Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the COVID-19 pandemic, which has led to a devastating global health crisis. The emergence of variants that escape neutralizing responses emphasizes the urgent need to deepen our understanding of SARS-CoV-2 biology. Using a comprehensive identification of RNA-binding proteins (RBPs) by mass spectrometry (ChIRP-MS) approach, we identify 107 high-confidence cellular factors that interact with the SARS-CoV-2 genome during infection. By systematically knocking down their expression in human lung epithelial cells, we find that the majority of the identified RBPs are SARS-CoV-2 proviral factors. In particular, we show that HNRNPA2B1, ILF3, QKI, and SFPQ interact with the SARS-CoV-2 genome and promote viral RNA amplification. Our study provides valuable resources for future investigations into the mechanisms of SARS-CoV-2 replication and the identification of host-centered antiviral therapies.
Keywords: severe acute respiratory syndrome coronavirus 2; comprehensive identification of RNA binding proteins by mass spectrometry, ChIRP-MS; SARS-CoV-2 RNA interactome; host RNA binding proteins; siRNA screen; host-dependency factors; SARS-CoV-2 infection inhibitors
Graphical abstract
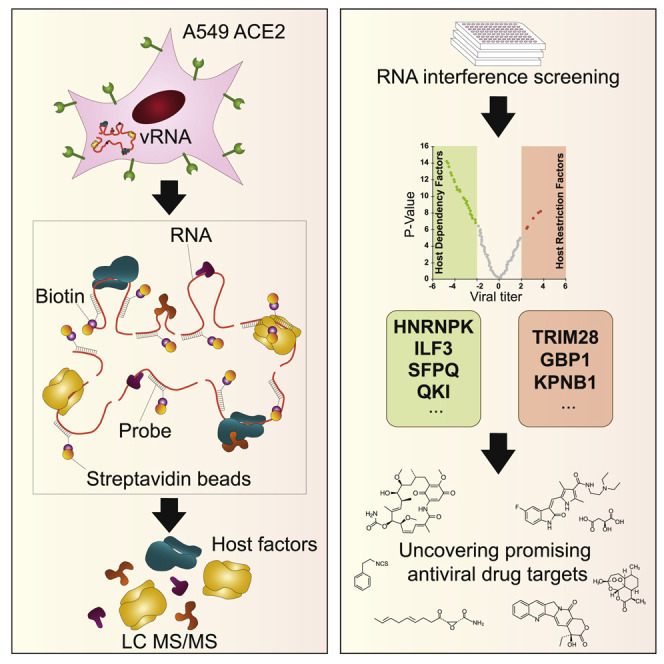
Labeau et al. combine a ChIRP-MS approach with a functional interrogation screen to provide an unbiased and comprehensive view of the host cell factors that interact with the viral genome and regulate SARS-CoV-2 infection. They show that these host dependency RNA-binding proteins (RBPs) represent pharmacological targets against SARS-CoV-2.
Introduction
Severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), which belongs to the Coronaviridae family, has been identified as the causative agent of the ongoing pandemic of coronavirus disease 2019 (COVID-19) (Zhu et al., 2020). One year after the emergence of SARS-CoV-2, several vaccines in Phase III clinical trials (Poland et al., 2020) received emergency approval. Most of these vaccines are primarily designed to elicit immune responses to the viral spike protein. However, several SARS-CoV-2 variants of concern (VOCs) have begun to emerge, which contain mutations in the spike protein that increase transmissibility and reduce susceptibility to natural or vaccine-induced neutralizing immunity (i.e., breakthrough infection) (Cele et al., 2021; Garcia-Beltran et al., 2021; Liu et al., 2021a, 2021b; Zhou et al., 2021). Thus, a deeper understanding of SARS-CoV-2 replication mechanisms is required to develop additional therapeutic strategies aimed at preventing the emergence of new SARS-CoV-2 variants with an increased rate of reinfection, transmissibility, and disease severity.
The genome of SARS-CoV-2 is composed of a single-stranded positive RNA with a length of approximatively 30 kb (Zhou et al., 2020). In the cytoplasm of infected cells, the viral RNA is translated by the host machinery in 2 overlapping replicase polyproteins that undergo subsequent viral protease-mediated processing to generate 16 non-structural proteins (NSPs) (de Wilde et al., 2018). These NSPs assemble and recruit poorly characterized host cell factors to form the replication and transcription complex (RTC), which localizes in a virus-induced network of double-membrane vesicles (DMVs) (Cortese et al., 2020; Eymieux et al., 2021). Within the RTC, the RNA-dependent-RNA polymerase (RdRp) complex uses the genomic RNA as a template to generate negative-strand intermediates that synthesize both progeny genomic and subgenomic viral RNAs (sgvRNAs) (Snijder et al., 2016). SgvRNAs are then translated in 4 structural (S, E, M, and N) and 7 accessory proteins (ORF3a-9b) (Kim et al., 2020). The viral genomic RNA associated with the nucleocapsid (N) forms the ribonucleocapsid, which together with the other structural proteins, drives the assembly of new viral particles that bud in the lumen of the endoplasmic reticulum-Golgi intermediate compartment (ERGIC).
Like all viruses, SARS-CoV-2 heavily relies on cellular proteins to accomplish its infectious life cycle. This host dependency represents a potential Achilles’ heel that could be exploited to develop new host-centered approaches to treat SARS-CoV-2 infection. In this context, affinity purification combined with mass spectrometry (MS) has allowed the identification of SARS-CoV-2 virus-host protein-protein interactions (PPIs) (Gordon et al., 2020a, 2020b; Li et al., 2020), while CRISPR-Cas9 phenotypic screens have provided valuable information about the host genes and cellular pathways important for the SARS-CoV-2 life cycle (Daniloski et al., 2020; Hoffmann et al., 2021; Schneider et al., 2020; Wang et al., 2021b; Wei et al., 2020). These works have led to the identification of potential cellular targets for drug repurposing, such as cholesterol homeostasis, the phosphatidylinositol 3-kinase (PI3K) complex or the sigma-1 and -2 receptors (Daniloski et al., 2020; Gordon et al., 2020a; Hoffmann et al., 2021; Wang et al., 2021b).
In this study, we undertook a comprehensive identification of RNA-binding protein by MS (ChIRP-MS) approach to establish a global map of the SARS-CoV-2 genome-associated host proteins. By combining these proteomics data with gene-silencing experiments, we identified a set of SARS-CoV-2 host dependency factors that affect infection and could be targetable for antiviral therapies.
Results and discussion
Characterization of the SARS-CoV-2 RNA interactome in human cells
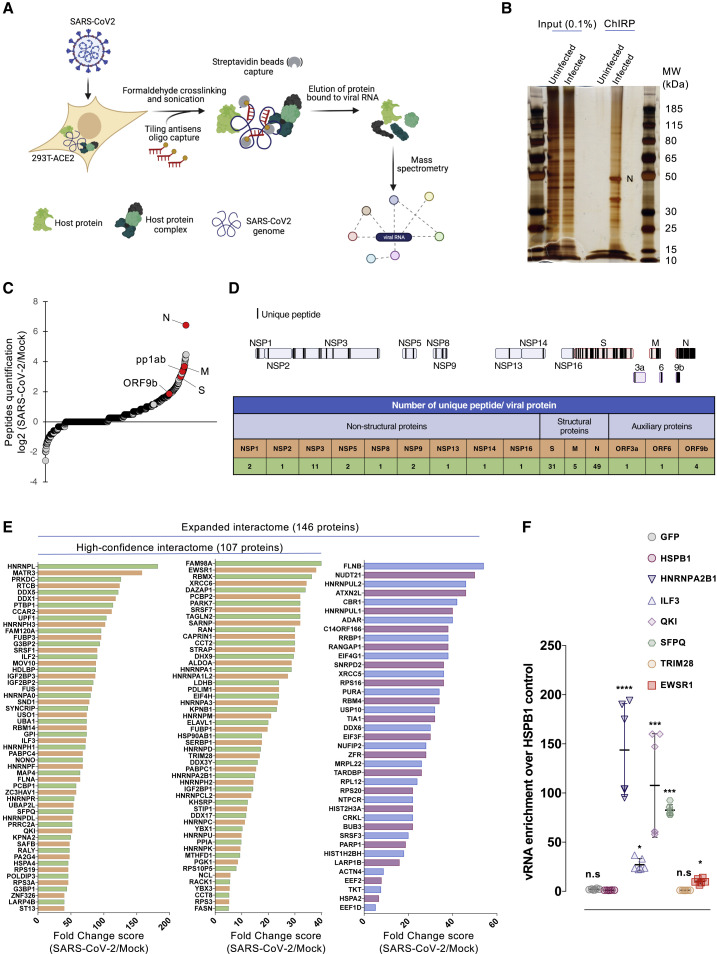
To characterize the host factors that interact with the SARS-CoV-2 genome during infection, we performed a ChIRP-MS (Figure 1A). Briefly, we challenged 293T cells stably expressing the human angiotensin-converting enzyme 2 (ACE2) receptor (293T-ACE2) with a primary clinical SARS-CoV-2 220/95 strain (EPI_ISL_469284) isolated by our laboratory in March 2020 (Onodi et al., 2021). Forty-eight hours later, SARS-CoV-2-infected and -uninfected cells were treated with formaldehyde to crosslink RNA with RNA-binding proteins (RBPs) to preserve ribonucleoprotein (RNP) complexes. To specifically capture SARS-CoV-2 RNA, we designed 129 biotinylated antisense probes (Table S1) complementary to the full-length viral RNA, and subsequently used streptavidin beads to pull down the RNPs. Co-immunoprecipitated proteins were separated by SDS-PAGE, visualized by silver staining (Figure 1B), and analyzed by MS (Table S2). Correlation experiments showed that our experimental replicates (n = 5) are highly consistent (Pearson’s correlation coefficient R > 0.97 between replicates). As expected, the SARS-CoV-2 N protein, which is the main viral RBP, was the most abundant viral protein (Figures 1B and 1C; Table S2). The replicase polyprotein pp1ab, as well as the structural M and S and the accessory ORF9b proteins were also significantly and reproducibly enriched in our experiments (Figures 1C and 1D; Table S3). We also identified 9 of the 16 pp1ab-derived NSPs (Figure 1D), which are part of the coronavirus replication/transcription complex (RTC) (V’kovski et al., 2019). Among them, NSP2–NSP16 play essential functions for RNA replication (V’kovski et al., 2021). NSP1 is known to interact both with the 40S ribosomal subunit to inhibit cellular translation, and the 5′ untranslated region (UTR) of the vRNA to promote translation (Schubert et al., 2020). The M protein associates with the N protein, within the ribonucleocapsid, to favor RNA genome packaging (Narayanan et al., 2000). M also orchestrates the assembly of viral particles at the ERGIC by interacting with the other structural proteins and ensures the retention of S within this compartment, and thereby its incorporation within the virion (Schoeman and Fielding, 2019). Thus, we speculate that enrichment of M and S may be a consequence of their association with the ribonucleocapsid during viral particle assembly. Finally, ORF9b has been shown to localize at the mitochondrial membrane and to antagonize the retinoic acid-inducible gene-I-mitochondrial antiviral-signaling protein (RIG-I-MAVS) antiviral signaling through its association with TOM70 (Gordon et al., 2020b; Jiang et al., 2020). Together, these data provide strong evidence that our ChIRP-MS approach reproducibly and specifically pulled down viral RNP complexes involved in different steps of the SARS-CoV-2 life cycle, from replication/transcription to particle assembly.
