Abstract
To investigate amoxicillin and metronidazole resistance of Helicobacter pylori, we compared putative resistance genes between resistant strains obtained in vitro and their sensitive parent strain. All metronidazole-resistant strains had rdxA mutations, and an amoxicillin-resistant strain had pbp1 and pbp2 mutations. By transforming PCR products of these mutated genes into antibiotic-sensitive strains, we showed that rdxA null mutations were sufficient for metronidazole resistance, while pbp1 mutations contributed to amoxicillin resistance of H. pylori.
Although most infections with Helicobacter pylori are asymptomatic, and some might even be beneficial for the host, the pathogen is usually eradicated with antibiotics such as amoxicillin and metronidazole in order to cure gastritis and peptic ulcer diseases (3, 4). Resistance against metronidazole is common among H. pylori strains, while there are only a few reports of amoxicillin-resistant H. pylori strains (6, 8; M. Guslandi, Letter, Lancet 353:241–242, 1999; A. A. van Zwet, C. M. Vandenbroucke-Grauls, J. C. E. Thijs, J. van der Wouden, M. M. Gerrits, J. G. Kusters, and C. M. Vandenbrouke-Grauls, Letter, Lancet 352:1595, 1998).
Metronidazole resistance of H. pylori was shown to be due to the mutational inactivation of rdxA (7, 11). In turn, a metronidazole-resistant strain (Mtzr) was rendered sensitive when complemented with the rdxA gene of a metronidazole-sensitive strain (Mtzs). It was concluded that RdxA functions as a metronidazole-reducing nitroreductase (11). Resistance against β-lactam antibiotics like amoxicillin is generally due to hydrolysis by a β-lactamase (5) or by mutational modification of the penicillin binding proteins (13).
To investigate the molecular basis for antibiotic resistance in H. pylori, we did an in vitro selection for amoxicillin and metronidazole resistance on the strains 503 (ATCC 43503) and 69A (69A and 888–0: clinical isolates obtained from R. Haas, Max-von Pettenkofer-Institut, Munich, Germany). In contrast to other investigators (12, 15), we performed the selection not on agar plates, but in liquid culture (brain heart infusion medium [BHI; Difco-BD Biosciences, Md.] plus 5% fetal calf serum [FCS; Eurobio, Les Ulis, France]) at 37°C with microaerobic incubation (5 to 6% O2, 8 to 10% CO2) in the presence of these antibiotics. The MIC was determined by inoculating logarithmically growing H. pylori cells with an optical density at 578 nm (OD578) of 0.04 in 10 ml of BHI–5% FCS in 50-ml cell culture flasks (Greiner) on a shaker incubator at 90 rpm. The MIC of metronidazole was defined as no OD578 increase in 10 to 14 days, and that of amoxicillin was defined as growth to an OD578 lower than 0.4, with no increase after the first 24 h of incubation. We obtained stable metronidazole resistance after three serial passages over the course of 8 to 10 days with increasing metronidazole concentration in the growth medium (from 2 μg/ml to 25 μg/ml). The metronidazole MIC for nine independently selected 69A/Mtzr and 503/Mtzr strains was >25 μg/ml, while that for strains 503 and 69A was <5 μg/ml (data not shown). These results correspond to the observation of metronidazole resistance developing de novo during the course of a typical antibiotic therapy (1). In vitro selection of amoxicillin-resistant H. pylori strains was done similarly. Since the amoxicillin MIC for strains 503 and 69A was 0.02 to 0.05 μg/ml, we began selection at an amoxicillin concentration of 0.01 μg/ml. After 11 passages and 35 days under the permanent selective pressure of amoxicillin, the amoxicillin MIC for strain 69A reached 0.5 to 1 μg/ml, and after 35 passages and 89 days, an amoxicillin MIC of 15 μg/ml for the highly resistant strain 69A/Amxr was obtained. Comparable selection results were obtained for the strain 503 (data not shown). In contrast to other Amxr strains described in the literature (9), the resistance was stable after cultivation in the absence of antibiotics and storage at −80°C. The induction of resistance was specific for the antibiotic used for selection. Amoxicillin-resistant strains remained sensitive to metronidazole and vice versa.
