INTRODUCTION
Surveillance of postoperative complications, including surgical site infections (SSI), can provide healthcare systems with reliable data to support continuous quality improvement (CQI) efforts.1, 2 For example, these data can allow healthcare systems to identify targets for CQI and conduct follow-up studies to determine the efficacy of interventions.3, 4 Several nationwide surveillance systems have been developed focusing on postoperative complications such as the American College of Surgeons National Surgical Quality Improvement Program (NSQIP) and the Centers for Disease Control National Healthcare Safety Network.1, 5, 6 While these programs provide high-quality clinical data to drive CQI efforts, both programs rely on manual chart abstraction to extract key clinical variables from the medical record limiting the scalability and generalizability across large healthcare systems.7
In 2008, the Health Information Technology for Economic and Clinical Health Act led to the rapid adoption of Electronic Healthcare Records (EHR) in US hospitals.8 By 2017 over 90% of hospitals had implemented an EHR most with advanced capabilities such as computerized provider order entry (CPOE), clinical decision support, and electronic provider documentation.9 The availability of electronic healthcare data in combination with artificial intelligence approaches, such as machine learning and natural language processing (NLP), has created opportunities to develop automated tools to support CQI efforts.10, 11
To demonstrate the value of electronic healthcare data in the surveillance of postoperative complications, Bucher, et. al. developed a machine learning approach using postoperative CPOE events for the detection of SSIs. Their approach demonstrated a sensitivity ranging from 62% to 88% and a specificity ranging from 72% to 92% compared to manual chart review for identification of SSIs in a single academic healthcare system.12 The use of structured EHR data (table-based data like CPOE events, laboratory results, vital signs, etc.) is inherently limited for surveillance of SSIs. These events are typically noted as a clinical diagnosis primarily documented in clinical unstructured text notes.13 To address this limitation, FitzHenry and Murff developed an NLP approach for the identification of SSIs in the VA Healthcare System. They published a sensitivity of 77% and a specificity of 63% compared to the VA Surgical Quality Improvement Program manual chart review.14, 15 While their study was the first to demonstrate the utility of NLP for surveillance of postoperative complications, the performance of their system is inherently limited. The methodology focused on a defined list of concepts in clinical text (e.g. “purulent”) and lacked a demonstration of generalizability outside the VA healthcare system.
In this paper, we address these limitations by presenting the development and validation of a portable NLP system capable of automated surveillance of SSI. We hypothesized that an NLP approach for surveillance of SSIs can be developed with high sensitivity and specificity, and the NLP system can be implemented across separate independent healthcare systems using different EHRs.
METHODS
Setting
The study population was drawn from two independent healthcare systems located in Utah that service the intermountain west: the University of Utah Health and Intermountain Healthcare. The University of Utah has maintained an EHR serviced by Epic since September 1, 2015. Intermountain Healthcare has maintained a separate EHR, Help2, locally serviced since 1996. The institutional review boards at each health care system approved the study, granting a waiver of informed consent for the use of patient healthcare data.
Participants
Patients were included in the study if their operative course underwent review by NSQIP-trained surgical clinical reviewers (SCR). The NSQIP methodology has been described previously.16 NSQIP SCRs select a random stratified set of patients undergoing surgical procedures and review all records for postoperative complications occurring within 30 days of the operative procedure.7 If a patient’s postoperative documentation is not complete in the EHR, attempts are made to contact the patient to complete 30-day follow-up. We included patients who were treated at one health care system from September 1, 2015 through August 30, 2017 as a training cohort and patients treated from September 1, 2017 through December 31, 2018 as an internal validation cohort. Patients who were treated at the other healthcare system from September 1, 2008 through June 30, 2017 were included as an external validation cohort.
Data Sources
We extracted the NSQIP reviewed cases for patient demographics, procedure characteristics, and outcomes for each cohort. The enterprise data warehouse at each healthcare system was queried using the NSQIP case identifiers for all electronic clinical text notes such as history and physical, operative reports, progress notes, nursing notes, radiology reports, and discharge summaries from the date of surgery to 30 days after the operative date. All patients who underwent NSQIP SCR review were included in the present study even if the complete documentation was not present for the entire 30-day postoperative period.
Reference Standard
We used the NSQIP SSI definition as the reference standard for the study. As the NSQIP definitions for SSI have changed over time, we used the NSQIP definitions of SSI from the 2017 Operations Manual.17 Each SSI was subsequently characterized as Superficial, Deep, or Organ Space according to standard NSQIP definitions. Each SSI was also characterized if an infection was present at the time of surgery (PATOS).
NLP Development
The description of our NLP architecture EasyCIE has been previously reported.18, 19 EasyCIE is a lightweight, rules-based NLP tool that supports quick and easy implementation of clinical information extractions. EasyCIE uses a set of highly optimized NLP components built on top of n-Trie20 (a fast rule processing engine), that includes a section detector, a sentence segmentater21, a named entity recognizer, a context detector22, a feature inferencer, a document inferencer23, and a patient inferencer.
EasyCIE builds a knowledge base during the training process with information extraction models (IEM) including a term mapping component, essentially a semantic representation of target concepts (“purulent”, “incision”, “abscess”, “Piperacillin,” etc.) and the corresponding contextual modifiers (i.e., affirmation, negation, temporality, anatomy, status of the infection, status of healing, purpose of treatment, and status of wound closure). Easy-CIE aggregates each IEM-term identified in a document in a rules-based manner to infer a document level classification.23 Subsequently, Easy-CIE aggregates documents along a temporal timeline to infer a patient-level classification.
