ABSTRACT
Partitioning systems ensure the stable inheritance of bacterial low-copy-number replicons, such as chromosomes, chromids, and megaplasmids. These loci consist of two genes encoding partition proteins A and B, and at least one parS centromere-like sequence. In chromids and megaplasmids, partitioning systems are often located in the vicinity of replication systems. An extreme example of this co-localization are alphaproteobacterial repABC replicons, where the partition (repAB) and replication (repC) genes form a single operon, with parS sequences usually positioned in close proximity to these genes. In this study, we characterized a more complex repABC system found in Paracoccus aminophilus (Rhodobacterales) megaplasmid pAMI4 (438 kb). Besides the repABC operon with a single parS site, this replicon has a 2-kb non-coding locus positioned 11.5 kb downstream of repC, which contains three additional parS repeats (3parS). We demonstrated that 3parS is bound by partition protein B in vitro and is essential for proper pAMI4 partitioning in vivo. In search of similar loci, we conducted a comparative analysis of parS distribution in other repABC replicons. This revealed different patterns of parS localization in Rhodobacterales and Rhizobiales. However, in both these taxonomic orders, parS sites are almost always located inside or close to the repABC operon. No other 3parS-like loci were found in the closest relatives of pAMI4. Another evolutionarily-independent example of such a locus was identified as a conserved feature in chromosome 2 of Allorhizobium vitis and related replicons.
IMPORTANCE The repABC replication/partitioning loci are widespread in extrachromosomal replicons of Alphaproteobacteria. They are evolutionarily diverse, subject to multi-layer self-regulation, and are responsible for the maintenance of different types of replicons, such as plasmids (e.g., Agrobacterium pTi and pRi tumorigenic and rhizogenic plasmids), megaplasmids (e.g., Sinorhizobium pSymA and pSymB) and essential chromids (e.g., secondary chromosomes of Agrobacterium, Brucella and Rhodobacter). In this study, we functionally analyzed an atypical partition-related component of repABC systems, the 3parS locus, found in the P. aminophilus megaplasmid pAMI4. We also identified parS centromere-like site distribution patterns in different groups of repABC replicons and found other unrelated 3parS-like loci, which had been overlooked. Our findings raise questions concerning the biological reasons for differential parS distribution, which may reflect variations in repABC operon regulation as well as different replication and partition modes of replicons belonging to the repABC family.
KEYWORDS: Alphaproteobacteria, partitioning system, plasmid evolution, plasmid segregation, repABC
INTRODUCTION
Partition of most bacterial chromosomes and large low-copy-number extrachromosomal replicons, including megaplasmids and chromids (often called secondary chromosomes), is dependent on ParAB-parS partitioning systems. These systems consist of a parAB operon, encoding an ATPase (ParA) and a DNA-binding protein (ParB), as well as palindromic sequences analogous to eukaryotic centromeres (parS) (1, 2).
The action of this system is performed in two steps: (i) ParB binding to the replicated parS sequences and (ii) ParA-driven separation of the ParB-parS complexes toward the opposite cell poles. The detailed mechanism of ParB binding to parS and its spread to adjacent DNA has recently been described (3).
Megaplasmid and chromid partitioning modules are often encoded in the vicinity of modules involved in plasmid replication. The most intricate examples of this co-existence are the repABC replicons found exclusively in Alphaproteobacteria. In this unique group of replicons, a single operon repABC encodes the proteins involved in both plasmid partition (RepA and RepB – homologs of ParA and ParB) and replication (RepC plasmid replication initiator). The repABC modules also contain the origin of replication (oriV), located inside the repC gene, as well as single or multiple parS sites, found at different locations within the module (4–7). The regulation of repABC expression occurs at many levels, including negative self-regulation of operon transcription by the RepA and RepB proteins, and modulation of repC transcript translation by a small regulatory ctRNA (RepE), encoded between repB and repC (8).
The repABC modules are also commonly found in extrachromosomal replicons of Paracoccus spp. (Rhodobacteraceae, Alphaproteobacteria), including P. aminophilus JCM 7686 – a methylotrophic strain isolated in Japan from a sample of soil polluted with dimethylformamide (DMF) (9–11). The multi-replicon genome of this strain contains eight extrachromosomal replicons (10). One of these, megaplasmid pAMI4 (438 kb), carries a repABC system, that, besides one parS centromere-like site localized in the classic manner in the repABC operon, may employ three additional parS repeats within a long (2.5 kb), low GC, non-coding region (henceforth referred to as the 3parS locus), located 11.5 kb downstream of repC. In this study, we demonstrated that these extra parS repeats are essential for the proper partitioning of pAMI4 and its stability in the host cell population. In addition, we conducted a bioinformatic analysis of a large set of repABC replicons from Rhodobacterales and Rhizobiales, which show different patterns of parS localization. Interestingly, we identified other 3parS-like loci that are evolutionarily unrelated to that of pAMI4.
RESULTS
Structure of the pAMI4 repABC module.
The pAMI4 repABC operon has a typical organization, with overlapping repA and repB genes (1-bp overlap) and a repC gene preceded by a 175-bp-long intergenic region (Fig. 1). This region contains a parS site (parSBC, 5′-GTTGACAGCTGTCAAC-3′), that matches the parS consensus sequence for repABC replicons, 5′-GTTNNCNGCNGNNAAC-3′, determined by Cevallos et al. (5).
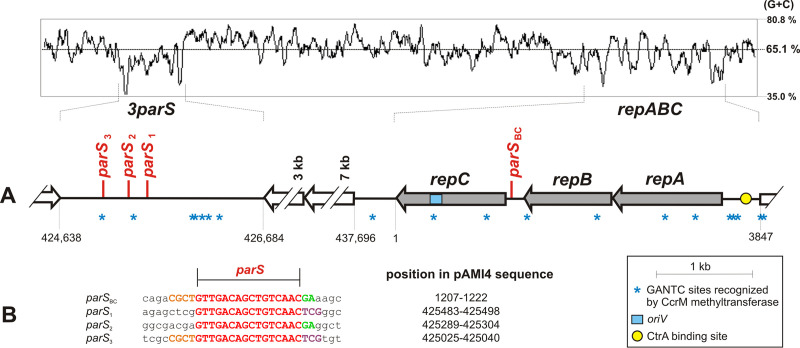
FIG 1.
