Abstract
Nanomaterials (NMs) are widely used in commercial and medical products, such as cosmetics, vaccines, and drug carriers. Exposure to NMs via various routes such as dermal, inhalation, and ingestion has been shown to gain access to the systemic circulation, resulting in the accumulation of NMs in the liver. The unique organ structures and blood flow features facilitate the liver sequestration of NMs, which may cause adverse effects in the liver. Currently, most in vivo studies were focused on NMs accumulation at the organ level and evaluation of the gross changes in liver structure and functions, however, cell-type-specific uptake and responses, as well as the molecular mechanisms at cellular levels linking the effects at organ levels are lagging. Herein, we systemically reviewed the diverse interactions of NMs with the liver, specifically on major liver cell types including Kupffer cells (KCs), liver sinusoidal endothelial cells (LSECs), hepatic stellate cells (HSCs), and hepatocytes as well as the detailed molecular mechanisms involved. In addition, we also reviewed the knowledge gained on nano-liver interactions that could facilitate the development of safer nanoproducts and nanomedicine.
Keywords: Nano-liver interaction, nanosafety, nanomedicine, cell-type-specific uptake, mode of action
Graphical Abstract
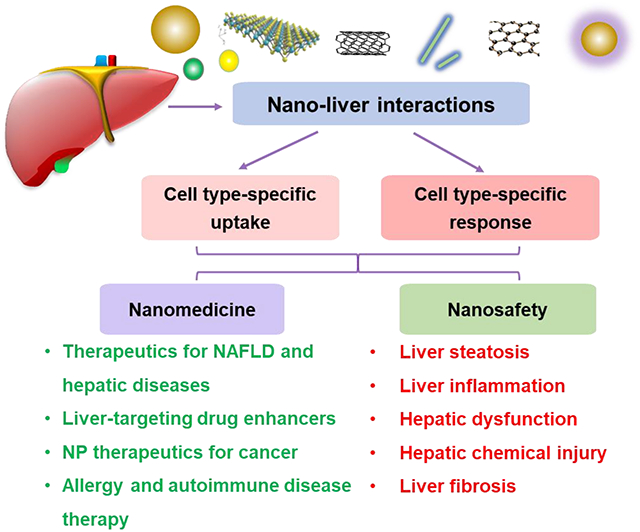
The widespread exposure of nanomaterials to the body and slow clearance during systemic circulation leads to nanomaterial liver accumulation. This induces cell-type-specific uptake and responses after exposure of representative NMs with various physicochemical properties to the liver and major liver cell types. Furtherly, understanding nano-liver interactions help to develop safer nanoproducts and nanomedicine.
1. Introduction
Nanomaterials (NMs) are engineered with sizes ranging from 1 to 100 nm in at least one dimension. They are designed to have unique chemical and physical properties that could substantially enhance the functional performance of a product compared to their bulk counterparts.[1–3] Since the launching of the National Nanotechnology Initiative (NNI), both NMs and nanoproducts have achieved rapid development in a variety of industrial, commercial, and medical fields that include bio-imaging, vaccines, and drug carriers (Figure 1). The functional advantages of NMs are derived from their unique physical and chemical properties.[1,4–6] Among all the NMs, metallic NMs including metal and metal oxides (MOx) nanoparticles (NPs), such as Ag and Au NPs, transition-metal oxides (TMOs, e.g., SiO2, Co3O4, and Mn2O3), and rare-earth oxides REOs (e.g., Gd2O3, La2O3, and Y2O3) NPs, are among the most produced NMs worldwide. Also, they are widely used in consumer products such as dietary supplements, fuel additives, cosmetics, bioimaging, and drug carriers, etc. due to their properties such as small size, high surface area, controlled particle shape as well as superior mechanical, electronic, optical, and magnetic properties.[7–9] For example, SiO2 NPs have been applied in the theranostic fields, including bioimaging and targeted drug delivery.[7] In addition, the popularity of carbon nanomaterials (CNMs) including zero-dimension fullerenes, one-dimensional carbon nanotubes (CNTs), and two-dimensional graphene-based nanoparticles (GBN) including graphene, graphene oxide (GO), and reduced graphene oxide (rGO) has been on the rise for their applications in battery, electronics, drug delivery, bio-sensing, bio-imaging, and tissue engineering due to their large surface area, diverse surface functional groups, excellent optical property, and efficient drug-loading capacity.[10–16] For example, an advanced NMs-based biosensing platform based on rGO has been developed to detect the coronavirus disease 2019 (COVID-19) antibodies within seconds.[17] Additionally, a form of nanostructured cellulose (nanocellulose), including cellulose nanocrystal (CNC) and cellulose nanofiber (CNF) has been increasingly considered for applications in papermaking, coatings, food, nanocomposite formulations and reinforcement, and biomedical fields (e.g., wound dressing) due to its biocompatibility, outstanding mechanical, chemical, and rheological properties.[18,19] Also, two-dimensional (2D) transition metal dichalcogenides (TMDs) (including MoS2 and BN) with high surface area and free surface energy levels are increasingly being used for commercial applications in energy generation, sensors, catalysis, electronics, and biomedicine fields.[13,20–22] Similarly, organic NPs (including lipid, liposome, polymer, micelle, dendrimer, and protein/peptide-based NPs) have been widely used in diagnosis, drug delivery, bioimaging, and cancer therapy because of their facile synthesis and chemical modification, self-assembly, biocompatibility, and biodegradability.[23–25] The global market value of NMs will be expected to reach US$ 125 billion marks by 2024.[26] The widespread production and use of NMs increase the possibility of human exposure via various exposure routes including dermal, inhalation, ingestion, and intravenous injection to NMs, raising significant public health concerns.[27,28]
Figure 1.
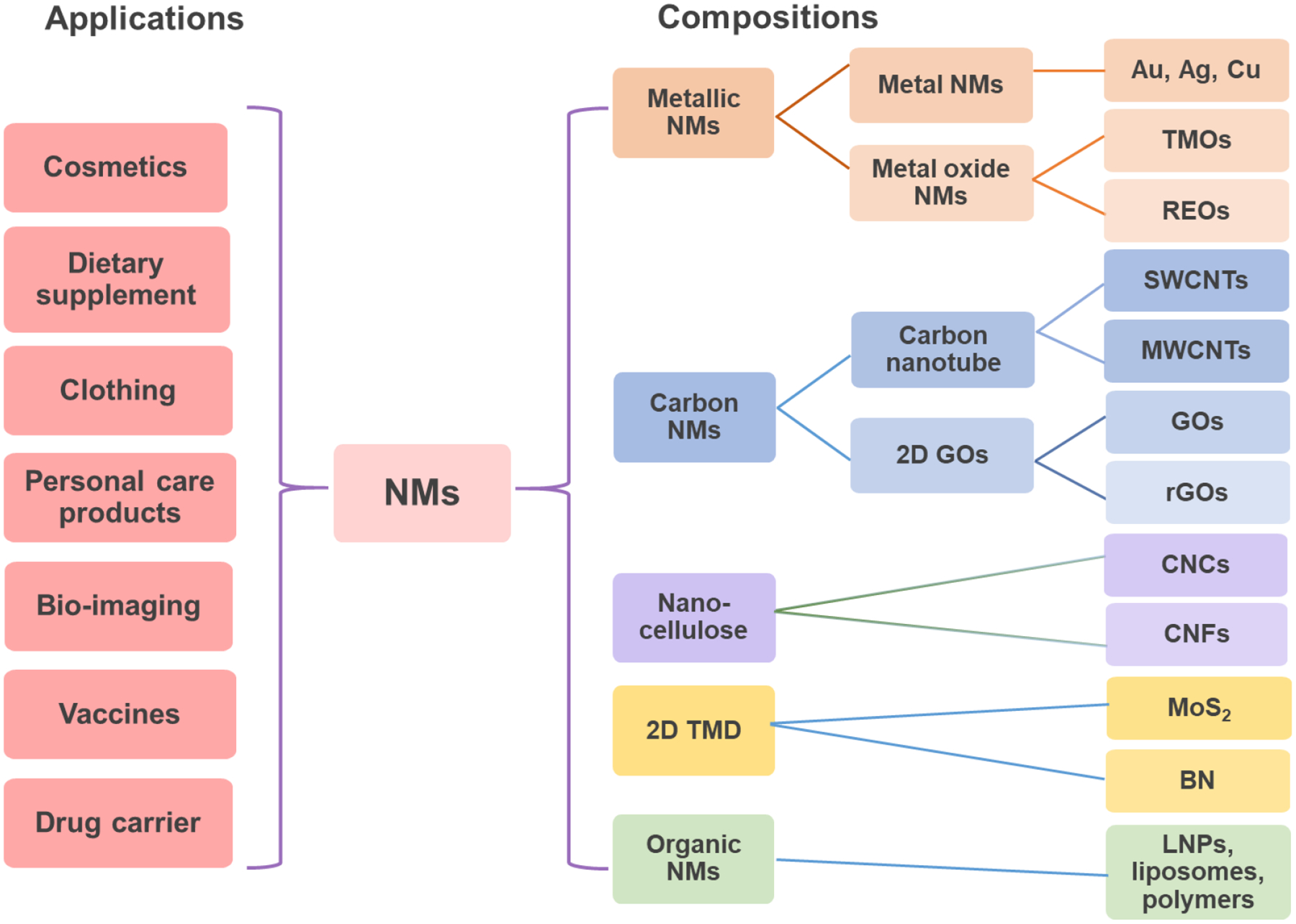
Major applications and compositions of nanomaterials (NMs) interacted with the liver. NMs have been widely applied in industrial, commercial, and medical fields, which include bio-imaging, vaccines, and drug carriers. NMs have diverse compositions, which include metallic NMs including metal (Ag and Au nanoparticles) and metal oxide (MOx) NMs including transition-metal oxides (TMOs, e.g., SiO2, Co3O4, and Mn2O3) and rare-earth oxides (REOs, e.g., Gd2O3, La2O3, and Y2O3) NPs, carbon NMs including one-dimensional carbon nanotubes (CNTs) and two-dimensional (2D) graphene-based NPs (graphene oxide, GO, and reduced graphene oxide, rGO), cellulose nanocrystal (CNC) and cellulose nanofiber (CNF), 2D transition metal dichalcogenide (TMD) including MoS2 and BN, and organic NMs including lipid NPs, liposomes, and polymer NPs.
While there are numerous comprehensive reviews on the environmental health impacts of NMs,[29–34] this current review is focused on the recent findings on NMs-induced effects in the liver. We focused on cell-type-specific interactions and the molecular mechanisms involved in the nano-liver interactions. In addition, we reviewed novel nanotherapeutics that could be developed based on the understanding of nano-liver interactions.
The liver is the largest solid organ and holds about 13% of the body’s blood supply at any given time, performing indispensable roles of metabolism, xenobiotic detoxification, glycogen storage, bile formation, protein synthesis, etc., maintaining the general homeostasis in the body.[35] The liver consists of 2 main lobes made up of 8 segments consisting of 1,000 lobules (small lobes). Each lobule is roughly hexagonal and is connected by the hepatic artery that delivers oxygenated blood from the general circulation, the portal vein that delivers deoxygenated blood from the small intestine containing nutrients, and the bile duct (canaliculi) that carries bile from the liver to be released into the gastrointestinal tract to facilitate digestion. These portal blood vessels branch into capillary-like structures named sinusoids, which receive blood from terminal branches of the hepatic artery and portal vein at the periphery of lobules and deliver it into central veins draining into the inferior vena cava (Figure 2).[36–40] Sinusoids are lined with liver sinusoidal endothelial cells (LSECs) and flanked by plates of hepatocytes, allowing larger molecules, including plasma proteins as well as small particles, to leave and reenter the bloodstream due to the presence of fenestrations in the LSEC layers. Liver cells could be divided into 2 major groups, the parenchymal hepatocytes, which account for about 60–80% of the total number of liver cells, and the non-parenchymal cells, which occupy approximately 20–40% of the total cells (Figure 3). The non-parenchymal cells consist of the LSECs, approximately 50% of the total number of non-parenchymal cells, liver resident macrophage designated Kupffer cells (KCs, approximately 20%), lymphocytes (approximately 25%), biliary cells (approximately 5%), and hepatic stellate cells (approximately 1–8%) that are found in the perisinusoidal space of Disse, a location between LSEC layer and hepatocytes. Similar to KCs, the intrahepatic lymphocytes are also present in the sinusoidal lumen, including T lymphocytes (approximately 63%), natural killer (NK) cells (approximately 31%), B lymphocytes (approximately 6%), and less than 1% of dendritic cells (DCs). T lymphocytes include the conventional T cells such as CD4+ T cells and CD8+ T cells, and the unconventional T cells, including natural killer T (NKT) cells, TCRγδ T cells, and others (Figure 3). The unique liver lobule architecture and position of cells play a pivotal role in liver functions.[37,41–44] The liver has been the primary target or target of secondary spread for NMs gaining access to the systemic circulation, e.g., extrapulmonary translocation after lung exposure, gastrointestinal absorption, intramuscular, and direct intravenous injection, which will lead to accumulation of NMs in the liver.[45–47] A thorough understanding of the liver effects and nano-liver interactions is needed.
Figure 2.

Structural and functional organization of the liver. A) Anatomy of the liver and its blood supply. The vessel (red) represents the hepatic artery that delivers oxygenated blood from the general circulation. The vessel (blue) represents the hepatic portal vein that delivers deoxygenated blood from the small intestine containing nutrients. The vessel (green) represents the bile duct that carries bile from the liver and gallbladder to the duodenum. B) Schematic of a liver lobule in a hexagonal shape with rows of hepatocytes radiating out from the central vein towards the portal triad. C) Schematic demonstrates the blood flow of the liver via the portal vein and hepatic artery through the sinusoids to the central vein. A lobule could be divided into three zones, zone 1 (periportal), zone 2 (transition zone), and zone 3 (pericentral) based on oxygen gradient from high to low. D) Schematic sinusoids that receive blood from terminal branches of the hepatic artery and portal vein at the periphery of lobules and drain into central veins (red arrow), and the bile ducts that carry bile from the liver and gallbladder to the duodenum (green arrow). Sinusoids are lined with endothelial cells and flanked by plates of hepatocytes. E) Schematic shows the cross-section of a liver lobule and the flow direction of blood and bile. F) Spatial map to demonstrate flow velocities within the virtual sinusoid network. The red and yellow colors indicate a greater flow velocity while the blue color represents a lower flow velocity. Color bar units indicate μm/s. Figures 2A–B are reproduced under terms of the CC-BY license.[35] Copyright 2018, Øie et al., published by De Gruyter; Figure 2C is reproduced with permission.[44] Copyright 2010, Nature Publishing Group; Figure 2D is reproduced under terms of the CC-BY license.[39] Copyright 2005, Frevert et al., published by PLOS; Figures 2E is courtesy of Bio Ninja (https://ib.bioninja.com.au/options/option-d-human-physiology/d3-functions-of-the-liver/liver-structure.html) and used with permission; Figure 2F is reproduced under terms of the CC-BY license.[40] Copyright 2018, Fu et al., published by PLOS.
Figure 3.

The major cell types in the liver. It includes the parenchymal hepatocytes, which occupy about 60–80% of the total number of liver cells, and the non-parenchymal cells occupying approximately 20–40% of the total number. In the non-parenchymal cell, it consists of the liver sinusoidal endothelial cells (approximately 50% of the total number of non-parenchymal cells), phagocytic Kupffer cells (approximately 20%), lymphocytes (approximately 25%), biliary cells (approximately 5%), and hepatic stellate cells (approximately 1–8%). In the lymphocytes, it includes the T lymphocytes (approximately 63%), natural killer (NK) cells (approximately 31%), B lymphocytes (approximately 6%), and less than 1% of dendritic cells (DCs). In the T lymphocytes, it contains the conventional T cells, including CD4+ T cells and CD8+ T cells, and the unconventional T cells, including natural killer T (NKT) cells, TCRγδ T cells, and others.
Studies have found that certain NMs including metal and MOx NPs (e.g., Au and TiO2) as well as CNTs and GOs could lead to hepatic toxicity.[48] Animal studies have shown NMs could induce liver injury as reflected by increases in blood biomarkers including aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (ALP), lactate dehydrogenase (LDH), etc., as well as by changes in liver histology.[49,50]. However, most in vivo studies are focused on the accumulation of the NMs at the organ level and the gross effects mentioned above.[8,9] It is not clear about the effects of NMs on individual cell types and how these cellular responses combine and contribute to the gross adverse outcomes. Furthermore, the diverse liver cell populations and specific metabolic zonation are vital to maintaining the hepatic structure and functions, and to understand the liver effects of NMs, we need to zoom in on the cellular levels.[37,41] However, most in vitro studies are typically focused on the NMs effects of an individual liver cell type. In addition, in vitro studies are generally done using primary hepatocytes or cell lines cultured in a flask without spatial context (3D) of the liver. Still, we have obtained much valuable information on the effects of NMs at the cellular level. For example, many NMs have been shown to induce oxidative stress in liver cells, which may contribute to their adverse effects on the liver.[51,52]
To gain a better understanding of the effects of NMs on the liver, a thorough examination of how NMs interact with the liver cells is required. Thus, we systemically reviewed the current state of our understanding of the effects of diverse NMs including metallic NMs, CNTs, 2D TMDs, GBNs, and nanocelluloses on the liver. We focus on hepatocytes, KCs, LSECs, and HSCs because they make up the vast bulk of the liver cell population, and these are the principal cell types that are responsible for liver function and diseases. We compiled information on cell type-specific uptake and cellular responses, as well as the molecular pathways involved in NMs-induced effects on these major liver cells. Also, we lay out how the knowledge on nano-liver interactions could be used to develop safer applications of NMs to treat diseases, including cancer, liver disorders and diseases, and diseases such as allergies and autoimmune diseases.
2. Liver pathological changes induced by NMs
It is estimated that 30–99% of administered NPs will accumulate and sequester in the liver after administration into the body.[49,53] The presence and accumulation of certain NMs in the liver have been shown to induce oxidative stresses that in turn interrupt the liver’s metabolism and homeostasis.[44,54] Although the liver possesses the ability to repair and restore sections of damaged tissue following acute injury, the prolonged exposure to NMs could impair the regenerative capabilities, inducing repetitive injury, provoking or worsening the liver condition as well as leading to chronic liver disease.[44,55,56] Many studies reported the adverse effects of NMs on the liver.[56,57] This includes that metallic NMs-induced liver disorders or liver damage, CNTs-induced hepatic steatosis and liver injury, GBNs-induced hepatic dysfunction or liver functional zonation changes, etc. (Figure 4).
Figure 4.

NMs able to induce or exacerbate liver disorders. For example, the silver, gold, and silicon NPs exacerbated the liver disorders including liver fibrosis and steatosis; Fe3O4, ZnO, and CuO NPs induced liver damage by triggering higher levels of liver enzyme release; heavy metal NMs including Cd, Ag, and ZnO aggravated hepatic chemical injury induced by environmental toxins; CNTs induced hepatic steatosis and liver injury, and graphene-based NMs induced hepatic dysfunction or liver functional zonation changes.
2.1. Metallic NMs induced liver steatosis and fibrosis
The imbalance between lipid storage and utilization will lead to supraphysiological triglyceride accumulation in hepatocytes, known as hepatic steatosis.[58] Many metallic NMs including metal NMs have been shown to induce hepatic steatosis due to the residual NMs in the liver. The 10 nm spherical Au NPs have been reported to induce hepatic steatosis in Wistar-Kyoto rats.[59] Also, 60 nm spherical Si NPs demonstrated increased hepatic steatosis via the TLR5-signaling pathway in mice or a zebrafish model.[60] Furthermore, Jia et al. demonstrated that Ag NPs at a safe dose in normal mice promoted the progression of fatty liver disease from steatosis to steatohepatitis in the overweighted mice. This disease progression was ascribed to the pro-inflammatory activation of KCs in the liver, enhancement of hepatic inflammation, and suppression of fatty acid oxidation.[61] Additional adverse effect assessment of other NMs in the liver is required for their safe application.
Liver fibrosis and cirrhosis are serious conditions of sustained wound healing in response to chronic liver injury caused by various factors including viral, cholestatic, and inflammatory diseases.[39] Chronic hepatic injury and liver fibrosis will also lead to hepatic carcinoma. Yu et al. reported SiO2 NPs induced oxidative damage- and hepatocyte apoptosis-activated transforming growth factor-β1 (TGF-β1)/Smad3 signaling pathway, which promoted the process of liver fibrosis (Table 1).[62] Also, the chronic treatment with silica NPs increased the expression of fibrosis-related genes, fibrosis-related markers, such as hydroxyproline, and the occurrence of periportal fibrosis in the liver.[63,64]
Table 1.
Summary of the liver pathologies induced by the representative NMs
| NMs | Characterization | Animal model | Liver responses | References |
|---|---|---|---|---|
| Au NPs | 10, 20 nm spherical | Wistar-Kyoto rats | Vacuolar to hydropic degeneration, cytoplasmic hyaline vacuolation, polymorphism, binucleation, karyopyknosis, karyolysis, karyorrhexis, steatosis, and necrosis in hepatocytes; inflammatory cell infiltration, Kupffer cells hyperplasia, central veins intima disruption, hepatic strands dilatation, and occasional fatty change with a loss of hepatic strand normal architecture | [59] |
| Ag NPs | 14 ± 4 nm, spherical shapes | Wistar albino rats | Dilation and display of red blood cells and cell debris in sinusoids; degradation in hepatic organelles such as mitochondria and nuclei; increase in lipid droplet accumulation and glycogen depletion in hepatocytes; nuclei abnormalities, vacuolization, increased lysosomes, and electron-dense bodies in Kupffer cells | [53] |
| Fe3O4 NPs | 20–30 nm | BLAB/c rats | Induction of higher ALT, AST, and ALP levels; central venous congestion; hypertrophy, regeneration, and necrosis in hepatocytes; increase in Kupffer cell numbers. | [69] |
| Fe2O3 NPs | 20 nm, spherical | Wistar rats | NP aggregation in the liver, induction of higher ALT level, increased hepatocytes necrosis, hepatic inflammation, interstitial congestion, and fatty degeneration around the central vein. | [67] |
| ZnO NPs | 50 nm, higher dissolution (>10%) | Kunming mice | Induction of higher ALT and AST levels, CAT and SOD activity, and MDA concentration; liver lesions, cell congestion, immune cell infiltration, and petechiae | [66] |
| TiO2 NPs | 10 nm, anatase | B6C3F1 mice | NP accumulation in the liver, induction of oxidative stress-induced DNA strand breaks or oxidative DNA adducts; upregulating metabolism pathways and genes involved in antioxidative stress, metal detoxifying enzymes, and metabolic enzymes | [56] |
| CuO or Cu2CO3(OH)2 | higher dissolution (>50%) | RjHan:WI rats | Increase in AST, ALT, LDH, free fatty acids, creatinine, iron, and potassium levels in the blood; decrease in ALP; induction in inflammation, vacuolization, and necrosis in hepatocytes; hypertrophy in Kupffer cells and hepatocytes | [70] |
| Si NPs and CdCl2 | 26.8 ± 0.9 nm with the unimodal distribution. | Kun Ming mice | Induction of significant hepatic oxidative stress, elevation in liver damage enzymatic markers, and severe damage in liver architecture | [50] |
| MWCNTs | CNTSmall D: 6–17 nm; L: 847 ± 102 nm; CNTLarge D: 24–138 nm; L: 4048 ± 366 nm |
C57BL/6 mice | Increase in binucleate hepatocyte number; induction of mild liver injury including microfoci of necrosis, eosinophilic necrosis of single hepatocytes, and hepatocytes with pyknotic nuclei; changes in hepatic inflammatory; causing granulomas | [75] |
| GOs | 90 to 290 nm, favorable stability and monodispersity | Zebrafish | Induction of liver dysfunction in zebrafish embryos, oxidative stress, immune cells, and inflammatory responses; reduction in the lipid metabolism; increase in the expression of immune genes | [77] |
| GOs | 10–20 nm, with abundant oxygen-containing functional groups | BALB/c male mice | A higher amount surrounding portal triad zones than the central vein zones, dysregulation of key signaling pathways governing liver zonation, and changes in liver functional zonation | [78] |
| Nanocellulose modified with oxalate ester | −50 mV to −10 mV | Albino rats | Increase in AST, ALT, myeloperoxidase, inflammation-related iNOS, and apoptosis-related Bax protein expression | [159] |
2.2. Induction of higher level of liver enzyme release by metallic NMs
AST is an enzyme found in the liver that helps metabolize amino acids and that is normally present in blood at low levels.[65] When the liver is damaged, AST is released into the bloodstream and the levels increase.[66] ALT is an enzyme that helps convert proteins into energy for the liver cells. Like AST, an increase in ALT levels may indicate liver damage and disease.[67] ALP, an enzyme in the liver, plays a vital role in breaking down proteins.[68] Higher-than-normal levels of ALP may indicate liver damage or disease, such as a blocked bile duct.[68] Also, the elevated levels of LDH in the liver may indicate liver damage. Metal-based NMs have been shown to induce the release of high levels of these liver enzymes. For example, the administration of a high dose of Fe3O4 NPs (150 or 300 μg/gr) in BLAB/c mice was associated with a significant elevation in liver enzymes, including AST, ALT, and ALP, together with the histopathological effects in the liver tissue.[69] The high iron accumulation or excessive ROS in the liver could be responsible for these effects. In addition, copper oxide (CuO) NPs and copper carbonate nanoparticles [Cu2CO3(OH)2 NPs] have been shown to induce increased AST, ALT, and LDH in male-specific pathogen-free rats (RjHan:WI) due to the shedding of toxic Cu ions from the NPs.[70]
2.3. Hepatic chemical injury exacerbated by metallic NMs
The xenobiotics including environmental pollutants, pesticides, chemicals, and pharmaceuticals were mainly metabolized in the liver. The accumulations of these xenobiotics together with NP deposition may show synergistic effects and induce greater damage to liver tissue. For example, cadmium is a heavy metal contaminant that can induce severe liver damage by the generation of excessive oxidative stress.[71] SiO2 NPs administered at safe levels together with cadmium chloride to mice have been shown to increase cadmium deposition in the liver, causing high oxidative stress, elevated liver enzyme release, and liver architecture damage.[50] Similarly, ZnO NPs co-administered with the xenobiotic compound, organophosphate dimethoate, increased hepatic deposition of zinc and dimethoate, resulting in increased liver oxidative stress and liver injury.[72] Non-alcoholic fatty liver disease (NAFLD) is a chronic liver condition that is characterized by the excessive fatty acid accumulation in hepatocytes without alcohol abuse.[73] Metal NMs including Au, Ag, and Si NPs have been found to worsen NAFLD through increased inflammation or hepatocellular damage in various mouse models.[9]
2.4. Liver injury and hepatic steatosis induced by CNTs
CNTs are cylindrical large molecules composed of a hexagonal arrangement of hybridized carbon atoms, which include single-walled carbon nanotubes (SWCNTs) and multiwall carbon nanotubes (MWCNTs). Recent studies have demonstrated that CNTs could induce liver injury in vivo (Table 1). For example, Zhang et al. reported that the exposure of adult mice to MWCNTs significantly reduced the weight of offspring mice (male and female) and disrupted the liver function as manifested by the accumulation of lipid droplets in hepatocytes, a sign of hepatic steatosis, and histopathological changes in the liver tissues.[74] Poulsen et al. found two MWCNTs with different physicochemical properties, small, entangled (CNTSmall, 0.8 ± 0.1 μm long) and large, thick (CNTLarge, 4 ± 0.4 μm long) MWCNTs, induced histological changes in the liver of C57BL/6 mice, including increased binucleate hepatocytes, induction of mild liver injury including microfoci of necrosis and eosinophilic necrosis of hepatocytes, and hepatic inflammation. CNTLarge induced higher effects than the small CNTsmall.[75] However, the mechanism of the size dependence is not clear.