NLP Rule Development
The NLP rules were developed initially using the NSQIP definitions for SSI as defined in the 2017 Operations Manual and Appendix.17, 18 We subsequently enriched the rules for synonyms using terms from the Unified Medical Language System (UMLS) from the National Library of Medicine.24 Using these rules, EasyCIE infers either SSI present or absent for each operative episode. To accomplish this inference, EasyCIE utilizes the operative report for each procedure to infer whether an SSI was PATOS, if the wound was closed or left open, and/or if the PATOS infection was healed within 30 days after surgery. We reviewed the rules manually iterating over the training cohort until all addressable errors were resolved. The final software package, EasyCIE-SSI, including the source code and SSI knowledgebase can be found at https://github.com/jianlins/EasyCIE_GUI.
Validation
After training was finalized, the performance of EasyCIE was evaluated blindly on both the internal and external validation cohorts without any rule modification.
Statistical Analysis
The analysis was performed using R v3.6 Statistical Software Package. We performed univariate analysis on the training, internal validation, and external validation cohort using the Pearson Chi-square test for discrete variables, and Student’s T-Test for continuous variables. The Bonferroni method was used to adjust for multiple comparisons on the univariate analysis.25 The performance of EasyCIE-SSI was evaluated for sensitivity, specificity, areas under the receiver operating curve (AUC), positive predictive value, negative predictive value, positive likelihood ratio, and negative likelihood ratio.26–28 A true positive occurred if EasyCIE-SSI inferred an SSI was present for the operative episode and the NSQIP SCR concluded any type of SSI was present, regardless of SSI depth. We used bootstrapping with 2000 iterations to obtain 95% confidence intervals for each performance metric.29 To evaluate the difference between the performance of EasyCIE-SSI between the internal and external validation cohorts, the Test of Two Proportions was used.30 We also performed an analysis of procedure subgroup characteristics including procedure types, wound classification, inpatient/outpatient status, and emergent procedures. A p-value of less than 0.05 was considered statistically significant.
RESULTS
A total of 21,784 operative events were included in the study and were divided into a training cohort (4574 events), an internal validation cohort (1850 events), and an external validation cohort (15360 events). (Table 1) In the internal validation cohort, compared to the training cohort, there were small significant differences in the rate of patients undergoing appendectomy (17% vs. 12%, p<0.001) and contaminated procedures (20% vs. 15%, p<0.001). In the external validation cohort, compared to the training cohort, there were small significantly higher rates of white patients (92% vs. 91%, p<0.001); and lower rates of patients with independent functional status (98% vs. 99%, p<0.001), steroid use (4% vs. 6%, p<0.001), and open wounds (3% vs. 6%, p<0.001). The external validation cohort also had significantly higher rates of appendectomy (17% vs. 14%, p<0.001) and vascular procedures (13% vs. 11%, p<0.001); and significantly lower rates of breast (9% vs. 11%, p<0.001), esophagus (3% vs 4%, p<0.001), and stomach (1% vs. 4%, p<0.001) procedures compared to the training cohort. In addition, in the external validation cohort, there were higher rates of emergent procedures (21% vs. 4%, p<0.001) and shorter operative duration (mean (SD): 99 (94) min vs. 120 (100) min, p<0.001).
TABLE 1.
Patient and Procedure Characteristics and Outcomes for Patients in Training, Internal Validation, and External Validation Cohorts.
| Training | Internal Validation | External Validation | |
|---|---|---|---|
| (n=4574) | (n=1850) | (n=15360) | |
| Patient Characteristics | |||
| Age (Mean Years, SD) | 53 (17) | 53 (17) | 53 (18) |
| Sex (male, %) | 2248 (49%) | 923 (50%) | 7536 (49%) |
| Race (%) | |||
| White | 4181 (91%) | 1679 (91%) | 14143 (92%) |
| Black or African American | 48 (1%) | 23 (1%) | 126 (1%) |
| Asian | 69 (2%) | 31 (2%) | 191 (1%) |
| Native Hawaiian or Other Pacific Islander | 30 (1%) | 12 (1%) | 147 (1%) |
| American Indian or Alaska Native | 64 (1%) | 28 (2%) | 59 (0%) |
| Hispanic Ethnicity (%) | 455 (10%) | 173 (9%) | 1382 (9%) |
| BMI (Mean, SD) | 29 (7.3) | 29 (6.7) | 29 (7.1) |
| Independent Functional Health Status (%) | 4518 (99%) | 1832 (99%) | 14986 (98%)* |
| Congestive Heart Failure (%) | 21 (0%) | 4 (0%) | 123 (1%) |
| Hypertension (%) | 1533 (34%) | 597 (32%) | 5421 (35%) |
| COPD (%) | 114 (2%) | 44 (2%) | 410 (3%) |
| Current Smoker (%) | 660 (14%) | 237 (13%) | 2236 (15%) |
| Dialysis (%) | 93 (2%) | 26 (1%) | 280 (2%) |
| Diabetes Mellitus (%) | 630 (14%) | 253 (14%) | 2123 (14%) |
| Steroid Use (%) | 284 (6%) | 139 (8%) | 639 (4%)* |
| Weight Loss (%) | 122 (3%) | 31 (2%) | 371 (2%) |
| Open Wound (%) | 262 (6%) | 101 (5%) | 506 (3%)* |