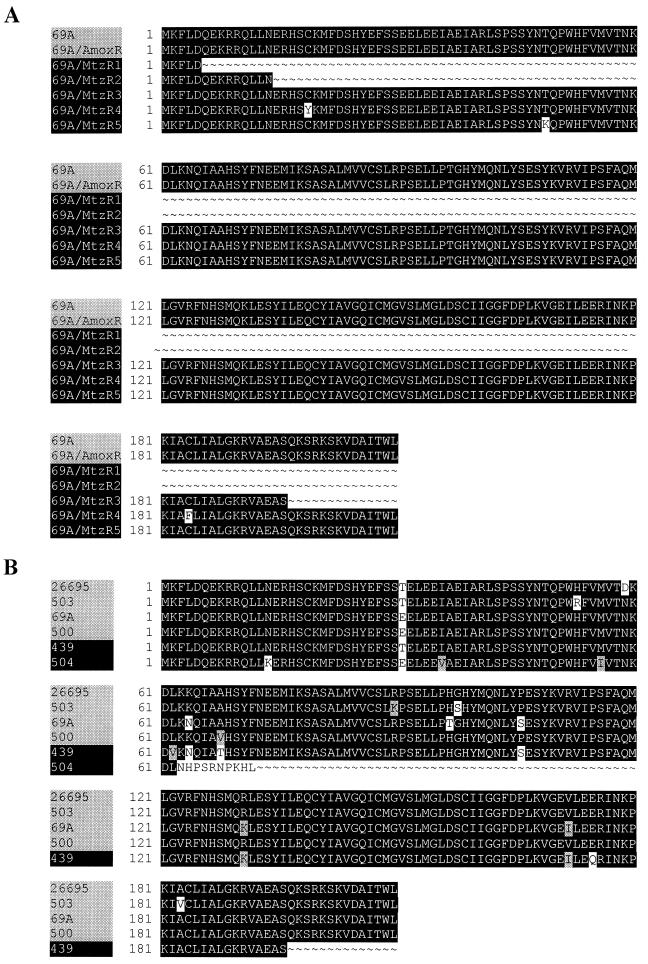
To investigate the molecular mechanisms for resistance, we sequenced putative resistance genes from 69A strains: five independently selected 69A/Mtzr strains and one 69A/Amxr strain. The rdxA gene of each 69A/Mtzr strain had at least one mutation compared to the copy of the wild-type 69A strain (Fig. 1A). In three cases, the mutations caused stop codons in the rdxA open reading frame, which is also common for metronidazole-resistant clinical isolates (Fig. 1B). In the remaining two strains, the mutations caused either one or two amino acid changes at positions conserved in metronidazole-sensitive strains (Fig. 1B). The 69A/Amxr strain had no rdxA mutation (Fig. 1A), but had four pbp1 mutations (S414R, Y484C, T541I, and P600T) and one pbp2 mutation (T498I). All of these mutations cause amino acid changes at positions conserved in the Amxs strains 26695 (16), J99 (2), and 69A (GenBank accession no. AF315503 and AF315504) and were located in the putative transpeptidase domains of the proteins (10).
FIG. 1.
Alignment of RdxA proteins from H. pylori 69A strains selected for antibiotic resistance (A) and those of H. pylori wild-type strains (B). (A) The strains 69A and 69A/Amxr were metronidazole sensitive, and the 69A/Mtzr1 to -5 strains were metronidazole resistant. (B) H. pylori strains 26695, 503, 69A, and 500 were metronidazole sensitive, and H. pylori strains 439 and 504 (ATCC 43504) were metronidazole resistant. A black background shows the conservation of amino acid residues. H. pylori amino acid sequences are given as follows: 26695, reference 16; strains 500 and 439, reference 11; strain 503, GenBank accession no. AF316109; strain 504, accession no. AF315501; and strain 69A, accession no. AF315502.
To prove that these mutations were indeed responsible for antibiotic resistance, we amplified these genes by PCR, purified the DNA by gel electrophoresis, and transformed it into antibiotic-sensitive strains. To do this, we used a simplified protocol for transformation of H. pylori, in which we added linear PCR fragments without an additional resistance marker to logarithmically growing H. pylori and then selected for transformants with amoxicillin or metronidazole, respectively. After transformation with the mutated rdxA genes from four 69A/Mtzr strains, bacteria of three metronidazole-sensitive H. pylori strains (26695, 69A, and 888–0) were rendered resistant (Table 1). Transformation with the mutated pbp1 gene from the 69A/Amxr strain rendered bacteria of these strains (26695, 69A, and 888–0) moderately amoxicillin resistant (MIC of 0.5 to 1 μg/ml). Transformation with the pbp2 gene from 69A/Amxr caused no amoxicillin resistance. The cotransformation of pbp1 and pbp2 did not show increased resistance compared to transformation with pbp1 alone (Table 1).
TABLE 1.