2.5. Liver function affected by graphene-based NMs
Graphene-based NMs including graphene and its derivatives, GO and rGO, are widely used in electronics, energy storage, and biomedicine fields due to their 2D structures.[14,17] Graphene-based NPs in the liver have been shown to induce adverse effects in the liver.[76] For example, GO exposure in larval and adult zebrafish could induce hepatic dysfunction mainly through the reactive oxygen species (ROS) and PPAR-α mediated innate immune signaling.[77] A recent study detailed the GO distribution and the zonation changes in liver lobules. Wu et al. found a higher GO accumulation surrounding portal triad zones than the central vein zones. Meanwhile, the liver zonation patterns also changed, such as changes in cytochrome P450 expression patterns and vital zonation-related genes involved in hepatocyte integrity and metabolism, leading to compromised hepatic functions. Furthermore, they identified dysregulation of key signaling pathways governing liver zonation, such as Wnt signaling and TET-dependent signaling as determined by RNA-Seq and DNA methylation sequencing analyses, which contributed to the GO-induced changes in the liver functional zonation.[78] There are also other types of GBNs, including graphene quantum dots (GQDs), few-layer graphene (FLG), and multilayered graphene (MLG) NMs.[76–78] Some of these graphene derivatives induced similar changes compared with GOs in the liver. For example, FLG- and carboxylated FLG could induce inflammatory changes and liver degeneration due to their accumulation in the liver for 90 days.[76]
There are limited studies focusing on the liver toxicity of NMs in humans. For example, Heringa et al. discovered TiO2 particles were present in 15 post-mortem human livers and spleens, with at least 24 percent of the nanoparticles smaller than 100 nm. Although we are still unclear about whether the presence of TiO2 could be directly linked to liver damage (i.e. liver edema and liver enzyme changes), the role of TiO2 particles cannot be ruled out.[79] More research on this topic in humans should be performed.
3. Interaction of NMs with the liver during systematic circulation
Why the liver is a major target of NMs? This aspect has been reviewed thoroughly by the Chan group.[49,80,81] The major point is that after the NMs gain access to the liver through blood circulation, the blood flow slows down about 1,000-fold, allowing the NMs to have sufficient time to interact with a variety of liver cells including KCs, LSECs, HSCs, and hepatocytes, and undergo cellular uptake.[81] Also, there is a concentration gradient of oxygen dropping from Zone 1 to 3 (Figure 2C), which may affect the cellular uptake of NMs by different cell types, however, this aspect has not been systemically explored. In addition, the exposure of NMs to the different cells is not homogeneous. This is because hepatocytes and HSCs are separated from the bloodstream by a layer of LSECs, which has a fenestration size (50 to 200 nm) that changes based on model animals and disease states, serving a sieve to filter out the NMs that are smaller than 200 nm to gain access to the space of Disse and have contact with HSCs and hepatocytes.[49] The NMs could pass through the hepatocyte layer and enter the bile canaliculi through transcytosis (Figure 5). The exposure of NMs to these liver cells may provoke the activation of various cellular uptake mechanisms including phagocytosis, receptor-mediated endocytosis, macropinocytosis, etc. depending on the NM physicochemical properties and specific cell types. Notably, some NMs may pass through the liver sinusoid and return to the systemic circulation via the central vein, which can lead to accumulation in other organs or tissue such as the spleen, lungs, tumor, etc., the NMs escaped or released from other organs or tissues could circulate back to the liver and get captured.[49,81] This recurring process could further increase the chance of NMs retention because the liver is so efficient in this aspect.
Figure 5.
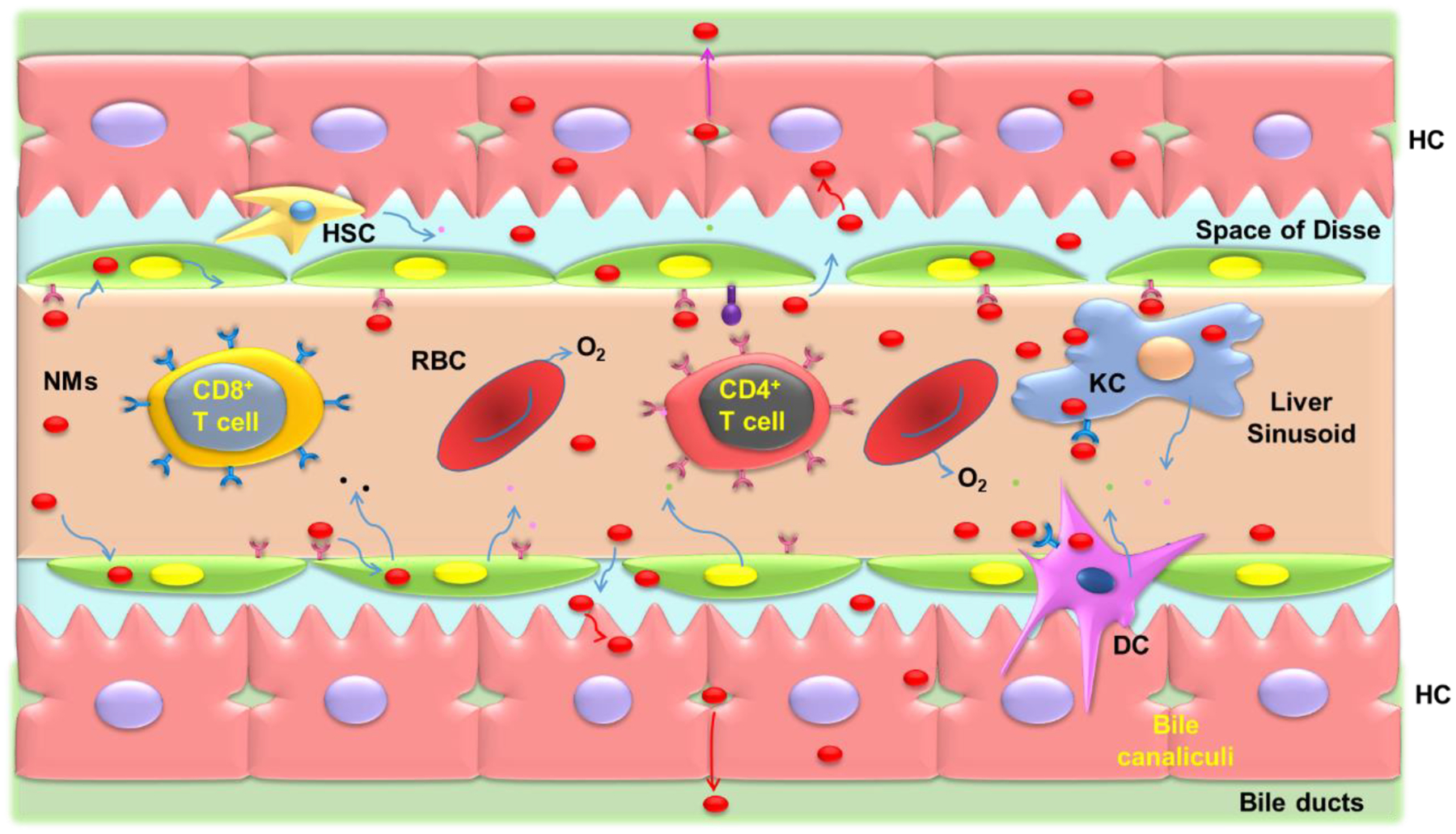
Interactions of NM uptake and elimination in the liver during systematic circulation after NMs exposure. As NMs move along the sinusoid, they will come into contact with T cells, Kupffer cells, sinusoidal endothelial cells, and DC cells. Depending on their physicochemical properties, NMs have better access through fenestrae to enter the space of Disse and contact with and hepatocytes. The smaller NMs may transcytose through the hepatocytes and enter the bile duct through bile canaliculi.
4. NMs properties and cell type-specific uptake in the liver
NMs have intrinsic properties including chemical composition, size, shape, surface chemistry, and hydrophobicity/hydrophilicity. However, when NMs encounter the biological/physiological fluid, they will assume additional properties by forming corona on the NMs surface, which will significantly impact their cellular uptake by liver cells.[3,49,82,83] The composition of the corona is influenced by NMs physicochemical properties (including size, shape, surface charge, hydrophilicity, etc.).[49,83,84] Additionally, the composition of the corona, e.g., proteins such as albumin, apolipoprotein E (ApoE), and IgG antibodies, will determine the NMs-cell interactions and cellular uptake mechanisms of liver cells once NMs are distributed in the liver. [85–93] Among the different liver cells, most studies are focused on the interactions of NMs with KCs, LSECs, HSCs, and hepatocytes due to their important roles in liver physiology and diseases. The unique liver architecture and location of these liver cells profoundly affect their interaction with NMs and cellular uptake, which will eventually determine the cellular responses (Figure 6).
Figure 6.
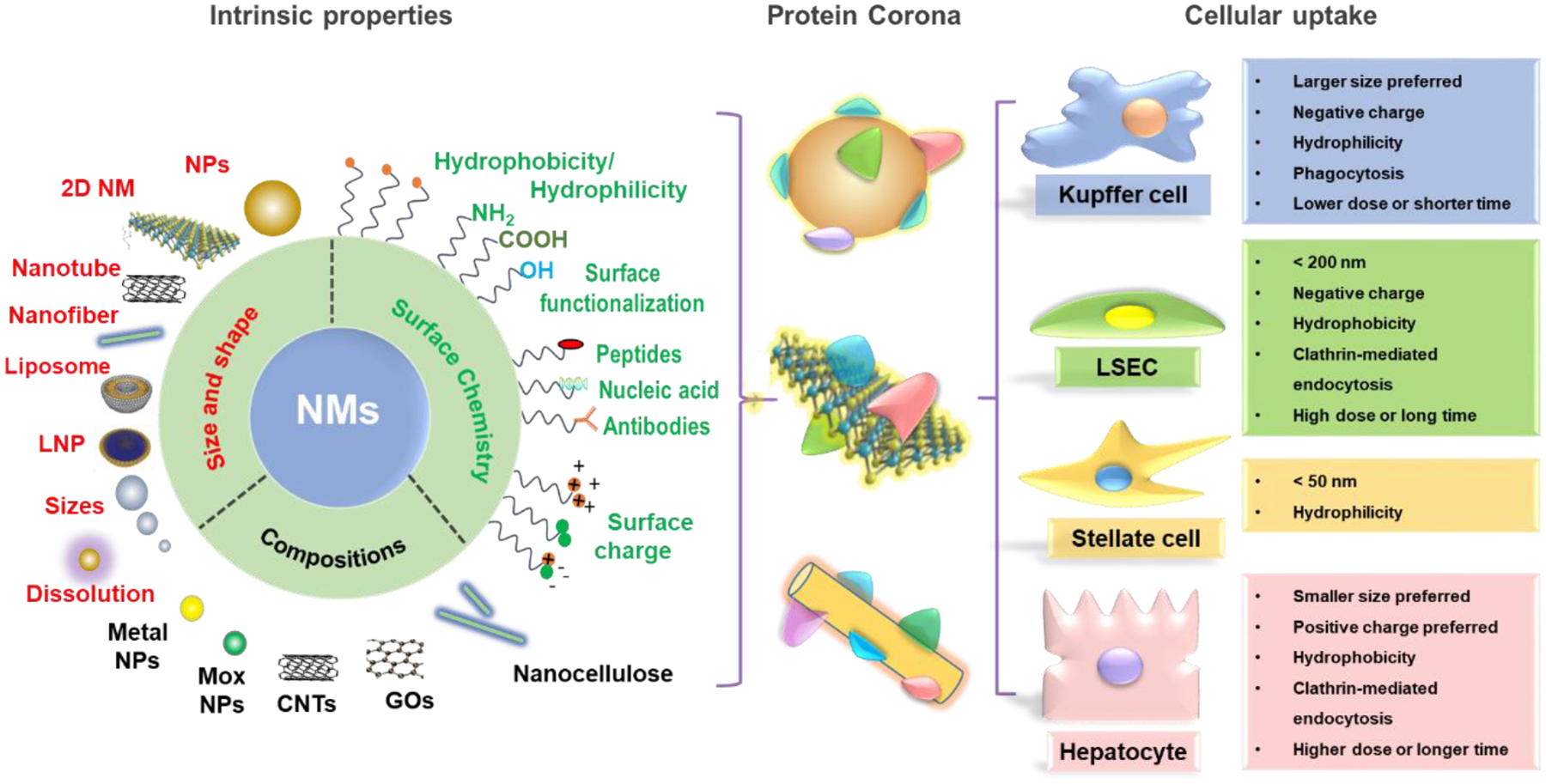
NMs intrinsic properties affect corona composition and cellular uptake in the liver. The major intrinsic properties of NMs include size, shape, surface chemistry, and composition, which will determine the corona composition and cellular uptake by the major liver cells. Larger size, negatively charged or hydrophilic NMs are preferentially swallowed by KCs via phagocytosis; NMs less than 200 nm or with negative surface charge or hydrophobicity tend to be taken up by endothelial cells through clathrin-mediated endocytosis with a high exposure dose or long time. NMs less than 50 nm or hydrophilic NMs could be captured by stellate cells. Smaller NMs with positive surface charge or hydrophobic NMs are preferentially taken up by hepatocytes through clathrin-mediated endocytosis.
4.1. Protein corona
The encounter of NMs with the biological environments will form a dynamic corona on NMs surfaces, including proteins, lipids, and certain sugar motifs.[85,86] The structure and composition of the corona are determined by the intrinsic physicochemical properties of NMs, the duration of exposure, and the nature of the physiological environment.[87–89] It has shown that the biomolecular corona regulated cellular recognition and penetration of NMs, and further influence their intracellular trafficking.[90,91] Corona proteins have been found to facilitate the binding of the opsonized particle to specific receptor molecules that are available in macrophages.[92] For example, Cao et al. found that the biodistribution of 2D MoS2 NMs in vivo was mediated by protein coronas, principally with ApoE, and the uptake by KCs is approximately 5.4- to 9.2-fold higher than that by hepatocytes.[93] In addition, Cai et al. found that the serum albumin corona on the surface of gold nanorods could confer a certain degree of stealth property to these NMs, and some nanorods escaped the clearance by KCs and entered the hepatocytes.[87] Moreover, Choi and co-workers also demonstrated the protein coronas on the surface of gold NPs reduced uptake by hepatocytes, possibly related to the increase in NP aggregation or the decrease in cell binding and subsequent transport.[94]
4.2. Size effects
The cellular uptake of NMs was size-dependent in many cell types.[95] After entry into the bloodstream, NPs larger than 150 nm in diameter could be preferentially captured by KCs and this uptake will enhance with an increase in particle size, although KCs are also able to take up NPs smaller than 150 nm.[96] The LSECs lining the liver sinusoids are proficient to take up NPs with sizes up to 200 nm size range by clathrin-mediated endocytosis.[43,49,96] Moreover, NPs with a smaller size than 50 nm in diameter could diffuse into the space of Disse through the fenestrations on the LSEC layer, and gain access to hepatocytes, and NMs on a scale of 10–20 nm could undergo rapid uptake by the hepatocytes.[49,96]
Large GOs with lateral sizes >500 nm were predominantly swallowed by KCs rather than LSECs or hepatocytes.[97] While the large-sized GOs induced significant lipid peroxidation and gasdermin-D (GSDMD)-mediated pyroptotic cell death in KCs, they did not induce as significant effects in LSECs or hepatocytes.[97] This size-dependent GO uptake was also supported by Zhang et al., where they showed the large GOs (500–2000 nm) taken up by KCs but only associated with the plasma membrane of hepatocytes in vivo.[98] Additionally, the uptake and subsequent cellular responses in KCs were in a size-dependent manner and the large size of GOs showed stronger effects than that of GOs with a size of 50–200 nm.[98] Furthermore, Lu and co-workers demonstrated that the larger graphene triggered membrane perturbation of RBCs and enhanced erythrophagocytosis by the KCs, resulting in higher liver toxicity in vitro and in vivo than smaller ones.[99]
4.3. Surface charge effects
In addition to the intrinsic NP size, the NP surface charge has also been reported to affect uptake by liver cell types. This is because of the differences in nanoparticle–cell membrane electrostatic interactions as well as protein adsorption to the NP surface.[49,100–102]
KCs and LSECs are efficient to interact with negatively-charged NPs because of the abundant expression of scavenger receptors binding to anionic NPs on the cell surfaces.[96,103] However, hepatocytes have been shown the most uptakes of positively-charged NPs rather than their negatively-charged counterparts.[96,103] Using fluorescently labeled mesoporous silica nanoparticles (MSNPs) in HepG2 and mice as models, Cheng et al. observed significant hepatocyte uptake of positively charged NPs, but not negatively charged ones.[104] They also demonstrated the negatively charged, but not positively charged, MSNPs were rapidly taken up by KCs in vivo and in vitro, respectively. The uptake of positively charged MSNPs by hepatocytes and negatively charged MSNPs by KCs was furtherly confirmed using transmission electron microscopy (TEM) of excised tissues and the accumulation of negatively charged MSNP in KCs induced significant adverse effects.[104]
4.4. Hydrophilicity effects
Generally speaking, NMs could be rapidly removed from circulation by components of the MPS, in particular, by the liver and spleen. Compared to hydrophilic NPs, hydrophobic NMs are more rapidly removed from circulation by KCs.[104] For example, studies found the polyethylene glycol (PEG) decoration on particle surface could escape uptake by KCs in the liver because their PEG chains partially blocked serum protein adsorption and reduced protein binding. This would minimize the uptake of NMs by KCs or increase uptake by hepatocytes.[96] Thus, this could explain that MWCNTs functionalized with PEG (MWCNTs–PEG) induced weaker ROS-mediated pro-inflammatory responses in KCs than carboxylated MWCNTs.[106] Lee et al. showed that silica NPs with hydrophobic surface modification were taken up by LSECs, while hydrophilic surface modification of silica NPs mainly was taken up by HSCs.[107] This could explain why hydrophobic rGO only has weaker effects in HSCs than hydrophilic GO under the same conditions.[108] Furthermore, Lee et al. demonstrated that in the hydrophobic-NP-treated liver, LSECs took up NPs the most (41%), followed by KCs (36%), HSCs (21%), and hepatocytes (2%); while in the hydrophilic-NP treated liver, KCs were the main cells taking up NPs (38%), followed by HSCs (29%), LSECs (29%), and hepatocytes (4%).[107].
4.5. Cellular uptake mechanism effects
It is well known that the induction of adverse effects by NMs is determined by their entry pathway and intracellular content.[84] The common mechanisms of how NMs enter the cells include phagocytosis and pinocytosis, which can be subcategorized into clathrin-mediated endocytosis, caveolae-mediated endocytosis, clathrin- and caveolae-independent endocytosis, and macropinocytosis.[84,97,109–111] Summarizing the cellular uptake mechanism of NMs in liver cells may shed light on understanding the differential actions of NMs in the liver. Many studies have demonstrated that NMs can be taken up by KCs via phagocytosis and hepatocytes via clathrin-mediated endocytosis.[97,109–111] Similarly, LSECs with high clathrin-coated pits per membrane unit are also proficient in taking up NMs by clathrin-mediated endocytosis.[112,113] Fortuna et al. showed that the agent to disturb the classical clathrin-mediated endocytosis did not interfere with retinol uptake in HSCs,[114] suggesting a different uptake mechanism involving in HSCs from clathrin-mediated endocytosis. Using various endocytosis inhibitors, including phagocytosis inhibitor wortmannin (WM), macropinocytosis inhibitor cytochalasin D (Cyto D), and clathrin-dependent endocytosis inhibitor Pitstop 2, Li et al. determined that only WM significantly inhibited the uptake of GO nanosheets labeled with fluorescein isothiocyanate (FITC-BSA-GOs) and 1 μm polystyrene beads (positive control), indicating that KCs take up GOs predominantly through phagocytosis; meanwhile, only Pitstop 2 reduced the GO uptake in LSECs and hepatocytes, suggesting the uptake of GOs in LSECs and Hepa 1–6 cells mainly by clathrin-mediated endocytosis.[97] The different uptake mechanisms in the liver cells are further confirmed using MoS2 and nanocellulose.[109,111] Also, Ouyang et al. showed that KCs were taking up NPs via receptor-mediated phagocytosis rather than clathrin- and caveolin-mediated endocytosis or micropinocytosis through localization of NPs by TEM and fluorescent imaging.[115]
The difference in uptake mechanisms of cells for NMs would determine their intracellular contents in the liver cells. Many studies also showed that NMs were differentially taken up by hepatic non-parenchymal cells, such as KCs, LSECs, and other cells, although the predominant component of liver cells is parenchymal hepatocytes.[97] For example, it has been shown that the phagocytic KCs played a major role in taking up graphene and removing quantum dots from circulation, whereas hepatocytes and LSECs contribute less compared to KCs.[81,99] Park et al. determined the relative distribution of polymeric poly(lactic-co-glycolic acid) (PLGA)-NPs among liver cells in vivo and found that KCs were the major cells that took up NPs, followed by LSECs, HSCs, and hepatocytes.[116] Also, Lee et al. found that in hydrophilic silica NP-treated mice, KCs mainly took up silica NPs (38%), followed by LSECs (29%) and HSCs (29%), and then hepatocytes (4%).[107] For other NMs, Zhang et al. found that the majority of GOs were actively swallowed by KCs and only a small amount of GOs was found inside hepatocytes in BALB/c mice.[98] Li et al. and Cao et al. also showed a much more uptake of MoS2 by KCs than other liver cell types, such as hepatocytes or LSECs, respectively.[93,111]
4.6. Dose and time effects
The exposure dose and time of NMs are important parameters in determining the NM biocompatibility profiles in vivo.[117,118] Many studies have also found that magnetic nanoparticles (MNPs) such as iron oxide NPs and citrate-coated manganese ferrite (Ci-MnFe2O4) NPs had distinct uptake profiles in the liver at different doses, as well as changing biodistribution profiles at different time points after injection.[117,118] Furthermore, some NMs including lipid nanoparticles (LNPs) were found to accumulate in KCs, LSECs, and hepatocytes in a time- and dose-dependent manner. [119,120] Shi et al. reported when LNP-siRNA (composed of CLinDMA:Cholesterol: PEG-DMG, 50:44:6 mol%) were administered at low doses (0.3 and 1 mg/kg), there was about 50% of LNP-siRNA delivered to KCs; however, at high doses (3 and 9 mg/kg), LNP-siRNA localization in hepatocytes increased to 74% at 3 mg/kg and 83% at 9 mg/kg, respectively, indicating uptake by KCs was reaching saturation, which could potentially allow more LNP–siRNA delivered to hepatocytes.[121] Furthermore, after 0.5 hr injection, LNPs were mainly localized in the space of Disse, followed by accumulation in hepatocytes after 2 hr injection. Interestingly, the siRNAs delivered to hepatocytes resulted in efficient gene silencing, while the delivery to KCs and LSECs was shown to be inactive.[122] Additionally, Sago et al. demonstrated a similar intrahepatic distribution of LNP-DNA barcode systems that are composed of the ionizable lipid D-Lin-MC3-DMA or cKK-E12 in vivo. At a dose of 0.3 mg/kg, LNP accumulation in KCs was higher than LSECs, while the trend was reversed at higher LNP doses of 1 mg/kg.[123]
In addition, many studies have shown that adjusting the physical size of organic NPs, PEG-lipid content, and incorporation of active targeting ligands can result in cell type-specific uptake in liver cell types. For example, Kim and colleagues found that mannose-incorporated LNPs with a hydrodynamic size larger than the fenestrae size were less uptake by hepatocytes, allowing for more selective uptake into LSECs.[124]
Taken together, these data show the cell type-specific uptake of NMs in the liver cells is mediated by the properties of NMs, protein corona, and the exposure dose and time. We here summarized a tendency of preferential uptake of NMs by the major liver cells based on limited reports (Figure 6). This includes NMs with a larger size, negative charge, or hydrophilicity are preferentially swallowed by KCs through phagocytosis at lower exposure dose or shorter time; NMs less than 200 nm or with negative surface charge or hydrophobicity tend to be taken up by endothelial cells through clathrin-mediated endocytosis at a high exposure dose or long time. The NMs less than 50 nm and hydrophilic NMs could be captured by stellate cells. Smaller NMs with positive surface charge or hydrophobic NMs are preferentially taken up by hepatocytes through clathrin-mediated endocytosis at a higher exposure dose or longer time. We need to recognize that cellular uptake of NMs is a complex and dynamic process, and cells could engage different cellular uptake pathways. More research is needed due to the diverse liver cell types and the complexity of cellular uptake mechanisms.