Distribution of parS sites in the pAMI4 genome. (A) Genetic organization of the DNA region containing repABC and the 3parS locus. Numbers below the diagram correspond to nucleotide positions in the pAMI4 sequence deposited in the GenBank database (NC_022049). Genes of the repABC module are marked in gray. The GC profile of the analyzed pAMI4 sequence is shown in the top panel. (B) Comparison of the identified parS sites (in red) in their original sequence context. Conserved nucleotides are shown in upper case letters. The colors orange, green, and violet indicate different types of conserved neighboring sequences.
The pAMI4 repABC operon carries a functional origin of replication as it was used for construction of a pAMI4 mini-derivative that replicates in Paracoccus spp. (10). Interestingly, 11.5 kb downstream of the repC gene, beyond two large open reading frames (ORFs) encoding hypothetical membrane proteins, we identified a long (2047 bp) intergenic region carrying three more parS repeats (parS1-3) (Fig. 1). These repeats are identical to parSBC, and some local sequence similarities are also found in their immediate vicinity (Fig. 1B). This 3parS region lacks any remnant genes and its nucleotide sequence has a higher AT content (42.5%) than the pAMI4 sequence as a whole (35.8%) or the oriV-containing repC gene (37.3%).
Further analysis of the DNA region containing repABC and the 3parS locus revealed the uneven distribution of GANTC sites recognized by the CcrM methyltransferase (Fig. 1) – a key epigenetic mediator in Alphaproteobacteria (12), which is equivalent to the gammaproteobacterial Dam methylase (13). This observation strongly suggests cell cycle dependent regulation of repABC operon transcription (three GANTC sites in the repABC promoter region) and an unknown type of regulation occurring in the 3parS region (five GANTC sites ∼500 bp from parS3) (the mean distribution of GANTC sites in the pAMI4 genome is one per 1132 bp) (Fig. 1). It is noteworthy that the intergenic region upstream of repABC also contains a putative CtrA-binding site (5′-CTAAN7TTAAC-3′), with one nucleotide difference compared to the consensus CtrA-binding site of Caulobacter crescentus (14). CtrA is a global regulator of many cell cycle-dependent genes in Alphaproteobacteria and a negative regulator of replication initiation of the C. crescentus chromosome (14). The ctrA gene is also found in the P. aminophilus genome (GenBank acc. no. AGT08327). The presence of a CtrA-binding site within the promoter region of a repABC operon was previously observed in the pTi plasmid of Agrobacterium tumefaciens (15).
Function of the 3parS locus in pAMI4 partition.
To confirm that the 3parS locus is involved in the partitioning of pAMI4 molecules, in vitro and in vivo analyses were performed to determine whether this region possesses the three basic properties typical for plasmid centromere-like sites: (i) expression of incompatibility behavior, (ii) interaction with the RepB protein, and (iii) the ability to ensure in cis stability of a plasmid in the presence of partition proteins. Furthermore, a Δ3parS mutant was obtained and the effect on pAMI4 stability and the positioning of a GFP-RepBpAMI4 fusion protein in the cells was investigated.
The parS sites are strong incompatibility determinants (inc) and therefore their introduction into the strain of origin may result in removal of the parental parS-containing plasmid from the bacterial cells (16). The entire 3parS locus was cloned into vector pBBR1MCS-5, and the resulting plasmid was introduced into P. aminophilus JCM 7686R (Rifr) by triparental mating. All tested Gmr Rifr transconjugants carrying the autonomous form of pBBRGm-3parS had lost pAMI4, which was verified by PCR with pAMI4-specific primers. This confirmed that the 3parS region exerts incompatibility toward pAMI4. At this stage, a P. aminophilus pAMI4-less strain, designated UW100, was obtained (lacking both pAMI4 and pBBRGm-3parS), for use as a convenient host for pAMI4-based mini-derivatives in subsequent experiments.
To confirm that 3parS interacts with RepBpAMI4, an in vitro electrophoretic mobility shift assay (EMSA) was performed. The pAMI4 repB gene was cloned into vector pET28b(+) in order to express a 6×His-RepBpAMI4 fusion protein. The purified protein was used in EMSAs with DNA fragments containing (i) parSBC of the repABC module (positive control), (ii) individual parS sites from the 3parS locus – parS1, parS2, parS3, (iii) the entire 3parS region, and (iv) a DNA fragment from vector pBGS18 (negative control). The 6×His-RepBpAMI4 protein was found to specifically bind parSBC and the entire 3parS region, as well as each of the single parS1-3 sequences. In the case of the fragment comprising the entire 3parS region, three complexes with different electrophoretic mobilities were formed in a protein concentration-dependent manner, most probably representing the binding of 6×His-RepBpAMI4 to one, two, or three parS sites within 3parS (Fig. 2A).
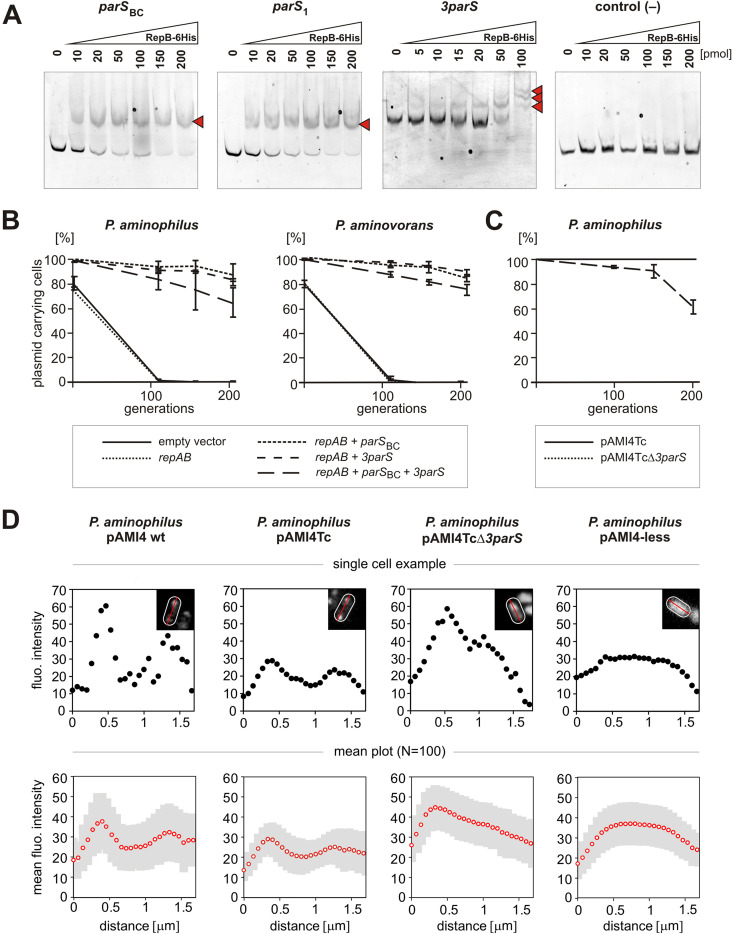
FIG 2.
Functional analysis of the 3parS locus. (A) Electrophoretic mobility shift assays of the 6His-RepBpAMI4 protein with pAMI4 parS sequences. The protein-DNA complexes are marked with red arrowheads. The tested DNA fragments of pAMI4, labeled with fluorescein, were incubated with increasing amounts of the 6His-RepBpAMI4 protein. The parS1, parS2 and parS3 sites showed the same pattern of 6His-RepBpAMI4 binding. Therefore, the result for only one of them (parS1) is presented. FAM-labeled DNA fragment of pBGS18 was used as a negative control. (B) Stability assays of test vector pABW3 (Kmr) containing different combinations of the components of the pAMI4 partitioning system (repAB, parSBC and the 3parS locus). P. aminophilus UW100 and P. aminovorans JCM 7685R transformed with different pABW3 constructs were grown under nonselective conditions. Retention of plasmids was determined by the percentage of kanamycin resistant cells at each time interval (100 colonies tested by replica plating, the plotted values are the means from triplicate assays and standard deviations are shown). (C) Stability assays of pAMI4Tc (control) and pAMI4TcΔ3parS in its natural host P. aminophilus during growth under non-selective conditions. Retention of plasmids was determined by the percentage of tetracycline resistant cells at each time interval (100 colonies tested by replica plating, the plotted values are the means from triplicate assays and standard deviations are shown). (D) Measurement of GFP-RepBpAMI4 fluorescence intensity profiles in various P. aminophilus strains with pJC43-GFP-RepB cultured to mid-exponential phase in LB with gentamicin and taurine. From left to right: P. aminophilus JCM 7686R carrying pAMI4 wt, pAMI4Tc or pAMI4TcΔ3parS, and P. aminophilus UW100 (pAMI4-less). The top row of plots show the typical patterns of fluorescence in cells expressing GFP-RepBpAMI4 (black dots). The fluorescence intensity signal was measured along the bacterial longitudinal axis (red line from one pole of the cell (outlined in white) to the other pole (inset pictures)) using ImageJ (Fiji) software. The bottom row of plots show the mean fluorescence intensity (empty red dots) and standard deviation (gray shaded area) over the length of the cells (N = 100). Enlarged microscopy images of P. aminophilus cells expressing GFP-RepBpAMI4 are presented in Fig. S1 in the supplemental material.
The stabilizing effect of the pAMI4 partitioning system on a test plasmid was examined in its natural host P. aminophilus strain UW100 and in a heterologous host P. aminovorans JCM 7685R (17). Several derivatives of vector pABW3 (Kmr; unstable in Paracoccus spp.) (4) were constructed for this analysis, containing (i) the repAB genes alone, (ii) repAB with its cognate parSBC, (iii) repAB with the 3parS region, and (iv) repAB with parSBC and 3parS. The obtained constructs were introduced into both strains by triparental mating. The stability of the different plasmids was tested by monitoring the proportion of kanamycin resistant cells during growth in liquid culture without antibiotic selection (Fig. 2B).
The results showed that 3parS fully compensates for the lack of parSBC in both bacterial hosts. However, the presence of 3parS and parSBC in the same plasmid was found to decrease the stabilizing effect compared to plasmids carrying only 3parS or only parSBC (Fig. 2B). A similar effect was observed in the case of the parS sequences of pSymA of Sinorhizobium meliloti 1021, a repABC replicon with six parS repeats upstream of repA. The first parS (counting from repA) stabilized a test plasmid carrying the repAB genes of pSymA, while addition of the other five sequences destabilized the system (18). It should be noted that results obtained in assays performed using small plasmids do not necessarily reflect the function of the tested parS sites in large natural replicons.
To test the function of 3parS in its original context, a Δ3parS strain was constructed in a pAMI4-derivative carrying a tetracycline resistance cassette (pAMI4Tc). The stability of the mutated plasmid was tracked during growth in LB medium for up to ∼200 generations, by monitoring the fraction of tetracycline resistant cells. The deletion of 3parS caused a significant drop in pAMI4 stability. At the final checkpoint, less than 70% of the cell population carried pAMI4TcΔ3parS, compared to 100% stability of pAMI4Tc (Fig. 2C).
Finally, to investigate the influence of 3parS on the formation of RepBpAMI4 foci, a plasmid for taurine-dependent expression of a GFP-RepBpAMI4 fusion protein was constructed and introduced into P. aminophilus JCM 7686R carrying pAMI4 (wt), pAMI4Tc or pAMI4TcΔ3parS, or strain UW100. Two green fluorescent foci were formed in the control strains P. aminophilus JCM 7686R carrying pAMI4 (wt) or pAMI4Tc, while in the pAMI4TcΔ3parS strain the GFP was much more dispersed, with a higher fluorescent signal on only one side of the cell. In the pAMI4-less strain UW100, the fluorescence was evenly distributed throughout the cell (Fig. 2D and Fig. S1 in the supplemental material).
Distribution of parS sites in repABC replicons.
To our knowledge, no other repABC replicon carrying an additional locus involved in partition has been described previously. Therefore, to identify loci similar to 3parS and to uncover its evolutionary origin, we analyzed the distribution of parS sites in diverse repABC replicons available in the NCBI database using two different data sets. The first data set, comprised of 115 complete plasmid genomes, represented the diversity of repABC replicons found in Rhodobacterales and Rhizobiales (Alphaproteobacteria) (Table S1 in the supplemental material). The analysis of this data set was to determine the variability of parS localization on a larger evolutionary scale without losing information about parS sites located distantly from the repABC operon (only complete plasmid sequences are included in the data set). The second data set represented the 100 closest relatives of pAMI4 (according to RepB similarity) identified in NCBI GenBank and is comprised mainly of shotgun sequencing contigs (Table S2).
These analyses identified different parS localization patterns in Rhodobacterales and Rhizobiales. In Rhodobacterales, the parS sequences tend to be localized in the intergenic region between repB and repC, and directly downstream of repC (Fig. 3, Fig. S2 in the supplemental material). Multiplication of the parS sequences located downstream of repC is frequently observed, with up to four parS repeats found in this region. These sequences are located not only in the intergenic region directly downstream of repC, but also within the following ORFs and intergenic regions. The interval between the end of the intergenic region downstream of repC and the more distant parS sequences is typically between ∼0.5 and ∼4 kb, with pAMI4 being an exception at 12 kb (Fig. 3). Interestingly, few putative parS sequences were identified at locations very distant (tens of kb) from repABC. However, in Rhodobacterales we did not find any long non-coding loci with multiple parS sites resembling 3parS.
FIG 3.
Distribution of putative parS sequences within repABC replicons from Rhodobacterales and Rhizobiales. Fifty representative replicons from Data set 1 (Table S1 in the supplemental material) are presented. In the left-hand panel, a neighbor joining tree of RepA-RepB concatenated protein sequences is shown. In the right-hand panel, the identified parS sequences are marked in red on diagrams of the repABC modules. These show the repA, repB, and repC genes (rectangles) and intergenic regions upstream of repA, between repA and repB, between repB and repC, and downstream of repC (bold lines). The intergenic/intragenic localization of putative parS sequences located outside the repABC modules (following the numbers giving the distance from the repABC module in kb) is not indicated. In some cases, putative parS sequences and/or the repC gene were absent. Replicons carrying long intergenic loci with parS repeats are in bold.
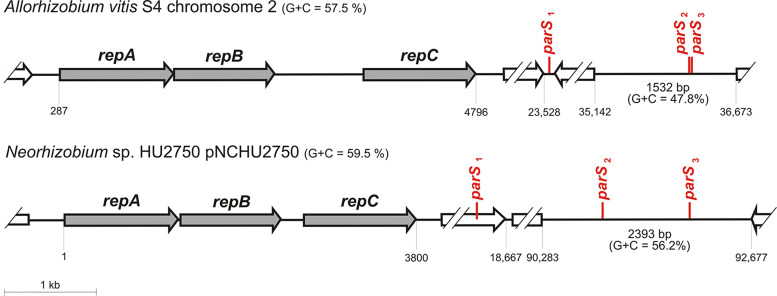
In Rhizobiales, the parS sequences tend to be localized in the intergenic region directly upstream of repA, and in the intergenic region between repA and repB (if these genes do not overlap), or at the 5′ end of repB (Fig. 3). There are some exceptions to this pattern, with the most interesting example present in chromosome 2 of Allorhizobium vitis S4 and its close relatives. These replicons do not have any parS sites located in the repABC operon. However, they possess a long (1.5-2.4 kb) non-coding AT-rich region located far downstream of repC (30-90 kb), containing two parS repeats (Fig. 4). This region resembles 3parS, but it has an independent evolutionary history. Interestingly, all replicons carrying this 3parS-like region, have an additional degenerate parS (1 nt different from other parS sites in the same replicon) located 15–20 kb downstream of repC in a short intergenic region (∼100 bp) or within a gene (Fig. 4, Table S2 in the supplemental material).
FIG 4.
Distribution of putative parS sites in Allorhizobium vitis S4 chromosome 2 (NC_011988.1) and Neorhizobium sp. HU2750 plasmid pNCHU2750 (CP030828.1).
DISCUSSION
Long inter-ORF regions are rare in bacteria and when they occur they often display regulatory functions. In this study, we identified and characterized such a region in P. aminophilus megaplasmid pAMI4. This locus, referred to as 3parS, carries three functional parS centromere-like sites recognized by the RepBpAMI4 protein, which are essential for pAMI4 stability. Additional unusual features of 3parS (high % AT compared to the whole plasmid and proximity to a region rich in GANTC sites, which are methylated by the CcrM methyltransferase, an important player in Alphaproteobacteria cell cycle regulation) indicate that this locus may be involved in other uncharacterized layers of pAMI4 maintenance regulation.
The role of the three parS locus is not immediately evident, as the repABC operon of pAMI4 contains another parS site, which could mediate the plasmid’s partition. However, we demonstrated that 3parS is required for proper maintenance of this plasmid. The single parS site located between repB and repC is not sufficient to act as a focus for RepBpAMI4 nor to sustain full stability of pAMI4 under the tested conditions. This indicates possible specialization of the parS sites into those that play a leading role in partition (the sites located in the 3parS region) and those performing another function (the site between repB and repC). Indeed, the latter parS site is located in a region where a gene encoding ctRNA RepE, a negative regulator of RepC translation, was identified in other repABC replicons (5). Thus, this site may participate in RepB-dependent regulation of RepE transcription, which might be a specific mechanism to modulate RepC expression.
Similar parS specialization might be common in other repABC replicons of Rhodobacterales. They frequently have the parS site in the repB-repC intergenic region, but also possess multiple parS repeats (two to four) located up to 4 kb downstream of repC (Fig. 3, Fig. S2 in the supplemental material). The pAMI4 3parS seems to have evolved from these latter sites, which were moved away from repC by an insertion. It is impossible to state whether such a new configuration was positively selected and conserved, as we lack information about the repABC systems that are very closely related to that of pAMI4 (the closest relatives of RepCpAMI4 in GenBank NCBI share only around 66% aa identity).
Interestingly, the parS sites located downstream of repC are found not only in the intergenic region directly after this gene. Many of them are located in subsequent intergenic regions or within ORFs (Fig. 3, Fig. S2 in the supplemental material). The most intriguing example of parS sequences located within ORFs are those identified in cas genes (cas3 – encoding ATP-dependent single-strand DNA translocase/helicase that degrades DNA as part of CRISPR based immunity; casA and casB – encoding CRISPR-associated proteins of the multi-subunit ribonucleoprotein Cascade complex (19)). This arrangement was identified in six contigs originating from six different species (Marinibacterium profundimaris, Frigidobacter mobilis, Tabrizicola sp. YIM 78059, Ruegeria intermedia, Jhaorihella thermophila and Paenimaribius caenipelagi; the contig numbers may be found in Tables S1 and S2), and seems to have evolved several times, as judged by RepB- and Cas3-based phylogenetic trees (compare Fig. S2 and Fig. S3 in the supplemental material). We hypothesize that as the presence of multiple parS sites located downstream of repC may be advantageous for repABC replicons of Rhodobacterales, they are positively selected even when present within coding sequences (they may arise in this location de novo or they may be present in incoming genes that already contain such sequences).
We observed another pattern of parS distribution in Rhizobiales, with parS sites located upstream of repA and in the repA/repB intergenic region, or at the 5′ end of repB. Specialized roles for parS sites at such locations have been proposed and tested previously, i.e., transcription regulation of the repABC operon for sequences located upstream of repA and transcription regulation of repC only for sequences located in the repA/repB region (5). It is noteworthy that repABC replicons of Rhizobiales are often large (>1 Mb) and fulfill important biological roles for their hosts, e.g., symbiotic megaplasmids, essential chromids. Especially in these largest repABC replicons (pSymA, Rhizobium rhizogenes K84 chromosome 2, Agrobacterium tumefaciens chromosome 2), we observed multiplication of parS upstream of repA and the absence of the parS site located in the repA/repB region, which may reflect specific regulatory issues important for the maintenance of such large DNA molecules. Interestingly, in this group of replicons, we identified a locus which resembles 3parS of pAMI4. It is a non-coding, AT-rich region containing two parS repeats located 30–90 kb downstream of repC, which is found in Allorhizobium vitis S4 chromosome 2 and related replicons. As the closest relatives of these replicons carry multiple parS sites upstream of repA, as do most repABC replicons in Rhizobiaceae, we suppose that this locus could have arisen by translocation of the repA upstream region to a position far downstream of repC.
The reasons for the different patterns of parS localization in repABC replicons of Rhodobacterales and Rhizobiales remain unclear. However, they may reflect differences in the features of these replicons in these two taxonomic orders. In Rhodobacterales, the repABC replicons appear to be smaller and less domesticated, while they are larger and better domesticated in Rhizobiales (and thus have more regulatory mechanisms to synchronize their replication and partition with the cell cycle) (20).
The 3parS and similar loci described in this study may only be interesting exceptions from the general parS localization patterns typical for Rhodobacterales and Rhizobiales. Nevertheless, their specific features (low % GC, CcrM binding sites in the case of 3parS of pAMI4) and evolutionary conservation (the case of A. vitis S4 chromosome 2 and its relatives) may lead to speculation that moving the parS function further from the repABC region is important for a specific mode of replication and partition, and regulation of these processes.
MATERIALS AND METHODS
Bacterial strains cultivation.
Paracoccus spp. were grown at 30°C on LB (or LA) supplemented with antibiotics (rifampicin, 50 μg/mL; kanamycin, 50 μg/mL; gentamicin, 40 μg/mL; tetracycline, 2 μg/mL), sucrose (11% wt/vol) or taurine (20 mM), as required. E. coli was grown at 37°C in LB (or LA) medium, supplemented with antibiotics (kanamycin 50 μg/mL; gentamicin 40 μg/mL; tetracycline, 20 μg/mL) and diaminopimelate (DAP) (0.3 mM), as required.
Genetic manipulations.
Routine DNA manipulation procedures were performed according to standard methods (21). Some plasmids were constructed by Gibson assembly (22). E. coli plasmid DNA was isolated using a Plasmid Mini isolation kit (A&A Biotechnology). P. aminophilus plasmid DNA was isolated using an alkaline lysis procedure (21). Bacterial genomic DNA was isolated using a GeneMATRIX Bacterial & Yeast Genomic DNA purification kit (EurX). Plasmid DNA was introduced into E. coli cells by the chemical transformation method of Kushner (23) and into Paracoccus spp. cells by bi- and triparental mating procedures (11). Genetic modifications of pAMI4 were performed by a gene replacement technique adapted to P. aminophilus (11, 24).
Bacterial strains used in this study.
E. coli DH5α (21) was used as a standard cloning strain and as a donor for triparental mating. E. coli β2163 (dapA) (25) was used as a cloning strain for ori R6K plasmid pDS132 and as a donor for biparental mating for gene replacement in P. aminophilus. E. coli BL21 (26) was used as a strain for 6×His-RepBpAMI4 recombinant protein overexpression. P. aminophilus JCM 7686R (27) was used as a strain carrying the wild type pAMI4 megaplasmid. P. aminovorans JCM 7685R (17) was used as a heterologous host in plasmid stability assays. P. aminophilus pAMI4Tc was constructed based on P. aminophilus JCM 7686R by inserting a tetracycline resistance gene (tetC) between 383337 and 383338 bp of pAMI4 in a location which is neutral for maintenance functions of the megaplasmid. P. aminophilus pAMI4TcΔ3parS was constructed based on P. aminophilus pAMI4Tc by deleting the whole 3parS region, between 424710 and 426551 bp of pAMI4, and inserting a kanamycin resistance gene (aph) in its place. P. aminophilus UW100 was constructed by removing pAMI4 from P. aminophilus JCM 7686 by incompatibility with 3parS delivered on pBBR1MCS-5 and subsequent loss of the vector by cultivation without the antibiotic pressure.
Plasmid used in this study.
pBBR1MCS-5 (28) was used as a vector to introduce 3parS (amplified pAMI4 fragment between 424627 and 426685 bp) to P. aminophilus. pBBR1MCS-3 (28) was used as a source of the tetC gene. pRK2013 (29) was used as a helper plasmid for triparental mating. pABW3 was used as an unstable vector for stability assays in P. aminophilus and P. aminovorans. Its four derivatives constructed for the stability assays carried (i) repABpAMI4 (amplified pAMI4 fragment between 1312 and 3696 bp), (ii) repABpAMI4 with parSBC (amplified pAMI4 fragment between 1184 and 3696 bp), (iii) repABpAMI4 ([same as in (i)] and 3parS (amplified pAMI4 fragment between 424627 and 426685 bp), and (iv) repABpAMI4 with parSBC [as in (ii)] and 3parS [as in (iii)]. pDS132 (ori R6K) (30) was used as a suicide plasmid with counterselection gene (sacB) for gene replacement in P. aminophilus. Its derivatives were used to insert the tetC gene into pAMI4 and to delete the 3parS locus. pDYI-Km (31) was used as a source of the aph gene. pET28b(+) (EMD Biosciences) was used as a plasmid for overexpression of the 6×His-RepBpAMI4 protein (repBpAMI4 gene was amplified with primers 5′-CATAAGCTTTCACCCATCGGACTTAGCCTTG-3′ and 5′-GGTCATATGGCACGCAAGGATCTTCTCAAAG-3′ and cloned into pET28b(+) using HindIII and NdeI). pUC18 (32) was used as a cloning vector of artificial DNA fragments carrying single parS sequences and the 3parS locus. pJC43 was used for expression of a GFP-RepBpAMI4 fusion protein from PtauAB promoter positively induced by taurine in the presence of TauR (33, 34). pZE1-GFP (35) was a source of the gfp gene.
Plasmid stability assays.
Stability of pABW3 and pAMI4 derivatives was tested by monitoring the percentage of antibiotic resistant bacteria (kanamycin for pABW3 derivatives and tetracycline for pAMI4 derivatives) in populations grown in liquid cultures without antibiotics, as described previously (36). Cultures of Paracoccus spp. for the stability assays (20 mL in 100 mL flasks) were refreshed at 24 h intervals (dilution 1:1000) and continued for up to approximately 200 generations (ca. 400 h, 2 h/generation). In selected time points, the cultures were plated on LA in appropriate dilutions to obtain single colonies, which were tested for the antibiotic resistance by replicas on LA and LA with kanamycin or tetracycline. All stability assays were performed in triplicate.
Overexpression and purification of recombinant 6×His-RepBpAMI4 protein.
The repB gene of pAMI4 was amplified by PCR and cloned into expression vector pET28b(+), to obtain pET28b+repB. RepBpAMI4 with an N-terminal 6×His tag was overexpressed in E. coli BL21(DE3)(pET28b+repB) and purified as described previously (37) with some modifications. A 1- L culture of the expression strain was grown to mid-log phase and production of the 6×His-RepBpAMI4 protein was induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.4 mM. After further incubation for 5 h at 28°C the cells were harvested by centrifugation and lysed in 15 mL of lysis buffer (50 mM sodium phosphate pH 8.0, 300 mM NaCl, 10 mM imidazole and 300 μL of 100 mg/mL lysozyme, supplemented with 1 mM PMSF and Protease Inhibitor Cocktail; Sigma). After sonication and centrifugation, the cleared lysate was incubated with 0.5 mL of Ni-NTA beads (Qiagen) for 30 min, with gentle shaking. The Ni-NTA resin was then given a series of washes with (i) 3 mL of W1 buffer (50 mM sodium phosphate pH 8.0, 300 mM NaCl, 10 mM imidazole), (ii) 2 mL of W2 buffer (50 mM sodium phosphate pH 8.0, 2 M NaCl, 10 mM imidazole), (iii) 16 mL of W3 buffer (50 mM sodium phosphate pH 8.0, 300 mM NaCl, 50 mM imidazole). The 6×His-tagged protein was eluted in 3 lots of 0.5 mL of elution buffer (50 mM sodium phosphate pH 8.0, 300 mM NaCl, 250 mM imidazole). The final concentration of the purified recombinant protein was estimated using the Bradford dye-binding method (38). Protein aliquots were frozen in liquid nitrogen and stored at −70°C.
Production of fluorescein (FAM)-labeled DNA fragments.
The 3parS DNA fragment of pAMI4 (519 bp) was amplified by PCR with primers 5′-ATCCCCGGGCTTATGGGATTCATCGCCGCTG-3′ and 5′-ATCCCCGGGCAAATCTGTCGCGAGCCAGC-3′. To create individual partitioning sites, pairs of complementary synthetic oligonucleotides containing single parS sequences (primers 5′-AATTACGCTGTTGACAGCTGTCAACGAAAG-3′ and 5′-GATCCTTTCGTTGACAGCTGTCAACAGCGT-3′ for parSBC; 5′-AATTCCGCTGTTGACAGCTGTCAACTCGTG-3′ and 5′-GATCCACGAGTTGACAGCTGTCAACAGCGG-3′ for parS1; 5′-AATTGACGAGTTGACAGCTGTCAACGAGGC-3′ and 5′-GATCGCCTCGTTGACAGCTGTCAACTCGTC-3′ for parS2; 5′-AATTGCTCGGTTGACAGCTGTCAACTCGGG-3′ and 5′-GATCCCCGAGTTGACAGCTGTCAACCGAGC-3′ for parS3) were mixed together at a molar ratio of 1:1. Diluted oligonucleotide mixtures (10 μmol of each) were incubated at 95°C for 10 min and then slowly cooled to room temperature. The annealed synthetic dsDNA fragments, as well as the 3parS region amplicon were cloned into the SmaI site of pBGS18. The resulting plasmids were used as templates in PCR with standard primers M13pUCf (5′-CCAGTCACGACGTTGTAAAACG-3′) and FAM-labeled M13pUCrFAM (5′-AGCGGATAACAATTTCACACAGG-3′). The same primer pair was used for the amplification of a 136-bp DNA fragment of pBGS18, which served as a negative control.
DNA binding assay.
DNA-protein complexes were detected using a gel electrophoresis mobility shift assay (EMSA). First, competitor DNA (salmon sperm DNA) was incubated with 0, 10, 20, 50, 100, 150 or 200 pM of purified 6×His-RepBpAMI4 protein for 10 min at room temperature in a 12 μL volume of binding buffer (40 mM Tris-HCl pH 8.0, 10 mM MgCl2, 1 mM dithiothreitol, 100 mM KCl, 10% BSA). Then, 1 pmol of each FAM-labeled DNA fragment was added to each variant (8 μL). Next, 4 μL of 50% glycerol were added to the reaction mixtures before loading on a gel. The complexes were separated by electrophoresis in 5% non-denaturing polyacrylamide (19:1 acrylamide:bisacrylamide) gels cast in 0.25 × TBE (22.5 mM Tris base, 22.5 mM boric acid, 0.5 mM EDTA, pH 8.0) and run in 0.5 × TBE. Fluorescent DNA-protein complexes were visualized with a GE Healthcare AI600 imager.
Fluorescence microscopy.
Plasmid pJC43-GFP-RepB was constructed for taurine-dependent expression of a GFP-RepBpAMI4 fusion protein and introduced from E. coli into P. aminophilus JCM 7686R carrying pAMI4, pAMI4Tc or pAMI4TcΔparS and strain UW100 lacking pAMI4. Overnight cultures of the strains carrying pJC43-GFP-RepB, grown on LB with gentamicin, were diluted 1:1000 in LB with gentamicin plus taurine (or without taurine for the negative control) and grown to mid-exponential phase (OD600 0.2) prior to imaging. Cells were transferred to 1.5% agarose-padded slides containing LB medium with gentamicin and taurine (or without taurine for the negative control). A coverslip was placed on top of the agarose pad and sealed with a vaseline:lanolin:paraffin mix (ratio 1:1:1) to prevent pad evaporation. Slides were placed under a Zeiss ApoTome inverted wide-field microscope. Bacterial cells and the GFP-RepBpAMI4 signal were imaged in the brightfield and the green channel respectively using a Plan Apo 63× objective (numerical aperture = 1.4, +optovar 1.6×) and a Hamamatsu sCMOS ORCA-Flash 4.0 v3 (Institut Pasteur Imaging Facility Imagopole). Images were taken using a 600 msec exposure. For each experiment, randomly selected images of 100 cells were analyzed to track the localization of the GFP-RepBpAMI4 fusion using ImageJ Fiji software (NIH) (39). The GFP fluorescent signal was only observed where taurine was added to the culture, and not in the non-induced controls.
Bioinformatic analyses.
Sequence similarity searches of the GenBank database were performed using online BLAST tools (https://blast.ncbi.nlm.nih.gov) (40). The GC profile of plasmid sequences was determined using Artemis software (41). Protein phylogenetic trees were generated using MEGA software (42–44) by the maximum likelihood method with 500 bootstrap replications or by the neighbor joining method with 1000 bootstrap replications (for the tree presented in Fig. 3, the neighbor joining method was selected for ease of presentation, as the maximum likelihood tree of the same set of sequences showed a more complicated topology for branches with poor statistical support. For comparison, the maximum likelihood tree is presented in Fig. S4 in the supplemental material. Putative parS sites within the repABC region (defined as the region including repA, repB and repC plus the adjacent intergenic regions) were identified as palindromic or almost palindromic (up to one nucleotide position disturbing the palindrome) sequences matching the repABC parS consensus (5′-GTTNNNNGCNNNNAAC-3′) (5). If such sequences were found in the repABC region, they were used to search the whole available plasmid sequence for additional putative copies of parS, with a tolerance of a one nucleotide change.
ACKNOWLEDGMENTS
This work was supported by the National Science Centre (NCN), Poland, on the basis of decision no. DEC-2013/09/B/NZ1/00133. J.C. was the recipient of a long-term post-doctoral fellowship from Roux-Cantarini foundation (Institut Pasteur). J.C. work was funded by the French Government's Investissement d'Avenir program Laboratoire d'Excellence Integrative Biology of Emerging Infectious Diseases [ANR-10-LABX-62- IBEID].
Footnotes
Supplemental material is available online only.
Contributor Information
Jakub Czarnecki, Email: jakub.czarnecki@pasteur.fr.
Dariusz Bartosik, Email: bartosik@biol.uw.edu.pl.
Maia Kivisaar, University of Tartu.
REFERENCES
- 1.Livny J, Yamaichi Y, Waldor MK. 2007. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J Bacteriol 189:8693–8703. 10.1128/JB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawalek A, Wawrzyniak P, Bartosik AA, Jagura-Burdzy G. 2020. Rules and exceptions: the role of chromosomal ParB in DNA segregation and other cellular processes. Microorganisms 8. 10.3390/microorganisms8010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soh YM, Davidson IF, Zamuner S, Basquin J, Bock FP, Taschner M, Veening JW, De Los Rios P, Peters JM, Gruber S. 2019. Self-organization of parS centromeres by the ParB CTP hydrolase. Science 366:1129–1133. 10.1126/science.aay3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartosik D, Szymanik M, Wysocka E. 2001. Identification of the partitioning site within the repABC-type replicon of the composite Paracoccus versutus plasmid pTAV1. J Bacteriol 183:6234–6243. 10.1128/JB.183.21.6234-6243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cevallos MA, Cervantes-Rivera R, Gutierrez-Rios RM. 2008. The repABC plasmid family. Plasmid 60:19–37. 10.1016/j.plasmid.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Pinto UM, Pappas KM, Winans SC. 2012. The ABCs of plasmid replication and segregation. Nat Rev Microbiol 10:755–765. 10.1038/nrmicro2882. [DOI] [PubMed] [Google Scholar]
- 7.Petersen J, Brinkmann H, Pradella S. 2009. Diversity and evolution of repABC type plasmids in Rhodobacterales. Environ Microbiol 11:2627–2638. 10.1111/j.1462-2920.2009.01987.x. [DOI] [PubMed] [Google Scholar]
- 8.MacLellan SR, Smallbone LA, Sibley CD, Finan TM. 2005. The expression of a novel antisense gene mediates incompatibility within the large repABC family of alpha-proteobacterial plasmids. Mol Microbiol 55:611–623. 10.1111/j.1365-2958.2004.04412.x. [DOI] [PubMed] [Google Scholar]
- 9.Urakami T, Araki H, Oyanagi H, Suzuki K, Komagata K. 1990. Paracoccus aminophilus sp. nov. and Paracoccus aminovorans sp. nov., which utilize N,N-dimethylformamide. Int J Syst Bacteriol 40:287–291. 10.1099/00207713-40-3-287. [DOI] [PubMed] [Google Scholar]
- 10.Dziewit L, Czarnecki J, Wibberg D, Radlinska M, Mrozek P, Szymczak M, Schluter A, Pühler A, Bartosik D. 2014. Architecture and functions of a multipartite genome of the methylotrophic bacterium Paracoccus aminophilus JCM 7686, containing primary and secondary chromids. BMC Genomics 15:124. 10.1186/1471-2164-15-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dziewit L, Czarnecki J, Prochwicz E, Wibberg D, Schluter A, Pühler A, Bartosik D. 2015. Genome-guided insight into the methylotrophy of Paracoccus aminophilus JCM 7686. Front Microbiol 6:852. 10.3389/fmicb.2015.00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton JR, Woodcock CB, Opot SB, Reich NO, Zhang X, Cheng XD. 2019. The cell cycle-regulated DNA adenine methyltransferase CcrM opens a bubble at its DNA recognition site. Nat Commun 10:4600. 10.1038/s41467-019-12498-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marinus MG. 1987. DNA methylation in Escherichia coli. Annu Rev Genet 21:113–131. 10.1146/annurev.ge.21.120187.000553. [DOI] [PubMed] [Google Scholar]
- 14.Laub MT, Chen SL, Shapiro L, McAdams HH. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci USA 99:4632–4637. 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon JE, Christie PJ. 2014. The Agrobacterium Ti plasmids. Microbiol Spectr 2. 10.1128/microbiolspec.PLAS-0010-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novick RP. 1987. Plasmid incompatibility. Microbiol Rev 51:381–395. 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Czarnecki J, Dziewit L, Puzyna M, Prochwicz E, Tudek A, Wibberg D, Schluter A, Pühler A, Bartosik D. 2017. Lifestyle-determining extrachromosomal replicon pAMV1 and its contribution to the carbon metabolism of the methylotrophic bacterium Paracoccus aminovorans JCM 7685. Environ Microbiol 19:4536–4550. 10.1111/1462-2920.13901. [DOI] [PubMed] [Google Scholar]
- 18.MacLellan SR, Zaheer R, Sartor AL, MacLean AM, Finan TM. 2006. Identification of a megaplasmid centromere reveals genetic structural diversity within the repABC family of basic replicons. Mol Microbiol 59:1559–1575. 10.1111/j.1365-2958.2006.05040.x. [DOI] [PubMed] [Google Scholar]
- 19.Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA. 2014. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci USA 111:6618–6623. 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fournes F, Val ME, Skovgaard O, Mazel D. 2018. Replicate once per cell cycle: replication control of secondary chromosomes. Front Microbiol 9:1833. 10.3389/fmicb.2018.01833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual 3rd ed Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 22.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 23.Kushner SR. 1978. An improved method for transformation of E. coli with ColE1 derived plasmids, p 17–23. In: Boyer HB, Nicosia S (ed), Genetic Engineering. Elsevier/North-Holland, Amsterdam, Netherlands. [Google Scholar]
- 24.Morton R, Hooykaas PJJ. 1995. Gene replacement. Molecular Breeding 1:123–132. 10.1007/BF01249697. [DOI] [Google Scholar]
- 25.Demarre G, Guérout AM, Matsumoto-Mashimo C, Rowe-Magnus DA, Marlière P, Mazel D. 2005. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol 156:245–255. 10.1016/j.resmic.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 27.Bartosik D, Baj J, Bartosik AA, Wlodarczyk M. 2002. Characterization of the replicator region of megaplasmid pTAV3 of Paracoccus versutus and search for plasmid-encoded traits. Microbiology (Reading) 148:871–881. 10.1099/00221287-148-3-871. [DOI] [PubMed] [Google Scholar]
- 28.Kovach ME, Phillips RW, Elzer PH, Roop RM, Peterson KM. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802. [PubMed] [Google Scholar]
- 29.Ditta G, Stanfield S, Corbin D, Helinski DR. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA 77:7347–7351. 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255. 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Dziewit L, Adamczuk M, Szuplewska M, Bartosik D. 2011. DIY series of genetic cassettes useful in construction of versatile vectors specific for Alphaproteobacteria. J Microbiol Methods 86:166–174. 10.1016/j.mimet.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 32.Norrander J, Kempe T, Messing J. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101–106. 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 33.Tett AJ, Rudder SJ, Bourdès A, Karunakaran R, Poole PS. 2021. Regulatable vectors for environmental gene expression in Alphaproteobacteria. Appl Environ Microbiol 78:7137–7140. 10.1128/AEM.01188-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mostafavi M, Lewis JC, Saini T, Bustamante JA, Gao IT, Tran TT, King SN, Huang Z, Chen JC. 2014. Analysis of a taurine-dependent promoter in Sinorhizobium meliloti that offers tight modulation of gene expression. BMC Microbiol 14:295. 10.1186/s12866-014-0295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Da Re S, Le Quéré B, Ghigo JM, Beloin C. 2007. Tight modulation of Escherichia coli bacterial biofilm formation through controlled expression of adhesion factors. Appl Environ Microbiol 73:3391–7403. 10.1128/AEM.02625-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czarnecki J, Dziewit L, Kowalski L, Ochnio M, Bartosik D. 2015. Maintenance and genetic load of plasmid pKON1 of Paracoccus kondratievae, containing a highly efficient toxin-antitoxin module of the hipAB family. Plasmid 80:45–53. 10.1016/j.plasmid.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Wawrzyniak P, Sobolewska-Ruta A, Zaleski P, Lukasiewicz N, Kabaj P, Kieryl P, Goscik A, Bierczynska-Krzysik A, Baran P, Mazurkiewicz-Pisarek A, Plucienniczak A, Bartosik D. 2019. Molecular dissection of the replication system of plasmid pIGRK encoding two in-frame Rep proteins with antagonistic functions. BMC Microbiol 19:254. 10.1186/s12866-019-1595-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 39.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–469. 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 44.Saitou N, Nei M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4. Download aem.00207-22-s0001.pdf, PDF file, 0.8 MB (834.6KB, pdf)
Table S1. Download aem.00207-22-s0002.xlsx, XLSX file, 0.05 MB (54.5KB, xlsx)
Table S2. Download aem.00207-22-s0003.xlsx, XLSX file, 0.05 MB (49.2KB, xlsx)