| Procedure Characteristics | |||
| Procedure Type (%) | |||
| Appendectomy | 536 (12%) | 314 (17%)* | 2583 (17%)* |
| Breast | 518 (11%) | 182 (10%) | 1366 (9%)* |
| Colon/Rectal | 826 (18%) | 394 (21%) | 2533 (16%) |
| Esophagus | 184 (4%) | 66 (4%) | 390 (3%)* |
| General Abdominal | 138 (3%) | 30 (2%) | 516 (3%) |
| Hepatobiliary | 448 (10%) | 184 (10%) | 1545 (10%) |
| Hernia | 806 (18%) | 327 (18%) | 3184 (21%) |
| Skin/Subcutaneous | 413 (9%) | 135 (7%) | 987 (6%)* |
| Stomach | 204 (4%) | 31 (2%)* | 205 (1%)* |
| Vascular | 499 (11%) | 186 (10%) | 2037 (13%)* |
| Outpatient Procedure (%) | 2378 (52%) | 993 (54%) | 7578 (49%) |
| Emergent Procedure (%) | 188 (4%) | 45 (2%) | 3224 (21%)* |
| Wound Classification (%) | |||
| Clean | 2349 (51%) | 873 (47%) | 7874 (51%) |
| Clean/Contaminated | 1143 (25%) | 425 (23%) | 3861 (25%) |
| Contaminated | 676 (15%) | 376 (20%)* | 2402 (16%) |
| Dirty/Infected | 406 (9%) | 176 (10%) | 1223 (8%) |
| Operative Duration (minutes, SD) | 120 (100) | 120 (110) | 99 (94)* |
| Hospital Length of Stay (days, SD) | 3.4 (9.5) | 3.1 (5.6) | 3.2 (6.2) |
| Outcomes | |||
| Superficial SSI (%) | 97 (2%) | 24 (1%) | 389 (3%) |
| Deep SSI (%) | 14 (0%) | 4 (0%) | 94 (1%) |
| Organ/Space SSI (%) | 155 (3%) | 47 (3%) | 252 (2%)* |
| SSI Present at the Time of Surgery (%) | 40 (1%) | 19 (1%) | 66 (0%) |
| Any SSI | 255 (6%) | 72 (4%) | 721 (5%) |
p<0.001 compared to Training Cohort; SD: Standard Deviation; BMI: Body Mass Index; COPD: Chronic Obstructive Pulmonary Disease
Within each cohort, the incidence of any SSI was 6% (training), 4% (internal validation), and 5% (external validation). (Table 1) There was a significantly lower rate of Organ/Space SSI in the external validation cohort compared to the training cohort (2% vs 3%, p<0.001). There were no significant differences in the incidence of any other SSI or SSI subtype between the training and internal or external validation cohorts.
The performance of EasyCIE-SSI for the detection of an SSI is shown in Table 2. For detection of any SSI, external validation (compared to internal validation) had a lower sensitivity (0.79 vs. 0.94, p=0.002), higher specificity (0.92 vs 0.88, p=0.01), and lower AUC (0.852 vs. 0.912, p<0.001). On subgroup analysis, for detection of Superficial SSI, there was a significantly lower sensitivity (0.71 vs. 0.92, p=0.047), higher specificity (0.90 vs 0.86, p=0.004), and lower AUC (0.805 vs 0.887, p<0.001) in the external validation cohort compared to the internal validation cohort. There was a significantly lower AUC for the detection of Deep SSI in external validation compared to internal validation (0.850 vs 0.925, p<0.001). There was no significant difference between internal and external validation in the performance of EasyCIE-SSI for the detection of Organ/Space SSI.
TABLE 2.
EasyCIE-SSI Performance in the Training, Internal and External Validation Cohorts
| Training | Internal Validation | External Validation | p-Value* | |
|---|---|---|---|---|
| Sensitivity | ||||
| Superficial SSI | 0.90 (0.82–0.95) | 0.92 (0.79–1.00) | 0.71 (0.66–0.75) | 0.047 |
| Deep SSI | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 0.81 (0.72–0.88) | 0.757 |
| Organ/Space SSI | 0.92 (0.88–0.96) | 0.96 (0.89–1.00) | 0.90 (0.86–0.94) | 0.334 |
| SSI PATOS | 1.00 (1.00–1.00) | 0.95 (0.84–1.00) | 0.88 (0.79–0.95) | 0.665 |
| Any SSI | 0.91 (0.88–0.95) | 0.94 (0.89–0.99) | 0.79 (0.75–0.82) | 0.002 |
| Specificity | ||||
| Superficial SSI | 0.86 (0.85–0.87) | 0.86 (0.84–0.87) | 0.90 (0.90–0.91) | 0.004 |
| Deep SSI | 0.85 (0.84–0.86) | 0.85 (0.83–0.87) | 0.89 (0.89–0.90) | 0.127 |
| Organ/Space SSI | 0.87 (0.86–0.88) | 0.87 (0.85–0.88) | 0.90 (0.90–0.90) | 0.443 |
| SSI PATOS | 0.85 (0.84–0.86) | 0.86 (0.84–0.87) | 0.89 (0.88–0.89) | 0.156 |
| Any SSI | 0.89 (0.88–0.90) | 0.88 (0.86–0.89) | 0.92 (0.92–0.92) | 0.012 |
| AUC | ||||
| Superficial SSI | 0.879 (0.848–00.91) | 0.887 (0.830–0.944) | 0.805 (0.782–0.827) | 0.008 |
| Deep SSI | 0.924 (0.919–00.93) | 0.925 (0.917–0.933) | 0.850 (0.810–0.890) | <0.0001 |
| Organ/Space SSI | 0.897 (0.876–00.92) | 0.913 (0.883–0.943) | 0.900 (0.882–0.919) | 0.484 |
| SSI PATOS | 0.926 (0.921–00.93) | 0.902 (0.849–0.954) | 0.884 (0.845–0.924) | 0.608 |
| Any SSI | 0.902 (0.884–00.92) | 0.912 (0.884–0.940) | 0.852 (0.837–0.868) | <0.0001 |
p-Value: comparison of EasyCIE-SSI performance on external validation cohort compared to internal validation cohort; SSI: Surgical Site Infection, PATOS: Present at the Time of Surgery, AUC: Area Under the Receiver Operating Curve
The value of a positive and negative EasyCIE-SSI result is shown in Table 3. Given the low prevalence of SSI, the positive predictive value of EasyCIE-SSI was low: 0.24 (95%CI: 0.22–0.27) for internal validation and 0.33 (95%CI: 0.31–0.34) for external validation. In external validation, compared to internal validation, there was a significant increase in both the positive likelihood ratio (PLR) (9.8 vs. 7.9, p<0.001) and negative likelihood ratio (NLR) (0.23 vs. 0.06, p<0.001).