Figure 1.
Purification of the SARS-CoV-2 RNA interactome
(A) Schematic illustrating the ChIRP-MS approach used to identify host factors bound to SARS-CoV-2 RNA.
(B) Proteins enriched by ChIRP from uninfected and SARS-CoV-2-infected cells were resolved by SDS-PAGE and visualized by silver staining.
(C) Quantification of the peptides ratio as the average of peptides count for a prey in SARS-CoV-2 divided by the average peptides count for the same prey in the control (zero counts are replaced by 1). Viral proteins are marked by red circles (n = 5 biological replicates).
(D) Top: Schematic representing unique peptide distribution along the enriched viral proteins. Bottom: Table showing the number of unique peptides for each viral protein. Data from 1 representative biological replicate are shown.
(E) Fold change (FC; SAINTexpress FC score) of the high-confidence and the expanded interactomes identified in our SARS-CoV-2 ChIRP-MS.
(F) vRIP assays in 293T-ACE2 cells transfected with plasmid encoding the indicated proteins bearing a hemagglutinin (HA) or FLAG tag and infected with SARS CoV-2. FLAG-HSPB1 and FLAG-GFP were used as negative controls. Enrichment of immunoprecipitated vRNA was calculated as 2(−ΔΔCt [normalized RIP/normalized HSPB1]. The data shown are means ± SDs of 2 biological replicates (with technical duplicates). Adjusted p values were calculated by the Kruskal-Wallis test with Benjamini, Krieger, and Yekutieli correction (n.s, non-significant; ∗p < 0.05, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001).
A view of the SARS-CoV-2 RNA-host protein interactome
We used the SAINTexpress scoring algorithm to compare the MS data from the 5 replicates (SARS-CoV-2 over Mock-infected cells) and defined a high-confidence interactome (Bayesian false discovery rate [BFDR] ≤0.05 and fold change [FC] ≥5; Table S3). We identified 107 human proteins that reproducibly associated with the SARS-CoV-2 RNA (Figure 1E). We also defined an expanded SARS-CoV-2 interactome (BFDR <0.1 and FC ≥5; Table S3) and identified 39 additional proteins. Next, we compared our high-confidence interactome with a SARS-CoV-2 interactome in which we have used SARS-CoV-2-infected cells without cross-link as negative control (Figure S1A; Tables S2 and S3). We found 78 common enriched factors and defined a high-confidence core interactome (Figures S1B and S1C). Because the majority of our high-confidence hits (78/107; 73%) were confirmed by a second approach, we could likely exclude obvious shortcomings in our enrichment calculation approach (SARS-CoV-2 over Mock-infected cells). To rule out biases resulting from infection-induced host proteome changes, we monitored the expression levels of selected hits before and after infection. We selected a panel of 9 proteins chosen among the top-ranked (e.g., G3BP1, HNRNPK, HNRNPA2B1, KHSRP, ILF3, NONO, and SFPQ) and lower-ranked (QKI, EWSR1) proteins in our MS analysis (Table S2) for which antibodies were available in our laboratory. As shown in Figure S1D, none of the studied proteins were upregulated upon SARS-CoV-2 infection of either 293T or A549 cells stably expressing ACE2.
Among the proteins included in the expanded interactome, 88% (128/146) were annotated by the Gene Ontology (GO) term “RNA binding” (GO: 0003723), suggesting a direct interaction with the viral genome. To test this hypothesis, we performed native viral RNA immunoprecipitation (vRIP) experiments on a subset of 5 selected factors (HNRNPA2B1, ILF3, QKI, SFPQ, and EWSR1). As negative controls, we included HSPB1, a heat shock protein not enriched in our ChIRP-MS, and the heterologous GFP protein, which unlikely binds RNA. We also added TRIM28, a protein for which no RNA-binding activity was reported, which likely was co-purified with vRNA in our ChIRP-MS through protein-mediated interaction. We confirmed that for the 5 proteins tested, there was a significant enrichment of vRNA relative to the negative controls (Figures 1F and S2A), whereas there was no vRNA enrichment in the TRIM28 sample, strongly pointing to the existence of specific interaction. These results were also confirmed in A549-ACE2 stably expressing the different proteins (Figures S2B and S2C). Altogether, these observations demonstrate that our ChIRP approach can identify proteins that associate with vRNA.
Next, we compared our RNA interactome with recently published datasets obtained by either RNA antisense purification coupled with MS (RAP-MS) or ChIRP-MS approaches in different SARS-CoV-2-infected cell lines (Flynn et al., 2021; Lee et al., 2021; Schmidt et al., 2020). We found that 36 (Schmidt et al.: 34.2%), 79 (Flynn et al.: 34.6%), and 74 (Lee et al.: 38.3%) SARS-CoV-2 RNA host factors identified in these studies were also enriched in our ChIRP-MS interactome (Table S4). Interestingly, we highlighted 58 common hits that defined a reproducible set of cellular factors binding SARS-CoV-2 RNA across 3 cell lines (293T, VeroE6, and Huh7.5; Figure S3). These encompass multiple RBPs involved in nucleic acid metabolism, including splicing, mRNA stability, and transcriptional regulation such as several heterogeneous nuclear RNPs (hnRNPs) (Geuens et al., 2016), the serine/arginine-rich splicing factor (SRSF) (Shepard and Hertel, 2009), poly(A) binding protein (PABP) (Mangus et al., 2003), and the insulin-like growth factor 2 mRNA binding protein (IGF2BP) (Bell et al., 2013). In our expanded interactome, we identified several components of the small 40S ribosome subunit (RPS3, RPS3A, RPS16, RPS19, and RPS20), the translation initiation factor 4F complex (eIF4G1, eIF3F, and eIF4H), and translation elongation factors (EEF1D and EEF2). In agreement with other published RNA-centric SARS-CoV-2 interactomes (Flynn et al., 2021; Kamel et al., 2021; Lee et al., 2021; Schmidt et al., 2020), the eukaryotic initiation factor 4E (eIF4E), the mRNA CAP binding protein, was not found. Although we cannot exclude the inability of RNA-centric methods to capture the vRNA-EIF4E complex, it is tempting to speculate that SARS-CoV-2 may either use an alternate CAP-dependent EIF4E-independent translation mechanism (de la Parra et al., 2018) to bypass host translation shutoff during the course of infection or switch from a CAP-dependent to a CAP-independent translation initiation, as proposed for dengue virus (Edgil et al., 2006). Chaperone proteins (HSP90AB1, HSPA2, CCT2, and CCT8) were also significantly enriched in our study. It is likely that they are indirectly associated with the vRNA either by contributing to nascent viral protein co-translational folding (Cabrita et al., 2016) or by interacting with viral NS proteins, as shown for dengue virus NS1 (Hafirassou et al., 2017) or hepatitis C virus NS5B or influenza PB2 viral polymerases (Fislová et al., 2010; Inoue et al., 2011). Characteristic RBPs involved in RNP granules such as paraspeckles (RBM14, NONO, MATR3, SFPQ) and stress granules (SGs) (G3BP1, G3BP2, TIA1, CAPRIN1, ATXN2L) (Nakagawa et al., 2018; Yang et al., 2020) were among the highest scoring candidates. Many paraspeckles-interacting proteins were also highly represented in our ChIRP-MS dataset (DAZAP1, EWSR1, FAM98A, FUS, RBMX, UBAP2L, as well as hnRNPA1, A1L2, H1, H3, K, R, UL1) (Naganuma et al., 2012). We also identified several components of the tRNA-splicing ligase complex (DDX1, FAM98A, RTCB, C14ORF166/hCLE), which is involved in XBP1 cytoplasmic splicing (Filipowicz, 2014), a key regulator of the unfolded protein response that is induced by coronavirus replication (Fung and Liu, 2014). Finally, we identified several proteins with antiviral functions among the hits (ZC3HAV1, MOV10, ILF3, ADAR). For instance, ZC3HAV1 is an interferon-stimulated gene (ISG) that directly binds the viral RNA and promotes viral RNA decay (Guo et al., 2007). MOV10 was recently shown to bind bunyavirus N protein to interfere with RNP assembly and RNA synthesis (Mo et al., 2020). It was also found to interact with coronaviruses N protein and to restrict Middle East respiratory syndrome (MERS)-CoV infection (Wang et al., 2021a). These proteins contribute to the host cell response to viral infection, and their enrichment strongly suggests that SARS-CoV-2 infection triggers an innate immunity response in 293T-ACE2 cells.