Transformation of mutated genes into antibiotic-sensitive H. pylori with metronidazole or amoxicillin selection mediuma
| H. pylori strain | Metronidazole concn (μg/ml) | Result for transformed geneb
|
Amoxicillin concn (μg/ml) | Result for transformed geneb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rdxA/Mtzr1 | rdxA/Mtzr3 | rdxA/Mtzr4 | rdxA/Mtzr5 | Control (no DNA added) | pbp1 | pbp2 | pbp1 + pbp2 | Control (no DNA added) | |||
| 69A | 5 | + | + | + | + | − | 0.2 | + | − | + | − |
| 69A | 25 | + | + | + | + | − | 0.5 | + | ND | + | − |
| 26695 | 5 | + | + | + | + | − | 0.2 | + | − | + | − |
| 26695 | 25 | + | + | + | + | − | 0.5 | + | ND | + | − |
| 888-0 | 5 | + | + | + | + | − | 0.2 | + | − | + | − |
| 888-0 | 25 | + | + | + | + | − | 0.5 | + | ND | + | − |
For transformation with mutated rdxA genes, the rdxA genes of the indicated 69A/Mtzr strains were amplified by PCR with primers 5′rdxA1 (ATGGGTTGCTGATTGTGGTTTATGG) and 3′rdxA2 (GCTTGAAAACACCCCTAAAAGAGCG) and purified by gel electrophoresis. For transformation, the DNA (250 to 500 ng) was added to logarithmically growing metronidazole-sensitive H. pylori strains. After 16 h of incubation, the bacteria were diluted in medium containing metronidazole to select for transformants. For transformation with mutated pbp1 and pbp2 genes, the mutated pbp genes of the H. pylori 69A/Amxr strain were amplified by PCR with 5′pbp1-1 primers (AATCAAGCGGTGAGTATCCTTGTGG), 3′pbp1-2 (CTACGGTTTCTAAACCCCTTTTACG), 5′pbp2-1 (GTTATAAGCGGTGGAATGAGTTGG), and 3′pbp2-2 (TGACGGCTTTTTATTCAAAACTTTGC), purified by gel electrophoresis, and transformed into logarithmically growing amoxicillin-sensitive H. pylori strains. Transformants were selected for by dilution in medium containing amoxicillin.
+, growth of bacteria after 3 to 5 days of incubation; −, no growth after 14 days of incubation; ND, not determined.
To exclude the possibility that antibiotic resistance after transformation was due to a different spontaneous mutation, we sequenced the rdxA genes from six transformed metronidazole-resistant strains and the pbp1 genes from two transformed amoxicillin-resistant strains. In all but one case, we found the same mutations as in the respective donor strain (Table 2). The only exception was observed for transformation with the rdxA gene of 69A/Mtzr4: The rdxA gene from 69A/Mtzr4 contained two mutations (C19Y and C184F), while H. pylori transformed with this gene acquired only the C19Y mutation (Table 2). We therefore do not know if the C184F mutation is involved in metronidazole resistance.
TABLE 2.
Sequences of rdxA and pbp1 genes from antibiotic-resistant H. pylori strains gained by transformation with mutated rdxA and pbp1 genes
| Donor strain | Gene | Amino acid change(s) in donor strain | Acceptor strain | Amino acid change(s) in acceptor strain |
|---|---|---|---|---|
| 69A/Mtzr1 | rdxA | 6 Q → stop | 69A | 6 Q → stop |
| 69A/Mtzr3 | rdxA | 197 Q → stop | 69A | 197 Q → stop |
| 69A/Mtzr4 | rdxA | 19 C → Y, 184C → F | 69A | 19 C → Y |
| 69A/Mtzr5 | rdxA | 49 T → K | 69A | 49 T → K |
| 69A/Mtzr4 | rdxA | 19 C → Y, 184C → F | 26695 | 19 C → Y |
| 69A/Mtzr5 | rdxA | 49 T → K | 26695 | 49 T → K |
| 69A/Amxr | pbp1 | 414 S → R, 484 Y → C, 541 T → I, 600 P → T | 69A | 484 Y → C, 541 T → I, 600 P → Ta |
| 69A/Amxr | pbp1 | 414 S → R, 484 Y → C, 541 T → I, 600 P → T | 26695 | 484 Y → C, 541 T → I, 600 P → Ta |
Codon 414 of pbp1 was not sequenced in these strains.