TABLE 3.
Value of EasyCIE-SSI Information in the Training, Internal and External Validation Cohorts
| Training | Internal Validation | External Validation | |
|---|---|---|---|
| Positive Predictive Value | |||
| Superficial SSI | 0.12 (0.11–0.13) | 0.08 (0.07–0.09) | 0.16 (0.15–0.17) |
| Deep SSI | 0.02 (0.02–0.02) | 0.01 (0.01–0.02) | 0.04 (0.04–0.05) |
| Organ/Space SSI | 0.20 (0.19–0.22) | 0.16 (0.14–0.18) | 0.13 (0.12–0.14) |
| SSI PATOS | 0.06 (0.05–0.06) | 0.06 (0.05–0.07) | 0.03 (0.03–0.04) |
| Any SSI | 0.33 (0.31–0.35) | 0.24 (0.22–0.27) | 0.33 (0.31–0.34) |
| Negative Predictive Value | |||
| Superficial SSI | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 0.99 (0.99–0.99) |
| Deep SSI | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) |
| Organ/Space SSI | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) |
| SSI PATOS | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) |
| Any SSI | 0.99 (0.99–1.00) | 1.00 (0.99–1.00) | 0.99 (0.99–0.99) |
| Positive Likelihood Ratio | |||
| Superficial SSI | 6.51 (6.46–6.56) | 6.52 (6.45–6.60) | 7.27 (7.21–7.30) |
| Deep SSI | 6.58 (6.56–6.61) | 6.68 (6.65–6.72) | 7.43 (7.37–7.48) |
| Organ/Space SSI | 7.28 (7.22–7.35) | 7.38 (7.31–7.45) | 9.03 (8.98–9.08) |
| SSI PATOS | 6.85 (6.81–6.89) | 6.69 (6.63–6.76) | 8.00 (7.94–8.06) |
| Any SSI | 8.39 (8.32–8.46) | 7.91 (7.84–7.98) | 9.83 (9.74–9.84) |
| Negative Likelihood Ratio | |||
| Superficial SSI | 0.12 (0.12–0.13) | 0.10 (0.09–0.11) | 0.32 (0.32–0.33) |
| Deep SSI | 0.00 (0.00–0.00) | 0.00 (0.00–0.00) | 0.22 (0.21–0.22) |
| Organ/Space SSI | 0.09 (0.09–0.09) | 0.05 (0.05–0.05) | 0.11 (0.11–0.11) |
| SSI PATOS | 0.00 (0.00–0.00) | 0.06 (0.06–0.07) | 0.14 (0.13–0.15) |
| Any SSI | 0.10 (0.09–0.10) | 0.06 (0.06–0.07) | 0.23 (0.23–0.24) |
SSI: Surgical Site Infection, PATOS: Present at the Time of Surgery, AUC: Area Under the Receiver Operating Curve
We next evaluated the performance of EasyCIE-SSI in several procedure subgroups. The differences in the performance of EasyCIE-SSI between the internal and external validation cohorts are show in Figure 1. Compared to the internal validation cohort, the sensitivity of EasyCIE-SSI for detection of SSI in the external validation cohort was significantly worse for hernia (absolute difference (95%CI): −17.3% (−33.7%– −1%)), and skin/soft tissue procedures (absolute difference (95%CI): −43.2% (−77.2% – −9.3%)). EasyCIE-SSI had a significantly lower sensitivity for detection of SSI in clean procedures (absolute difference (95%CI): −26.6% (−42.0% – −11.2%)), and outpatient procedures (absolute difference (95%CI): −31.7% (−46.2% – −17.3%)) in external validation compared to internal validation.
FIGURE 1.