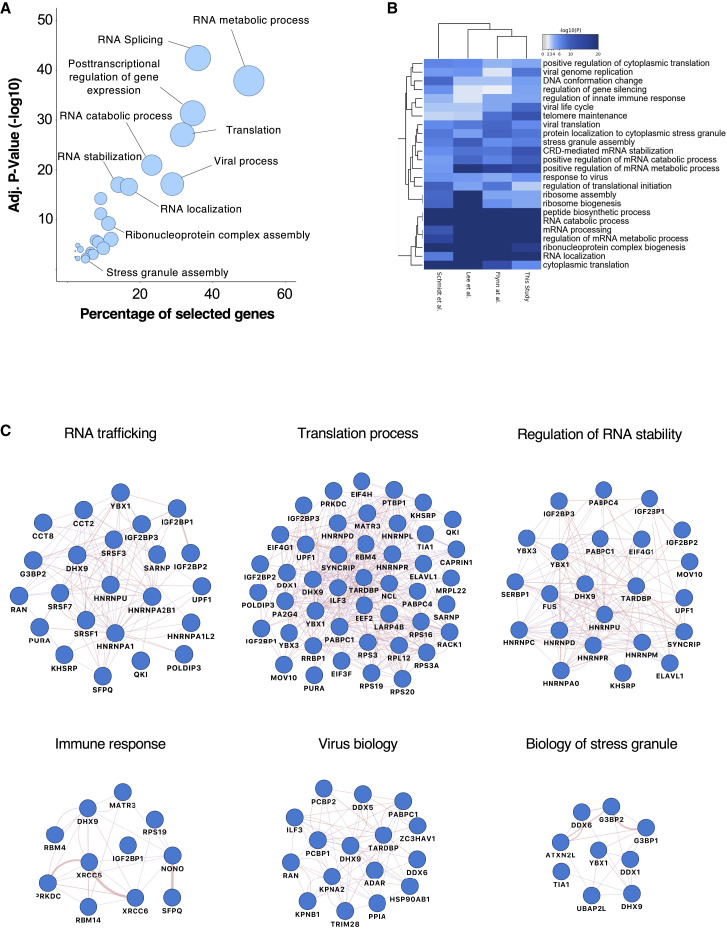
Functional analyses of the vRNA-interacting proteins revealed more than 150 statistically enriched GO terms, which clustered into 23 functional groups (Figure 2A) that are shared by the 3 other published SARS-CoV-2 RNA interactome studies (Figure 2B). Among them, we found host molecules that participate in RNA metabolic process, translation, RNA stabilization, RNP complex assembly, viral process, and innate immune response (Figures 2A and 2C). Based on its predominant nuclear localization and its role in mRNA maturation, the significant enrichment of the RNA splicing GO term (p = 3.55 × 10−43) was unexpected. Pre-mRNA splicing is critical for mRNA nucleo-cytoplasmic export, thereby contributing to the regulation of gene expression (Reed and Hurt, 2002). We infer that SARS-CoV-2 RNA may sequester host RBPs involved in splicing to hamper cellular mRNA nuclear export, thus favoring the cytoplasmic translation of viral RNAs. This hypothesis is consistent with recent findings showing that SARS-CoV-2 NPS16 disrupts mRNA splicing to antagonize innate immunity at a post-transcriptional level (Banerjee et al., 2020).
Figure 2.
Biological analysis of the SARS-CoV-2 RNA interactome
(A) GO enrichment analysis of SARS-CoV-2 RNA interactome proteins. Circles represent the enriched function for an annotated ontology term and circle size corresponds to the number of enriched proteins within that term.
(B) Heatmap of shared GO:biological processes enriched across all SARS-CoV-2 RNA interactomes. Horizontally, biological functions have been clustered according to their enrichment significance among cohorts. Vertically, cohorts have been clustered according to their functional enrichment similarities.
(C) Selected biological process networks of SARS-CoV-2 interactome proteins.
Finally, we asked whether the vRNA-binding cellular factors identified in our study also interact with viral proteins. By examining a reference coronavirus interactome (Perrin-Cocon et al., 2020), we found that 81 (55%) of the 146 hits identified by our ChIRP-MS approach are known interactors of 1 or several coronaviruses viral proteins (Figure S4). This suggests that many of the proteins identified in our SARS-CoV-2 interactome could interact with viral proteins within RNP complexes.
Functional interrogation of the SARS-CoV-2 RBP identified in our ChIRP-MS approach
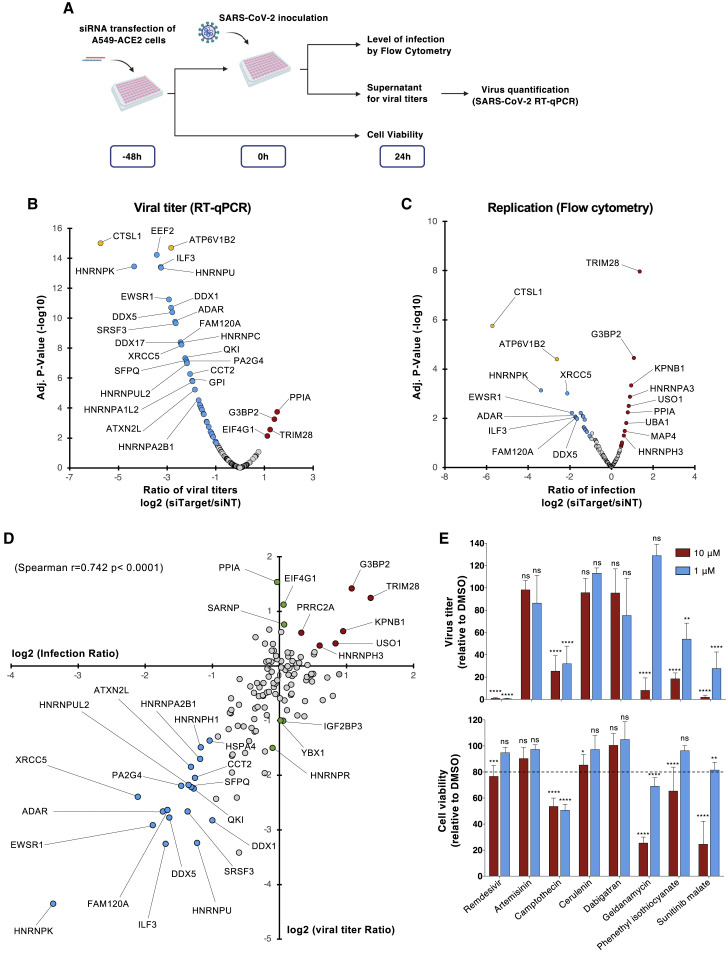
To further pinpoint the function of the identified host factors in SARS-CoV-2 infection, we analyzed the consequences of silencing their expression by RNA interference (RNAi) on virus replication. We designed a library of small interfering RNA (siRNA) pools targeting 138 RBPs identified in our study and performed a loss-of-function screen in the human lung epithelial cell line A549 stably expressing ACE2. siRNA targeting the cathepsin L protease (CTSL) and the vacuolar ATPase subunit ATP6V1B2, 2 host molecules important for SARS-CoV-2 viral entry (Daniloski et al., 2020), were used as positive controls, while a non-targeting siRNA pool (siNT) was used as negative control. siRNA transfected cells were challenged with SARS-CoV-2 for 24 h and viral infection was assessed either by quantifying the intracellular accumulation of the N protein by flow cytometry (viral replication) or the vRNA released in the supernatant of infected cells by qRT-PCR (release of viral particles) (Figure 3A). All readouts were normalized to the siNT-transfected cells. Transfected cells were also monitored for cell viability, and the siRNA with cytotoxic effects (below 75% of cell viability in control) were removed from the analysis (7%, 9/138). As expected, siRNA-mediated depletion of either CTSL or ATP6V1B2 resulted in a strong decrease in infection in both assays (Figures 3B and 3C). qRT-PCR experiments revealed that 45 siRNAs reduced viral particle release by at least 50%, whereas 4 siRNAs increased it by 200% (Figure 3B). Flow cytometry studies showed that 21 siRNAs decreased the percentage of infected cells by more than 50%, while 12 enhanced it by 150% (Figure 3C). By intersecting the data from both assays, we identified 25 proteins that affected viral infection by acting either as host dependency factors (HDFs) (19/25) or host restriction factors (HRFs) (6/25) (Figure 3D). This contrasts with the data recently published by Flynn et al. (2021) showing that the majority of SARS-CoV-2 RBPs identified in their study has an antiviral function. It is likely that RNP complexes are dynamically regulated and that some RBPs could function as pro- or antiviral host factors at different stages of the viral life cycle. We observed a high correlation coefficient (r = 0.742) between our qRT-PCR and flow cytometry datasets indicating that most of the identified proteins are required for genome replication (Figure 3D), and only very few act at a late step (assembly/particle egress) of the SARS-CoV-2 life cycle (Figure 3D, green circles). Interestingly, several HDFs such as ILF3, HNRNPU, HNRNPK, FAM120A, and QKI were predicted to bind the 5′- or 3′-UTR of SARS-CoV-2 RNA (Sun et al., 2021). Others, including SFPQ, DDX5, EWSR1, HNRNPK, ADAR, and PA2G4 were previously reported to promote the replication of other RNA viruses (Garcia-Moreno et al., 2019; Gélinas et al., 2011; Landeras-Bueno et al., 2011; Li et al., 2013; Oakland et al., 2013; Thompson et al., 2020; Zhou et al., 2019a), supporting their role in SARS-CoV-2 replication. Interestingly, ILF3 was shown to participate in the antiviral type I interferon response by promoting the expression of IFNB1 and ISGs during host translational shutoff (Watson et al., 2020). QKI was shown to repress host interferon response by downregulating MAVS expression at a post-transcriptional level (Liao et al., 2020). This suggests that SARS-CoV-2 has evolved several mechanisms to dampen the type I interferon response by targeting specific RBPs.
Figure 3.
Functional interrogation of the SARS-CoV-2 RNA interactome and compounds screening
(A) Schematic illustrating the loss-of-function screen procedure.
(B and C) A549-ACE2 cells were transfected with an arrayed siRNA library and challenged with SARS-CoV-2 (MOI 0.05) for 24 h.
(B) Yield of viral particles released in the supernatant of infected cells was quantified by qRT-PCR and normalized to the siNT-transfected cells. The data shown are the means of 2 biological replicates (with technical duplicates).
(C) Viral replication was assessed by flow cytometry using anti-N protein monoclonal antibody (mAb) and normalized to the siNT-transfected cells. The data shown are the means of 2 biological replicates (with technical duplicates). Adjusted p values were calculated by 1-way ANOVA with Benjamini and Hochberg correction. Host dependency factors are marked in blue and host restriction factors in are marked in red. Positive controls (CTSL1 and ATP6V1B2) are highlighted in yellow.
(D) Intersection of the data obtained from N protein quantification by flow cytometry and viruses released in the supernatant of infected cells by qRT-PCR. The data shown are the means of 2 biological replicates (with technical duplicates). Host dependency factors are marked in blue and host restriction factors are marked in red.