Our findings independently confirm previous results (7, 11, 14) and expand their data in the sense that not only stop codons and extensive deletions in the rdxA open reading frame cause metronidazole resistance, but so do single amino acid changes. We have proven that two alterations of the RdxA protein, C19Y and T49K, have the same effect on the phenotype of H. pylori (Mtzs→Mtzr) as rdxA null mutations and conclude that they severely affect RdxA function. The same is true for deletion of the C-terminal 14 amino acids of RdxA. By systematically transforming rdxA genes with single mutations into metronidazole-sensitive H. pylori strains by the simplified protocol, we have illustrated how it would be possible to identify additional residues essential for RdxA function. We have also shown that pbp1 mutations can affect amoxicillin resistance, but are not sufficient for the high-level amoxicillin resistance of 69A/Amxr. This indicates that mutations in more than one gene are probably required to render H. pylori amoxicillin resistant. This could explain the many cycles necessary for the in vitro selection of amoxicillin-resistant H. pylori strains and their low occurrence in vivo.
Acknowledgments
The work described here was supported by a grant from the German Ministry of Education and Research (BMBF; BEO/22 031 0777).
We thank Klaus Bensch and Wolfram Steinhilber for discussions.
REFERENCES
- 1.Adamek R J, Suerbaum S, Pfaffenbach B, Opferkuch W. Primary and acquired Helicobacter pylori resistance to clarithromycin, metronidazole, and amoxicillin—influence on treatment outcome. Am J Gastroenterol. 1998;93:386–389. doi: 10.1111/j.1572-0241.1998.00386.x. [DOI] [PubMed] [Google Scholar]
- 2.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 3.Blaser M J. Not all Helicobacter pylori strains are created equal: should all be eliminated? Lancet. 1997;349:1020–1022. doi: 10.1016/S0140-6736(96)09133-7. [DOI] [PubMed] [Google Scholar]
- 4.Blaser M J. Helicobacter pylori and gastric diseases. Br Med J. 1998;316:1507–1510. doi: 10.1136/bmj.316.7143.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 6.Debets-Ossenkopp Y J, Herscheid A J, Pot R G, Kuipers E J, Kusters J G, Vandenbroucke-Grauls C M. Prevalence of Helicobacter pylori resistance to metronidazole, clarithromycin, amoxicillin, tetracycline and trovafloxacin in The Netherlands. J Antimicrob Chemother. 1999;43:511–515. doi: 10.1093/jac/43.4.511. [DOI] [PubMed] [Google Scholar]
- 7.Debets-Ossenkopp Y J, Pot R G J, van Westerloo D J, Goodwin A, Vandenbroucke-Grauls C M J E, Berg D E, Hoffman P S, Kusters J G. Insertion of mini-IS605 and deletion of adjacent sequences in the nitroreductase (rdxA) gene cause metronidazole resistance in Helicobacter pylori NCTC11637. Antimicrob Agents Chemother. 1999;43:2657–2662. doi: 10.1128/aac.43.11.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dore P M, Piana A, Carta M, Atzei A, Are B M, Mura I, et al. Amoxicillin resistance is one reason for failure of amoxicillin-omeprazole treatment of Helicobacter pylori infection. Aliment Pharmacol Ther. 1998;12:635–639. doi: 10.1046/j.1365-2036.1998.00350.x. [DOI] [PubMed] [Google Scholar]
- 9.Dore P M, Graham D Y, Sepulveda A R. Different penicillin-binding protein profiles in amoxicillin-resistant Helicobacter pylori. Helicobacter. 1999;4:154–161. doi: 10.1046/j.1523-5378.1999.99310.x. [DOI] [PubMed] [Google Scholar]
- 10.Goffin C, Ghuysen J M. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten S J, Berg D E, Hoffman P S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 12.Sörberg M, Hanberger H, Nilsson M, Björkman A, Nilsson L E. Risk of development of in vitro resistance to amoxicillin, clarithromycin, and metronidazole in Helicobacter pylori. Antimicrob Agents Chemother. 1998;42:1222–1228. doi: 10.1128/aac.42.5.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spratt B G. Resistance to antibiotics mediated by target alterations. Science. 1994;264:388–393. doi: 10.1126/science.8153626. [DOI] [PubMed] [Google Scholar]
- 14.Tankovic J, Lamarque D, Delchier J-C, Soussy C-J, Labigne A, Jenks P J. Frequent association between alteration of the rdxA gene and metronidazole resistance in French and North African isolates of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44:608–613. doi: 10.1128/aac.44.3.608-613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenney J H, Maack R W, Chippendale G R. Rapid selection of organisms with increasing resistance on subinhibitory concentrations of norfloxacin in agar. Antimicrob Agents Chemother. 1983;23:188–189. doi: 10.1128/aac.23.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]