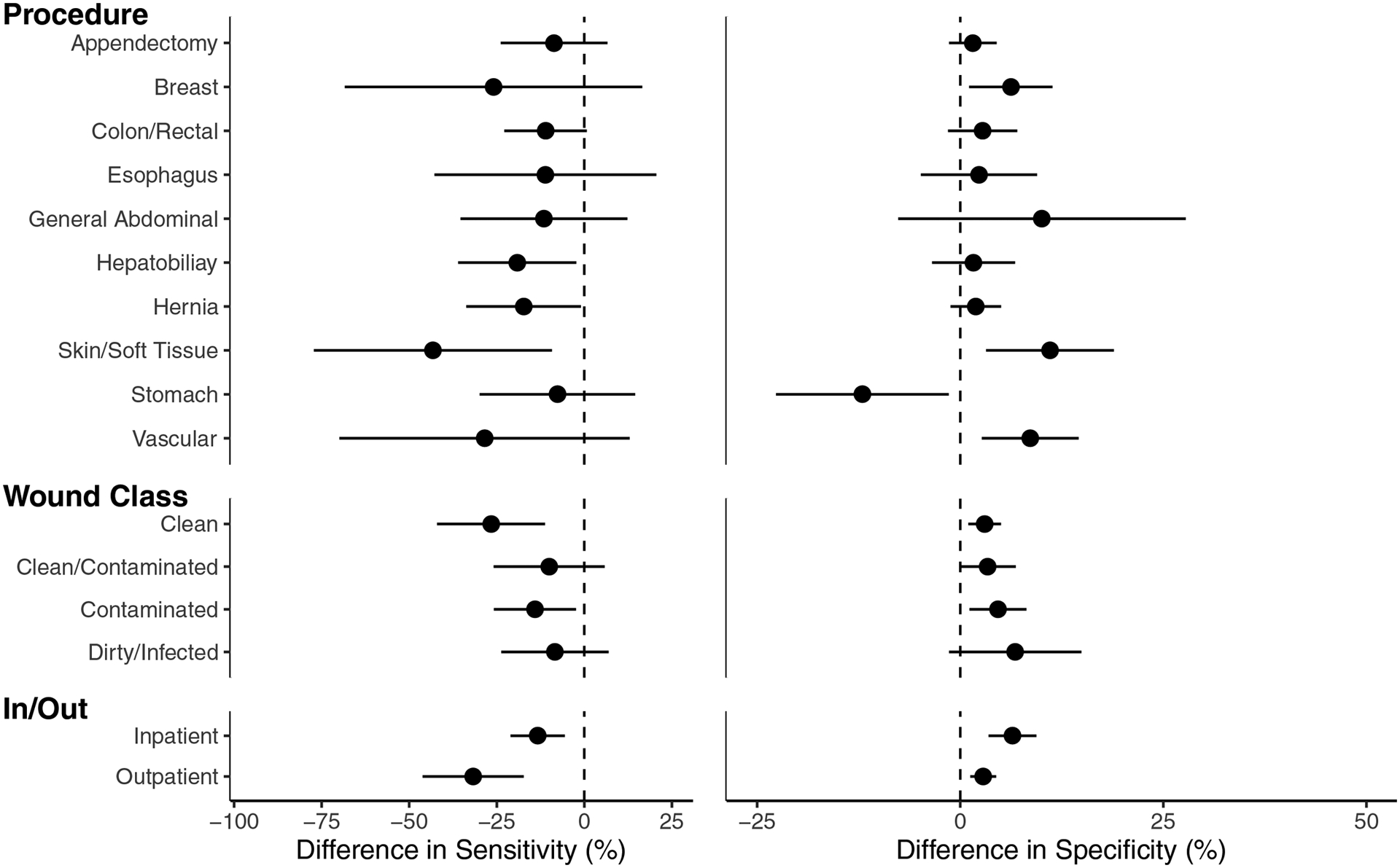
Subgroup analysis of the difference in sensitivity (left) and specificity (right) of EasyCIE-SSI between the external validation cohort compared to the internal validation cohort by procedure characteristics. The error bars represent 95% Confidence Interval for each point estimate and the dotted line represents 0 or no difference between external and internal validation. Sensitivity was uniformly decreased in External Validation, with characteristics whose error bar excludes 0, for example, Clean Procedures, representing statistically significant decreases.
We next analyzed the performance errors in EasyCIE-SSI compared to the reference standard NSQIP review (Table 4). We chose to focus on false-negative errors given the importance of missing an SSI identified on NSQIP review. In the training, internal validation, and external validation cohort 68%, 50%, and 56% respectively, of the false-negative errors were due to the documentation not being present in electronic format, respectively. These were most commonly due to outside documentation obtained from the NSQIP SCR patient contact. Other causes of errors associated with the NLP pipeline included errors in named entity recognition, local inferencing, document inferencing, and patient inferencing.
TABLE 4.
False Negative Error Analysis of EasyCIE-SSI in the internal and External Validation Cohorts.
| Error Type | Cohort | Description | Example | |
|---|---|---|---|---|
| Internal Validation (n=4) | External Validation (n=155) | |||
| Documentation Not Present | 2 (50%) | 88 (56.8%) | The documentation needed to identify an SSI was not available electronically. | Patient treated at non-index facility post discharge, |
| Named Entity Recognition | 20 (12.9%) | The local terminology was not in the dictionary required to identify key terms related to SSI. | Misspelling (i.e. “prulent”), Anatomic name not present in Training (i.e. “Morrison’s Pouch”) | |
| Local Inference | 1 (25%) | 33 (21.3%) | Sentence/Paragraph inferencing rules made an incorrect conclusion in aggregating the concepts. | Concluded SSI was historical due to phrase “patient has a history of an SSI” |
| Document Inference | 10 (6.5%) | Document level inferencing rules made an incorrect conclusion aggregating the sentences and paragraphs. | Antibiotics mentioned separately in document and not associated with SSI symptoms to conclude SSI Present | |
| Patient Inference | 1 (25%) | 4 (2.6%) | Patient level inferencing rules made an incorrect conclusion aggregating the documents conclusions | Infection PATOS not identified as resolved prior to Organ Space SSI development. |
DISCUSSION
In this paper, we present the development and validation of a natural language processing approach, EasyCIE-SSI, for automated surveillance of surgical site infections. There are several novel and significant findings of our work. First, SSI surveillance can be achieved with high sensitivity (94% in internal validation), specificity (86% in internal validation), and distinction (AUC 0.912 in internal validation). Second, EasyCIE is portable across EHRs and can be implemented in health care systems without the need for significant retraining. Lastly, the post-discharge surveillance of SSI continues to remain a challenge especially for clean, skin/soft tissue, and outpatient procedures despite the benefits of natural language processing.