(E) A549-ACE2 were infected with SARS-CoV-2 (MOI 0.05) in the continuous presence of compounds (10 and 1 μM). Viruses released in supernatant were quantified 24 hpi by qRT-PCR (top panel). Cell viability was assessed in parallel (bottom panel). The data shown are the means ± SDs of 3 biological replicates (with technical duplicates). Significance was calculated using a 2-way ANOVA statistical test with Dunnett’s multiple comparisons test (n.s, not significant; ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
We then looked at the DrugBank, ChEMBL, DGIdb (Drug-Gene Interaction Database), and PanDrugs databases and identified 274 small molecules targeting 30 vRNA-binding proteins. Among them, we selected camptothecin (XRCC5), cerulenin (FASN), dabigatran (HNRNPC), geldanamycin (HSP90AB1), phenethyl isothiocyanate (HNRNPK), sunitinib malate (EWSR1), and artemisinin (an antimalarial interacting with multiple RBPs), which were predicted to interact with HDF having a log2 (titer ratio) ≤ 0.5 in our siRNA screen. To test their effect on infection, A549-ACE2 cells were incubated with each compound at a concentration of 10 and 1 μM and then challenged with SARS-CoV-2. We used equivalent amounts of DMSO as negative control and similar concentrations of remdesivir, an inhibitor SARS-CoV-2 replication (Wang et al., 2020), as positive control. We monitored both the release of progeny viruses in the supernatant and cell viability 24 h post-infection. Of all of the compounds tested, we observed a significant inhibitory effect for phenethyl isothiocyanate (40% inhibition) and sunitinib malate (80% inhibition) at a concentration of 1 μM without toxicity (Figure 3E). These data suggest that the host factors identified in our ChIRP-MS study could represent promising antiviral drug targets.
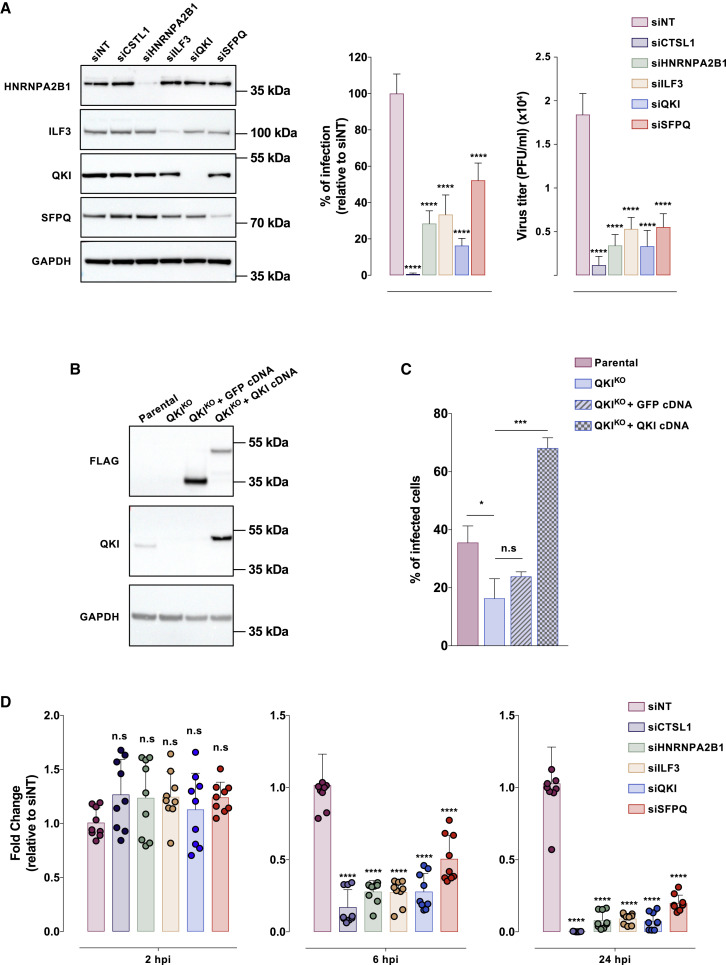
HNRPNA2B1, ILF3, QKI, and SFPQ mediate SARS-CoV-2 replication
Among the HDF identified, we were particularly interested in ILF3 and QKI (for which limited information on viral infection are available), as well as HNRNPA2B1 and SFPQ, which are likely able to associate with SARS-CoV-2 vRNA (Figures 1F and S2C). Consistent with our screen, siRNA-mediated depletion of HNRPNA2B1, ILF3, QKI, or SFPQ (Figure 4A, left panel) significantly reduced the percentage of infected cells and progeny viruses release (Figure 4A, center and right panels). To reinforce these data, we generated QKI knockout (QKIKO) cells using the CRISPR-Cas9 technology and performed infection assays (Figure 4B). We found that QKIKO cells were approximately 2-fold less permissive than parental cells, while trans-complementation with the human QKI cDNA rescued the infection (Figure 4C). Of note, our efforts to generate KO cells for the other selected HDFs (HNRPNA2B1, ILF3, SFPQ) were unsuccessful, suggesting that these genes are essential for the survival of A549 cells. To dissect the viral steps where these host factors are required, we infected A549-ACE2 cells knocked down for the expression of HNRPNA2B1, ILF3, QKI, and SFPQ, and quantified vRNA at different time points (Figure 4D). We did not observe any significant differences in vRNA levels in all of the conditions at 2 hpi, suggesting that these proteins are not involved in viral particle uptake. In contrast, we observed a large reduction in vRNA levels at 6 hpi, which increased at 24 hpi. Since in A549-ACE2, the beginning of vRNA amplification occurred between 4 and 6 hpi (data not shown), it is likely that HNRPNA2B1, ILF3, QKI, and SFPQ are involved in vRNA replication. Nonetheless, because SARS-CoV-2 vRNA translation and replication are tight, intricate steps, we could not rule out a role of some of these proteins in vRNA translation.
Figure 4.
HNRNPA2B1, ILF3, QKI, and SFPQ mediate SARS-CoV-2 replication
(A) A549-ACE2 cells were reverse transfected with the indicated siRNA pool. In 48 h, protein knockdown was ascertained by western blot (left panel) and cells were challenged with SARS-CoV-2 (MOI 0.01) for 24 h. Viral replication was assessed by flow cytometry using anti-N protein mAb and normalized to the siNT-transfected cells (center panel); the yield of viral particles released in the supernatant of infected cells was quantified by qRT-PCR (right panel). The data shown are the means ± SDs of 3 biological replicates (with technical triplicates). Significance was calculated using 1-way ANOVA statistical test with Dunnett’s multiple comparisons test.
(B) Immunoblot with anti-FLAG or anti-QKI mAb in parental, QKIKO A549-ACE2 cells, and QKIKO cells trans-complemented with GFP or QKI cDNA.
(C) Indicated cells were inoculated with SARS-CoV-2 (MOI 0.01). Infection was quantified 24 h later by flow cytometry using anti-N protein mAb. The data shown are the means ± SDs of 3 biological replicates (with technical duplicates). Significance was calculated by the Kruskal-Wallis test with Dunnett’s multiple comparisons test.
(D) A549-ACE2 cells were treated as in (A). At the indicated time points, cells were treated with trypsin to remove cell surface-bound viral particles and viral RNA was quantified by qRT-PCR. The data shown are the means ± SDs of 3 biological replicates (with technical triplicates). The p values were calculated by 1-way ANOVA with Dunnett’s multiple comparisons test (n.s, non-significant; ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
In conclusion, our data provide a landscape of functional interactions that the SARS-CoV-2 RNA genome establishes with the host cell during infection. We provide robust evidence showing that SARS-CoV-2 interacts with several cellular RBPs to facilitate viral replication, some of which could represent promising targets for antiviral intervention. All of this information constitutes a valuable resource for future investigations into the mechanism of SARS-CoV-2 replication and the identification of antiviral drugs to manage the emergence of future variants of concern or new human coronavirus(es).