5. Transformation and metabolic processes of NMs in the liver
After liver accumulation, NMs are taken up by diverse liver cells with distinct functions and cellular programs. In most studies, the common cellular endpoints include cell viability, oxidative stress, DNA damage, and pro-inflammatory effects.[51,52] However, a less researched aspect is how the NMs change once they are inside the cells, which include subcellular localization (e.g., phagosomes, endosomes, and lysosomes), intracellular or intra-organellar transformation/degradation, and metabolization of the degradation products (Figure 7).
Figure 7.
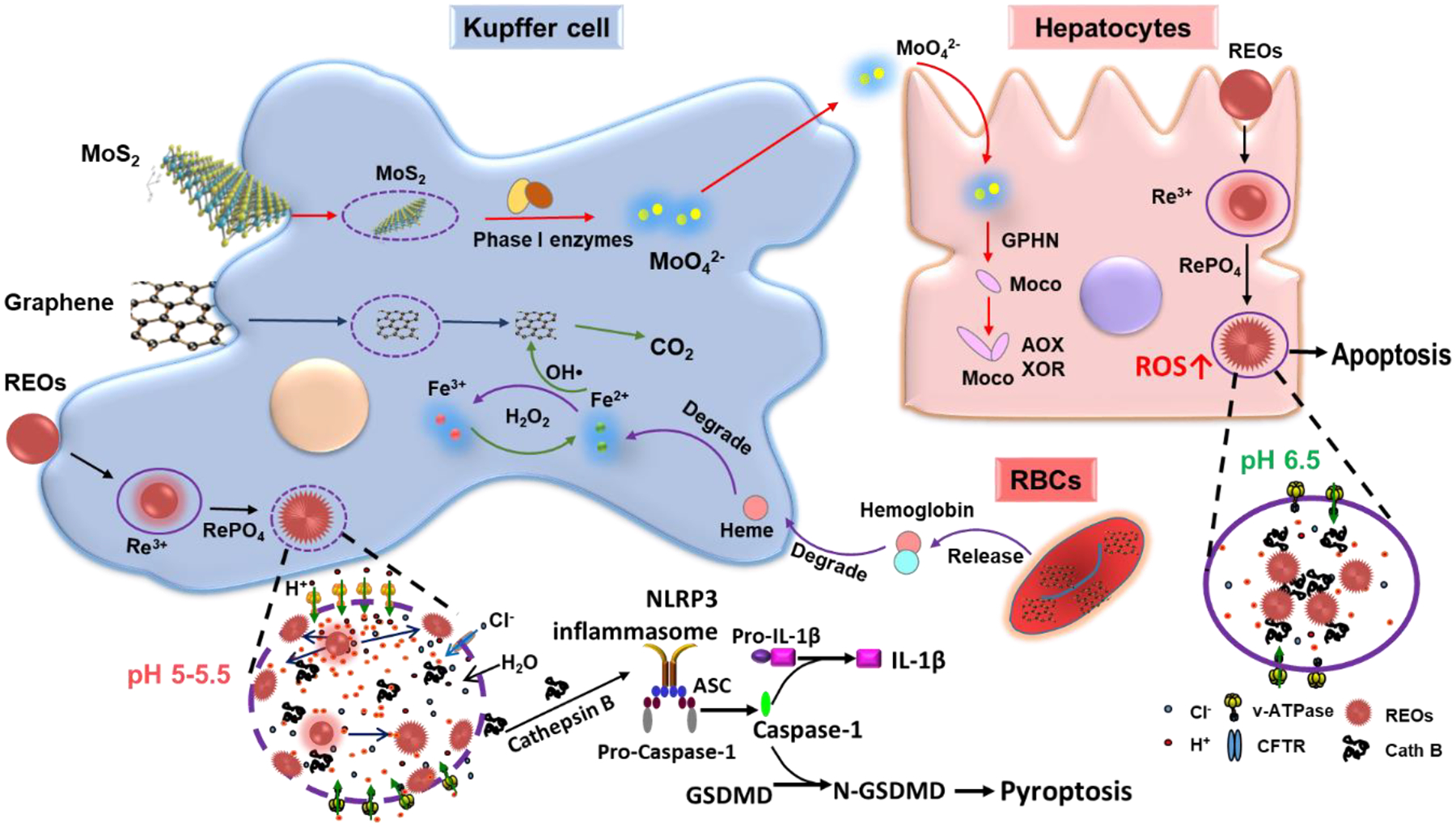
Schematic to demonstrate the transformation and metabolic processes of NMs in the liver. For example, MoS2 degrade into MoO42− by phase I enzymes in KCs, which can be used for biosynthesis of molybdenum cofactors (Moco) in hepatocytes.[93] Few layer graphene is degraded into CO2 by the OH• generated through the Fenton reaction in KCs, which originated from the degradation of released hemoglobin from the damaged RBCs by graphene into hemes, and the differential transformation of REOs in KCs and hepatocytes due to different levels of acidification in the phagolysosomes of macrophages (pH 5–5.5) vs hepatocytes (pH 6.5). The intense lysosomal acidification in KCs is driven by v-ATPase on lysosomal membranes, creating a high concentration of protons near the lysosomal membrane, driving the transformation of REOs and the formation of sea urchin structures on the lysosomal membrane by stripping phosphate groups from the phospholipids, leading to lysosomal membrane damage, NLRP3 inflammasome activation and pyroptosis in KCs. The same transformation also happens in hepatocytes, however, only in the interior of lysosomes, which will not lead to lysosomal damage. The Figure is reproduced with permission.[99] Copyright 2021, American Chemical Society.
5.1. NMs degradation and biotransformation through dissolution
Dissolution is involved in the degradation and transformation of inorganic NMs in the liver due to the low pH environments in lysosomes, a main target of NMs after cellular uptake. [7,105] We found that phagocytosed REOs could dissolve in the acidic environment of lysosomes in KCs. Because rare-earth ions (RE3+) have a high binding affinity with phosphates that is abundant in physiological media and membrane composed of phospholipids, which prefers to form needle-like crystalline structures.[7] As the dissolution and growth of needle-like structures are happening at the same time, this leads to morphological transformation from sphere to sea urchin-shaped structures as well as a change in chemical composition from oxides to rare-earth phosphate (REPO4).[7] This biotransformation process resulted in the depletion of phosphate groups from phospholipids and enzymes, leading to lysosomal damage as manifested by the loss of lysosomal proteins (e.g., cathepsin B). The released cathepsin B could trigger the NLR family pyrin domain containing 3 (NLRP3) inflammasome and caspase-1 activation, which could induce GSDMD-mediated pyroptosis in KCs. In hepatocytes, we found that REOs could also transform into sea urchin-shaped structures in the lysosomes, however, it did not lead to lysosomal damage. A possible explanation is the differences in the acidification levels in the phagolysosomes of macrophages (pH 5–5.5) vs hepatocytes (pH ≈6.5). The transformation of REOs was found to be pH-dependent, they exhibit a higher dissolution rate at pH 5.5 than pH 6.5. In addition, pH 5.5 requires an intense lysosomal acidification process that is driven by v-ATPase on the lysosomal membrane by pumps protons into the organelle, which forms to proton gradient that a high concentration of protons is present near the lysosomal membrane. This leads to rapid REO dissolution and transformation near the lysosomal membranes, resulting in the formation of sea urchin structures on the lysosomal membrane by directly stripping phosphates from the phospholipids. The transformation process is much slower in the lysosomes of hepatocytes, leading to the formation of sea urchin structures in the lysosomal fluids, but not on the lysosomal membranes, leaving the membranes intact (Figure 7).[7] As for transformation in vivo, it has been shown that the thiol-capped CdSe were degraded to release free Cd ions, which accumulated in the liver with a half-life of 15–30 years and induced adverse effects male Sprague Dawley rats.[125], Guo et al. found that ZnO NPs dissolved in the liver of rats, and the amount of free Zn ions increased over time after exposure, it only started to decrease on day 5. Furthermore, zinc accumulation in the liver induced oxidative stress and affected the energy metabolism pathways [126]
5.2. NMs degradation and metabolization mediated by enzymes
The elucidation of the degradation and metabolization of NMs is important to understand their physiological fate and evaluate their safety profiles. In cells, phase I and phase II enzymes are the main proteins for biotransformation and metabolization in the liver. Specifically, the phase I enzymes are responsible for phase I metabolism that is oxidation, reduction, and hydrolysis reactions, while phase II enzymes are responsible for phase II metabolism that is conjugation reactions.[127] There are few studies on NMs metabolism in the liver, however, progress has been made recently.
Cao et al. found that MoS2 NPs were accumulated in the liver and mainly sequestered by KCs, where they could be chemically degraded from MoS2 into MoO42− by phase I cytochrome P450 enzymes in liver microsomes. MoS2 nano-complexes were degraded through dissolution and the dissolved Mo element could be released from KCs and utilized for the biosynthesis of molybdenum cofactors (Moco) in the hepatocytes (Figure 7), leading to enhancement in enzymatic activities of the main molybdoflavoenzymes, including aldehyde oxidase and xanthine oxidoreductase, which could metabolize many anticancer drugs and generate nitric oxide, an important factor for tumor progression.[93,128,129] Similarly, Yang et al. found that CNTs could be degraded by ROS generated in primary KCs from rats.[130] The degradation process was driven by peroxidases including horseradish peroxidase as well as myeloperoxidase (MPO). After the internalization of CNTs, the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex assembles at the phagolysosomal membrane after cellular uptake, which transfers electrons to oxygen for the formation of superoxide anions (oxidative burst). These superoxide anions are further catalyzed by superoxide dismutase for the formation of H2O2 that is turned into hydroxyl radical (OH•) by the Fenton reaction. Then the generated OH• radicals attack CNT defects and unsaturated carbon bonds on the sidewalls of CNTs for the generation of carboxylic acids, creating holes in the graphitic structure.[130,131] The roles of phase enzymes in the biotransformation process of NMs in the liver were also supported by Lu et al.[99] They found few-layer graphene was taken up by liver cells, especially the KCs, after intravenous injection. At the same time, graphene could induce membrane perturbation on red blood cells (RBCs), which induced enhanced erythrophagocytosis of the damaged RBCs by KCs.[99] This triggered the degradation of hemoglobin into hemes and a rise of iron concentrations in KCs, which triggers the Fenton reaction to generate OH• (Figure 7). Similar to CNTs, OH• could attack the defects on graphenes in KCs, surprisingly, leading to the generation of CO2.[99] For organic NMs accumulated in the liver, liposomes were found to be degraded by serum proteins during blood circulation, lipases in cells, or metabolized by the body, and polymeric NPs were reported to be degraded into constituent monomeric units and dissociated polymer chains.[49]
5.3. NMs chemical modifications in a physiological environment
Certain NMs are subject to chemical modifications in the reductive physiological environment. For example, GOs could be readily reduced in the reduction environment in cells, including the presence of GSH, thiol-group containing proteins and enzymes, and unsaturated lipids, leading to the conversion of epoxy and carbonyl groups to phenolic groups in macrophages.[132–134] This transformation can change the properties of GOs, leading to reduced colloidal stability and agglomeration, which results in a significant reduction in cellular uptake of GOs by scavenging macrophages. Interestingly, in lung fluids Gamble’s solution, the transformation enhanced the layer-by-layer aggregation of GOs that led to the precipitation of GOs, causing a reduction in interactions with cells. On the contrary, GO transformation in artificial lysosomal fluid (ALF) enhanced the adhesion of large sheet-like GO aggregates on the plasma membrane.[134] Similar phenomenons could also happen in the liver, and, further detailed studies are required to explore the transformation and metabolic process of NMs in the liver.
6. NMs clearance from the liver
Sequestration by the liver and cellular uptake by liver cells are balanced by NMs clearance through MPS, renal, and hepatobiliary systems,[135] which are critical in determining the dose and biopersistence of NMs in the liver as well as their effects. When NMs circulate in the blood to reach organs or tissues, including MPS, kidney, and liver, the majority of non-biodegradable NMs are more likely to be taken up and retained long-term in the MPS for months to years, while the biodegradable NMs can be broken down, disassembled, and metabolized.[136,137] For NMs less than the glomerular filtration size limit (~5.5 nm), the majority of them will be filtered out by the kidney and excreted through urine rapidly, ranging from hours to days.[138] NMs larger than 5.5 nm will stay in the blood circulation and accumulation in the liver and other organs. For the liver, NMs could be cleared from the liver by hepatobiliary clearance. Evidence has shown that hepatocytes could excrete NMs in bulk via emptying of lysosomal contents into the bile,[139] which are transited through bile ducts and stored in the gallbladder, eventually released to the gastrointestinal tract and eliminated in feces (Figure 8).[49] This clearance route happens at a relatively slow pace ranging from hours to weeks. In addition, the size, shape, composition, and surface chemistry of NMs determine the efficiency of the hepatobiliary clearance pathway.[49,140–142] For example, the mesoporous silica NPs with a low aspect ratio of 1.5 were found to have a higher hepatobiliary clearance compared to those with a high aspect ratio of 5.[49,142]
Figure 8.
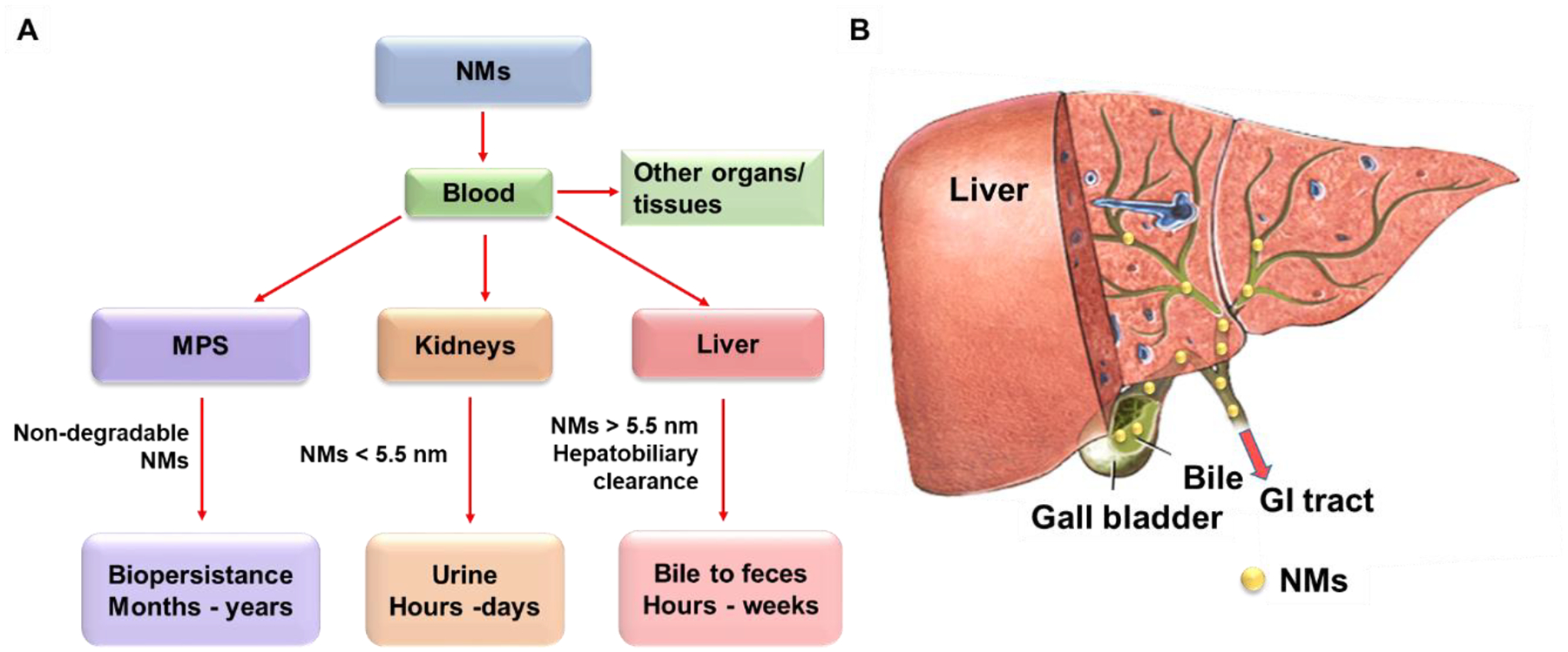
Main pathways for NMs clearance. A) NMs are cleared through hepatobiliary, renal, and mononuclear phagocyte systems. NMs circulate in the blood to reach organs or tissues, including MPS, liver, and kidneys. The non-degradable NMs are more likely to be taken up and retained by the MPS for months to years. The NMs less than 5.5 nm are cleared from the kidneys by renal clearance and eliminated in feces within hours to days. The NMs larger than 5.5 nm can be cleared from the liver by hepatobiliary clearance within hours to weeks. B) The hepatobiliary clearance is performed through interactions among the hepatic ducts, bile, gallbladder, common bile duct, duodenum, gastrointestinal tract, and feces. Figure 8 is reproduced with permission.[49] Copyright 2016, Elsevier B.V.
The slow hepatobiliary clearance contributes to the accumulation of NMs in the liver for long periods, which will enhance NMs uptake by liver cells. For example, after exposure of TiO2, CeO2, or carbon black to C57BL/6 mice, these NMs were all found to translocate and accumulate in the liver, where they remained within the liver tissue even at 180 days.[8] Liver accumulation of NMs and cellular uptake by different liver cells will induce cell type-specific responses.
7. Cell type-specific responses and molecular mechanisms to NMs exposure
The cellular uptake level of NMs by liver cells has been shown to induce differential cellular responses. For example, we demonstrated that the uptake of GOs in KCs was significantly more than that in LSECs and hepatocytes. This leads to a prominent plasma membrane lipid peroxidation and adverse effects in KUP5 cells but not in LSECs or hepatocytes. Furthermore, in graphene-treated mice by intravenous injection, we observed higher graphene uptake in KCs than hepatocytes. The difference in cellular uptake led to differential cellular responses, while KCs showed a higher iron content as a result of taking up and degradation of damaged red blood cells (RBCs) by graphenes, hepatocytes showed little iron content changes even though they represent a main iron storage site in the body.[99] This lead to the generation of OH• by Fenton reaction and degradation of graphenes into CO2 in KCs but not in hepatocytes as described above.[99] In addition, studies found that CNCs and aggregated MoS2 could induce lysosomal damage, cathepsin B release, NLRP3 inflammasome and caspase-1 activation, and interleukin-1β (IL-1β) production KCs due to the uptake of NMs by phagocytosis, but not in the non-phagocytotic cells including hepatocytes or LSECs.[109,111] Below we summarized the current knowledge on the cellular responses of NMs in major liver cell types, including KCs, LSECs, HSCs, and hepatocytes.
7.1. Effects of NMs on KCs
The Kupffer cell, constitution ~10% of all liver cells and 80–90% of all the residential macrophages in the body, is a major component of the MPS and plays a major role in the phagocytosis of NMs exposed at sinusoidal blood capillary barriers, modulation of innate immune responses, and endotoxin removal.[42,137,143–145] KCs serve as the first line of defense for NMs by phagocytic removal in the liver, which has a profound impact on liver functions.[137,144]
7.1.1. Metallic NMs induce oxidative stress-mediated inflammatory responses and apoptosis
For metallic NMs, different chemical compositions determined their cellular responses in KCs, as listed in Table 2. Wang et al. detailed that the shedding of toxic ions by Ag, CuO, and ZnO NPs induced mitochondrial reactive oxygen species (mtROS) production and oxidative stress and, leading to GSH depletion, heme oxygenase-1 (HO-1) expression, and the caspases 3 and 7-mediated apoptotic cell death in KCs.[105] V2O5 is another interesting example, in addition to oxidative stress and caspases 3/7- mediated apoptosis, which is induced by dissolved V5+ ions, it could induce NLRP3 inflammasome and caspase-1 activation, leading to IL-1β production, which is attributed to the interference of membrane Na+/K+ ATPase activity by V5+ions. Na+/K+ ATPase inhibition leads to the cease of K+ pumping into cells, which causes a decrease in the intracellular K+ levels due to the K+ leakage through the constitutive potassium leak channels.[105] A drop in intracellular K+ concentrations by potassium efflux or leakage is a trigger for NLRP3 inflammasome activation as discussed above. Furthermore, Mirshafiee et al. showed the pro-oxidative TMOs (e.g., Mn2O3 and Co3O4) induced the activation of caspases 3/7 in KCs, resulting in apoptotic cell death because of their conduction band energy overlaps with biological redox potential, which causes electron transfer to TMOs from biological molecules, triggering oxidative stress in cells (Figure 9).[7] In addition, Cho et al. showed that the uptake of 13 nm PEG-coated AuNPs in primary KCs played an important role in inducing oxidative stress, inflammation, and apoptosis in the liver of BALB/c mice.[146]
Table 2.