Natural language processing has been used to identify postoperative complications in several previous studies. A study by Murff, et. al., developed an NLP tool, Post-Operative Event Monitor (POEM), for automated surveillance of several postoperative complications, including SSI, in the VA healthcare system.14, 15 In their approach, they electronically parsed clinical notes for medical terminology and mapped to SNOMED CT concepts. They subsequently created rules using a combination of SNOMED CT concepts to detect the presence or absence of a surgical site infection compared to the VA Surgical Quality Improvement Program Review. Their sensitivity and specificity for the detection of surgical site infections was 77% and 63%, respectively. Given that their study used a validation cohort from the same health care system, the appropriate comparison to the current study would be comparing to the internal validation cohort, which demonstrated improved sensitivity and specificity of 94% and 88%.
Several reasons contribute to our improved performance compared to previous studies. First, we utilized an expanded vocabulary for SSI detection including signs, symptoms, treatments, and medications related to SSIs. Second, we utilized information located in the operative report, including inferring for evidence of infection present at the time of surgery (PATOS) and inferring based on the status of the wound closure. This information is key in identifying subsequent events in the patient’s postoperative course. For example, if the wound was not closed at the time of the initial operation, a superficial infection at the surgical site will not be considered as an SSI until the wound is documented to be closed. Lastly, we integrated temporal information across the patient’s postoperative course. Integrating temporal information improves NLP accuracy by inferring the status of an SSI if there is conflicting information documented multiple notes.31 For example, if the patient had an intrabdominal abscess at the time of the operation then, understanding when a reference to intrabdominal abscess refers to the original abscess which has resolved or a newly developed organ/space infection is necessary to minimize false-positive errors.
We demonstrated the portability of our approach by implementing EasyCIE-SSI across two independent healthcare systems that utilize different electronic health care records. Portability across healthcare systems is feasible because EasyCIE-SSI is directly interacting with the database using a general-purpose NLP-database schema and utilizes free-text clinical notes commonly available in enterprise data warehouses of healthcare systems.32 Therefore, our approach can easily be implemented locally in health care systems or through remote services utilizing Fast Healthcare Interoperability Resources (FHIR).33 However, there are key differences in the performance of EasyCIE-SSI between the internal and external validation cohorts. Overall, we observed a 15% decrease in Sensitivity and a 4% increase in Specificity between our internal and external validation cohorts. Given NSQIP definitions have changed over time, the performance of EasyCIE-SSI in the external validation is likely the lower bound of performance as the reference standard labels were based on an older definition of SSI by NSQIP. We observed larger decreases in the Sensitivity of EasyCIE-SSI for Superficial SSI (−20%), Clean Procedures (−26%), Skin/Soft Tissue Procedures (−43%), and Outpatient Procedures (−31%).
To get a better understanding of the reasons for the differences between the Sensitivity of EasyCIE-SSI in the internal and external validation cohorts, we performed an error analysis of all the false negative errors occurring in the validation cohorts. The most common reason for a false negative was the documentation necessary to diagnose an SSI did not exist in the enterprise data warehouse of the health care system. The lack of documentation accounted for over 50% of the false-negative errors in both the internal and external validation cohorts. We intentionally did not exclude these patients from analysis as we aimed to demonstrate the performance of an NLP approach for SSI surveillance in a “real world” setting where post-procedural documentation may be limited. If the patients with a lack of documentation were removed from our analysis, the Sensitivity of EasyCIE-SSI on the internal and external validation cohorts increases to 97% and 89%, respectively. Other false-negative errors were due to technical aspects of the NLP pipeline, including named entity recognition (NER) errors, local, document, and patient inferencing errors. NER errors could be mitigated with additional training data before NLP implementation. For example, hospital antibiotic formulations can be anticipated and added to the rules dictionary to minimize named entity recognition errors. The inferencing errors represent the complexities of clinical language and remain general NLP challenge.22, 31 However, NLP systems can be designed to minimize false positive or false negative errors to maximize sensitivity or specificity.
The lack of documentation also explains the observed differences between the likelihood ratios between our internal and external validation cohorts. In external validation, both the PLR (9.83 vs 7.91) and NLR increased (0.23 vs 0.06), compared to internal validation. Due to the higher number of patients who lacked post-discharge documentation in external validation, there are fewer false positives and more false negatives in the external validation cohort. However, given the low prevalence of SSIs, the decrease in false positives was greater than the increase in false negatives. Therefore the value of a positive result increases (increase PLR), while the value of a negative result decreases (increased NLR) in external validation.