Limitations of the study
This study provides a valuable resource to improve our understanding of the molecular interactions that SARS-CoV-2 establishes with the infected cells. However, our ChIRP-MS was performed in a cell line that is not derived from the lung; thus, the vRNA-host protein interactions shown in this study could not fully cover the whole interactions that take place during natural infection. It will be interesting to extend this study to more physiological cellular models, such as primary cells or cells derived from nasopharyngeal or lung epithelium, to derive an overall view of the SARS-CoV-2 interactome.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Human/Rat/Hamster ACE2 | Biotechne | Cat #MAB9332 |
| Mouse monoclonal SARS-CoV/SARS-CoV-2 Nucleocapsid | Sino Biological | Cat #40143-MM05; RRID:AB_2827977 |
| Alexa Fluor® 647 AffiniPure Donkey Anti-Mouse IgG (H + L) | Jackson ImmunoResearch | Cat #715-605-150; RRID:AB_2340862 |
| Monoclonal ANTI-FLAG M2 antibody | Sigma Aldrich | Cat #F1804; RRID:AB_262044 |
| Rabbit monoclonal HA-Tag (C29F4) | Cell Signaling Technology | Cat #3724S; RRID:AB_1549585 |
| Rabbit polyclonal GAPDH (0411) Antibody | SantaCruz Biotechnology | Cat #sc-47724; RRID:AB_627678 |
| Peroxidase AffiniPure Donkey anti-Rabbit IgG | Jackson ImmunoResearch | Cat #711-035-152; RRID:AB_10015282 |
| Rabbit anti-mouse Immunoglobulins/HRP | Dako | Cat #P0260; AB_2636929 |
| Rabbit monoclonal anti-ILF3 | Abcam | Cat #Ab 92355; RRID:AB_2049804 |
| Rabbit monoclonal anti-QKI | Abcam | Cat #Ab 126742; RRID:AB_11129508 |
| Rabbit monoclonal anti-HNRNPA2B1 | Abcam | Cat #Ab 259894 |
| Rabbit monoclonal anti-SFPQ | Abcam | Cat #Ab 177149 |
| Bacterial and virus strains | ||
| SARS-CoV-2 isolate 220_95 | Isolated from patient (Onodi et al., 2021) | EPI_ISL_469284 |
| Chemicals, peptides, and recombinant proteins | ||
| 32% Paraformaldehyde (formaldehyde) aqueous solution | Electron Microscopy Sciences | Cat #15714 |
| DMSO | Sigma-Aldrich | Cat #D5879 |
| Triton X-100 | Sigma-Aldrich | Cat #T9284 |
| Artemisinin | Sigma-Aldrich | Cat #361593 |
| (S)-(+)-Campthotecin | Sigma-Aldrich | Cat #C9911 |
| Cerulenin | Sigma-Aldrich | Cat #C2389 |
| Dabigatran | Sigma-Aldrich | Cat #SML2370 |
| Geldanamycin from Streptomyces hygroscopicus | Sigma-Aldrich | Cat #G3381 |
| Phenethyl isothiocyanate | Sigma-Aldrich | Cat #253731 |
| Remdesivir | Selleckchem | Cat #SE-S8932 |
| Sunitinib malate | Sigma-Aldrich | Cat #PZ0012 |
| MyOne C1 beads | Thermo Fisher Scientific | Cat #65001 |
| 50 mM D-biotin | Thermo Fisher Scientific | Cat #B20656 |
| SUPERase-in | Thermo Fisher Scientific | Cat #AM2696 |
| NA-Deoxycholate | Sigma Merck | Cat #30970 |
| N-Laurosylsarcosine sodium salt | Sigma Merck | Cat #L7414 |
| EDTA ULTROL grade | Sigma Merck | Cat #324504 |
| Trichloroacetic acid | Sigma Merck | Cat #T0699 |
| Proteinase K | Qiagen | Cat #19133 |
| TRizol LS Reagent | Thermo Fisher Scientific | Cat #10296028 |
| Dynabeads Protein G | Thermo Fisher Scientific | Cat #10004D |
| 3XFLAG peptide | Sigma Merck | Cat #F4799 |
| HA peptide | Sigma Merck | Cat #11666975001 |
| GlycoBlue Coprecipitant | Thermo Fisher Scientific | Cat #AM9515 |
| Lipofectamine RNAiMax Transfection Reagent | Thermo Fisher Scientific | Cat #13778150 |
| Lipofectamine 3000 Transfection Reagent | Thermo Fisher Scientific | Cat #L3000015 |
| Bolt 4–12% Bis-Tris plus | Thermo Fisher Scientific | Cat #NW04122BOX |
| Bolt 10% Bis-Tris plus | Thermo Fisher Scientific | Cat #NW00100BOX |
| 2X Laemmli Sample Buffer | Biorad | Cat #1610737 |
| Critical commercial assays | ||
| CellTiter-Glo® 2.0 Assay | Promega | Cat #G9242 |
| Profection® Mammalian Transfection System | Promega | Cat #E1200 |
| Luna® Universal One-Step RT-qPCR Kit | New England Biolabs | Cat #E3005E |
| Maxima First Strand cDNA Synthesis Kit | Thermo Fisher Scientific | Cat #K1671 |
| Experimental models: Cell lines | ||
| Lenti-X 293T | Takara Bio | Cat #632180 |
| Lenti-X 293T-ACE2 | This manuscript | N/A |
| A549 | ATCC | Cat #CCL-185 |
| A549-ACE2 | This manuscript | N/A |
| Vero E6 | ATCC | Cat #CRL-1586 |
| Oligonucleotides | ||
| ChIRP tilling oligos, see Table S1 | This paper | N/A |
| E_Sarbeco Forward: ACA GGT ACG TTA ATA GTT AAT AGC GT | Corman et al., (2020) | N/A |
| E_Sarbeco Reverse: ATA TTG CAG CAG TAC GCA CAC A | Corman et al., (2020) | N/A |
| EWSR1 HA Forward: TAA ACT ATG AAT TCA GGA TGG CGT CCA CGG ATT ACA GTA CCT ATA GCC AAG CTG CAG CGC AGC AGG GC | This paper | N/A |
| EWSR1 HA Reverse: ATA GTT TAG CGG CCG CTC AAG CGT AAT CTG GAA CAT CGT ATG GGT ATC CTC CAG CGG CCG AGT AGG GCC GAT CTC TGC GC | This paper | N/A |
| ACE2 Forward: CCG CTC GAG CGG GCC ATG TCA AGC TCT TCC TGG CTC | This paper | N/A |
| ACE2 Reverse: ATA AGA ATG CGG CCG CTT AAA AGG AGG TCT GAA CAT CAT CAG TG | This paper | N/A |
| ON-TARGETplus Human ILF3 siRNA - SMARTpool | Horizon Discovery | Cat #L-012442-00 |
| ON-TARGETplus Human HNRNPA2B1 siRNA -SMARTpool | Horizon Discovery | Cat #L-011690-01 |
| ON-TARGETplus Human QKI siRNA - SMARTpool | Horizon Discovery | Cat #L-024905-01 |
| ON-TARGETplus Human SFPQ siRNA - SMARTpool | Horizon Discovery | Cat #L-006455-00 |
| ON-TARGETplus Human CTSL siRNA SMART Pool | Horizon Discovery | Cat #L-005841-00 |
| ON-TARGETplus Non-targeting Pool | Horizon Discovery | Cat #D-001810-10-05 |
| QKI sgRNA targeting sequence: GGATCTTCAACCACCTCGAG | This paper | N/A |
| Recombinant DNA | ||
| pLVX-EF1alpha-IRES-Puro | Takara Bio | Cat #631988 |
| pLVX-ACE2 | This manuscript | N/A |
| MGC Human EWSR1 Sequence-Verified cDNA | Horizon Discovery | Cat #MHS6278-202759357 |
| 3xFlag SFPQ_pUC57-mini Plasmid | Genscript | N/A |
| 3xFlag QKI_pUC57 Plasmid | Genscript | N/A |
| 3xFlag ILF3_pUC57-Mini Plasmid | Genscript | N/A |
| 3xFlag HNRNPA2B1_pUC57 Plasmid | Genscript | N/A |
| 3xFlag HSPB1_pUC57 Plasmid | Genscript | N/A |
| 3xFlag GFP_pUC57 Plasmid | Genscript | N/A |
| 3xFlag TRIM28_pUC57-mini Plasmid | Genscript | N/A |
| Software and algorithms | ||
| Attune NxT | Thermo Fisher Scientific | Version 2.6 |
| FlowJo | Tree Star | Version 10.0.8 |
| Prism | GraphPad Software | Version 7.0d |
| Serial Cloner | SerialBasics | Version 2.6.1 |
| ClueGo | Cytoscape | Version 2.5.7 |
| Genamania | Cytoscape | Version 3.5.2 |
| Other | ||
| Attune NxT | Thermo Fisher Scientific | N/A |
| TriStar2 LB 942 | BERTHOLD Technologies | N/A |
| LightCycler 480 | Roche | N/A |
Resource availability
Lead contact
Further information and requests or resources and reagents should be directed to and will be fulfilled by the lead contact, Laurent Meertens (laurent.meertens@inserm.fr).
Materials availability
All the plasmids generated in this study, the 293T-ACE2 and A549-ACE2 are available on request.
Experimental model and subject details
Cell lines
Lenti-X 293T cells (human embryonic kidney cells), A549 cells (Human alveolar basal epithelial carcinoma), and Vero E6 cells (African green monkey kidney cells) were maintained in Dulbecco Modified Eagle Medium (DMEM; Invitrogen Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S) and 1% GlutaMAX (Life Technologies). All cell lines were periodically tested negative for mycoplasma contamination prior to use in experiments.
Method details
Virus preparation and titration
SARS-CoV-2 strain 220_95 (EPI_ISL_469284) was isolated from nasopharyngeal swab specimens collected at Service de Virologie (Hospital Saint Louis, Paris) (Onodi et al., 2021). Virus was propagated on Vero E6 in DMEM-2% (DMEM supplemented with 2% FBS, 1% P/S, 1% GlutaMAX, and 25 mM Hepes). Viruses were passaged three times before being used for experiments. For the last passage, viruses were purified through a 20% sucrose cushion by ultracentrifugation at 80,000 × g for 2 h at 4°C. Pellets were resuspended in TNE1X pH7.4 (Tris 50 mM, NaCl 100mM, EDTA 0.5 mM), aliquoted and stored at −80°C.
Viruses titer was determined by plaque assays in Vero E6 cells and expressed as PFU/mL. Cells were incubated for one hour with 10-fold serial dilution of viral stocks. The inoculum was then replaced with Avicel 2.4% mixed at equal volume with DMEM supplemented with 4% FBS, 2% Glutamax, 50mM MgCl2, 0.225% of NaHCO3, and incubated 3 days at 37°C. Then, Vero E6 cells were washed twice with phosphate-buffered saline (PBS) and stained with an aqueous solution containing 1% formaldehyde, 0.5% crystal violet, and 0.89% sodium chloride, for 15 min at room temperature. Vero E6 cells were then washed with water to visualize lysis plaques.
Generation of cell lines stably expressing ACE2
The human ACE2 coding DNA sequence (CDS) was amplified by RT-PCR from pLenti-ACE2 plasmid (a gift from Olivier Schwartz, Virus and Immunity, Institut Pasteur) using the ACE2 forward and reverse primers. Amplification product was cloned into pLVX-IRES-ZsGreen1 vector using XhoI/NotI restriction enzymes. The ACE2 protein-expressing lentiviral vectors pseudotyped with vesicular stomatitis virus glycoprotein G (VSV-G) were generated by transfecting lenti-X 293T (5 × 106) cells with pLVX-ACE2-ZsGreen, psPAX2, and pVSV-G (4:3:1 ratio) using calcium phosphate transfection (Promega) following the manufacturer’s instructions. Supernatants were harvested 48 h after transfection, cleared by centrifugation, and filtered. Lentiviral vectors were purified as describe above (see ‘virus preparation and titration’ section). 293T and A549 cells (2 × 105) were seeded in 6 well plate and transduced by spinofection at 1,000 × g for 2 h at 33°C. Cell populations were sorted based on GFP expression with a BD FACSAria II (Becton Dickinson) with FACSDiva 6.1.2 software (Becton Dickinson).
Flow cytometry analysis of ACE2 cell surface expression
Cells were detached with 2.5 mM EDTA in PBS and incubated with anti-human ACE2 mAb (5 mg/mL) in 100 mL of PBS with 0.02% NaN3 and 5% FBS for 1 h at 4°C. Following the primary staining, cells were washed and stained with incubated with the Alexa 647-conjugated goat anti-mouse IgG (Jackson ImmunoResearch) for 30 min at 4°C. Acquisition was performed on an Attune NxT Flow Cytometer (Thermo Fisher Scientific) and analysis was done by using FlowJo software (Tree Star).
Comprehensive identification of RNA binding proteins by mass spectrometry
ChIRP-M/S was performed as previously described by Ooi and colleagues (Ooi et al., 2019). SARS-CoV-2 probes were designed using the online tool (https://www.biosearchtech.com/stellaris-designer), with a repeat masking setting of 5 and even coverage of the whole viral genome (1 oligos every 200 bp). Oligos were synthetized with 3′ biotin-TEG modification at Eurofins Genomics. The 129 oligos were resuspended at 100 μM and mixed at equal volume to prepare the probes pool.
293T cells stably expressing ACE2 were seeded at 9 × 106 cells per T150 flask (3 flasks per condition) and inoculated with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.001 or mock-treated, aiming for 50–60% of infected cells as monitored by flow cytometry (see “viral infection quantification assays” section) using a fraction of cells collected at 48 hpi. The media was removed 48 h later and the cells were rinsed once with PBS, trypsinized and pooled for each condition. Cells were centrifugated at 290 × g for 5 min and pellets were washed twice with PBS. The cells were resuspended with 35 mL of PBS containing 3% formaldehyde and incubated under agitation for 30 min at room temperature (RT). Of note, all the buffers used throughout the ChIRP experiments were freshly prepared. Cross-linking was terminated by adding glycine to a final concentration of 0.125 mM for 5 min at RT. Cross-linked cells were centrifugated at 1,000 × g for 5 min, washed once with PBS, weighted, then snap frozen and kept at −80°C until used. Cells were resuspended in 1 mL of lysis buffer (50 mM Tris-HCl, pH7.0, 10 mM EDTA, 1% SDS supplemented with SUPERase-IN and protease inhibitor) per 100 mg of cell pellet. The lysates were sonicated using a Diagenode Bioruptor. At this stage, 10 μL of lysates were kept as sample inputs. Lysates were precleared by adding 30 μL of washed MyOne C1 beads per ml of lysate and rocked for 30 min at 37°C. Beads were removed from the lysates using a magnetic stand and 2 mL of ChIRP hybridization buffer (750 mM NaCl, 1% SDS, 50 mM Tris-HCl pH 7.0, 1 mM EDTA, 15% formamide supplemented with 1 mM PMSF, protease inhibitor and SUPERase-in) and 1.5 μL of probes pool were added for every ml of lysate. After mixing, hybridization took place for 16h at 37°C under rotation. Then, 100 μL of washed MyOne C1 beads per microliter of probes pool were added to each sample and incubated for 45 min at 37°C. Magnetic beads were collected on a magnetic stand and washed 5 times with 1 mL of washing buffer (2× SSC, 0.5% SDS, add 1 mM PMSF) for 5 min at 37°C. To eluate the enriched ribonucleoprotein complexes, the beads were incubated with 300 μL of biotin elution buffer (12.5 mM D-biotin, 7.5mM Hepes pH 7.5, 75 mM NaCl, 1.5 mM EDTA, 0.15% SDS, 0.075% sarkosyl and 0.02% Na-Doexycholate) for 20 min at RT under rotation and then 15 min at 65°C with shaking. Eluate was collected in a fresh tube and the beads subject to elution again. The two eluents were pooled (600 μL) and 150 μL of trichloroacetic acid (25% of the total eluate volume) was added, mixed vigorously, and let overnight at 4°C for precipitation. The next day, the samples were centrifugated at 20,000 × g for 45 min at 4°C. Pellets were washed once with ice-cold acetone and spun at 20,000 × g for 5 min at 4°C. The acetone supernatant was removed, the tubes centrifuged briefly to carefully remove the residual acetone and the pellets were left to air-dry on the bench top. The pellets were solubilized in 30 μL of 2X Laemmli buffer with 20 mM DTT and boiled at 95°C for 30 min with a periodic mixing (every 5 min) to reverse the cross-linking.
One-tenth of the protein samples and their corresponding inputs (pre-diluted 1/10) were size-separated on a bolt 4–12% Bis-Tris plus (Thermo Fisher Scientific) and stained with the silver stain kit (Thermo Fisher Scientific) as per manufacturer’s instructions. The remaining samples were size-separated on a bolt 10% Bis-Tris plus and stained with the Imperial protein stain (Thermo Fisher Scientific) according to manufacturer’s instructions. For each of the 5 ChIRP experiments one gel slice from the SARS-CoV-2 and mock-infected samples were cut from the SDS-page. Gel slices from 5 independent replicates were sent for mass spectrometry analysis at Taplin Biological Mass Spectrometry Facility (Harvard Medical School) as previously described (Hafirassou et al., 2017).
High-confidence RNA binding protein scoring
ChIRP-M/S data were analyzed using the online server (https://reprint-apms.org/) to run SAINTexpress (Teo et al., 2014). Total peptides count from each of the five technical replicates, including both mock- and SARS-CoV-2-infected cells, were used as quantitative values to run the analysis. To determine a core high-confidence interactome, we applied stringent filtering criteria with BFDR ≤0.05 and a fold change ≥5. We also defined an expanded interactome by loosen the BFDR requirement (BFDR <0.1). Data shown in Figure 1E correspond to the fold change scores (FC) calculated by SAINTexpress from the average mean of spectral counts.
Identification of known interactors of coronaviruses in the ChIRP dataset
A recently published meta-analysis by Perrin-Cocon et al. was used as a reference for known interactions between coronavirus and host proteins (Perrin-Cocon et al., 2020). In this report, the literature was manually curated to assemble a dataset of 1,311 coronavirus-host interactions encompassing 1,140 distinct cellular proteins. Host proteins in the ChIRP dataset overlapping with those present in this reference interactome were considered as previously reported or « known » interactors of coronaviruses. They were displayed together with the interacting-coronavirus proteins as an interaction network using Cytoscape (Shannon et al., 2003).
siRNA screen assay
An arrayed ON-TARGETplus SMARTpool siRNA library targeting 138 of the 146 RNA-interacting proteins significantly enriched in our ChIRP-M/S was purchased from Horizon Discovery. SiRNA targeting 8 genes were not assessed because either no predesigned siRNA pools were available or proteins were not considered as candidates in our first analysis. These proteins are: hnRNP C-like 2, histone H2B type1-H, histone H3.2, putative 40S ribosomal protein S10-like, HSPA2, EEF1D, TKT and ACTN4. A549 cells stably expressing ACE2 were seeded in 24-well plate format (viral infection) or a 96-well plate (viability) and then were reverse transfected in duplicate with 30 nM of siRNA using the Lipofectamine RNAiMAX reagent (Invitrogen) according to manufacturer’s instruction. 48 h post-transfection, cells seeded in a 24-well format were challenged with SARS-CoV-2 at a MOI of 0.05 in DMEM containing 5% FBS. After three hours at 37°C, virus inoculum was removed and replace with fresh medium. 24 h post-infection, cells and 200 μL of supernatants were collected for N protein staining and viral RNA quantification as described in “Viral infection quantification assays”. Throughout the screening assay, siRNA-transfected cells seeded in 96-well format were incubated for 72 h at 37°C and used to assess cell viability.
Drug treatment and SARS-CoV-2 infection
Using DGIdb, DrugBank, ChEMBL, and PanDrugs databases, we identified 274 compounds targeting 30 RBPs. Compounds were first filtered to select molecules described either as an inhibitor or with the highest interaction score. Finally, we selected the compounds targeting RBPs with a log2 (titer ratio) ≤ 0.5 in our siRNA screen.
Cells grown in 24-well plates were treated with the indicated compound concentrations for two hours prior infection and compounds were maintained throughout the course of infection. Cells treated with equivalent concentration of DMSO were used as control. The cells were challenged with SARS-CoV-2 at a MOI of 0.05 for 3 h, then washed once with PBS and incubated with fresh media. 24 h post-infection, cells and 200 μL of supernatants were collected for N protein staining and viral RNA quantification, as described in “Viral infection quantification assays”. In parallel cells seeded in 96-well format were incubated 24 h with the drugs to assess cell viability.
Viral infection quantification assays
For infection quantification by flow cytometry analysis, 24 h post-infection cells were trypsinized and fixed with 2% (v/v) paraformaldehyde (PFA) diluted in PBS for 15 min at room temperature. Cells were incubated for 45 min at 4°C with 1 μg/mL of the anti-N mAb diluted in permeabilization flow cytometry buffer (PBS supplemented with 5% FBS, 0.5% (w/v) saponin, 0.1% sodium azide). After washing, cells were incubated with 1 μg/mL of Alexa Fluor 647-conjugated goat anti-mouse IgG diluted in permeabilization flow cytometry buffer for 30 min at 4°C. Acquisition was performed on an Attune NxT Flow Cytometer (Thermo Fisher Scientific) and analysis was done by using FlowJo software (Tree Star). To assess infectious viral particle release during infection by RT-qPCR, viruses were first inactivated by incubating the supernatants v/v with 1% Triton X-100 (Sigma) in PBS for 30 min under agitation at RT. Yields of viral RNA were quantified by real-time qPCR by using SARS-CoV-2 specific primers targeting the E gene with the Luna® Universal One-Step RT-qPCR Kit (New England Biolabs) in a LightCycler 480 thermocycler (Roche) according to the manufacturer’s protocol. The number of viral genomes is expressed as PFU equivalent/mL and was calculated by performing a standard curve with a similarly treated supernatant from a viral stock with a known titer as described by Gordon et al. (Gordon et al., 2020b).
Kinetic of infection by qPCR assay
A549-ACE2 cells, plated on 6 well plates and reverse transfected with the indicated siRNA, were inoculated with SARS-CoV-2 (MOI of 0.01). At indicated time point cells were washed twice with PBS, incubated with trypsin 0.25% for 5 min at 37°C to remove cells surface bound particles, and total RNA was extracted using the RNeasy plus mini kit (Qiagen) according to manufacturer’s instruction. cDNAs were generated from 400 ng total RNA by using the Maxima First Strand Synthesis Kit following manufacturer’s instruction (Thermo Fisher Scientific). Amplification products were incubated with 1 Unit of RNAse H for 20 min at 37°C, followed by 10 min at 72°C for enzyme inactivation, and diluted 10-fold in DNAse/RNAse free water. Real time quantitative PCR was performed using a Power Syber green PCR master Mix (Fisher Thermo Scientific) on a Light Cycler 480 (Roche). The primers used for qPCR were the E_Sarbeco primers for viral RNA quantification (Corman et al., 2020), and Quantitect primers for GAPDH were purchased from Qiagen. The relative expression quantification was performed based on the comparative threshold cycle (CT) method, using GAPDH as endogenous reference control.