The effects of NMs on Kupffer cells
| NMs | Characterization | Cell type | Cellular responses | References |
|---|---|---|---|---|
| Ag NPs | 20 nm, spherical shapes | KUP5 | Oxidative stress generation, mtROS production, GSH depletion, HO-1 expression, and the initiation of caspases 3 and 7 mediated apoptotic cell death | [105] |
| CuO NPs | 60 nm, higher dissolution (>10%) | KUP5 | Oxidative stress generation, mtROS production, GSH depletion, HO-1 expression, the release of TNF-α, and the initiation of caspases 3 and 7 mediated apoptotic cell death | [105] |
| ZnO NPs | 50 nm, higher dissolution (>10%) | KUP5 | Oxidative stress generation, mtROS production, GSH depletion, HO-1 expression, the release of TNF-α, and the initiation of caspases 3 and 7 mediated apoptotic cell death | [105] |
| V2O5 NPs | 400 nm, higher dissolution (>50%) | KUP5 | Dissolved ions-induced caspases 3/7 mediated apoptosis as well as interference in membrane Na+/K+ ATPase activity, NLRP3 inflammasome and caspase-1 activation, and IL-1β release | [105] |
| Au NPs | 13 nm, PEG-coating | Primary mouse Kupffer cell | Cellular uptake, induction of inflammation and apoptosis | [146] |
| Aggregated MoS2 | Large agglomerates, low dissolution | KUP5 | Phagocytosis, induction of mtROS and caspases 3/7 mediated apoptosis, lysosomal damage, cathepsin B release, NLRP3 inflammasome as well as caspase-1 activation, and IL-1β release | [111] |
| MoS2 dispersed in Pluronic F87 | T: 3.5 ± 1.9 nm, S: 56 ± 28 nm, high dissolution |
KUP5 | Dissolved ions-induced mtROS and caspases 3/7 mediated apoptosis | [111] |
| GOs | T: 1–4 nm; S: 10 to 2000 nm |
KUP5 | Phagocytosis, NADPH oxidase mediated plasma membrane lipid peroxidation, PLC activation, calcium flux, mtROS generation, NLRP3 inflammasome activation, caspase-1 activation, IL-1β production, and GSDMD-mediated pyroptosis | [97] |
| GOs | T: 1 nm; S: 50 to 2000 nm |
Primary mouse Kupffer cell | Cellular uptake, TLR-4 activation, macrophage polarization, and secretion of IL-1β and TNF-α, activated via NF-κB signaling pathway | [98] |
| Gd2O3 NPs | 43.8 ± 15.8 nm, cubic | KUP5 | Cellular uptake, lysosomal damage, NLRP3 inflammasome activation, caspase 1 activation, cell swelling, membrane blebbing, IL-1β release, increase in membrane permeability, and GSDMD-mediated pyroptosis | [7] |
| Y2O3 NPs | 32.7 ± 8.1, cubic | Primary human Kupffer cell | Cellular uptake, cell swelling and membrane blebbing, IL-1β release, cell death | [7] |
| SiO2 NPs | 20 nm, pyrolytic (fumed) silica | KUP5 | Surface membrane perturbation, K+ efflux, NLRP3 inflammasome and caspase-1 activation, and induction of GSDMD-mediated pyroptosis | [105] |
| CNTs | D: 3–7 nm L:100–400 nm |
Primary rat Kupffer cells | Cellular uptake, ROS generation, caspases 3/7 activation, drop of total glutathione level, time-dependent cytotoxicity | [130] |
| CNCs | 100–750 nm | KUP5 | Phagocytosis, mtROS generation, caspases 3/7 activation, apoptosis, lysosomal damage, cathepsin B release, NLRP3 inflammasome and caspase 1 activation, and IL-1β production | [109] |
| NaYF4:18%Yb, 2%Er | 40 nm | Primary mouse Kupffer cell | Induction of enlarged autolysosomes, pro-death autophagy, and liver toxicity | [160] |
Figure 9.
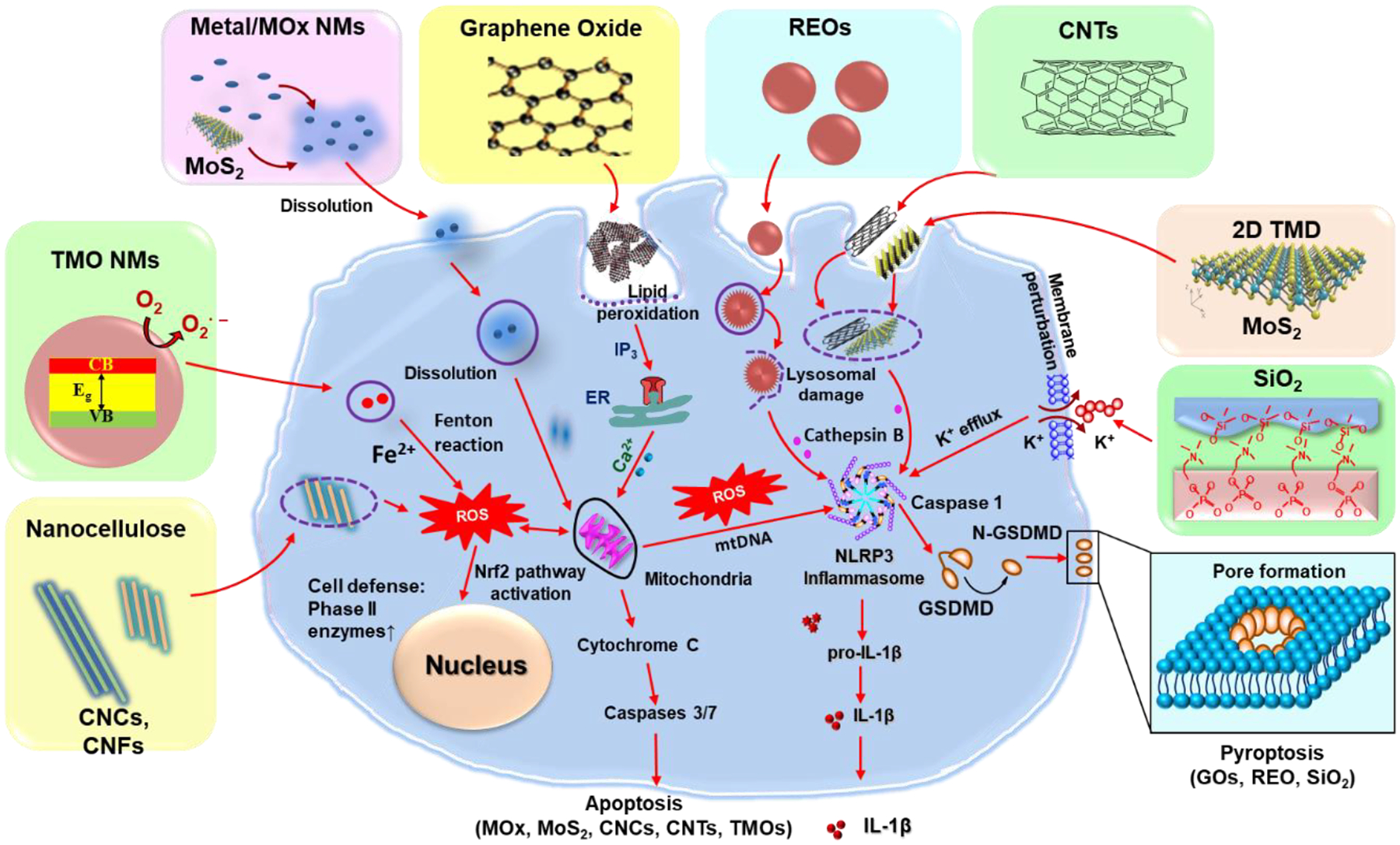
NMs-induced various cellular responses in KCs. This includes the metal or metal oxide (MOx) or transition-metal oxide (TMO) NMs, e.g., Ag, CuO, Co3O4, or Mn2O3, induce apoptosis due to their dissolution and shedding of toxic ions, bandgap energy, and oxidative stress; REOs (e.g., Gd2O3, La2O3, and Y2O3) and GOs induced pyroptosis in KCs. For REOs, the transformation from sphere to sea urchin-shaped and the formation of rare-earth phosphate (REPO4) structures on the lysosomal membrane, where RE(III) ions strip phosphate from the phospholipids on a lysosomal membrane and induce lysosomal damage, cathepsin B release, leading to NLRP3 inflammasome activation and GSDMD-mediated pyroptosis; the phagocytized GOs-induced NADPH oxidase activation and lipid peroxidation, triggering PLC activation that leads to calcium flux, mitochondrial ROS generation, and NLRP3 inflammasome activation, resulting in IL-1β production as well as subsequent pyroptosis; for fumed SiO2, the activation of NLRP3 inflammasome is involved in the pathway premised on K+ efflux resulting from the plasma membrane perturbation after SiO2 binding. Moreover, 2D TMD, CNCs, and CNTs induce ROS-mediated apoptosis and inflammatory responses in KCs after their internalization.
7.1.2. 2D MoS2 induces apoptotic inflammatory responses and cell death
MoS2, a representative of 2D TMD, consists of a molybdenum sheet bonded on both sides by sulfur layers.[147,148] It is generally considered safe and it has been used to develop drug carriers due to its large surface area and biocompatibility, however, recent data showed that it could lead to cytotoxicity.[93,149,150] Similar to metallic NMs, the dissolution of MoS2 is associated with cellular responses. Li et al. reported that the dissolution and the release of hexavalent Mo(VI) were responsible for the generation of oxidative stress, activation of caspases 3/7, and apoptotic cell death in KCs. Additionally, the phagocytosis of the aggregated form (MoS2-Agg) could trigger lysosomal damage, cathepsin B release, and NLRP3 inflammasome and caspase-1 activation, leading to IL-1β and IL-18 production in KCs. Despite the caspase-1 activation, KCs did not undergo pyroptosis. Instead, similar to V2O5 NPs,[105] MoS2-Agg induced apoptosis, this is because of the earlier onset of caspase 3/7 activation, while the caspase-1 activation happened later.[111] Cao et al. also showed the sequestration of protein-coated 24.5 nm MoS2@HSA nanocomplexes by the KCs and the long-term accumulation of MoS2 nanodots in the mouse liver. In addition, MoS2 was oxidized and degraded in the KCs, and the dissolved molybdenum ions were chemically transformed from Mo(IV) to Mo(VI) and used for biosynthesis of molybdenum cofactors (Moco) in hepatocytes as we discussed above.[93] Furthermore, Yu et al. have demonstrated the induction of pro-inflammatory and apoptotic responses (e.g., IL-1β, IL-6, and AIF gene) in the livers of adult zebrafish by chitosan-functionalized MoS2 micro-sheets.[150] Thus, much attention should be paid to the adverse effects induced by MoS2 NMs for developing safer nanoapplications.
7.1.3. GOs and MOx NMs-induced inflammatory responses and GSDMD-mediated pyroptosis
GO is an oxidized graphene derivative with a large surface area, high flexibility, and excellent dispersibility, which are useful for biomedical applications.[12] GOs have been shown to accumulate and induce toxicity to liver cells.[31,78] For example, Li et al. found that GOs were taken up into KCs through phagocytosis, which triggered NADPH oxidase-mediated plasma membrane lipid peroxidation, inducing the activation of phospholipase C (PLC) to cleave phosphatidylinositol 4,5-bisphosphate (PIP2) into diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3). IP3 mobilized intracellular calcium stores and induced mtROS generation when mitochondria absorb excessive or sustained calcium release from the endoplasmic reticulum (ER). Mitochondrial damage by mtROS could lead to mtDNA release and global oxidative stress in cells, activating NLRP3 inflammasome and caspase-1, leading to IL-1β release and cleavage of GSDMD. The resulting N-terminal GSDMD fragments could translocate to the plasma membrane, where oligomerization leads to pore formation, cellular swelling, and cell death (Figure 9).[97,151–153] It also showed that the lateral size of GOs played a key role in GO-induced pyroptosis, where the large-sized GOs showed strong effects than small ones.[97] Furthermore, Zhang et al. showed that GO was taken up by KCs in mouse liver sections, which induced ROS production, TLR-4 activation, and macrophage polarization, and secretion of pro-inflammatory cytokines IL-1β. In addition, GO-L (500–2000 nm) induced higher IL-1β production by primary KCs and in the liver homogenates compared to GO-S (50–200 nm).[98] Furthermore, Li et al. found GO could induce pyroptosis in various macrophages cell lines in addition to KCs, suggesting GO-induced pyroptosis could be a universal feature for all macrophages.[97]
In addition, Mirshafiee et al. detailed the REOs-induced pyroptosis in KCs. As we discussed above, REOs (e.g., Gd2O3, La2O3, and Y2O3) undergo biotransformation in KCs, which induce lysosomal damage, and NLRP3 inflammasome and caspase-1 activation. Caspase-1 will cleave GSDMD and trigger pyroptosis in KCs (Figure 9). Similar to GOs, REOs-induced pyroptosis was also confirmed in multiple macrophage cell lines, suggesting it is a universal feature.[7]
Different from GOs and REOs, Wang et al. reported another mechanism of NMs-induced pyroptosis. Fumed SiO2 does not undergo cellular uptake, instead, they bind to the plasma membrane due to their chain-like structures. The binding leads to membrane perturbation and potassium (K+) efflux because fumed silica NPs have abundant silanol groups on the particle surface that are capable of generating ROS, which leads to K+ efflux and NLRP3 inflammasome activation, eventually resulting in pyroptosis as discussed above. (Figure 9).[105]
7.1.4. CNTs and nanocellulose induced ROS-mediated inflammatory responses and apoptosis
CNTs have been one of the most studied NMs due to their high tensile strength, large surface area, and high conductivity, however, their fiber-like structure raised concerns on the potential hazards to humans because of the similarity to asbestos.[15,16] Many studies reported that both SWCNTs and MWCNTs could be taken up into KCs through phagocytosis, triggering oxidative stress. This in turn induced inflammatory cytokines, such as IL-1β, interleukin-6 (IL-6), and tumor necrosis factor-α (TNF-α) as well as apoptosis. (Table 2).[106,130,154] The mechanism involves the cellular uptake, lysosomal damage, and activation of the NLRP3 inflammasome (Figure 9). The receptors responsible for CNT uptake have also been identified. Singh et al. found that the cellular uptake of CNTs in a murine macrophage RAW264.7 cell line was mainly mediated by scavenger receptors and charge-dependent, although non-scavenger receptor-mediated mechanisms could also be engaged at low CNT concentrations.[106]
The surface properties of CNTs play a major role in cellular responses. Higher surface carboxyl density leads to reduced cytotoxicity of MWCNTs while hydrophobic surface increases ROS production.[154] Surface modification of MWCNTs with biocompatible polymers could reduce these responses and subsequent adverse effects.[155] Zhang et al. found that the phagocytosis of MWCNTs functionalized with polyethylene glycol (MWCNTs–PEG) induced weaker ROS-mediated pro-inflammatory responses in the model for liver KCs, RAW 264.7 cell line, compared with those MWCNTs functionalized with carboxylation (MWCNTs–COOH).[106] As we mentioned above, CNTs can be degraded by KCs, thus the adverse effects of CNTs will decrease as well. For example, Yang et al. reported that CNTs in RAW 264.7 and primary rat KCs were degraded by approximately 25–30% within the first 4 days after uptake. Meanwhile, the generation of ROS was attenuated and glutathione levels were recovered as CNTs degradation occurred.[130]
Nanocellulose has been widely used to develop drug carriers, 3D cell culture, antimicrobial materials, and tissue repair and regeneration because they are generally considered biocompatible.[18,156–158] However, nanocellulose as drug carriers have been reported to accumulate in the liver and induce adverse effects. For example, the nanocellulose modified with oxalate ester (NCD) has been reported to induce significant hepatotoxicity with the elevated AST, ALT, myeloperoxidase, inflammation-related iNOS, and apoptosis-related Bax protein expression in the liver of rats.[159] The mechanism of hepatoxicity at the cellular level has been also reported. Li et al. found that the phagocytosis of CNCs by KCs induced ROS production in mitochondria, caspases 3/7 activation, and apoptotic cell death (Figure 9). Although the phagocytosed CNCs also triggered lysosomal damage and cathepsin B release, triggering NLRP3 inflammasome and caspase-1 activation, they did not induce pyroptosis. Similar to V2O5 and MoS2 mentioned above, this is due to the differences in the activation time in which CNCs induced early onset of caspase 3/7 activation while caspase-1 was activated at a later time.[109] Furthermore, the length of nanocellulose played a vital role in the effects in KCs. Shorter nanocellulose (CNCs) can induce stronger apoptotic cell death than longer nanocellulose (CNFs), and the CNC with ~280 nm length (CNC-2) showed the most significant effects due to the highest cellular uptake in KCs.[109].
7.1.5. Upconversion NPs-induced autophagy
Zhu et al. found that rare-earth-based upconversion nanoparticles (UCNPs) including NaYF4:18%Yb, 2%Er induced the enlarged autolysosomes and pro-death autophagy in primary mouse KCs as well as liver toxicity in mice, which was likely due to the induction of autophagic cell death by ER stress in KCs (Table 2). They also revealed the inhibition of KC autophagy may constitute a novel strategy for abrogating NM-elicited liver toxicity.[160] Li et al. also showed rare earth materials, a major component of UCNPs were also able to induce autophagosome accumulation in macrophages by disruption of autophagy flux.[161] The mechanism of rare earth-induced autophagosome accumulation involves interference in the fusion of autophagosomes with lysosomes, in part because of the biotransformation process as we discussed above, which has negative impacts on lysosomal alkalization and phosphoprotein/enzyme function. The result of inhibition on autophagy flux results in the accumulation of activated NLRP3 inflammasome in cells, which leads to exaggerated IL-1β production. In addition, the biotransformation of UCNPs affected its optical properties, resulting in a reduction of the fluorescence intensity. Surface coating of UCNPs with phosphonate could reduce the dissolution, biotransformation, and NLRP3 inflammasome activation as well as maintain the optical property of UCNPs.[162]
7.2. Effects of NMs on LSECs
LSECs, making up 21 % of the total number of liver cells, are also an important part of the MPS.[112,113] Although the LSECs make up only about 3% of the total liver cell volume, their surface in a normal adult human liver is about 210 m.[112] LSECs have a high capacity of clathrin-mediated endocytic activity, which plays a central role in the clearance of blood-borne waste and innate immunity.[43,163,164] LSECs could take up NMs through active endocytosis into cells.[81]
To date, there are only a few studies on the effects of NMs on LSECs (Table 3). Among the studies, Nishimori et al. demonstrated that 70 nm silica NPs increased ALT levels in the liver and induced LESCs-mediated liver injury in a dose-dependent manner.[165] Li et al. found that GOs induced cytotoxicity in the immortalized SV40-transformed mouse LSECs only at a high concentration (100 μg/mL), although these cells were able to take up small-sized GOs by clathrin-mediated endocytosis.[97] The reason for the insensitivity of LSECs to GOs compared to KCs is still not clear. Additionally, Tee et al. showed that TiO2 NPs were rapidly internalized into the human hepatic sinusoidal endothelial cells (HHSECs), increasing endothelial permeability and inducing cellular shrinkage. This transient endothelial leakiness was induced by a reduction of Akt activation in liver sinusoids. However, TiO2 NPs-induced endothelial leakiness was not accompanied by significant oxidative stress or a decrease in cell viability.[166] More studies need to be performed on LSECs in vitro and in vivo.
Table 3.
The effects of NMs on sinusoidal endothelial cells (SECs)
| NMs | Characterization | Cell type | Cellular responses | References |
|---|---|---|---|---|
| Si NPs | 70 nm | Primary liver sinusoidal endothelial cells | Increase in ALT and induction of liver injury | [63] |
| GOs | T: 1–4 nm; S: 10 to 2000 nm |
Liver sinusoidal endothelial cells | Cellular uptake, induction of cytotoxicity at a high concentration (100 μg/mL) | [97] |
| TiO2 NPs | 21 nm | Human hepatic sinusoidal endothelial cells (HHSECs) | Cellular uptake, induction of transient leakiness by reducing the activation of Akt, increase in endothelial permeability, morphological changes, cellular shrinkage | [166] |
7.3. Effects of NMs on HSCs
HSCs, representing 5–8% of the total number of liver cells, are mesenchymal cells in the space of Disse.[167] Upon liver damage, HSCs are activated by differentiating into myofibroblasts, migrate to the site of wounding, and begin depositing extracellular matrix (ECM), which are pivotal for the development of liver fibrosis.[9,167] When NMs reach the space of Disse after passing through the fenestration of the LSEC layer in the liver, they will come into contact with HSCs, which may induce cell activation, leading to liver fibrosis or other adverse effects.
There are limited studies on the effects of NMs on HSCs. For metal NMs, Sun et al. found that the uptake of Ag NPs in primary rat HSCs induced significant alterations of cell morphology, including rupture of the cell membrane, swelling of cell and cell organelle, and formation of microvesicles. This also leads to the inhibitions on the production of matrix metallopeptidase (MMP)-2 and MMP-9, and induction of apoptosis or necrosis in a size- and dose-dependent manner, with the smaller NPs showing stronger effects at higher concentrations.[168] Additionally, Osmond-McLeod et al. reported that ZnO NPs induced significant cytotoxicity in primary human HSCs through an initial activation of cellular stress and injury responses, followed by dysregulation of the transcriptome, alterations in cellular function, and induction of apoptosis or senescence.[169] These responses may be mediated by the phosphorylated c-JUN and p38 proteins and the activated caspase 3/7. Expectedly, the presence of surface coatings reduced these adverse effects due to reduced dissolution. For carbon NMs, Chen et al. reported that GO and rGO flakes showed growth inhibition on human HSCs at a high concentration (> 31.25 μg/mL).[108] The mechanism of the cellular response is unclear. (Table 4)
Table 4.
The effects of NMs on hepatic stellate cells (HSCs)
| NMs | Characterization | Cell type | Cellular responses | References |
|---|---|---|---|---|
| Ag NPs | 10 or 30–50 nm, PVP-coating | Primary rat HSC | Morphology alterations, reduction in cell viability, induction of apoptosis or necrosis, inhibition of the production of MMP-2 and –9 | [168] |
| ZnO NPs | W: 44 ± 2 nm; L: 73 ± 4 nm, powders | Primary human HSC | Activation of cellular stress and protection responses, followed by dysregulation of the transcriptome, alterations in cellular function, and induction of apoptosis | [169] |
| GO | H: ~ 1.2 nm, flake | Human HSC | A growth inhibition at a concentration of 15.62 μg/mL | [108] |
| rGO | H: ~ 2.0 nm, well dispersed, flake | Human HSC | A growth inhibition at a high concentration (> 31.25 μg/mL) | [108] |
7.4. Effects of NMs on hepatocytes
Hepatocytes, making up more than 90% of the total liver volume and as high as 60–80% of the total number of liver cells, perform important roles in protein synthesis, metabolic, endocrine, secretory, and detoxification functions.[46,49] When NMs cross the LSEC layer through the fenestrations to reach the space of Disse in the liver, they will come into contact with hepatocytes (Figure 10).[81]
Figure 10.
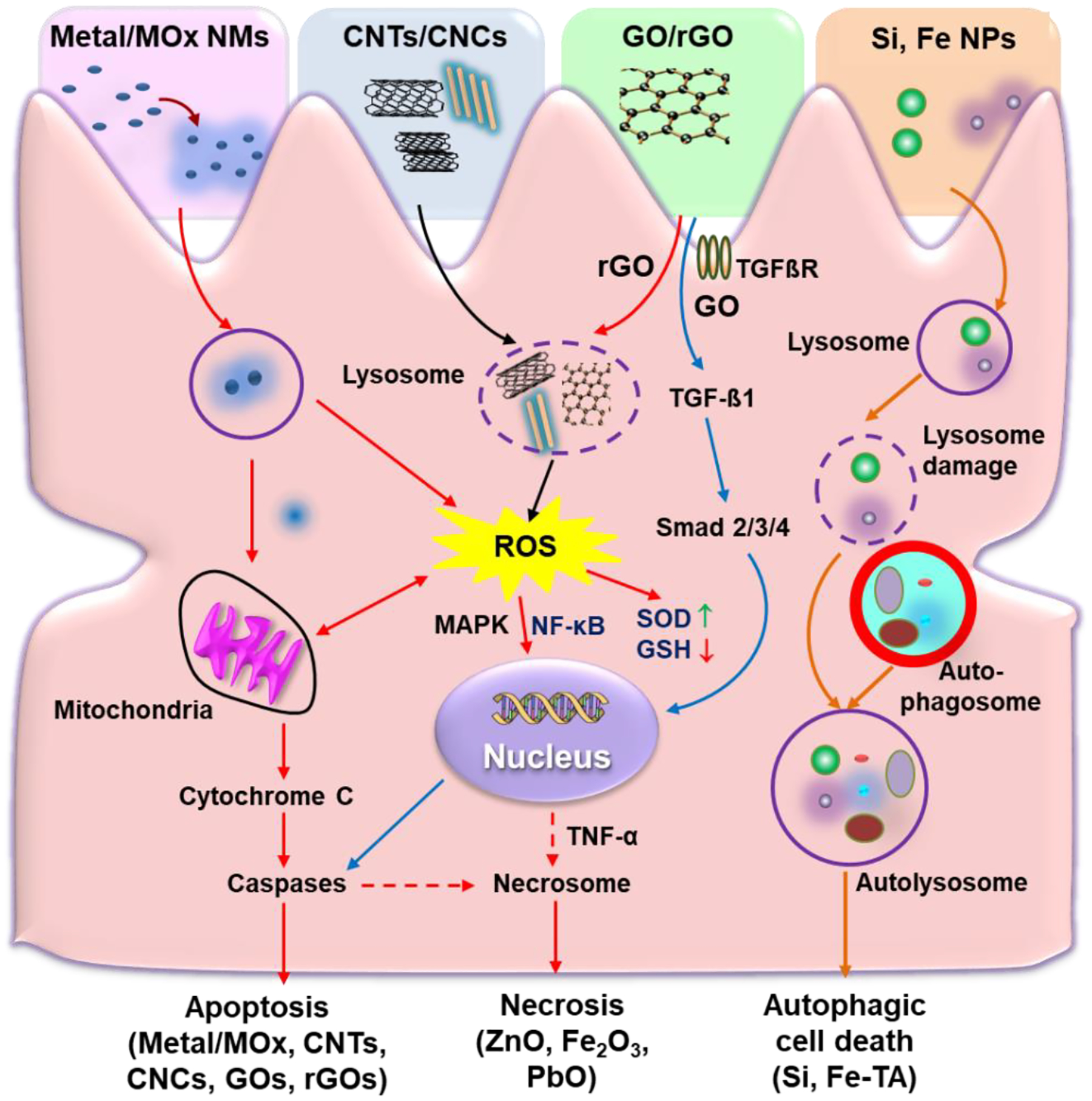
NMs-induced cellular responses in hepatocytes. Metallic NMs (e.g., Au, Ag, and ZnO NPs) could induce oxidative stress-mediated inflammatory responses, apoptosis, or necrosis; CNTs or CNCs could induce oxidative stress-mediated inflammatory responses and apoptotic cell death; rGOs could induce apoptosis via the nuclear factor kappa B (NF-κB) and oxidative stress pathways, while GOs induce apoptosis via the TGFβ1-mediated signaling pathway; Si and Fe-TA NPs could induce autophagic cell death.
7.4.1. Metallic NMs induce oxidative stress-mediated inflammatory responses and cell death
Many studies have shown that metallic NMs induced significant effects in hepatocytes, including oxidative stress-mediated cell death and inflammatory responses (Table 5). For example, Gaiser et al. reported that the uptake of Ag NPs could affect hepatocyte function by reducing albumin release, which was highly cytotoxic to hepatocytes following the in vivo exposures of female Wistar rats and in vitro exposures of human hepatocyte cell line C3A. Importantly, they also observed the similar effects of Ag NPs on the increase of IL-8/macrophage inflammatory protein 2, IL-1RI, and TNF-α expression in both models.[170] The uptake of Au NPs in the liver also provoked protein carbonylation, lipid peroxidation, DNA damage, and time-dependent adverse effects without any inflammatory responses in the HepG2 cell or Wistar rat model.[171] Wang et al. showed that the exposure of Cu NPs to primary hepatocytes dramatically decreased cell viability due to the oxidative stress-mediated apoptotic and necrotic cell death.[172]
Table 5.
The effects of NMs on hepatocytes
| NMs | Characterization | Cell type | Cellular responses | References |
|---|---|---|---|---|
| Ag NPs | 20 nm, spherical | Hepa 1–6 cell line | Oxidative stress generation, mtROS production, GSH depletion, HO-1 expression, and initiation of caspases 3 and 7 mediated apoptotic cell death | [105] |
| Ag NPs | 20 nm | C3A cell line | Cellular uptake, inflammatory mediator expression, increase in IL-8/macrophage inflammatory protein 2, IL-1RI, and TNF-α expression | [170] |
| Au NPs | 10–60 nm, citrate-stabilization, monodisperse | HepG2 cell line | protein carbonylation, lipid peroxidation, DNA damage, overproduction of free radicals and ROS | [171] |
| Cu NPs | 100 ±35 nm, spherical and aggregation | Primary hepatocytes of E.coioides | Intracellular ROS generation, antioxidative enzymatic defense systems alteration, induction of apoptosis and necrosis | [172] |
| CuO NPs | 60 nm, dissolution (>10%) | Hepa 1–6 cell line | Oxidative stress generation, mtROS production, GSH depletion, HO-1 expression, the release of TNF-α, and the initiation of caspases 3 and 7 mediated apoptotic cell death | [105] |
| Gd2O3 | 43.8 ± 15.8 nm, cubic | Hepa 1–6 cell line | Cellular uptake, caspases 3/7 activation, induction of apoptotic cell death | [7] |
| Mn2O3 | 51.5 ± 7.3, tetragonal | Hepa 1–6 cell line | Cellular uptake, cellular shrinking, caspases 3/7 activation, induction of apoptotic cell death | [7] |
| TiO2 NPs | 50 nm, spherical or rod-like crystal structure | Primary rat hepatocytes | Oxidative stress, intracellular ROS, morphological changes in mitochondria and substantial loss in the fusion process, disruption of the mitochondrial dynamics | [173] |
| V2O5 NPs | 400 nm, higher dissolution (>50%) | Hepa 1–6 cell line | Dissolved ions-induced caspases 3/7 mediated apoptosis | [105] |
| ZnO NPs | 47–106 nm, spherical/rhomboid/rod-shaped | Catfish primary hepatocytes, HepG2 cell line | Cellular uptake, ROS-induced cell death, and damages to the cell and mitochondrial membranes | [174] |
| ZnO NPs | 50 nm, dissolution (>10%) | Hepa 1–6 cell line | Oxidative stress generation, mtROS production, GSH depletion, HO-1 expression, and the initiation of caspases 3 and 7 mediated apoptotic cell death | [105] |
| Fe–TA NPs | 3–5 nm, tannic complexes | HepG2 cell line | Cellular uptake, autophagosomes formation, increase in LC3 expression, induction in autophagic cell death | [178] |
| SWCNTs | 1000–2000 nm with a diameter range of 1.0–6.0 nm | HepG2 cell line | Elevation in ROS level, reduction in cellular metabolic activity, perturbation of cell cycle, induction of apoptotic cell death | [181] |
| MWCNTs | D: 5–35 L:700–3000; D: 6–20 L: 700–4000 | C3A and primary hepatocyte | Cellular uptake, IL-8 production, cytotoxicity | [182] |
| GO | T: 6 nm; H: 20 nm; S:40 nm | HepG2 cell line | Cellular uptake, oxidative stress generation, NADPH oxidase-dependent ROS formation, high deregulation of antioxidant/DNA repair/apoptosis-related genes, DNA damage, TGFβ1 mediated signaling, cytotoxicity | [184] |
| rGO | T: 7 nm; H: 23 nm; S:40 nm | HepG2 cell line | DNA damage, oxidative stress, ROS generation, induction of innate immune response through TLR4–NFkB pathway, apoptosis | [184] |
| CNC | ~280 nm | Hepa 1–6 cell line | Cellular uptake through clathrin-mediated endocytosis, mtROS generation, caspases 3/7 activation, apoptosis | [109] |
Similar to metal NMs, metal oxide NMs could induce adverse effects on hepatocytes.[173] For example, Wang et al. reported that CuO, ZnO, and V2O5 NPs underwent dissolution, leading to shedding of >10% of the ions and ROS production, inducing caspases 3/7-mediated apoptosis in Hepa 1–6 cells.[105] Both the uptake of TMOs (e.g., Mn2O3) and REOs (e.g., Gd2O3) in Hepa 1–6 also induced cellular shrinking, caspases 3/7 activation, and apoptotic cell death.[7] In addition, NMs induced oxidative stress-mediated apoptosis in hepatocytes is generalizable in many hepatocytes including primary rat hepatocytes, catfish primary hepatocytes, HepG2 cells, and Hepa 1–6 cells,[174] as listed in Table 5. Furthermore, several studies have reported the inflammation in the liver and the necrosis in hepatocytes under the treatment of Fe2O3, PbO, or ZnO NPs in animals.[67,175–177]
7.4.2. Metallic NMs induce autophagic cell death
Metallic NMs could also induce autophagy in hepatocytes. For example, Saowalak et al. found iron(III)-tannic complexes (Fe–TA NPs) induced autophagosomes formation (double-membrane vesicle within the cells), increased LC3 expression, a specific marker for the early stage of autophagy, and triggered autophagic cell death.[178] However, the normal rat hepatocytes (AML12) had less uptake of Fe–TA NPs induced autophagy but no cell death. The reason is still not clear. Similarly, silica NPs were found to induce autophagy dysfunction and autophagic cell death in HepG2 cells or L-02 normal human hepatic cell lines through lysosomal impairment and inhibition of autophagosome degradation.[179,180] Further studies are needed to clarify the detailed mechanism of autophagic cell death in hepatocytes.
7.4.3. Carbon NMs induce inflammatory responses and apoptosis
Many studies have been shown that CNTs induced significant adverse effects on hepatocytes. Yuan et al. found the oxidized SWCNTs with a length of 1000–2000 nm and a diameter of 1–6 nm triggered oxidative stress and interfered with intracellular metabolism, protein synthesis, and cytoskeletal systems in HepG2 cells. They also found that the oxidized SWCNTs elevated ROS levels, perturbed cell cycles, leading to a significant increase in apoptotic cell death likely mediated by the p38 signaling pathway and the activation of the nuclear factor kappa B (NF-κB) pathway.181] Kermanizadeh et al. found that MWCNTs with different lengths and diameters were both uptakes by the primary human hepatocytes and C3A cell line, resulting in IL-8 production and cytotoxicity at a higher concentration (>5 μg/cm2). Furthermore, a higher level of cytochrome P450 activity was found in the untreated primary cells compared with the C3A cell line.[182] For inflammatory response, Ji et al. demonstrated the Tween-80-dispersed MWCNTs (T-MWCNTs) induced mitochondria destruction, oxidative damage, gene expression change of CYP450 and TNF-α, and hepatotoxicity in mice.[183] These effects may be mediated by the P450 pathway and NF-κB signaling pathway.
The adverse effects of GOs and rGOs on hepatocytes have been shown in many studies. Chatterjee et al. reported that after exposure to HepG2 cells, GOs with an average thickness of 6 nm, a height of 20 nm, and lateral size distribution of 40 nm induced NADPH oxidase-dependent ROS formation, oxidative stress, DNA damage, deregulation of antioxidant/DNA repair/apoptosis-related genes, and cell death. The subsequent global gene expression and pathway analysis demonstrated that the GO effects were attributed to TGFβ1-mediated signaling.[184] In addition, they also found that the hydrophobic rGO was mostly adsorbed on the cell surface without internalization. This induced higher oxidative stress, stronger DNA damages, and apoptosis, etc.[184] The innate immune response through TLR4–NFκB pathway might be responsible for these cellular responses.
7.4.4. Nanocellulose induced oxidative stress and apoptosis
Although nanocellulose is generally considered to be biocompatible (no or low cytotoxicity), recent studies reported that nanocellulose materials could induce adverse effects in hepatocytes. It has been reported that nanocrystalline celluloses could induce the loss of cell viability in rainbow trout hepatocytes.[185] Also, Li et al. described the length-dependent cellular uptake and cellular responses of nanocellulose in Hepa 1–6 cells. The ~280 nm length CNC showed the highest uptake by the mouse hepatocyte cell line through clathrin-mediated endocytosis, inducing significantly higher mtROS generation and caspases 3/7-mediated apoptotic cell death.[109] This information provides useful information for the safe application of nanocellulose materials.
We summarized the cell-type-specific responses induced by various NMs in liver cells in Figure 11. This includes the higher uptake of NMs through phagocytosis by KCs, which could induce apoptosis or pyroptosis depending on the particle types. LSECs generally have lower NMs uptake through endocytosis, showing no significant cell death. However, HSCs also showed lower NMs uptake through endocytosis, which could lead to apoptosis. Similarly, hepatocytes also showed lower uptake of NMs by endocytosis, which could engage diverse cell death mechanisms including apoptosis, necrosis, and autophagic cell death.
Figure 11.
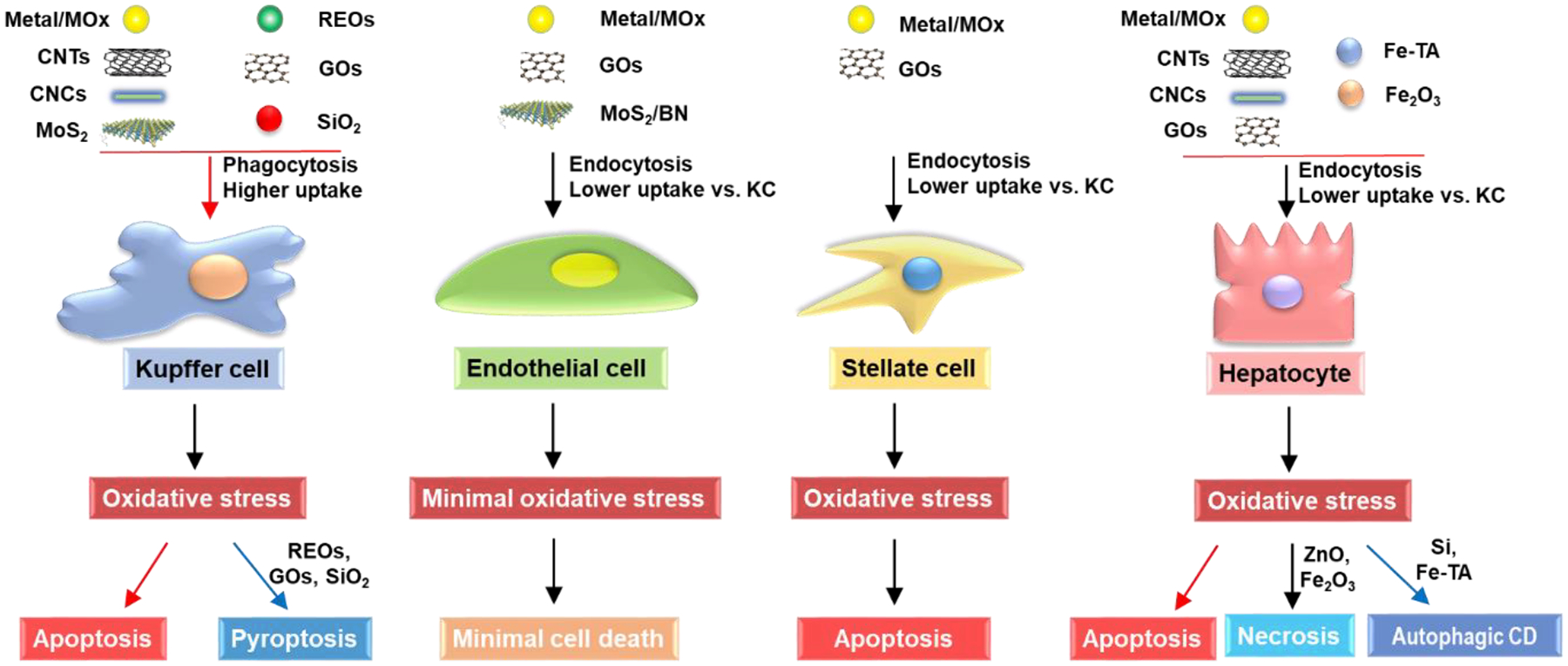
Cell type-specific responses induced by various NMs in the liver cells. High-level uptake of metal, MOx, CNTs, CNCs, MoS2, etc., and REOs, GOs, SiO2, etc., through phagocytosis, could induce apoptosis or pyroptosis in KCs, respectively. Low-level uptake of metal, MOx, GOs, MoS2, BN, etc., through endocytosis, did not induce significant cell death in LSECs. However, the lower uptake of metal, MOx, GOs, etc., through endocytosis, has been shown to induce apoptosis in stellate cells. Similarly, hepatocytes have lower uptake of metal, MOx, CNTs, CNCs, GOs, Fe-TA, etc. through endocytosis, which could induce cell death via apoptosis, necrosis, or autophagic cell death.
In general, most organic NPs including liposomes and polymeric NPs are considered to be biocompatible with low liver toxicity.[25] Although these NPs are also accumulated in the liver, they are readily degraded in the MPS or other tissues and organs. The safety concern is mainly from the payload, e.g., the burst release of drugs causing severe liver toxicity.[186] The impact of these organic NPs on the adverse effects of model drugs has only been recorded in a few cases.[122] In recent years, researchers have developed hybrid materials that have organic and inorganic components. For example, lipid bilayer-coated mesoporous silica nanoparticles (LB-MSNP) as drug carriers showed a much slower rate of payload release in the liver, which dramatically reduced the liver toxicity induced by liposomes carrying irinotecan, a key anticancer drug for pancreatic ductal adenocarcinoma (PDAC).[187]
8. Utilization of nano-liver interaction to treat diseases
Understanding the cell type-specific uptake and responses as well as the molecular mechanisms in the liver will facilitate the development of safer NMs applications. This includes treating liver conditions by using the NMs’ liver accumulation tendency to improve cancer and other diseases’ treatment (Figure 12).
Figure 12.
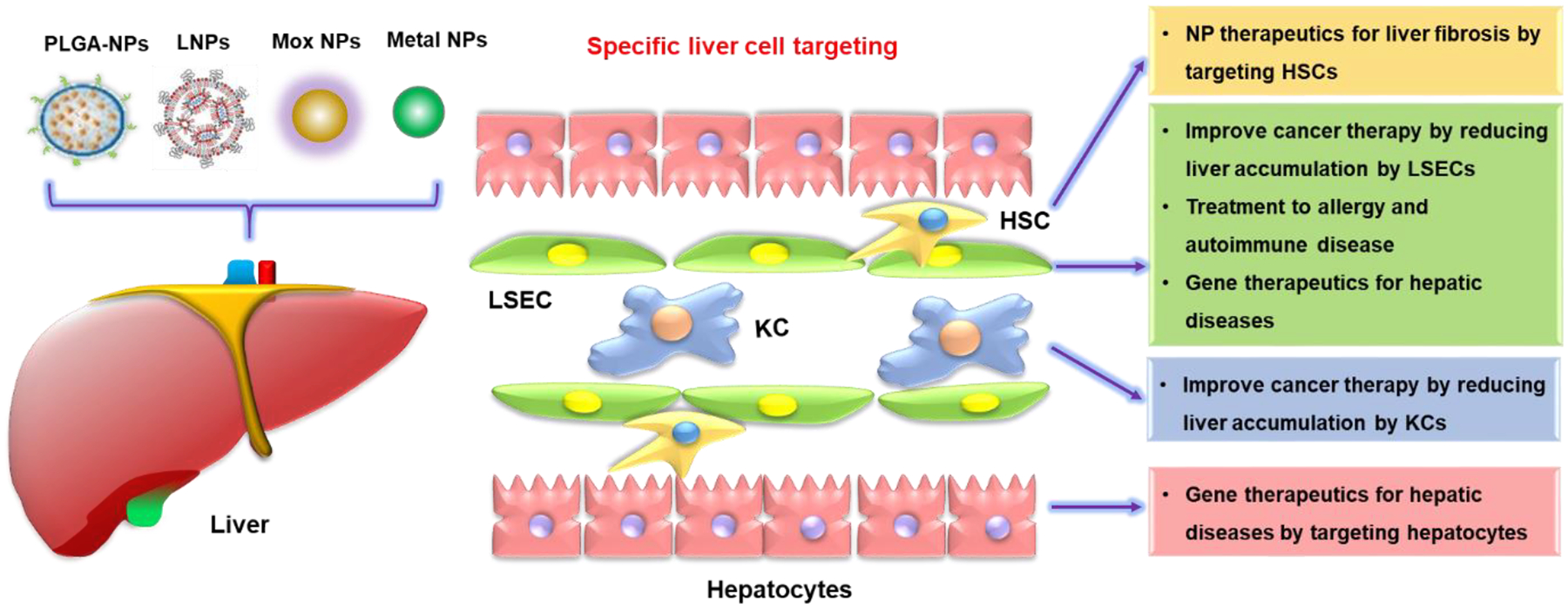
Schematic to demonstrate the utilization of nano-liver interactions to treat diseases based on cell-type-specific uptake or cellular response behaviors in the liver. This includes utilizing LNPs to treat liver fibrosis by targeting HSCs, utilizing metal NPs to improve cancer therapeutic efficacy by reducing LSEC or KC uptake, utilizing PLGA-NPs to treat allergy and autoimmune diseases by targeting LSEC, as well as using LNPs to improve gene therapy efficacy for hepatic diseases by targeting LSECs or hepatocytes. The Figure is reproduced with permission.[220] Copyright 2016, Elsevier B.V.
8.1. Treating liver conditions by the utilization of NMs liver accumulation
Many NMs accumulate in the liver, which is beneficial to develop nanomaterials as a drug to treat or improve liver conditions.[122,188] This includes the utilization of NMs to treat liver diseases including liver cancer, NAFLD, and hepatic fibrosis as well as enhancing drug bioavailability and biocompatibility through liver-targeting LNP delivery systems (Table 6).
Table 6.
The utilization of nano-liver interactions to treat diseases
| NMs | Methods | Models | Effects | References |
|---|---|---|---|---|
| AuNPs | Administering more than 1 trillion nanoparticles that is above the threshold to reduce uptake of drugs in macrophages | 4T1 tumor-bearing BALB/c mice | Reduction in the uptake rates, liver clearance, and prolonged circulation; enhancement in nanoparticle tumor penetration, delivery to the tumor cell population, and therapeutic efficacy independently of the active drug dose | [115] |
| ZnO NPs | Intravenous (i.v.) administration of NPs for the liver accumulation | Diethylnitrosamine (DEN)-induced hepatocellular carcinoma (HCC) in Wistar rats | Reduction in the elevated serum levels of HCC‐related tumor markers alpha-fetoprotein and alpha‐l‐fucosidase and the apoptotic marker caspase‐3. | [189] |
| Ag, Au, and Ag/Au alloy NPs | Administration of NPs for the liver accumulation | DEN-induced HCC in Sprague Dawley (SD) rats | Significant reduction (~45 to 65%) in tumor and the presence of BAX antibodies with up to immunoreactive (3+) level in the nanoparticle-treated animals | [190] |
| CeO2 NPs | Intravenously treating with NPs for the liver accumulation | Methionine and choline-deficient diet (MCDD) experimental model of NAFLD in Wistar rats | Reduction in the size and content of hepatocyte lipid droplets and the hepatic concentration of triglyceride- and cholesterol ester-derived FAs; expressing genes involved in cytokine, adipokine, and chemokine signaling pathways | [194] |
| ZnO NPs | Intraperitoneal (i.p.) injection NPs for the liver accumulation | High-fat-diet fed [HFD] obese C57BL/6 mice | significantly decreased HFD-induced hepatic steatosis and peripheral insulin resistance through hepatic SIRT1-LKB1-AMPK | [197] |
| TiO2 NPs | Internalization of nanomaterials in hepatic stellate cells | LX-2 cell line | Uptake, expression of Col-I and α-SMA, up-regulation of MMPs, down-regulation of TIMPs, regulation of EMT genes, and inhibition of hepatic fibrosis | [198] |
| SiO2 NPs | Internalization of nanomaterials in hepatic stellate cells | LX-2 cell line | Uptake, inhibition on cellular fibrosis, proteolytic breakdown of collagen, inhibition on adhesion and migration profiles of TGF-β, and potential treatment for liver fibrosis | [198] |
| PLGA-NPs attached ApoBP | Intravenous injection | LSECs and the antigen-specific tolerance in a murine anaphylaxis model | Endocytic uptake, induction of comparable immunotolerance in allergic airway disease and anaphylaxis as nanoparticle-delivering pharmaceuticals | [211] |
| LNP-mediated CRISPR-Cas9 delivery system | Intravenous injection to mice at a dose of 1.0, 2.0, and 3.0 mg/kg in total RNA.to target liver hepatocytes of mice | Wild-type C57BL/6 mice, aged 6 to 8 wk | A median gene editing rate of 38.5%, a corresponding 65.2% reduction of a target protein, and subsequent regulation of hypercholesterolemia. | [218] |
8.1.1. Treating liver cancer by NMs
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, which is more common in people who drink excessive alcohol or have an accumulation of fat in the liver.[188] A large number of metallic NMs have been reported to suppress HCC. For instance, ZnO NPs were found to inhibit the in vitro growth of the HCC cells as well as confer protective effects against diethylnitrosamine (DEN)-induced HCC in vivo by attenuating the elevated serum levels of HCC-related tumor markers alpha-fetoprotein and alpha-l-fucosidase and the apoptotic marker caspase-3.[189] Similarly, Au, Ag, and Ag-Au alloy NPs also can protect against DEN-induced HCC in rats through a reduction in tumor volume, blood hepatic biochemical markers, and improvement in liver architecture possibly via regulating oxidative stress by the NF-κB pathway.[190,191] Also, the low-density lipoprotein–docosahexaenoic acid (LDL-DHA) NPs have been demonstrated to selectively trigger ferroptosis in both human HCC cells and rats by lipid peroxidation, depletion of glutathione, and inactivation of the lipid antioxidant glutathione peroxidase-4 (GPX4).[192] Although these nanoparticles have shown anti-cancer effects, the application for cancer therapy is still a long shot due to the limitations on delivery, specificity, efficacy, etc.
8.1.2. Treating NAFLD and hepatic fibrosis by NMs
NAFLD is the most common form of chronic liver disease, affecting about one-quarter of the population in the United States.[73] The therapeutic effects of NMs on NAFLD have been reported.[9] For example, CeO2 NPs are strong antioxidants and capable of scavenging ROS in NAFLD-afflicted hepatocytes. Studies also found that CeO2 NPs could reduce fatty acid content by stimulating fatty acid (FA) metabolism in vitro,[193] and induce a reduction in NAFLD markers like steatosis, inflammation, and portal hypertension, lipid droplet size and content, FA concentrations, and the expression of NAFLD-signaling pathways in vivo.[194–196] In addition, ZnO NPs showed potential in the treatment of high-fat-diet fed (HFD)-induced hepatic steatosis and peripheral insulin resistance.[197] For example, Dorge et al. reported that ZnO NPs decreased high-fat-diet fed (HFD)-induced hepatic steatosis and peripheral insulin resistance through hepatic SIRT1-LKB1-AMPK which restricted SREBP-1c within the cytosol limiting its transcriptional ability in obese mice or HepG2 cells.[197]
In addition, Peng et al. showed TiO2 and SiO2 could block liver fibrosis and the associated phenotypes through their effects on the LX-2 human stellate cell line. They also found the internalized TiO2 and SiO2 NPs significantly suppressed the expression of α-smooth muscle actin (α-SMA) and the deposition of collagen I (Col-I) induced by TGF-β. The up-regulation of MMP-13 and down-regulation of tissue inhibitors of MMPs (TIMP-1) further improved the degradation of ECM, leading to a decrease in adhesion of TGF-β-activated LX-2 cells.[198] Furthermore, these NPs could reduce the migration of TGF-β-activated LX-2 cells.[198] Their results showed that TiO2 and SiO2 could potentially be used to treat hepatic fibrosis.
8.1.3. Treating liver diseases by organic nanoparticles
LDL-DHA NPs have been demonstrated to selectively trigger ferroptosis in both HCC cells and rats.[192] LNPs have received much attention recently in light of their successful use in COVID-19 vaccines to deliver the mRNA of spike proteins. LNPs have also been used as drug carriers due to advantages including controlled drug release, targeting ability, increased drug stability, and safety profiles.[199] It has shown that the LNP delivery systems could enhance drug bioavailability as well as increase its biocompatibility in the liver. For example, the triptolide-loaded solid lipid nanoparticle (TP-SLN) was found to enhance the anti-inflammatory activity of triptolide while avoiding triptolide-induced adverse effects in the liver, including oxidative stress, lipid peroxidation, DNA damage, ALT and AST release, and apoptotic/necrotic cell death.[200]
8.2. Improving therapeutic efficacy by liver cell-type-specific uptake
Various organic NMs including liposome and polymer-based nanoparticle carriers have shown promise in the targeted delivery of therapeutics (drugs, proteins, and nucleic acids) for the treatment of liver diseases, including hepatitis B virus (HBV) infection, liver fibrosis, and HCC.[188] This section is focused on the recent findings on cell-type-specific targeting of nanoparticle therapeutics for cancer and other diseases (Figure 12).
8.2.1. Improving the cancer therapeutic efficacy by reducing the liver accumulation
Although NMs have been widely used as drug carriers for the treatment of cancer, only limited numbers of nanomedicines have advanced to clinical use due to the low delivery efficiency (less than 0.7%) of these NMs to target sites.[6,201] This is ascribed to sequestration or clearance of NMs by the MPS, including KCs in the liver. One widely used approach is the use of PEG on a nanoparticle surface to reduce uptake by MPS and the liver.[202] For example, Doxil is the FDA-approved doxorubicin encapsulated in a liposome. The presence of PEG on the surface of the liposomal carrier has been shown to extend blood circulation time while reducing MPS or RES uptake, which is also called stealth nanoparticles.[95,203] This technology has resulted in a large number of nanoparticle formulations encapsulating active molecules, with high target efficiency and activity.[202,203] However, even in the presence of PEG, there is still substantial liver accumulation of NPs, which could explain the low efficacy of clinical anti-cancer formulations such as Caelyx®, Myocet®, and Onivyde®.[115] Chan et al. proposed that it is possible to reduce the accumulation of nanocarriers by saturation of the KCs by using a decoy nanoparticle first.[115] For example, Ouyang et al. showed that NP doses beyond the available binding site threshold in KCs could reduce liver clearance and prolong circulation time. They showed that administering sufficient NPs (more than 1 trillion) could overcome the threshold of KC uptake in the liver, resulting in enhanced tumor accumulation, which enhances the therapeutic efficacy in 4T1 tumor-bearing BALB/c mice.[115] The effectiveness of this strategy requires further testing.
8.2.2. Treating allergy and autoimmune diseases by targeting the natural tolerogenic liver microenvironment
The liver is maintaining an immune tolerant environment to prevent inflammation and allergic responses induced by the constant bombardment of bacterial materials such as LPS and foreign proteins from food sources coming from the gut to the portal vein.[44,112,163] In addition to the well-recognized roles of KCs, HSCs, and hepatocytes in immune tolerance, LSECs are increasingly recognized to be a major player.[43,80,164,204] LSECs have been shown to have a higher endocytotic activity than any other cell type and they can rapidly endocytose NMs through several families of endocytotic receptors: stabilin-1, stabilin-2, scavenger receptors, mannose receptor, and FcγRIIb2.[163,205] Although much of the clearance of blood-borne NMs was previously attributed to KCs, in recent decades it has become clear that LSECs and KCs play a complementary role in this process. LSECs are also increasingly recognized as a major antigen-presenting cell type in the liver, which can capture and present foreign antigens through the major histocompatibility complex (MHC)-II complex, and release TGF-β as well as IL-10 to direct the differentiation of naive T-cells into regulatory T-cells (Tregs) that can suppress antigen-specific immune responses.[43,206] Designing the tolerogenic NPs that introduce antigens into natural tolerogenic environments, including LSECs, can be used to accomplish antigen-specific immune tolerance in allergic and autoimmune diseases, including food allergy, anaphylaxis, asthma, rheumatoid arthritis, systemic lupus erythematosus, and type 1 diabetes (Figure 13).[207–210] For example, Liu et al. reported that the intravenous injected polymeric PLGA-NPs decorated with apolipoprotein B peptide (ApoBP) ligand could deliver the antigens or epitopes to LSECs through endocytic uptake, inducing the generation of Foxp3+ regulatory T cells (Treg), which is a powerful T cell that induces antigen-specific immune tolerance in allergic lung inflammation and anaphylaxis models.[206,211] The use of the polymeric NP platform targeting LSECs could also be possible for treating autoimmune diseases, which requires future research.
Figure 13.
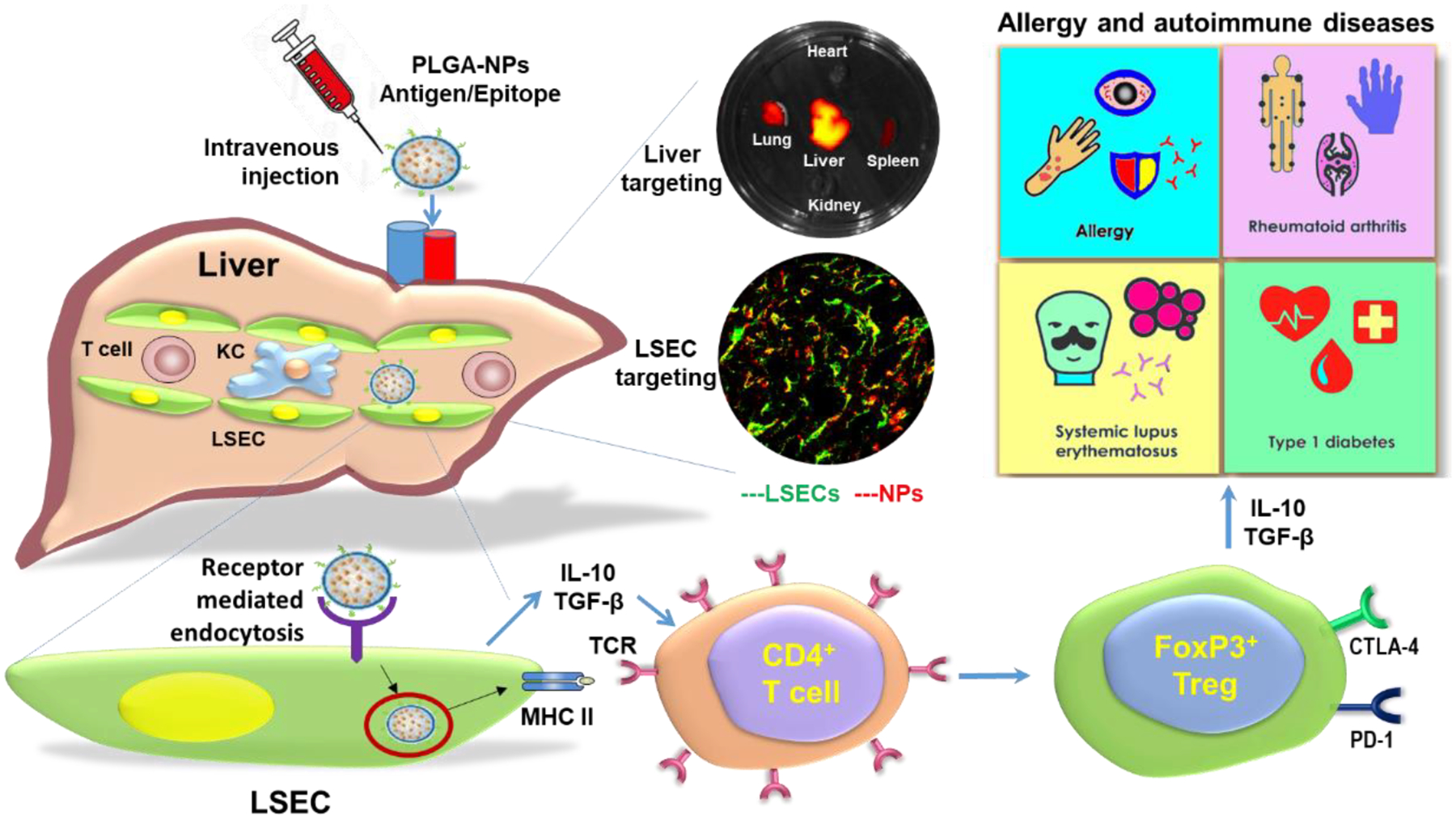
Schematic to demonstrate the treatment of allergy and autoimmune diseases by targeting natural tolerogenic LSEC. The intravenous injected PLGA-NPs attached ApoBP ligand delivering the antigens are epitopes to LSECs in the liver through endocytic uptake. Antigen processing and presentation to naive CD4+ T-cells are capable to generate Foxp3+ Tregs that are recruited to the site of pathology, where they exert immunosuppressive effects on allergy and autoimmune diseases, including allergy, rheumatoid arthritis, systemic lupus erythematosus, and type 1 diabetes. The middle panel shows the liver targeting and LSEC targeting determined by the representative ex vivo IVIS images and confocal images, respectively. The Figure is reproduced with permission.[206] Copyright 2020, American Chemical Society.
8.2.3. Alleviating liver fibrosis by targeting HSCs
The HSCs play a vital role in the process of liver fibrosis.[212] The activated hepatic stellate cells (aHSCs) are the main effector cells for liver fibrosis through the ECM production in response to chronic liver injuries.[167,213–215] Hydrophilic NMs less than 200 nm can be captured by HSCs. One study showed that 130~150 nm hydrophilic lipid–protamine–hyaluronic acid (LPH) NPs modified with aHSC-targeting aminoethyl anisamide (AEAA) and encapsulated the relaxin gene and miR-30a-5p mimic could preferentially target activated HSCs in the fibrotic liver. It has been also shown to achieve synergistic antifibrosis effects in models of mouse liver fibrosis with the decreased α-SMA expression and collagen content in the liver.[216] This provides a proof-of-concept approach to the alleviation of liver fibrosis.
8.2.4. Improving gene therapy efficacy by targeting hepatocytes or LSECs using LNPs
Genome editing technologies have emerged as a potentially powerful therapeutic tool to treat diseases with a genetic etiology.[122] However, the application of this technology has been limited by the technical challenge of achieving safe, effective, and specific in vivo delivery of the genes such as the CRISPR-Cas9 genome editing components. LNPs play a vital role in delivering messenger RNA (mRNA), short interfering RNA (siRNA), DNA, or gene-editing complexes, which provide opportunities to treat hepatic diseases through silencing pathogenic genes, expressing therapeutic proteins, or correcting genetic defects.[204] The LNP systems containing mRNA have been shown to target the liver as a bioreactor and produce therapeutic proteins, such as monoclonal antibodies and hormones following i.v. administration.[217] The recently developed LNP-mediated CRISPR-Cas9 delivery system based on nano-liver interactions specifically delivered Cas9 mRNA and target sgRNA to liver hepatocytes of wild-type mice, resulting in a median editing rate of 38.5% and a corresponding 65.2% reduction of a target protein.[218] Importantly, the adverse effects in the liver or off-target mutagenesis were not observed in this system, offering a clinically viable approach for liver-specific delivery of genome editing tools. Furthermore, similar to previous nano-liver interactions, Akinc et al. investigated the mechanism of cellular uptake of LNPs by hepatocytes, they reported the adsorption of serum ApoE on the surface of LNPs was a major effector facilitating the intracellular delivery of LNPs into hepatocytes through low-density lipoprotein (LDL) receptors.[219] Also, the NP size was found to be heavily influenced by the gene delivery efficacy of LNPs during blood circulation.[220] It has been shown that the LNPs with a size less than 80 nm can efficiently pass through liver fenestrae and LNPs with ionizable cationic lipid pKa value around 6.4 and near-neutral surface charge could help to prevent sequestration by the MPS, and avoid immune stimulation and adverse effects.[122,124]
In addition to delivering to hepatocytes that are often associated with diseases in metabolism and endocrine dysfunctions, other liver cell types including LSECs that are closely related to liver fibrosis and cirrhosis can also be targeted by LNPs.[65,112,221,222] For example, the larger LNPs that cannot penetrate through the fenestrae could be delivered to LSECs and induce the target gene silencing. Furthermore, the size and composition of LNPs were important for endothelium-specific delivery.[223] However, the development of LNP formulations for selective delivery of RNA into certain types of liver cells remains the main challenge. Recently, Kim et al. successfully showed a liver cell type-specific delivery and demonstrated the targeted delivery of RNA into hepatocytes and LSECs by controlling the size of LNPs, PEG-lipid content, and incorporation of active targeting ligands.[124] This cell type-specific delivery of RNA by LNPs may shed light on developing new gene therapies.
While substantial progress has been made in the delivery of nucleic acid, knowledge regarding the mechanism of LNP-mediated adverse effects in vivo has been limited. For example, Sato et al. reported that the hepatotropic LNP induced liver effects following the intravenous injection of a high dose, which could be attributed to the accumulation of the LNPs in LSECs. They found induction of the pro-inflammatory cytokines, and neutrophil recruitment, and neutrophilic inflammation. Furthermore, maximization of the hepatocyte-specificity of the LNPs by modification with both GalNAc ligands and PEGs enhances the safety without any loss in delivery efficiency. As a result, a single injection of the newly designed LNPs was found to show strong and durable inhibitory effects on HBV without any sign of the adverse effect.[224] Furthermore, Jackson et al. identified platelet-activating factor (PAF) release from KCs as a critical mediator of siRNA nanocarrier toxicity, and PAF receptor inhibition could serve as an effective approach for increasing the tolerated dose of siRNA nanocarriers.[225] This indicates the potential to develop safe nanoproducts and nanomedicine by the utilization of nano-liver interactions.
In 2018, the U.S. Food and Drug Administration had approved Onpattro® (Patisiran), a lipid nanoparticle-based siRNA drug that acts by inhibiting the synthesis of the transthyretin (TTR) protein in hepatocytes in the liver, to treat the peripheral nerve disease (polyneuropathy) resulted from the hereditary transthyretin-mediated amyloidosis (hATTR) in adult patients.[217,226] The success of Onpattro® opened new avenues for the clinical translation of LNP nanomedicines containing nucleic acid-based drugs to enable many novel therapeutics based on silencing or expressing target genes.
9. Conclusions and perspectives
Widespread use of NMs in commercial products and biomedicine will lead to increased human exposure, resulting in the potential accumulation of NMs in the liver. Thus a good understanding of nano-liver interactions will be critical for developing safer nanoapplications. Although it is still in its infancy, substantial evidence has been accumulated. We showed a comprehensive review of this issue including the unique liver structures and functions, diverse cell types, cellular uptake, transformation, clearance, and effects, as well as the molecular mechanisms involved induced by representative NMs with different physicochemical properties. We highlighted the cell type-specific uptake and responses after NMs exposure to the liver. We can use this knowledge on nano-liver interactions to develop safer and more effective NMs for nanoproducts and biomedical applications such as treating cancer and liver disorders.
Still much needs to be done on understanding the nano-liver interactions at the cellular levels. Our understanding of the interactions in vivo is hindered by the complexity of the liver structure, function, and diverse cell types. In addition, the current animal dominant toxicity testing approach is time-consuming, prohibitive in cost, and logistically impossible to perform considering the number of nanomaterials is well over 4500 by Nanowerk without counting additional functionalizations.[227–229] Furthermore, considering the 3Rs-replacement, reduction, and refinement in animal research, we have to develop reliable in vitro models that are based on the mechanism of action, which have been shown to be more predictive of in vivo outcomes. To speed up the discovery and establish a predictive paradigm, high-throughput screening (HTS) should be used and bioinformatics will facilitate the development of nano structure-activity relationships (nano-SARs) because there are so many NMs with diverse physicochemical properties.[228,229] Currently, 2D hepatocyte cell culture, a single layer of cells attached to a plastic surface and submerged in the cell culture medium, is still widely used for liver toxicity evaluations. However, hepatocytes under 2D cell culture will rapidly lose their original cell morphology and function, and using these cells for NMs screening will not reflect the hepatocyte responses in vivo. Moreover, the liver tissue is composed of diverse cell types with unique spatial distribution, the single-cell type 2D culture will not reflect the complexity of cell-cell interactions in the liver. The emerging 3D cell culture comprises single or multiple cell types (e.g., KCs, LSECs, HSCs, and hepatocytes) in the form of liver spheroids or liver microtissues could maintain the hepatocyte function for longer times and mimic the smallest functional unit of the liver.[230–232] The 3D liver spheroids provide a morphological and functional analog to liver tissue and are better in vitro safety screening models to comprehensively study the effects of NMs and more accurately predict liver responses to NMs in vivo. However, this is not to say 3D spheroid could replace in vivo animal testing, the idea is that when certain NMs are identified to have potential adverse effects in vitro, limited but focused animal studies should be performed to validate the results obtained using 3D spheroids.
Most of the studies on the liver is focusing on known mechanisms or common endpoints such as cell viability, oxidative stress, inflammation, biomarkers, histology, etc., which is not an unbiased approach to gain new insights. Recent technological advances in high throughput multi-omics such as single-cell RNA sequencing (scRNASeq) provide revolutionary opportunities to build a comprehensive and clearer map of the mechanisms of NMs effects in a global unbiased manner at an unprecedented single-cell resolution.[233,234] This map will enable us to identify new pathways or mechanisms of toxicity.[235,236] Another example is metabolomics, which is a powerful analytical methodology for studying a broad range of metabolites in the context of physiological stimuli or disease states,[237] may aid in the understanding of direct cellular phenotypes induced by NMs and lead to new biomarker discovery that is complementary to genomics, transcriptomics, and proteomics.[235,238] In addition, liquid chromatography with mass spectrometry (LC-MS)-based metabolomics has been shown to be highly sensitive to detect subtle metabolomic changes when functional cellular assays show no significant difference.[239] These technological advancements will facilitate our understanding of nano-liver interactions.
Currently, although there are many in vitro and in vivo studies that have evaluated the potential adverse effects of NMs on the liver,[33,34,240] the comparison among the different reports is often difficult due to the differences in NMs preparations, physicochemical characterizations, experimental procedures, biomarker measurements, and pathological endpoints, even leading to conflicting results.[241] For example, BN and MoS2 have been shown to induce adverse effects in human HepG2 hepatocytes.[242] However, Li et al. and Sobańska et al. did not observe any impairment to cell growth and survival of hepatocytes, even after high-dose exposures over prolonged periods.[243,244] Thus, the data reproducibility and reliability have to be improved in both nanosafety and nanomedicine research to ensure the accuracy, reliability, reproducibility, and intercomparison of experimental data. The application of reporting standards, research guidelines, international standards, and minimal characterization checklists is urgently needed.
Additionally, the liver is a complex organ, we need to understand the in vivo behavior of NMs better, including biodistribution, biotransformation, and bioavailability, for a well-established clinical trial.[245–249] We know much information about intravenously injected NPs, often functionalized by PEG to have colloidal stability in the blood, could accumulate in the liver in a high amount. However, we know little about the NMs entering through other routes including skin, gastrointestinal tract, the lung, or intramuscular delivery. Although there are reports claiming liver accumulation after the NMs gain access to the systemic circulation by passing through these physiological barriers, the properties, dose, cellular uptake mechanism, biotransformation, clearance, bioavailability, and effects of these NMs on the liver have not been elucidated and more research is needed in this area.
Our understanding of nano-liver interactions will benefit the development of nanoapplications. We have already begun to see the development of therapeutics based on the progress in this area. For example, the design of nanoparticles targeting specific liver cell types to treat liver disorders,[206,211,216–218,221–226] increased nano-drug delivery to tumors by reducing liver accumulation,[95,115,203] and taking advantage of the natural immune tolerant environment in the liver to induce the generation of antigen-specific regulatory T cells to treat allergy and autoimmune diseases.[206,211] We could expect a better understanding of nano-liver interactions with more effort, which will facilitate the safer development of NMs for nanoapplications.
Acknowledgments
The research reported in this publication was supported by the National Key Research and Development Program of China (2021YFA1200902), the Nanotechnology Health Implications Research (NHIR) Consortium of the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number (U01ES027237), the start-up funding support from the National Center for Nanoscience and Technology, Chinese Academy of Sciences (CAS) (E1763911), and the Science Fund for Creative Research Groups of the National Natural Science Foundation of China (11621505). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Biographies

Jiulong Li is an Associate Professor at the National Center for Nanoscience and Technology (NCNST) in China. He received his degrees Ph.D. in biophysics from Zhejiang University and gained postdoctoral training at the Center of Environmental Implications of Nanotechnology (UC CEIN), California NanoSystems Institute at UCLA. His research interests focus on the nano-bio interface, the structure-activity relationship of nanomaterials, and the nano-liver interactions for the safe application of nanomaterials.

Chunying Chen is a Principal Investigator at CAS Key Laboratory for Biomedical Effects of Nanomaterials and Nanosafety in the National Center for Nanoscience and Technology of China. Prof. Chen received her Bachelor’s degree in Chemistry (1991) and her Ph.D. degree in Biomedical Engineering from Huazhong University of Science and Technology (1996). Prof. Chen provides input to WHO, ISO, and OECD working groups investigating the health effects of nanomaterials. Her research interests mainly include nanotoxicology, nanomedicine, the nano–bio interfaces, the transformation and fate of nanomaterials in biological systems, and therapies for malignant tumors using theranostic nanomedicine systems and vaccine nanoadjuvants using nanomaterials.

Tian Xia is an Associate Professor in the Division of NanoMedicine, Department of Medicine at UCLA. He studies nanomedicine, which includes the development of nanomaterials for medical applications and research on nanosafety. For nanomedicine, he developed AlOOH and nanocellulose for vaccine adjuvants, tolerogenic liver-targeting PLGA nanoparticles for allergic and autoimmune diseases, and GO as antimicrobial to combat antibiotic resistance. For nanosafety, he used predictive toxicology and high throughput screening to establish toxicological profiles for over 100 different nanomaterials covering major material categories including carbonaceous materials (fullerene and carbon nanotubes), 2D materials (graphene, GO, and MoS2), metallic, silica, and III–V materials.
Contributor Information
Jiulong Li, CAS Key Laboratory for Biomedical Effects of Nanomaterials and Nanosafety, CAS Center for Excellence in Nanoscience, National Center for Nanoscience and Technology, Beijing 100190, P.R. China.
Chunying Chen, CAS Key Laboratory for Biomedical Effects of Nanomaterials and Nanosafety, CAS Center for Excellence in Nanoscience, National Center for Nanoscience and Technology, Beijing 100190, P.R. China.
Tian Xia, Center of Environmental Implications of Nanotechnology (UC CEIN), California NanoSystems Institute, Division of NanoMedicine, Department of Medicine, University of California, Los Angeles, CA 90095, USA.
References
- [1].Baig N, Kammakakam I, Falath W, Materials Advances 2021, 2, 1821. [Google Scholar]
- [2].Smith BR, Gambhir SS, Chem. Rev 2017, 117, 901. [DOI] [PubMed] [Google Scholar]
- [3].Valsami-Jones E, Lynch I, Science 2015, 350, 388. [DOI] [PubMed] [Google Scholar]
- [4].Hochella M, Mogk DW, Ranville J, Allen IC, Luther GW, Marr LC, Mcgrail BP, Murayama M, Qafoku NP, Rosso KM, Science 2019, 363, 1414. [DOI] [PubMed] [Google Scholar]
- [5].Khin MM, Nair AS, Babu VJ, Murugan R, Ramakrishna S, Energ. Environ. Sci 2012, 5, 8075. [Google Scholar]
- [6].Pelaz B, Alexiou C, Alvarez-Puebla RA, Alves F, Andrews AM, Ashraf S, Balogh LP, Ballerini L, Bestetti A, Brendel C, Bosi S, Carril M, Chan WCW, Chen C, Chen X, Chen X, Cheng Z, Cui D, Du J, Dullin C, Escudero A, Feliu N, Gao M, George M, Gogotsi Y, Grünweller A, Gu Z, Halas NJ, Hampp N, Hartmann RK, Hersam MC, Hunziker P, Jian J, Jiang X, Jungebluth P, Kadhiresan P, Kataoka K, Khademhosseini A, Kopeček J, Kotov NA, Krug HF, Lee DS, Lehr C, Leong KW, Liang X, Ling Lim M, Liz-Marzán LM, Ma X, Macchiarini P, Meng H, Möhwald H, Mulvaney P, Nel AE, Nie S, Nordlander P, Okano T, Oliveira J, Park TH, Penner RM, Prato M, Puntes V, Rotello VM, Samarakoon A, Schaak RE, Shen Y, Sjöqvist S, Skirtach AG, Soliman MG, Stevens MM, Sung H, Tang BZ, Tietze R, Udugama BN, Vanepps JS, Weil T, Weiss PS, Willner I, Wu Y, Yang L, Yue Z, Zhang Q, Zhang Q, Zhang X, Zhao Y, Zhou X, Parak WJ, ACS Nano 2017, 11, 2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mirshafiee V, Sun B, Chang CH, Liao Y, Jiang W, Jiang J, Liu X, Wang X, Xia T, Nel AE, ACS Nano 2018, 12, 3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yao Y, Zang Y, Qu J, Tang M, Zhang T, Int. J. Nanomed 2019, 14, 8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Boey A, Ho HK, Small 2020, 16, 2000153. [DOI] [PubMed] [Google Scholar]
- [10].Georgakilas V, Tiwari JN, Kemp KC, Perman JA, Bourlinos AB, Kim KS, Zboril R, Chem. Rev 2016, 116, 5464. [DOI] [PubMed] [Google Scholar]
- [11].Bottari G, Herranz MÁ, Wibmer L, Volland M, Rodríguez-Pérez L, Guldi DM, Hirsch A, Martín N, D’Souza F, Torres T, Chem. Soc. Rev 2017, 46, 4464. [DOI] [PubMed] [Google Scholar]
- [12].Reina G, González-Domínguez JM, Criado A, Vázquez E, Prato M, Chem. Soc. Rev 2017, 46, 4400. [DOI] [PubMed] [Google Scholar]
- [13].Hua Z, Hui-Ming C, Peide Y, Chem. Soc. Rev 2018, 47, 10. [Google Scholar]
- [14].Kurapati R, Kostarelos K, Prato M, Bianco A, Adv. Mater 2016, 28, 6052. [DOI] [PubMed] [Google Scholar]
- [15].Wang P, Nie X, Wang Y, Li Y, Ge C, Zhang L, Wang L, Bai R, Chen Z, Zhao Y, Chen C, Small 2013, 9, 3799. [DOI] [PubMed] [Google Scholar]
- [16].Harrison BS, Atala A, Biomaterials 2007, 28, 344. [DOI] [PubMed] [Google Scholar]
- [17].Ali MA, Hu C, Jahan S, Yuan B, Saleh MS, Ju E, Gao S, Panat R, Adv. Mater 2021, 33, 2006647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Thomas B, Raj MC, Athira KB, Rubiyah MH, Sanchez C, Chem. Rev 2018, 118, 11575. [DOI] [PubMed] [Google Scholar]
- [19].Moon RJ, Martini A, Nairn J, Simonsen J, Youngblood J, Chem. Soc. Rev 2011, 40, 3941. [DOI] [PubMed] [Google Scholar]
- [20].Chhowalla M, Shin HS, Eda G, Li LJ, Loh KP, Zhang H, Nat. Chem 2013, 5, 263. [DOI] [PubMed] [Google Scholar]
- [21].Li X, Wang X, Zhang J, Hanagata N, Wang X, Weng Q, Ito A, Bando Y, Golberg D, Nat. Commun 2017, 8, 13936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen Y, Fan Z, Zhang Z, Niu W, Li C, Yang N, Chen B, Zhang H, Chem. Rev 2018, 118, 6409. [DOI] [PubMed] [Google Scholar]
- [23].Zhang S, Geryak R, Geldmeier J, Kim S, Tsukruk VV, Chem. Rev 2017, 117, 12942. [DOI] [PubMed] [Google Scholar]
- [24].Zhao N, Yan L, Zhao X, Chen X, Li A, Zheng D, Zhou X, Dai X, Xu F, Chem. Rev 2019, 119, 1666. [DOI] [PubMed] [Google Scholar]
- [25].Fang F, Li M, Zhang J, Lee C, ACS Materials Letters 2020, 2, 531. [Google Scholar]
- [26].Wiseguyreports.com, Nanotechnology Market 2019. Global Industry-Key Players, Size, Trends, Opportunities, Growth Analysis and Forecast to 2024, 2019, https://www.wiseguyreports.com/reports/3702298-global-healthcare-nanotechnology-market-growth-status-and-outlook-2019-2024.
- [27].a) Pietroiusti A, Stockmann-Juvala H, Lucaroni F, Savolainen K, WIREs Nanomed. Nanobio 2018, 10, e1513. [DOI] [PubMed] [Google Scholar]; b) Huo L, Chen R, Zhao L, Shi X, Bai R, Long D, Chen F, Zhao Y, Chang Y, Chen C, Biomaterials 2015, 61, 307. [DOI] [PubMed] [Google Scholar]
- [28].Donaldson K, Schinwald A, Murphy F, Cho W, Duffin R, Tran L, Poland C, Accounts Chem. Res 2013, 46, 723. [DOI] [PubMed] [Google Scholar]
- [29].Lu X, Zhu Y, Bai R, Wu Z, Qian W, Yang L, Cai R, Yan H, Li T, Pandey V, Liu Y, Lobie PE, Chen C, Zhu T, Nat. Nanotechnol 2019, 14, 719. [DOI] [PubMed] [Google Scholar]
- [30].Wang Y, Cai R, Chen C, Accounts Chem. Res 2019, 52, 1507. [DOI] [PubMed] [Google Scholar]
- [31].Guiney LM, Xiang W, Tian X, Nel AE, Hersam MC, ACS Nano 2018, 12, 6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bondarenko O, Mortimer M, Kahru A, Feliu N, Javed I, Kakinen A, Lin S, Xia T, Song Y, Davis TP, Lynch I, Parak WJ, Leong DT, Ke PC, Chen C, Zhao Y, Nano Today 2021, 39, 101184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sharifi S, Behzadi S, Laurent S, Laird Forrest M, Stroeve P, Mahmoudi M, Chem. Soc. Rev 2012, 41, 2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang Y, Bai Y, Jia J, Gao N, Li Y, Zhang R, Jiang G, Yan B, Chem. Soc. Rev 2014, 43, 3762. [DOI] [PubMed] [Google Scholar]
- [35].øie CI, Mönkemöller V, Hübner W, Schüttpelz M, Mao H, Ahluwalia BS, Huser TR, Mccourt P, Nanophotonics 2018, 7, 575. [Google Scholar]
- [36].Gebhardt R, Pharmacol. Therapeut 1992, 53, 275. [DOI] [PubMed] [Google Scholar]
- [37].Ben-Moshe S, Itzkovitz S, Nat. Rev. Gastro. Hepat 2019, 16, 395. [DOI] [PubMed] [Google Scholar]
- [38].Donne R, Saroul-Aïnama M, Cordier P, Celton-Morizur S, Desdouets C, Nat. Rev. Gastro. Hepat 2020, 17, 391. [DOI] [PubMed] [Google Scholar]
- [39].Frevert U, Engelmann S, Zougbédé S, Stange J, Ng B, Matuschewski K, Liebes L, Yee H, PLoS Biol 2005, 3, e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fu X, Sluka JP, Clendenon SG, Dunn KW, Wang Z, Klaunig JE, Glazier JA, PLoS One 2018, 13, e198060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Teutsch HF, Hepatology 2005, 42, 317. [DOI] [PubMed] [Google Scholar]
- [42].Kazankov K, Jørgensen SMD, Thomsen KL, Møller HJ, Vilstrup H, George J, Schuppan D, Grønbæk H, Nat. Rev. Gastro. Hepat 2019, 16, 145. [DOI] [PubMed] [Google Scholar]
- [43].Shetty S, Lalor PF, Adams DH, Nat. Rev. Gastro. Hepat 2018, 15, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Thomson AW, Knolle PA, Nat. Rev. Immunol 2010, 10, 753. [DOI] [PubMed] [Google Scholar]
- [45].Trefts E, Gannon M, Wasserman DH, Curr. Biol 2017, 27, R1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gissen P, Arias IM, J. Hepatol 2015, 63, 1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Guo H, Feng C, Zhang W, Luo Z, Zhang H, Zhang T, Ma C, Zhan Y, Li R, Wu S, Abliz Z, Li C, Li X, Ma X, Wang L, Zheng W, Han Y, Jiang J, Nat. Commun 2019, 10, 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Walser T, Limbach LK, Brogioli R, Erismann E, Flamigni L, Hattendorf B, Juchli M, Krumeich F, Ludwig C, Prikopsky K, Rossier M, Saner D, Sigg A, Hellweg S, Günther D, Stark WJ, Nat. Nanotechnol 2012, 7, 520. [DOI] [PubMed] [Google Scholar]
- [49].Zhang YN, Poon W, Tavares AJ, Mcgilvray ID, Chan WCW, Control J. Release 2016, 240, 332. [DOI] [PubMed] [Google Scholar]
- [50].Guo M, Xu X, Yan X, Wang S, Gao S, Zhu S, J. Hazard. Mater 2013, 260, 780. [DOI] [PubMed] [Google Scholar]
- [51].Golbamaki N, Rasulev B, Cassano A, Marchese Robinson RL, Benfenati E, Leszczynski J, Cronin MTD, Nanoscale 2015, 7, 2154. [DOI] [PubMed] [Google Scholar]
- [52].Nel A, Xia T, Madler L, Li N, Science 2006, 311, 622. [DOI] [PubMed] [Google Scholar]
- [53].Loeschner K, Hadrup N, Qvortrup K, Larsen A, Gao X, Vogel U, Mortensen A, Lam HR, Larsen EH, Part. Fibre Toxicol 2011, 8, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ryter SW, Cloonan SM, Choi AMK, Mol. Cells 2013, 36, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Friedrich-Rust M, Poynard T, Castera L, Nat. Rev. Gastro. Hepat 2016, 13, 402. [DOI] [PubMed] [Google Scholar]
- [56].Li Y, Yan J, Ding W, Chen Y, Pack LM, Chen T, Mutagenesis 2017, 32, 33. [DOI] [PubMed] [Google Scholar]
- [57].Szabo G, Petrasek J, Nat. Rev. Gastro. Hepat 2015, 12, 387. [DOI] [PubMed] [Google Scholar]
- [58].Gluchowski NL, Becuwe M, Walther TC, Farese RV, Nat. Rev. Gastro. Hepat 2017, 14, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Abdelhalim MAK, Jarrar BM, Lipids Health Dis 2011, 10, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Duan J, Liang S, Feng L, Yu Y, Sun Z, Int J Nanomedicine 2018, 13, 7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Jia J, Li F, Zhou H, Bai Y, Liu S, Jiang Y, Jiang G, Yan B, Environ. Sci. Technol 2017, 51, 9334. [DOI] [PubMed] [Google Scholar]
- [62].Yu Y, Duan J, Li Y, Li Y, Jing L, Yang M, Wang J, Sun Z, Int. J. Nanomed 2017, 12, 6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Nishimori H, Kondoh M, Isoda K, Tsunoda S, Tsutsumi Y, Yagi K, Eur. J. Pharm. Biopharm 2009, 72, 626. [DOI] [PubMed] [Google Scholar]
- [64].van der Zande M, Vandebriel RJ, Groot MJ, Kramer E, Herrera Rivera ZE, Rasmussen K, Ossenkoppele JS, Tromp P, Gremmer ER, Peters RJ, Hendriksen PJ, Marvin HJ, Hoogenboom RL, Peijnenburg AA, Bouwmeester H, Part. Fibre Toxicol 2014, 11, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Burra P, Best Pract Res Clin Gastroenterol 2013, 27, 553. [DOI] [PubMed] [Google Scholar]
- [66].Kong T, Zhang S, Zhang C, Zhang J, Yang F, Wang G, Yang Z, Bai D, Zhang M, Wang J, Zhang B, Biol. Trace Elem. Res 2019, 189, 478. [DOI] [PubMed] [Google Scholar]
- [67].Sadeghi L, Babadi VY, Espanani HR, Bratislava Medical Journal 2015, 116, 373.26084739 [Google Scholar]
- [68].Kunutsor SK, Apekey TA, Khan H, Atherosclerosis 2014, 236, 7. [DOI] [PubMed] [Google Scholar]
- [69].Parivar K, Malekvand Fard F, Bayat M, Alavian SM, Motavaf M, Iran. Red Crescent Me 2016, 18, e28939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].De Jong WH, De Rijk E, Bonetto A, Wohlleben W, Stone V, Brunelli A, Badetti E, Marcomini A, Gosens I, Cassee FR, Nanotoxicology 2019, 13, 50. [DOI] [PubMed] [Google Scholar]
- [71].Bernhoft RA, Grant H, Hansen DK, Sci. World J 2013, 2013, 394652. [Google Scholar]
- [72].Yan X, Xu X, Guo M, Wang S, Gao S, Zhu S, Rong R, Environ. Toxicol 2017, 32, 1202. [DOI] [PubMed] [Google Scholar]
- [73].Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ, Nat. Med 2018, 24, 908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zhang H, Chen R, Shao Y, Wang H, Liu Z, Toxicol. Res.-UK 2018, 7, 809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Poulsen SS, Saber AT, Mortensen A, Szarek J, Wu D, Williams A, Andersen O, Jacobsen NR, Yauk CL, Wallin H, Halappanavar S, Vogel U, Toxicol. Appl. Pharm 2015, 283, 210. [DOI] [PubMed] [Google Scholar]
- [76].Fadeel B, Bussy C, Merino S, Vázquez E, Flahaut E, Mouchet F, Evariste L, Gauthier L, Koivisto AJ, Vogel U, Martín C, Delogu LG, Buerki-Thurnherr T, Wick P, Beloin-Saint-Pierre D, Hischier R, Pelin M, Candotto Carniel F, Tretiach M, Cesca F, Benfenati F, Scaini D, Ballerini L, Kostarelos K, Prato M, Bianco A, ACS Nano 2018, 12, 10582. [DOI] [PubMed] [Google Scholar]
- [77].Xiong G, Deng Y, Liao X, Zhang JE, Cheng B, Cao Z, Lu H, Nanotoxicology 2020, 14, 667. [DOI] [PubMed] [Google Scholar]
- [78].Wu Y, Feng W, Liu R, Xia T, Liu S, ACS Nano 2020, 14, 877. [DOI] [PubMed] [Google Scholar]
- [79].Heringa MB, Peters RJB, Bleys RLAW, van der Lee MK, Tromp PC, van Kesteren PCE, van Eijkeren JCH, Undas AK, Oomen AG, Bouwmeester H, Part. Fibre. Toxicol 2018, 15, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Chen YY, Syed AM, Macmillan P, Rocheleau JV, Chan WCW, Adv. Mater 2020, 32, 1906274. [DOI] [PubMed] [Google Scholar]
- [81].Tsoi KM, Macparland SA, Ma X, Spetzler VN, Echeverri J, Ouyang B, Fadel SM, Sykes EA, Goldaracena N, Kaths JM, Conneely JB, Alman BA, Selzner M, Ostrowski MA, Adeyi OA, Zilman A, Mcgilvray ID, Chan WCW, Nat. Mater 2016, 15, 1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Roduner E, Chem. Soc. Rev 2006, 35, 583. [DOI] [PubMed] [Google Scholar]
- [83].Liu Y, Workalemahu B, Jiang X, Small 2017, 13, 1701815. [DOI] [PubMed] [Google Scholar]
- [84].Albanese A, Tang PS, Chan WCW, Annu. Rev. Biomed. Eng 2012, 14, 1. [DOI] [PubMed] [Google Scholar]
- [85].Liu X, Xie X, Jiang J, Lin M, Zheng E, Qiu W, Yeung I, Zhu M, Li Q, Xia T, Meng H, Adv. Funct. Mater 2021, 2104487. [Google Scholar]
- [86].Cai R, Chen C, Adv. Mater 2018, 1805740. [DOI] [PubMed] [Google Scholar]
- [87].Cai H, Ma Y, Wu Z, Ding Y, Zhang P, He X, Zhou J, Chai Z, Zhang Z, NanoImpact 2016, 3–4, 40. [Google Scholar]
- [88].Walkey CD, Chan WCW, Chem. Soc. Rev 2012, 41, 2780. [DOI] [PubMed] [Google Scholar]
- [89].Gao J, Zeng L, Yao L, Wang Z, Yang X, Shi J, Hu L, Liu Q, Chen C, Xia T, Qu G, Zhang X, Jiang G, Nano Today 2021, 39, 101161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ge C, Tian J, Zhao Y, Chen C, Zhou R, Chai Z, Arch. Toxicol 2015, 89, 519. [DOI] [PubMed] [Google Scholar]
- [91].Westmeier D, Chen C, Stauber RH, Docter D, Eur J Nanomed 2015, 7, 153. [Google Scholar]
- [92].Gao H, Shi W, Freund LB, P. Natl. Acad. Sci. U S A 2005, 102, 9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Cao M, Cai R, Zhao L, Guo M, Wang L, Wang Y, Zhang L, Wang X, Yao H, Xie C, Cong Y, Guan Y, Tao X, Wang Y, Xu S, Liu Y, Zhao Y, Chen C, Nat. Nanotechnol 2021, 16, 708. [DOI] [PubMed] [Google Scholar]
- [94].Choi K, Riviere JE, Monteiro-Riviere NA, Nanotoxicology 2017, 11, 64. [DOI] [PubMed] [Google Scholar]
- [95].Walkey CD, Olsen JB, Guo H, Emili A, Chan WCW, J. Am. Chem. Soc 2012, 134, 2139. [DOI] [PubMed] [Google Scholar]
- [96].Wang H, Thorling CA, Liang X, Bridle KR, Grice JE, Zhu Y, Crawford DHG, Xu ZP, Liu X, Roberts MS, J. Mater. Chem. B 2015, 3, 939. [DOI] [PubMed] [Google Scholar]
- [97].Li J, Wang X, Mei K, Chang CH, Jiang J, Liu X, Liu Q, Guiney LM, Hersam MC, Liao Y, Meng H, Xia T, Nano Today 2021, 37, 1010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zhang Y, Ma C, Wang Z, Zhou Q, Sun S, Ma P, Lv L, Jiang X, Wang X, Zhan L, Nanoscale 2020, 12. [Google Scholar]
- [99].Lu K, Dong S, Xia T, Mao L, ACS Nano 2021, 15, 396. [DOI] [PubMed] [Google Scholar]
- [100].Yang Y, Yuan S, Zhao L, Wang C, Ni J, Wang Z, Lin C, Wu M, Zhou W, Mol. Pharmaceut 2015, 12, 644. [DOI] [PubMed] [Google Scholar]
- [101].Lee M, Yang J, Jung HS, Beack S, Choi JE, Hur W, Koo H, Kim K, Yoon SK, Hahn SK, ACS Nano 2012, 6, 9522. [DOI] [PubMed] [Google Scholar]
- [102].Zhang H, Xia T, Meng H, Xue M, George S, Ji Z, Wang X, Liu R, Wang M, France B, Rallo R, Damoiseaux R, Cohen Y, Bradley KA, Zink JI, Nel AE, ACS Nano 2011, 5, 2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Poelstra K, Prakash J, Beljaars L, Control J. Release 2012, 161, 188. [DOI] [PubMed] [Google Scholar]
- [104].Cheng S, Li F, Souris JS, Yang C, Tseng F, Lee H, Chen C, Dong C, Lo L, ACS Nano 2012, 6, 4122. [DOI] [PubMed] [Google Scholar]
- [105].Wang X, Chang CH, Jiang J, Liu X, Li J, Liu Q, Liao YP, Li L, Nel AE, Xia T, Small 2020, 16, e2000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zhang T, Tang M, Kong L, Li H, Zhang T, Xue Y, Pu Y, J. Hazard. Mater 2015, 284, 73. [DOI] [PubMed] [Google Scholar]
- [107].Lee AR, Nam K, Lee BJ, Lee SW, Baek SM, Bang JS, Choi SK, Park SJ, Kim TH, Jeong KS, Lee DY, Park JK, Int. J. Mol. Sci 2019, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Chen K, Ling Y, Cao C, Li X, Chen X, Wang X, Mater Sci Eng C Mater Biol Appl 2016, 69, 1222. [DOI] [PubMed] [Google Scholar]
- [109].Li J, Wang X, Chang CH, Jiang J, Liu Q, Liu X, Liao Y, Ma T, Meng H, Xia T, Small 2021, 17, 2102545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Linares J, Matesanz MC, Vila M, Feito MJ, Gonçalves G, Vallet-Regí M, Marques PAAP, Portolés MT, ACS Appl. Mater. Inter 2014, 6, 13697. [DOI] [PubMed] [Google Scholar]
- [111].Li J, Guiney LM, Downing JR, Wang X, Chang CH, Jiang J, Liu Q, Liu X, Mei K, Liao Y, Ma T, Meng H, Hersam MC, Nel AE, Xia T, Small 2021, 17, e202101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Poisson J, Lemoinne S, Boulanger C, Durand F, Moreau R, Valla D, Rautou P, J. Hepatol. 2016, 66, 212. [DOI] [PubMed] [Google Scholar]
- [113].Sørensen KK, Simonsantamaria J, Mccuskey RS, Smedsrød B, Compr. Physiol 2015, 5, 1751. [DOI] [PubMed] [Google Scholar]
- [114].Fortuna VA, Martucci RB, Trugo LC, Borojevic R, J. Cell. Biochem 2003, 90, 792. [DOI] [PubMed] [Google Scholar]
- [115].Ouyang B, Poon W, Zhang Y, Lin ZP, Kingston BR, Tavares AJ, Zhang Y, Chen J, Valic MS, Syed AM, Macmillan P, Couture-Senécal J, Zheng G, Chan WCW, Nat. Mater 2020, 19, 1362. [DOI] [PubMed] [Google Scholar]
- [116].Park J, Utsumi T, Seo Y, Deng Y, Satoh A, Saltzman WM, Iwakiri Y, Nanomed-Nanotechnol 2016, 12, 1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Quini CC, Próspero AG, Calabresi MFF, Moretto GM, Zufelato N, Krishnan S, Pina DR, Oliveira RB, Baffa O, Bakuzis AF, Miranda JRA, Nanomed-Nanotechnol 2017, 13, 1519. [DOI] [PubMed] [Google Scholar]
- [118].Jain TK, Reddy MK, Morales MA, Leslie-Pelecky DL, Labhasetwar V, Mol. Pharmaceut 2008, 5, 316. [DOI] [PubMed] [Google Scholar]
- [119].Paunovska K, Da Silva Sanchez AJ, Sago CD, Gan Z, Lokugamage MP, Islam FZ, Kalathoor S, Krupczak BR, Dahlman JE, Adv. Mater 2019, 31, 1807748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Paunovska K, Gil CJ, Lokugamage MP, Sago CD, Sato M, Lando GN, Gamboa Castro M, Bryksin AV, Dahlman JE, ACS Nano 2018, 12, 8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Shi B, Keough E, Matter A, Leander K, Young S, Carlini E, Sachs AB, Tao W, Abrams M, Howell B, Sepp-Lorenzino L, J. Histochem. Cytochem 2011, 59, 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Witzigmann D, Kulkarni JA, Leung J, Chen S, Cullis PR, van der Meel R, Adv. Drug Deliver. Rev 2020, 159, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Sago CD, Krupczak BR, Lokugamage MP, Gan Z, Dahlman JE, Cell. Mol. Bioeng 2019, 12, 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Kim M, Jeong M, Hur S, Cho Y, Park J, Jung H, Seo Y, Woo HA, Nam KT, Lee K, Lee H, Science Advances 2021, 7, f4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Arslan Z, Ates M, Mcduffy W, Agachan MS, Farah IO, Yu WW, Bednar AJ, J. Hazard. Mater 2011, 192, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Guo Z, Luo Y, Zhang P, Chetwynd AJ, Qunhui Xie H, Abdolahpur Monikh F, Tao W, Xie C, Liu Y, Xu L, Zhang Z, Valsami-Jones E, Lynch I, Zhao B, Environ. Int 2020, 136, 105437. [DOI] [PubMed] [Google Scholar]