The organization of the healthcare systems included in the study can explain the large increase in the lack of documentation observed in external validation. The healthcare system for the internal evaluation cohort is a vertically integrated healthcare system with employed physicians and providers.34 The external validation cohort is a horizontally integrated healthcare system with both employed and affiliated physicians and providers. In the vertically integrated healthcare system, all the providers use the same electronic documentation for postoperative care. In the horizontally integrated healthcare system, the providers have the option of using the EHR of the healthcare system for outpatient documentation. Our findings demonstrate an NLP approach to SSI surveillance is limited in the detection of post-discharge events outside of vertically integrated healthcare systems. Given the majority of post-discharge readmissions, after surgical procedures are due to new-onset postoperative complications, alternative strategies should be investigated to improve post-discharge surveillance.35
The implications of adopting an NLP approach for automated SSI surveillance should be considered with caution for quality assessment programs. Given the low prevalence of SSIs, the positive predictive value of EasyCIE-SSI was predictably low (24%–33%), and the negative predictive value predictably near perfect (greater than 99%) in both cohorts. In addition, the PLR and NLR both increased in external validation. Therefore, after the implementation of EasyCIE at a new healthcare system without any additional training, a negative result could be trusted. However, a positive result would still require manual chart review for confirmation of SSI. Given the high sensitivity and high negative predictive value of our NLP approach, NSQIP SCRs could safely restrict their review to positively flagged patients after an NLP evaluation. This option potentially could lead to a decrease in chart review burden, allowing an expanded surveillance scope of cases across a healthcare system at a lower cost. Also, our approach could easily be extended to other common postoperative complications including urinary tract infection, pneumonia,36 venous thromboembolism, and pulmonary embolism.22
There are several limitations to the present study. First, the data were abstracted from two independent healthcare systems in a single geographic region. We cannot account for variation in our NLP performance across geographic regions due to differences in clinical language and treatments between healthcare systems. However, the benefit of a rules-based NLP system is that it can easily be adapted to language and terminology differences between healthcare systems with a small amount of training data. Second, the definition of SSI was based on NSQIP methodology. The performance EasyCIE for SSI surveillance based on other definitions of SSI, such as the NHSN definition, is unknown. Likely, small variations in the definitions of operative episodes and an SSI would lead to the diminished performance of EasyCIE-SSI compared to other surveillance systems. We limited our definition of operative episodes to procedures defined by NSQIP methodology. It is not generalizable at this time to other procedures including orthopedic and neurosurgical procedures. Lastly, we included only clinical text notes and did not include any structured data fields in our surveillance approach, such as laboratory values, microbiology reports, and medication administration. This information was only included if documented in the clinical notes. Additional research studies are needed to determine if the inclusion of these structured data fields in combination with NLP would improve the overall performance of our surveillance approach.
CONCLUSION
We report the development and validation of a portable natural language processing approach for surveillance of postoperative surgical site infections. We demonstrate that surveillance of surgical site infections can be achieved with high sensitivity and specificity in two independent healthcare systems. We observe an NLP approach is limited in the surveillance of some post-discharge events particularly among skin and soft tissue procedures. Since the tool’s NPV is near perfect, using an NLP approach would allow a healthcare system to focus their chart review resources solely on NLP-positive cases, saving considerable effort and cost in the review process. This approach can easily be integrated into the NSQIP methodology at independent healthcare systems and can be generalized to other postoperative complications.
ACKNOWLEDGMENTS
The support and resources from the Center for High-Performance Computing at the University of Utah are gratefully acknowledged.
FUNDING/SUPPORT
This research was supported by grant 1K08HS025776 from the Agency for Healthcare Research and Quality (Dr. Bucher). The computational resources used were partially funded by the NIH Shared Instrumentation Grant 1S10OD021644-01A1.
ROLE OF THE FUNDER/SPONSOR
The Agency for Healthcare Research and Quality had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
CONFLICT OF INTEREST DISCLOSURE
Dr. Chapman reported consulting for IBM and serving on the Scientific Advisory Board of Health Fidelity. These companies had no role in the study. No other disclosures are reported.
ACCESS TO DATA AND DATA ANALYSIS
Dr. Bucher had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis
REFERENCES
- 1.Ko CY, Hall BL, Hart AJ, et al. The American College of Surgeons National Surgical Quality Improvement Program: achieving better and safer surgery. Jt Comm J Qual Patient Saf 2015; 41(5):199–204. [DOI] [PubMed] [Google Scholar]
- 2.Hu QL, Liu JY, Hobson DB, et al. Best Practices in Data Use for Achieving Successful Implementation of Enhanced Recovery Pathway. J Am Coll Surg 2019; 229(6):626–632.e1. [DOI] [PubMed] [Google Scholar]
- 3.Classen DC, Griffin FA, Berwick DM. Measuring Patient Safety in Real Time: An Essential Method for Effectively Improving the Safety of Care. Ann Intern Med 2017; 167(12):882–883. [DOI] [PubMed] [Google Scholar]
- 4.McGee MF, Kreutzer L, Quinn CM, et al. Leveraging a Comprehensive Program to Implement a Colorectal Surgical Site Infection Reduction Bundle in a Statewide Quality Improvement Collaborative. Ann Surg 2019; 270(4):701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ju MH, Ko CY, Hall BL, et al. A comparison of 2 surgical site infection monitoring systems. JAMA Surg 2015; 150(1):51–7. [DOI] [PubMed] [Google Scholar]