Viral RNA immunoprecipitation (vRIP)
The human EWSR1 coding DNA sequence (CDS) was amplified by PCR from MGC Human EWSR1 cDNA plasmid (Horizon Discovery) using the EWSR1-HA forward and reverse primers. Amplification product was cloned into pLVX-EF1alpha-IRES-Puro vector using EcoRI/NotI restriction enzymes. Plasmids expressing FLAG-tagged version of HNRNPA2B1, ILF3, QKI, SFPQ, HSPB1, GFP and TRIM28 cloned in pUC57 vector were purchased from GenScript, and subcloned into pLVX-EF1alpha-IRES-Puro vector using EcoRI/NotI restriction enzymes, with the exception of SFPQ (EcoRI/BamHI). For A549-ACE2 transductions, lentiviral particles production and spinofection, were performed as described above for ACE2-expressing cells generation.
293T cells stably expressing ACE2 were seeded in 10 cm2 dishes and transfected with 20 μg of plasmid using the Lipofectamine 3000 reagent (Thermo Fisher Scientific) according to manufacturer’s instruction. 24 h post-transfection, cells were challenged with SARS-CoV-2 at a MOI of 0.1. Thirty hours post-infection, cells were lysed for 30 min at 4°C in IP lysis buffer (Thermo Fisher Scientific) supplemented with 1% Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific) and 0.5% SUPERase-in, then cleared by centrifugation for 30 min at 13,000 × g. For each condition, 1 mg of total proteins was transferred in a new tube in a 900 μL final volume for IP, from which 2 × 20 μL (2%) were kept as input for RNA extraction and immunoblot. Protein G magnetic beads (30 μL per conditions) were washed twice with 1 mL of IP Lysis buffer supplemented with 1% Triton and 0.01% SDS. Beads were resuspended in IP lysis buffer (100 μL per conditions) and incubated with anti-HA or anti-FLAG antibodies (3 μg per conditions) for one hour at room temperature under rotation. After 5 washes with IP lysis buffer, the beads were resuspended in IP lysis buffer (50 μL per conditions) supplemented with 1% Halt™ Protease and Phosphatase Inhibitor Cocktail and 0.5% SUPERase-in. One mg of total proteins was incubated 3 h at 4°C with 50 μL with the magnetic beads. Beads were washed five times with RIP buffer (KCl 150 mM, Tris-HCl 25 mM, EDTA 5mM, IPGAL 0.5%, 1% protease and phosphatase inhibitor and 0.5% SUPERase-in). Proteins were eluted twice (2 × 50 μL) with either HA (400 mg/mL) or 3XFLAG (200 mg/mL) peptides for 30 min at room temperature. Twenty microliters of the eluate were kept for the immunoblot, whilst the remaining eluate and 20 μL of the input were treated with proteinase K for 30 min at 50°C. Immunoprecipitated RNA was purified using Trizol (Qiagen) following the manufacturers recommendations.
For immunoblot, 20 μL of the inputs and eluates were resuspended in LDS Sample Buffer 4X (Thermo Fisher Scientific) containing 25 mM dithiothreitol (DTT) and were separated by SDS PAGE. Immunoblotting was performed as previously described (Hafirassou et al., 2017) with an anti-HA rabbit monoclonal antibody (Cell Signaling) or an anti-FLAG M2 mouse monoclonal antibody (Sigma).
For vRNA quantification, reverse transcription was performed on 2 μL of input and immunoprecipitated RNA as described above. Real time quantitative PCR was performed as described above with the E_Sarbeco primers. All our analyses were performed as previously described (Marmisolle et al., 2018) using the ΔΔCT method. Briefly, we normalized each RIP fractions’ Ct to the corresponding input fraction Ct average for the same qPCR assay (ΔCt [normalized RIP]) to account for sample preparation differences. Then we adjusted the normalized RIP fraction Ct value for the normalized WT Ct value (ΔΔCt = ΔCt [normalized RIP]- ΔCt [normalized HSPB1 negative control]). Finally, we performed a linear conversion of the ΔΔCt (2ˆ[-ΔΔCt]) to calculate the fold change over the HSPB1 protein binding to vRNA.
Gene editing and trans-complementation experiments
sgRNA targeting QKI were designed using the CRISPOR software (http://crispor.org). Sequences for all the sgRNAs are listed in the key resources table. The sgRNAs were cloned into the plasmid lentiCRISPR v2 (Addgene) according to the recommendations of members of the Zhang laboratory. A549-ACE2 gene-edited cells were generated as previously described (Meertens et al., 2019). Gene-edited cells were transduced with the FLAG-GFP and FLAG-QKI cDNA cloned in the lentiviral vector pLVX-EF1alpha-IRES-Puro.
Cell viability assay
Cell viability and proliferation were assessed using the CellTiter-Glo 2.0 Assay (Promega). Briefly, cells were plated in 96-well plates (5 × 103). At specific times, 100 μL of CellTiter-Glo reagent mixed v/v with PBS were added to each well. After 10 min incubation, 90 μL from each well were transferred to an opaque 96-well plate (Cellstar, Greiner bio-one) and luminescence was measured on a TriStar2 LB 942 (Berthold) with 0.1 s integration time.
Quantification and statistical analysis
Statistical analyses
Graphical representation and statistical analyses were performed using Prism7 software (GraphPad Software). Unless otherwise stated, results are shown as means +/− standard deviation (SD) from at least 2 independent experiments in duplicates. Differences were tested for statistical significance using One-way or two-way ANOVA with multiple comparisons post-test.
Protein functional enrichment was performed in Cytoscape (Version. 3.8.2). Gene Ontology (GO) Biological process were enriched with the ClueGO (Bindea et al., 2009) extension (Version. 2.5.7) using a two-sided hypergeometric test with Bonferroni correction. Adjusted p-values < 0.5 were considered as significant. p-values and percentage of represented genes per GO terms were computed to select the most enriched functional families. This was illustrated with ggplot2 package (Version. 3.3.2) in R Studio interface (Version. 1.2.1335). Most representative terms were represented in networks using the Cytoscape’s Genemania extension (Version. 3.5.2). Comparison of functional enrichment between our datasets and previous studies was performed in Metascape (Zhou et al., 2019b). GO Biological process with p-value < 0.01, enrichment score >1.5 and overlap = 4 were selected.
Acknowledgments
This work was supported by the French government's Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (grant no. ANR-10-LABX-62-IBEID), the Fondation pour la Recherche Médicale (grant no. FRM-EQU202003010193), the Agence Nationale de la Recherche (ANR/FRM Flash COVID project IDISCOVR and ANR-20-CO11-0004 project FISHBP), and University Paris Cité (Plan de Soutien Covid-19: RACPL20FIR01-COVID-SOUL). The authors thank Alessia Zamborlini for critical reading of the manuscript. L.M. and A.A. dedicate this work to the memory of Professor Jean-Louis Virelizier (Unité d’Immunologie Virale, Institut Pasteur, Paris) and Professor Renaud Mahieux (Ecole Normale Supérieure, Lyon, France), who left us during the SARS-CoV-2 epidemic.
Author contributions
L.M. and A.A. conceived of the project; L.M. supervised the project. L.M. performed the ChIRP experiments and analyzed the data with the help of M.P. and P.-O.V. A.L.-U., P.-O.V., and V.S. performed the bioinformatic analysis. L.B.-M. and L.M. produced the SARS-CoV-2 virus stocks used in this study. P.-O.V. and V.L. analyzed the viral protein-RBP protein interactions. A.L. and L.M. performed the siRNA screen and the vRIP assays. A.L.-U. screened the compounds. A.L.-U., L.F.-S., and L.M. performed the knockdown and KO experiments. L.M., P.-O.V., A.L.-U., and A.A. wrote the manuscript, with input of all of the authors. A.A. and L.M. secured the funding for the study.
Declaration of interests
The authors declare no competing interests.
Published: April 26, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110744.
Supplemental information
Data and code availability
-
•
The datasets of the mass spectrometry analyses are provided in supplemental files (Table S2). Additional Supplemental Items are available from Mendeley Data at https://data.mendeley.com/datasets/9pmrdxdpyg/1.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.
References
- Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339.e21. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J.L., Wächter K., Mühleck B., Pazaitis N., Köhn M., Lederer M., Hüttelmaier S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol. Life Sci. 2013;70:2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.-H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrita L.D., Cassaignau A.M.E., Launay H.M.M., Waudby C.A., Wlodarski T., Camilloni C., Karyadi M.-E., Robertson A.L., Wang X., Wentink A.S., et al. A structural ensemble of a ribosome–nascent chain complex during cotranslational protein folding. Nat. Struct. Mol. Biol. 2016;23:278–285. doi: 10.1038/nsmb.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Gazy I., Jackson L., Hwa S.-H., Tegally H., Lustig G., Giandhari J., Pillay S., Wilkinson E., Naidoo Y., et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese M., Lee J.-Y., Cerikan B., Neufeldt C.J., Oorschot V.M.J., Köhrer S., Hennies J., Schieber N.L., Ronchi P., Mizzon G., et al. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe. 2020;28:853–866.e5. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T.X., Wessels H.-H., Hoagland D.A., Kasela S., Legut M., Maniatis S., Mimitou E.P., Lu L., Geller E., et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2020;184:92–105.e16. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgil D., Polacek C., Harris E. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J. Virol. 2006;80:2976–2986. doi: 10.1128/JVI.80.6.2976-2986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymieux S., Rouillé Y., Terrier O., Seron K., Blanchard E., Rosa-Calatrava M., Dubuisson J., Belouzard S., Roingeard P. Ultrastructural modifications induced by SARS-CoV-2 in Vero cells: a kinetic analysis of viral factory formation, viral particle morphogenesis and virion release. Cell. Mol. Life Sci. 2021;78:3565–3576. doi: 10.1007/s00018-020-03745-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W. Making ends meet: a role of RNA ligase RTCB in unfolded protein response. EMBO J. 2014;33:2887–2889. doi: 10.15252/embj.201490425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fislová T., Thomas B., Graef K.M., Fodor E. Association of the influenza virus RNA polymerase subunit PB2 with the host chaperonin CCT. J. Virol. 2010;84:8691–8699. doi: 10.1128/JVI.00813-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn R.A., Belk J.A., Qi Y., Yasumoto Y., Wei J., Alfajaro M.M., Shi Q., Mumbach M.R., Limaye A., DeWeirdt P.C., et al. Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions. Cell. 2021;184:2394–2411.e16. doi: 10.1016/j.cell.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:296. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St. Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno M., Noerenberg M., Ni S., Järvelin A.I., González-Almela E., Lenz C.E., Bach-Pages M., Cox V., Avolio R., Davis T., et al. System-wide profiling of RNA-binding proteins uncovers key regulators of virus infection. Mol. Cell. 2019;74:196–211.e11. doi: 10.1016/j.molcel.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gélinas J.-F., Clerzius G., Shaw E., Gatignol A. Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA-activated protein kinase. J. Virol. 2011;85:8460–8466. doi: 10.1128/JVI.00240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuens T., Bouhy D., Timmerman V. The hnRNP family: insights into their role in health and disease. Hum. Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., Jureka A.S., Obernier K., Guo J.Z., Batra J., et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370:eabe9403. doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Ma J., Sun J., Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl. Acad. Sci. U S A. 2007;104:151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafirassou M.L., Meertens L., Umaña-Diaz C., Labeau A., Dejarnac O., Bonnet-Madin L., Kümmerer B.M., Delaugerre C., Roingeard P., Vidalain P.-O., et al. A global interactome map of the dengue virus NS1 identifies virus restriction and dependency host factors. Cell Rep. 2017;21:3900–3913. doi: 10.1016/j.celrep.2017.11.094. [DOI] [PubMed] [Google Scholar]
- Hoffmann H.-H., Sánchez-Rivera F.J., Schneider W.M., Luna J.M., Soto-Feliciano Y.M., Ashbrook A.W., Le Pen J., Leal A.A., Ricardo-Lax I., Michailidis E., et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2021;29:267–280.e5. doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Aizaki H., Hara H., Matsuda M., Ando T., Shimoji T., Murakami K., Masaki T., Shoji I., Homma S., et al. Chaperonin TRiC/CCT participates in replication of hepatitis C virus genome via interaction with the viral NS5B protein. Virology. 2011;410:38–47. doi: 10.1016/j.virol.2010.10.026. [DOI] [PubMed] [Google Scholar]
- Jiang H., Zhang H., Meng Q., Xie J., Li Y., Chen H., Zheng Y., Wang X., Qi H., Zhang J., et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel W., Noerenberg M., Cerikan B., Chen H., Järvelin A.I., Kammoun M., Lee J.Y., Shuai N., Garcia-Moreno M., Andrejeva A., et al. Global analysis of protein-RNA interactions in SARS-CoV-2-infected cells reveals key regulators of infection. Mol. Cell. 2021;81:2851–2867.e7. doi: 10.1016/j.molcel.2021.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeras-Bueno S., Jorba N., Pérez-Cidoncha M., Ortín J. The splicing factor proline-glutamine rich (SFPQ/PSF) is involved in influenza virus transcription. PLoS Pathog. 2011;7:e1002397. doi: 10.1371/journal.ppat.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee Y., Choi Y., Son A., Park Y., Lee K.-M., Kim J., Kim J.-S., Kim V.N. The SARS-CoV-2 RNA interactome. Mol. Cell. 2021;81:2838–2850.e6. doi: 10.1016/j.molcel.2021.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Ge L., Li P., Wang Y., Sun M., Huang L., Ishag H., Di D., Shen Z., Fan W., et al. The DEAD-box RNA helicase DDX5 acts as a positive regulator of Japanese encephalitis virus replication by binding to viral 3′ UTR. Antivir. Res. 2013;100:487–499. doi: 10.1016/j.antiviral.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Guo M., Tian X., Wang X., Yang X., Wu P., Liu C., Xiao Z., Qu Y., Yin Y., et al. Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med. 2020;12:99–112.e7. doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao K.-C., Chuo V., Fagg W.S., Bradrick S.S., Pompon J., Garcia-Blanco M.A. The RNA binding protein Quaking represses host interferon response by downregulating MAVS. RNA Biol. 2020;17:366–380. doi: 10.1080/15476286.2019.1703069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236.e13. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F.-W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the omicron variant of SARS-CoV-2. Nature. 2021;602:678–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Mangus D.A., Evans M.C., Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmisolle F.E., García M.L., Reyes C.A. RNA-binding protein immunoprecipitation as a tool to investigate plant miRNA processing interference by regulatory proteins of diverse origin. Plant Methods. 2018;14:9. doi: 10.1186/s13007-018-0276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meertens L., Hafirassou M.L., Couderc T., Bonnet-Madin L., Kril V., Kümmerer B.M., Labeau A., Brugier A., Simon-Loriere E., Burlaud-Gaillard J., et al. FHL1 is a major host factor for chikungunya virus infection. Nature. 2019;574:259–263. doi: 10.1038/s41586-019-1578-4. [DOI] [PubMed] [Google Scholar]
- Mo Q., Xu Z., Deng F., Wang H., Ning Y.-J. Host restriction of emerging high-pathogenic bunyaviruses via MOV10 by targeting viral nucleoprotein and blocking ribonucleoprotein assembly. PLoS Pathog. 2020;16:e1009129. doi: 10.1371/journal.ppat.1009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma T., Nakagawa S., Tanigawa A., Sasaki Y.F., Goshima N., Hirose T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Yamazaki T., Hirose T. Molecular dissection of nuclear paraspeckles: towards understanding the emerging world of the RNP milieu. Open Biol. 2018;8:180150. doi: 10.1098/rsob.180150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakland T.E., Haselton K.J., Randall G. EWSR1 binds the hepatitis C virus cis-acting replication element and is required for efficient viral replication. J. Virol. 2013;87:6625–6634. doi: 10.1128/JVI.01006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodi F., Bonnet-Madin L., Meertens L., Karpf L., Poirot J., Zhang S.-Y., Picard C., Puel A., Jouanguy E., Zhang Q., et al. SARS-CoV-2 induces human plasmacytoid predendritic cell diversification via UNC93B and IRAK4. bioRxiv. 2021 doi: 10.1101/2020.07.10.197343. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi Y.S., Majzoub K., Flynn R.A., Mata M.A., Diep J., Li J.K., van Buuren N., Rumachik N., Johnson A.G., Puschnik A.S., et al. An RNA-centric dissection of host complexes controlling flavivirus infection. Nat. Microbiol. 2019;4:2369–2382. doi: 10.1038/s41564-019-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Parra C., Ernlund A., Alard A., Ruggles K., Ueberheide B., Schneider R.J. A widespread alternate form of cap-dependent mRNA translation initiation. Nat. Commun. 2018;9:3068. doi: 10.1038/s41467-018-05539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin-Cocon L., Diaz O., Jacquemin C., Barthel V., Ogire E., Ramière C., André P., Lotteau V., Vidalain P.-O. The current landscape of coronavirus-host protein-protein interactions. J. Transl. Med. 2020;18:319. doi: 10.1186/s12967-020-02480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland G.A., Ovsyannikova I.G., Kennedy R.B. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396:1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- Schmidt N., Lareau C.A., Keshishian H., Ganskih S., Schneider C., Hennig T., Melanson R., Werner S., Wei Y., Zimmer M., et al. The SARS-CoV-2 RNA–protein interactome in infected human cells. Nat. Microbiol. 2020;6:339–353. doi: 10.1038/s41564-020-00846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Luna J.M., Hoffmann H.-H., Sánchez-Rivera F.J., Leal A.A., Ashbrook A.W., Le Pen J., Ricardo-Lax I., Michailidis E., Peace A., et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2020;184:120–132.e14. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.-A., Leibundgut M., Thiel V., Mühlemann O., Ban N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard P.J., Hertel K.J. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Decroly E., Ziebuhr J. In: Advances in Virus Research. Ziebuhr J., editor. Academic Press; 2016. Chapter three - the nonstructural proteins directing coronavirus RNA synthesis and processing; pp. 59–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Li P., Ju X., Rao J., Huang W., Ren L., Zhang S., Xiong T., Xu K., Zhou X., et al. In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs. Cell. 2021;184:1865–1883.e20. doi: 10.1016/j.cell.2021.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo G., Liu G., Zhang J., Nesvizhskii A.I., Gingras A.-C., Choi H. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics. 2014;100:37–43. doi: 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M.G., Dittmar M., Mallory M.J., Bhat P., Ferretti M.B., Fontoura B.M., Cherry S., Lynch K.W. Viral-induced alternative splicing of host genes promotes influenza replication. Elife. 2020;9:e55500. doi: 10.7554/eLife.55500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P., Gerber M., Kelly J., Pfaender S., Ebert N., Braga Lagache S., Simillion C., Portmann J., Stalder H., Gaschen V., et al. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. Elife. 2019;8:e42037. doi: 10.7554/eLife.42037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Sola I., Enjuanes L., Zuñiga S. MOV10 helicase interacts with coronavirus nucleocapsid protein and has antiviral activity. mBio. 2021;12:e0131621. doi: 10.1128/mBio.01316-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P., et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. 2021;184:106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson S.F., Bellora N., Macias S. ILF3 contributes to the establishment of the antiviral type I interferon program. Nucleic Acids Res. 2020;48:116–129. doi: 10.1093/nar/gkz1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Alfajaro M.M., DeWeirdt P.C., Hanna R.E., Lu-Culligan W.J., Cai W.L., Strine M.S., Zhang S.-M., Graziano V.R., Schmitz C.O., et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell. 2020;184:76–91.e13. doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Snijder E.J., Kikkert M., van Hemert M.J. Host factors in coronavirus replication. Curr. Top Microbiol. Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Mathieu C., Kolaitis R.-M., Zhang P., Messing J., Yurtsever U., Yang Z., Wu J., Li Y., Pan Q., et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell. 2020;181:325–345.e28. doi: 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Wu F., Han J., Qi F., Ni T., Qian F. Exploitation of nuclear protein SFPQ by the encephalomyocarditis virus to facilitate its replication. Biochem. Biophys. Res. Commun. 2019;510:65–71. doi: 10.1016/j.bbrc.2019.01.032. [DOI] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The datasets of the mass spectrometry analyses are provided in supplemental files (Table S2). Additional Supplemental Items are available from Mendeley Data at https://data.mendeley.com/datasets/9pmrdxdpyg/1.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.