- [127].Swinney R, Hsu S, Tomlinson G, J. Invest. Med 2006, 54, 303. [DOI] [PubMed] [Google Scholar]
- [128].Li H, Cui H, Kundu TK, Alzawahra W, Zweier JL, J. Biol. Chem 2008, 283, 17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Hille R, Hall J, Basu P, Chem. Rev 2014, 114, 3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Yang M, Zhang M, Nakajima H, Yudasaka M, Iijima S, Okazaki T, Int. J. Nanomed 2019, 14, 2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Elgrabli D, Dachraoui W, Ménard-Moyon C, Liu XJ, Bégin D, Bégin-Colin S, Bianco A, Gazeau F, Alloyeau D, ACS Nano 2015, 9, 10113. [DOI] [PubMed] [Google Scholar]
- [132].Kotchey GP, Allen BL, Vedala H, Yanamala N, Kapralov AA, Tyurina YY, Klein-Seetharaman J, Kagan VE, Star A, ACS Nano 2011, 5, 2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Kurapati R, Russier J, Squillaci MA, Treossi E, Ménard-Moyon C, Del Rio-Castillo AE, Vazquez E, Samorì P, Palermo V, Bianco A, Small 2015, 11, 3985. [DOI] [PubMed] [Google Scholar]
- [134].Qi Y, Liu Y, Xia T, Xu A, Liu S, Chen W, NPG Asia Mater 2018, 10, 385. [Google Scholar]
- [135].Mardinoglu A, Boren J, Smith U, Uhlen M, Nielsen J, Nat. Rev. Gastro. Hepat 2018, 15, 365. [DOI] [PubMed] [Google Scholar]
- [136].Poon W, Zhang Y, Ouyang B, Kingston BR, Wu JLY, Wilhelm S, Chan WCW, ACS Nano 2019, 13, 5785. [DOI] [PubMed] [Google Scholar]
- [137].Sadauskas E, Wallin H, Stoltenberg M, Vogel U, Doering P, Larsen A, Danscher G, Part. Fibre Toxicol 2007, 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Soo Choi H, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, Bawendi MG, Frangioni JV, Nat. Biotechnol 2007, 25, 1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Renaud G, Hamilton RL, Havel RJ, Hepatology 1989, 9, 380. [DOI] [PubMed] [Google Scholar]
- [140].Longmire M, Choyke PL, Kobayashi H, Nanomedicine-UK 2008, 3, 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Balasubramanian SK, Jittiwat J, Manikandan J, Ong C, Yu LE, Ong W, Biomaterials 2010, 31, 2034. [DOI] [PubMed] [Google Scholar]
- [142].Huang X, Li L, Liu T, Hao N, Liu H, Chen D, Tang F, ACS Nano 2011, 5, 5390. [DOI] [PubMed] [Google Scholar]
- [143].Dixon LJ, Barnes M, Tang H, Pritchard MT, Nagy LE, Compr. Physiol 2013, 3, 785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Bilzer M, Roggel F, Gerbes AL, Liver Int 2006, 26, 1175. [DOI] [PubMed] [Google Scholar]
- [145].Hume David A, Curr. Opin. Immunol 2006. [DOI] [PubMed] [Google Scholar]
- [146].Cho W, Cho M, Jeong J, Choi M, Cho H, Han BS, Kim SH, Kim HO, Lim YT, Chung BH, Jeong J, Toxicol. Appl. Pharm 2009, 236, 16. [DOI] [PubMed] [Google Scholar]
- [147].Yadav V, Roy S, Singh P, Khan Z, Jaiswal A, Small 2019, 15, e1803706. [DOI] [PubMed] [Google Scholar]
- [148].Zhou W, Zou X, Najmaei S, Liu Z, Shi Y, Kong J, Lou J, Ajayan PM, Yakobson BI, Idrobo JC, Nano Lett 2013, 13, 2615. [DOI] [PubMed] [Google Scholar]
- [149].Wang S, Li K, Chen Y, Chen H, Ma M, Feng J, Zhao Q, Shi J, Biomaterials 2015, 39, 206. [DOI] [PubMed] [Google Scholar]
- [150].Yu Y, Yi Y, Li Y, Peng T, Lao S, Zhang J, Liang S, Xiong Y, Shao S, Wu N, RSC Adv 2018, 8, 17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J, Nature 2016, 535, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F, Nature 2015, 526, 660. [DOI] [PubMed] [Google Scholar]
- [153].Bergsbaken T, Fink SL, Cookson BT, Nat. Rev. Microbiol 2009, 7, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Singh RP, Das M, Thakare V, Jain S, Chem. Res. Toxicol 2012, 25, 2127. [DOI] [PubMed] [Google Scholar]
- [155].Gupta R, Mehra NK, Jain NK, Pharm. Res.-Dordr 2014, 31, 322. [DOI] [PubMed] [Google Scholar]
- [156].Shahriari-Khalaji M, Zarkesh M, Nozhat Z, Curr Pharm Des 2021, 27. [DOI] [PubMed] [Google Scholar]
- [157].Lin N, Dufresne A, Eur. Polym. J 2014, 59, 302. [Google Scholar]
- [158].Dufresne A, Mater. Today 2013, 16, 220. [Google Scholar]
- [159].Otuechere CA, Adewuyi A, Adebayo OL, Ebigwei IA, Hum. Exp. Toxicol 2019, 39, 1538216903. [DOI] [PubMed] [Google Scholar]
- [160].Zhu S, Zhang J, Zhang L, Ma W, Man N, Liu Y, Zhou W, Lin J, Wei P, Jin P, Zhang Y, Hu Y, Gu E, Lu X, Yang Z, Liu X, Bai L, Wen L, Adv. Healthc. Mater 2017, 6, 1601252. [DOI] [PubMed] [Google Scholar]
- [161].Li R, Ji Z, Qin H, Kang X, Sun B, Wang M, Chang CH, Wang X, Zhang H, Zou H, Nel AE, Xia T, ACS Nano 2014, 8, 10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Li R, Ji Z, Dong J, Chang CH, Wang X, Sun B, Wang M, Liao Y, Zink JI, Nel AE, Xia T, ACS Nano 2015, 9, 3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Gracia-Sancho J, Caparrós E, Fernández-Iglesias A, Francés R, Nat. Rev. Gastro. Hepat 2021, 18, 411. [DOI] [PubMed] [Google Scholar]
- [164].Knolle PA, Wohlleber D, Cell. Mol. Immunol 2016, 13, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Nishimori H, Kondoh M, Isoda K, Tsunoda S, Tsutsumi Y, Yagi K, Eur. J. Pharm. Biopharm 2009, 72, 496. [DOI] [PubMed] [Google Scholar]
- [166].Tee JK, Ng LY, Koh HY, Leong DT, Ho HK, Int. J. Mol. Sci 2019, 20, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Tsuchida T, Friedman SL, Nat. Rev. Gastro. Hepat 2017, 14, 397. [DOI] [PubMed] [Google Scholar]
- [168].Sun X, Wang Z, Zhai S, Cheng Y, Liu J, Liu B, Mol. Med. Rep 2013, 8, 1365. [DOI] [PubMed] [Google Scholar]
- [169].Osmond Mcleod MJ, J. Nanomed. Nanotechnol 2014, 05, 1000232. [Google Scholar]
- [170].Gaiser BK, Hirn S, Kermanizadeh A, Kanase N, Fytianos K, Wenk A, Haberl N, Brunelli A, Kreyling WG, Stone V, Toxicol. Sci 2013, 131, 537. [DOI] [PubMed] [Google Scholar]
- [171].Lopez-Chaves C, Soto-Alvaredo J, Montes-Bayon M, Bettmer J, Llopis J, Sanchez-Gonzalez C, Nanomed-Nanotechol 2018, 14, 1. [DOI] [PubMed] [Google Scholar]
- [172].Wang T, Chen X, Long X, Liu Z, Yan S, PLoS One 2016, 11, e149484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Natarajan V, Wilson CL, Hayward SL, Kidambi S, PLoS One 2015, 10, e134541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Wang Y, Aker WG, Hwang H, Yedjou CG, Yu H, Tchounwou PB, Sci. Total Environ 2011, 409, 4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Dumková J, Smutná T, Vrlíková L, Le Coustumer P, Večeřa Z, Dočekal B, Mikuška P, apka L, Fictum P, Hampl A, Buchtová M, Part. Fibre Toxicol 2017, 14, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Esmaeillou M, Moharamnejad M, Hsankhani R, Tehrani AA, Maadi H, Environ. Toxicol. Phar 2013, 35, 67. [DOI] [PubMed] [Google Scholar]
- [177].Askri D, Ouni S, Galai S, Chovelon B, Arnaud J, Sturm N, Lehmann SG, Sakly M, Amara S, Sève M, Food Chem. Toxicol 2019, 127, 173. [DOI] [PubMed] [Google Scholar]
- [178].Saowalak K, Titipun T, Somchai T, Chalermchai P, Sci. Rep.-UK 2018, 8, 6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Yu Y, Duan J, Yu Y, Li Y, Liu X, Zhou X, Ho K, Tian L, Sun Z, J. Hazard. Mater 2014, 270, 176. [DOI] [PubMed] [Google Scholar]
- [180].Wang J, Yu Y, Lu K, Yang M, Li Y, Zhou X, Sun Z, Int. J. Nanomed 2017, 12, 809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Yuan J, Gao H, Sui J, Duan H, Chen WN, Ching CB, Toxicol. Sci 2012, 126, 149. [DOI] [PubMed] [Google Scholar]
- [182].Kermanizadeh A, Gaiser BK, Ward MB, Stone V, Nanotoxicology 2012, 7, 1255. [DOI] [PubMed] [Google Scholar]
- [183].Ji Z, Zhang D, Li L, Shen X, Deng X, Dong L, Wu M, Liu Y, Nanotechnology 2009, 20, 445101. [DOI] [PubMed] [Google Scholar]
- [184].Chatterjee N, Eom H, Choi J, Biomaterials 2014, 35, 1109. [DOI] [PubMed] [Google Scholar]
- [185].Kovacs T, Naish V, O’Connor B, Blaise C, Gagné F, Hall L, Trudeau V, Martel P, Nanotoxicology 2010, 4, 255. [DOI] [PubMed] [Google Scholar]
- [186].Meng H, Nel AE, Adv. Drug Deliver. Rev 2018, 130, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Liu X, Situ A, Kang Y, Villabroza KR, Liao Y, Chang CH, Donahue T, Nel AE, Meng H, ACS Nano 2016, 10, 2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Li L, Wang H, Ong ZY, Xu K, Ee PLR, Zheng S, Hedrick JL, Yang Y, Nano Today 2010, 5, 296. [Google Scholar]
- [189].Hassan HFH, Mansour AM, Abo-Youssef AMH, Elsadek BEM, Messiha BAS, Clin. Exp. Pharmacol. P 2017, 44, 235. [DOI] [PubMed] [Google Scholar]
- [190].Shanmugasundaram T, Radhakrishnan M, Gopikrishnan V, Kadirvelu K, Balagurunathan R, Nanoscale 2017, 9, 16773. [DOI] [PubMed] [Google Scholar]
- [191].Singh D, Yadav E, Kumar V, Verma A, Curr. Drug Deliv 2020, 18, 634. [DOI] [PubMed] [Google Scholar]
- [192].Ou W, Mulik RS, Anwar A, Mcdonald JG, He X, Corbin IR, Free Radical Bio. Med 2017, 112, 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Parra-Robert M, Casals E, Massana N, Zeng M, Perramon M, Fernandez-Varo G, Morales-Ruiz M, Puntes V, Jimenez W, Casals G, Biomolecules 2019, 9, 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Carvajal S, Perramón M, Oró D, Casals E, Fernández-Varo G, Casals G, Parra M, González B De La Presa, Ribera J, Pastor Ó, Morales-Ruíz M, Puntes V, Jiménez W, Sci. Rep.-UK 2019, 9, 12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Kobyliak N, Virchenko O, Falalyeyeva T, Kondro M, Beregova T, Bodnar P, Shcherbakov O, Bubnov R, Caprnda M, Delev D, Sabo J, Kruzliak P, Rodrigo L, Opatrilova R, Spivak M, Biomed. Pharmacother 2017, 90, 608. [DOI] [PubMed] [Google Scholar]
- [196].Rocca A, Moscato S, Ronca F, Nitti S, Mattoli V, Giorgi M, Ciofani G, Nanomed-Nanotechnol 2015, 11, 1725. [DOI] [PubMed] [Google Scholar]
- [197].Dogra S, Kar AK, Girdhar K, Daniel PV, Chatterjee S, Choubey A, Ghosh S, Patnaik S, Ghosh D, Mondal P, Nanomed-Nanotechnol 2019, 17, 210. [DOI] [PubMed] [Google Scholar]
- [198].Peng F, Tee JK, Setyawati MI, Ding X, Yeo HLA, Tan YL, Leong DT, Ho HK, ACS Appl. Mater. Inter 2018, 10, 31938. [DOI] [PubMed] [Google Scholar]
- [199].Mehnert W, Mäder K, Adv. Drug Deliver. Rev 2012, 64, 83. [DOI] [PubMed] [Google Scholar]
- [200].Mei Z, Li X, Wu Q, Hu S, Yang X, Pharmacol. Res 2005, 51, 345. [DOI] [PubMed] [Google Scholar]
- [201].Shi J, Kantoff PW, Wooster R, Farokhzad OC, Nat. Rev. Cancer 2017, 17, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Liu LY, Ma X, Ouyang B, Ings DP, Marwah S, Liu J, Chen AY, Gupta R, Manuel J, Chen X, Gage BK, Cirlan I, Khuu N, Chung S, Camat D, Cheng M, Sekhon M, Zagorovsky K, Abdou Mohamed MA, Thoeni C, Atif J, Echeverri J, Kollmann D, Fischer S, Bader GD, Chan WCW, Michalak TI, Mcgilvray ID, Macparland SA, ACS Nano 2020, 14, 4698. [DOI] [PubMed] [Google Scholar]
- [203].Zhou H, Fan Z, Li PY, Deng J, Arhontoulis DC, Li CY, Bowne WB, Cheng H, ACS Nano 2018, 12, 10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Knolle PA, Gerken G, Immunol. Rev 2000, 174, 21. [DOI] [PubMed] [Google Scholar]
- [205].Mousavi SA, Sporstøl M, Fladeby C, Kjeken R, Barois N, Berg T, Hepatology 2007, 46, 871. [DOI] [PubMed] [Google Scholar]
- [206].Liu Q, Wang X, Liu X, Kumar S, Gochman G, Ji Y, Liao Y, Chang CH, Situ W, Lu J, Jiang J, Mei K, Meng H, Xia T, Nel AE, ACS Nano 2019, 13, 4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Sabatos-Peyton CA, Verhagen J, Wraith DC, Curr. Opin. Immunol 2010, 22, 609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Vickery BP, Scurlock AM, Jones SM, Burks AW, J. Allergy Clin. Immun 2011, 127, 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Stabler CL, Li Y, Stewart JM, Keselowsky BG, Nat. Rev. Mater 2019, 4, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Pozsgay J, Szekanecz Z, Sármay G, Nat. Rev. Rheumatol 2017, 13, 525. [DOI] [PubMed] [Google Scholar]
- [211].Liu Q, Wang X, Liu X, Liao Y, Chang CH, Mei K, Jiang J, Tseng S, Gochman G, Huang M, Thatcher Z, Li J, Allen SD, Lucido L, Xia T, Nel AE, ACS Nano 2021, 15, 1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Hoffmann C, Djerir NEH, Danckaert A, Fernandes J, Roux P, Charrueau C, Lachagès A, Charlotte F, Brocheriou I, Clément K, Aron-Wisnewsky J, Foufelle F, Ratziu V, Hainque B, Bonnefont-Rousselot D, Bigey P, Escriou V, Sci. Rep.-UK 2020, 10, 3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Cai X, Wang J, Wang J, Zhou Q, Yang B, He Q, Weng Q, Pharmacol. Res 2020, 155, 104720. [DOI] [PubMed] [Google Scholar]
- [214].Yin C, Evason KJ, Asahina K, Stainier DY, J. Clin. Invest 2013, 123, 1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Zhang C, Yuan W, He P, Lei J, Wang C, World J. Gastroentero 2016, 22, 10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Hu M, Wang Y, Liu Z, Yu Z, Guan K, Liu M, Wang M, Tan J, Huang L, Nat. Nanotechnol 2021, 16, 466. [DOI] [PubMed] [Google Scholar]
- [217].Akinc A, Maier MA, Manoharan M, Fitzgerald K, Jayaraman M, Barros S, Ansell S, Du X, Hope MJ, Madden TD, Mui BL, Semple SC, Tam YK, Ciufolini M, Witzigmann D, Kulkarni JA, van der Meel R, Cullis PR, Nat. Nanotechnol 2019, 14, 1084. [DOI] [PubMed] [Google Scholar]
- [218].Qiu M, Glass Z, Chen J, Haas M, Jin X, Zhao X, Rui X, Ye Z, Li Y, Zhang F, Xu Q, Proc. Natl. Acad. Sci. U S A 2021, 118, e2020401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN, Jayaraman M, Rajeev KG, Cantley WL, Dorkin JR, Butler JS, Qin L, Racie T, Sprague A, Fava E, Zeigerer A, Hope MJ, Zerial M, Sah DW, Fitzgerald K, Tracy MA, Manoharan M, Koteliansky V, Fougerolles AD, Maier MA, Mol. Ther 2010, 18, 1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Chen S, Tam YYC, Lin PJC, Sung MMH, Tam YK, Cullis PR, J. Control. Release 2016, 235, 236. [DOI] [PubMed] [Google Scholar]
- [221].Wang K, Shang F, Chen D, Cao T, Wang X, Jiao J, He S, Liang X, J. Nanobiotechnol. 2021, 19, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Krawczyk M, Müllenbach R, Weber SN, Zimmer V, Lammert F, Nat. Rev. Gastro. Hepat 2010, 7, 669. [DOI] [PubMed] [Google Scholar]
- [223].Khan OF, Zaia EW, Yin H, Bogorad RL, Pelet JM, Webber MJ, Zhuang I, Dahlman JE, Langer R, Anderson DG, Angew. Chem 2014, 53, 14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Sato Y, Matsui H, Yamamoto N, Sato R, Munakata T, Kohara M, Harashima H, J. Control. Release 2017, 266, 216. [DOI] [PubMed] [Google Scholar]
- [225].Jackson MA, Patel SS, Yu F, Cottam MA, Glass EB, Hoogenboezem EN, Fletcher RB, Dollinger BR, Patil P, Liu DD, Kelly IB, Bedingfield SK, King AR, Miles RE, Hasty AM, Giorgio TD, Duvall CL, Biomaterials 2021, 268, 120528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].van der Meel R, Nat. Nanotechnol 2020, 15, 253. [DOI] [PubMed] [Google Scholar]
- [227].Nel AE, Lutz MD, Darrell V, Tian X, Hoek EMV, Ponisseril S, Fred K, Vince C, Mike T, Nat. Mater 2009, 8, 543.19525947 [Google Scholar]
- [228].Meng H, Xia T, George S, Nel AE, ACS Nano 2009, 3, 1620. [DOI] [PubMed] [Google Scholar]
- [229].Nel A, Xia T, Meng H, Wang X, Lin S, Ji Z, Zhang H, Accounts Chem. Res 2012, 46, 164609029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Khetani SR, Bhatia SN, Nat. Biotechnol 2008, 26, 120. [DOI] [PubMed] [Google Scholar]
- [231].Kermanizadeh A, Berthing T, Guzniczak E, Wheeldon M, Whyte G, Vogel U, Moritz W, Stone V, Part. Fibre Toxicol 2019, 16, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Llewellyn SV, Niemeijer M, Nymark P, Moné MJ, van de Water B, Conway GE, Jenkins GJS, Doak SH, Small 2021, 17, 2006055. [DOI] [PubMed] [Google Scholar]
- [233].Potter SS, Nat. Rev. Nephrol 2018, 14, 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA, Mol. Cell 2015, 58, 610. [DOI] [PubMed] [Google Scholar]
- [235].Shin TH, Lee DY, Lee H, Park HJ, Jin MS, Paik M, Manavalan B, Mo J, Lee G, BMB Rep 2018, 51, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Costa PM, Fadeel B, Toxicol. Appl. Pharm 2016, 299, 101. [DOI] [PubMed] [Google Scholar]
- [237].Idle JR, Gonzalez FJ, Cell Metab 2007, 6, 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Fröhlich E, J. Nanobiotechnol. 2017, 15, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Cui L, Wang X, Sun B, Xia T, Hu S, ACS Nano 2019, 13, 13065. [DOI] [PubMed] [Google Scholar]
- [240].Yan X, Sedykh A, Wang W, Yan B, Zhu H, Nat. Commun 2020, 11, 2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Leong HS, Butler KS, Brinker CJ, Azzawi M, Conlan S, Dufés C, Owen A, Rannard S, Scott C, Chen C, Dobrovolskaia MA, Kozlov SV, Prina-Mello A, Schmid R, Wick P, Caputo F, Boisseau P, Crist RM, Mcneil SE, Fadeel B, Tran L, Hansen SF, Hartmann NB, Clausen LPW, Skjolding LM, Baun A, ågerstrand M, Gu Z, Lamprou DA, Hoskins C, Huang L, Song W, Cao H, Liu X, Jandt KD, Jiang W, Kim BYS, Wheeler KE, Chetwynd AJ, Lynch I, Moghimi SM, Nel A, Xia T, Weiss PS, Sarmento B, Das Neves J, Santos HA, Santos L, Mitragotri S, Little S, Peer D, Amiji MM, Alonso MJ, Petri-Fink A, Balog S, Lee A, Drasler B, Rothen-Rutishauser B, Wilhelm S, Acar H, Harrison RG, Mao C, Mukherjee P, Ramesh R, Mcnally LR, Busatto S, Wolfram J, Bergese P, Ferrari M, Fang RH, Zhang L, Zheng J, Peng C, Du B, Yu M, Charron DM, Zheng G, Pastore C, Nat. Nanotechnol 2019, 14, 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Ahankari S, Paliwal P, Subhedar A, Kargarzadeh H, ACS Nano 2021, 15, 3849. [DOI] [PubMed] [Google Scholar]
- [243].Sobańska Z, Domeradzka-Gajda K, Szparaga M, Grobelny J, Stpnik M, Toxicol. in Vitro 2020, 68, 104931. [DOI] [PubMed] [Google Scholar]
- [244].Li H, Tay RY, Tsang SH, Zhen X, Teo EHT, Small 2016, 11, 6491. [DOI] [PubMed] [Google Scholar]
- [245].Chen C, Li Y, Qu Y, Chai Z, Zhao Y, Chem. Soc. Rev 2013, 42, 8266. [DOI] [PubMed] [Google Scholar]
- [246].Matter MT, Li J, Lese I, Schreiner C, Bernard L, Scholder O, Hubeli J, Keevend K, Tsolaki E, Bertero E, Bertazzo S, Zboray R, Olariu R, Constantinescu MA, Figi R, Herrmann IK, Advanced Science 2020, 7, 2000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Guo Z, Zhang P, Chakraborty S, Chetwynd AJ, Abdolahpur Monikh F, Stark C, Ali-Boucetta H, Wilson S, Lynch I, Valsami-Jones E, P. Natl. Acad. Sci. U S A 2021, 118, e2105245118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Foldbjerg R, Jiang X, Miclăuş T, Chen C, Autrup H, Beer C, Toxicol. Res.-UK 2015, 4, 563. [Google Scholar]
- [249].Balfourier A, Luciani N, Wang G, Lelong G, Ersen O, Khelfa A, Alloyeau D, Gazeau F, Carn F, P. Natl. Acad. Sci. U S A 2020, 117, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]