- 6.West N, Eng T. Monitoring and reporting hospital-acquired conditions: a federalist approach. Medicare Medicaid Res Rev 2014; 4(4):E1–E16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiloach M, Frencher SK Jr., Steeger JE, et al. Toward robust information: data quality and inter-rater reliability in the American College of Surgeons National Surgical Quality Improvement Program. J Am Coll Surg 2010; 210(1):6–16. [DOI] [PubMed] [Google Scholar]
- 8.Joseph S, Sow M, Furukawa MF, et al. HITECH spurs EHR vendor competition and innovation, resulting in increased adoption. Am J Manag Care 2014; 20(9):734–40. [PubMed] [Google Scholar]
- 9.Adler-Milstein J, Jha AK. HITECH Act Drove Large Gains In Hospital Electronic Health Record Adoption. Health Aff (Millwood) 2017; 36(8):1416–1422. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto DA, Rosman G, Rus D, et al. Artificial Intelligence in Surgery: Promises and Perils. Ann Surg 2018; 268(1):70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanauer DA, Englesbe MJ, Cowan JA Jr., et al. Informatics and the American College of Surgeons National Surgical Quality Improvement Program: automated processes could replace manual record review. J Am Coll Surg 2009; 208(1):37–41. [DOI] [PubMed] [Google Scholar]
- 12.Bucher BT, Ferraro JP, Finlayson SRG, et al. Use of Computerized Provider Order Entry Events for Postoperative Complication Surveillance. JAMA Surg 2019; 154(4):311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman AB, Mowery DL, Swords DS, et al. Detecting Evidence of Intra-abdominal Surgical Site Infections from Radiology Reports Using Natural Language Processing. AMIA Annu Symp Proc 2017; 2017:515–524. [PMC free article] [PubMed] [Google Scholar]
- 14.FitzHenry F, Murff HJ, Matheny ME, et al. Exploring the frontier of electronic health record surveillance: the case of postoperative complications. Med Care 2013; 51(6):509–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murff HJ, FitzHenry F, Matheny ME, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA 2011; 306(8):848–55. [DOI] [PubMed] [Google Scholar]
- 16.Hall BL, Hamilton BH, Richards K, et al. Does surgical quality improve in the American College of Surgeons National Surgical Quality Improvement Program: an evaluation of all participating hospitals. Ann Surg 2009; 250(3):363–76. [DOI] [PubMed] [Google Scholar]
- 17.American College of Surgeons. NSQIP Operations Manual. 2017.
- 18.Shi J, Liu S, Pruitt LCC, et al. Using Natural Language Processing to improve EHR Structured Data-based Surgical Site Infection Surveillance. AMIA Annu Symp Proc 2019; 2019:794–803. [PMC free article] [PubMed] [Google Scholar]
- 19.Bucher BT, Shi J, Pettit RJ, et al. Determination of Marital Status of Patients from Structured and Unstructured Electronic Healthcare Data. AMIA Annu Symp Proc 2019; 2019:267–274. [PMC free article] [PubMed] [Google Scholar]
- 20.Shi J, Hurdle JF. Trie-based rule processing for clinical NLP: A use-case study of n-trie, making the ConText algorithm more efficient and scalable. J Biomed Inform 2018; 85:106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi J, Mowery D, Doing Harris K, et al. RuSH: a Rule-based Segmentation Tool Using Hashing for Extremely Accurate Sentence Segmentation of Clinical Text. AMIA Annual Symposium. Chicago, IL, 2016. [Google Scholar]
- 22.Chapman BE, Lee S, Kang HP, et al. Document-level classification of CT pulmonary angiography reports based on an extension of the ConText algorithm. J Biomed Inform 2011; 44(5):728–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi J, Mowery D, Zhang M, et al. Extracting Intrauterine Device Usage from Clinical Texts Using Natural Language Processing. Healthcare Informatics (ICHI), 2017 IEEE International Conference on: IEEE, 2017. pp. 568–571. [Google Scholar]
- 24.Scuba W, Tharp M, Mowery D, et al. Knowledge Author: facilitating user-driven, domain content development to support clinical information extraction. J Biomed Semantics 2016; 7(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaffer JP. Multiple Hypothesis Testing. Annual Review of Psychology 1995; 46(1):561–584. [Google Scholar]
- 26.McGee S Simplifying likelihood ratios. Journal of general internal medicine 2002; 17(8):646–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982; 143(1):29–36. [DOI] [PubMed] [Google Scholar]
- 28.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011; 12(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Austin PC, Tu JV. Bootstrap Methods for Developing Predictive Models. The American Statistician 2004; 58(2):131–137. [Google Scholar]
- 30.Newcombe RG. Interval estimation for the difference between independent proportions: comparison of eleven methods. Statistics in Medicine 1998; 17(8):873–890. [DOI] [PubMed] [Google Scholar]
- 31.Chu D, Dowling JN, Chapman WW. Evaluating the effectiveness of four contextual features in classifying annotated clinical conditions in emergency department reports. AMIA Annu Symp Proc 2006; 2006:141–5. [PMC free article] [PubMed] [Google Scholar]
- 32.Wu ST, Kaggal VC, Dligach D, et al. A common type system for clinical natural language processing. J Biomed Semantics 2013; 4(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandel JC, Kreda DA, Mandl KD, et al. SMART on FHIR: a standards-based, interoperable apps platform for electronic health records. J Am Med Inform Assoc 2016; 23(5):899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans Jenna M, Baker Ross G, Berta W, et al. The Evolution Of Integrated Health Care Strategies. Annual Review of Health Care Management: Revisiting The Evolution of Health Systems Organization, Vol. 15: Emerald Group Publishing Limited; 2014:pp. 125–161. [DOI] [PubMed] [Google Scholar]
- 35.Merkow RP, Ju MH, Chung JW, et al. Underlying reasons associated with hospital readmission following surgery in the United States. JAMA 2015; 313(5):483–95. [DOI] [PubMed] [Google Scholar]
- 36.Dublin S, Baldwin E, Walker RL, et al. Natural Language Processing to identify pneumonia from radiology reports. Pharmacoepidemiology and drug safety 2013; 22(8):834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Dr. Bucher had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis


