Abstract
We tested the hypothesis that divergent genetic merit for fertility of dairy cows is due to aberrant reproductive neuroendocrine function. The kisspeptin status of non-pregnant cows of either positive (POS) or negative (NEG) breeding values (BVs) for fertility was studied in three groups (n = 8), based on their previous post-partum period: POS cows, which had spontaneous ovarian cycles (POS-CYC) and NEG cows, which either cycled (NEG-CYC) or did not cycle (NEG-NONCYC). Ovarian cycles were synchronized, blood samples were taken to define endocrine status, and the animals were slaughtered in an artificial follicular phase. The brains and the pituitary glands were collected for quantitative polymerase chain reaction (qPCR) and in situ hybridization of hypothalamic GNRH1, Kiss1, TAC3, and PDYN and pituitary expression of LHB and FSHB. Gonadotropin releasing hormone (GnRH) and kisspeptin levels were quantified in snap frozen median eminence (ME). GNRH1 expression and GnRH levels in the ME were similar across groups. Kiss1 expression in the preoptic area of the hypothalamus was also similar across groups, but Kiss1 in the arcuate nucleus was almost 2-fold higher in POS-CYC cows than in NEG groups. TAC3 expression was higher in POS-CYC cows. The number of pituitary gonadotropes and the level of expression of LHB and FSHB were similar across groups. We conclude that the lower levels of Kiss1 and TAC3 in NEG cows with low fertility status and may lead to deficient GnRH and gonadotropin secretion.
Keywords: kisspeptin, hypothalamus, fertility, bovine
Introduction
Delayed resumption of estrus post-partum and reduced fertility in dairy cows is influenced by many factors including increased metabolic demand for milk production, negative energy balance, immune dysfunction and inflammation, and mineral deficiency and associated metabolic disorders [1, 2]. This is notwithstanding genetic factors [3]. Fertility has decreased across time, such as shown in a UK study, comparing data for the period between 1975 and 1982 with that from 1995–1998 [4]; the rate of pregnancy to first service in untreated cows fell from 65% in the earlier period to 43% in the later period. The low heritability of reproductive traits [5] exacerbates efforts to correct the problem, but certain traits of moderate-high heritability, such as age at puberty, may be worthy of consideration [6].
Reproduction is driven by the brain, through the secretion of gonadotropin releasing hormone (GnRH), which stimulates the synthesis and secretion of the gonadotropins, luteinizing hormone (LH) and follicle stimulating hormone (FSH) from the pituitary gonadotropes. Anovulation in the post-partum dairy cow is associated with reduced gonadotropin secretion [7], but there are no available data on the secretion of GnRH in such cows. In spite of this, methods to improve fertility in the transition dairy cow have been developed, particularly the OvSynch procedure, which involves timed sequential treatment with prostaglandin and GnRH [8] in various iterations [9]. This is predicated upon the likelihood that post-partum anestrus is due to deficient GnRH secretion.
GnRH secretion is regulated by a number of brain systems and the feedback effects of gonadal steroids [10]. In particular, kisspeptin is a hypothalamic neuropeptide that stimulates GnRH secretion and mediates sex steroid feedback effects on GnRH secretion [11]. Kisspeptin cells are located in the preoptic area (POA) and the arcuate nucleus (ARC) of the ungulate brain including the bovine [12, 13]. Although not detailed in the bovine, studies in the ewe show that the ARC Kisspeptin cells mediate negative feedback effects of gonadal steroids, estrogen, and progesterone [14]. The positive feedback effect of estrogen, which causes the preovulatory GnRH/LH surge, is initiated in the ARC Kisspeptin cells in ewes [15] and is facilitated by the kisspeptin cells in the POA of the ewe [16]. In addition to production of kisspeptin, the kisspeptin cells of the ARC of the ewe and other species also produce dynorphin and neurokinin B [17]. These ARC cells have been referred to as KNDY cells; it has been shown that dynorphin inhibits and neurokinin B stimulates kisspeptin production/secretion by these cells [18]. In turn, the combined function of these three neuropeptides controls the pulsatile secretion of GnRH [18]. In particular, kisspeptin cells in the ARC project into the median eminence (ME) in sheep to drive pulsatile release of GnRH at this level [19]. Further evidence of the role of ARC Kisspeptin neurons driving pulsatile GnRH secretion is found in a recent study in mice, whereby conditional knockout of ARC Kiss1 was shown to cause marked reduction in pulsatile secretion of LH [20]. Mutations in the kisspeptin gene (Kiss1) and its receptor may cause hypogonadotropic hypogonadism in some species, although the phenotype is variable [21]. Nevertheless, Kisspeptin is regarded as an essential factor in the stimulation of pulsatile GnRH secretion from the hypothalamus [22].
Genetic merit for fertility in dairy cows in New Zealand is based on a fertility BV (FBV), which takes account of eight phenotypic traits and is expressed as the calving rate (%) in the first 42 days after the start of the seasonal calving period [23]. Using this predictor, a custom-bred herd of Holstein-Friesian female dairy cattle with either high (+5%) or low (−5%) FBV has been established [24–26]; these are referred to as POS or NEG animals [26]. In particular, Meier et al. [26] show that NEG animals have delayed resumption of estrus post-partum. These two divergent cohorts provide an excellent opportunity to identify factors that relate directly to fertility. Relatively little is known about the hypothalamo-pituitary axis of dairy cows in relation to fertility, although the fact the parturition-to-conception period can be reduced with “OvSynch” suggests a deficiency in GnRH/gonadotropin secretion in low fertility cows. As indicated above, “OvSynch” involves sequential treatment with GnRH and a prostaglandin F2α analog [9, 27]. In addition, ovulation can be advanced in the post-partum period by administration of Senktide, which acts via NK3R receptors to activate Kisspeptin cells [28]. This suggests that the activation of Kisspeptin cells, leading to increased GnRH secretion, may mitigate poor fertility in the post-partum dairy cow. In the present study, we quantified hypothalamic Kiss1 and GNRH1 and GnRH and Kisspeptin peptide levels in NEG and POS cows. Because of the role that Neurokinin (NKB encoded by the gene TAC3) and Dynorphin (encoded by PDYN) play in the regulation of Kisspeptin secretion [29, 30], expression in the ARC of genes for these peptides was also quantified. In addition, the function of the pituitary gonadotropes was measured. We tested the hypothesis that divergent genetic merit for fertility traits (FBVs) of dairy cows is due to aberrant function of key neuroendocrine factors.
Materials and methods
Ethics
The experiment was approved prior to commencement by the Ruakura Animal Ethics Committee (Hamilton, New Zealand) and was conducted at Ruakura, New Zealand (37.78°S, 175.28°E) in May 2018.
Animal experimentation
Primiparous, non-pregnant Holstein-Friesian cows were selected from a research herd established to compare performance of animals with POS and NEG genetic merit for fertility based on the FBV, as previously described [24–26] and detailed in Supplementary S1. These cows were approximately 34 months old, had not become pregnant during the prior seasonal artificial breeding period between October and December 2017 and had completed their first lactation at the commencement of the study in May–June 2018. Typically, these cows have similar BVs for milk volume, milk fat percentage, and milk protein percentage [26]. POS FBV cows have a score of approximately +5, and NEG FBV cows have a value of approximately −5. Initially, 39 cows were selected in three groups as follows:
POS FBV cows (POS-CYC; n = 13) that displayed estrous cycles throughout the first 6 weeks of the 2017 seasonal breeding period;
NEG FBV cows (NEG-CYC; n = 11) that displayed estrous cycles during the first 6 weeks of the 2017 seasonal breeding period;
NEG FBV cows (NEG-NONCYC; n = 16) that did not display estrous cycles within 6 weeks of the 2017 seasonal breeding period
The experimental timeline is presented in Figure 1. In May–June 2018, approximately 45 weeks after calving, the cows were administered 2 i.m. injections of 500 μg Cloprostenol (Ovuprost®, Bayer New Zealand Ltd) 10 h apart on Day 1 of the study and were observed for signs of estrus for 30 min twice-daily over the following 6 days (detailed in Supplementary S1). Transrectal ultrasound (5–15 MHz probe—SonoScope S6V, Euromed Medical Systems) was performed on Day 7 to determine presence or absence of a corpus luteum (CL), signifying ovulation following the Cloprostenol treatment. Then, eight cows from each of the three groups described above were selected, based on genetic merit for FBV, body weights and body condition score BVs, as detailed in Supplementary S1 (Table 1), to undergo further synchronization of estrous cycles prior to slaughter and tissue collection. All cows in these three experimental groups had CL and had displayed estrus within the previous 7 days (following the above treatment). The POS-CYC group had higher (P < 0.001) FBV than the other two groups (Table 1). Although bodyweights were similar, body condition score BVs were lower (P < 0.05) in the NEG-CYC group than the POS-CYC group and tended to be lower (P = 0.06) in the NEG-NONCYC group than in the POS-CYC group. The post-partum anoestrus interval (PPAI) for the NEG-NONCYC cows was longer than that of the POS-CYC (P < 0.01) and the NEG-CYC cows (P < 0.05) (Table 1).
Figure 1.
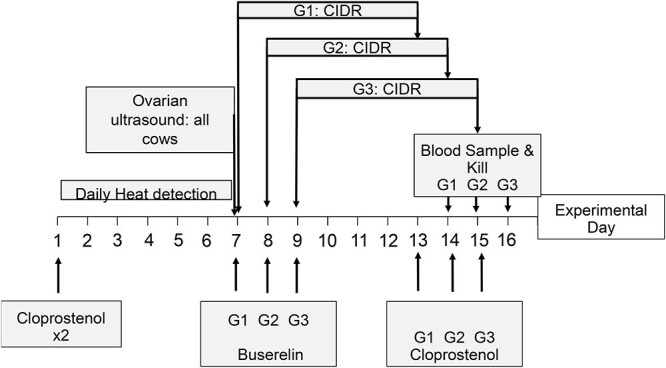
Timeline of treatments and slaughter. The cows of NEG and POS cohorts were divided into three groups (G1, G2, and G3) for treatment and for slaughter, so that the collection of brains was within a 1-hour period. The assignment to groups balanced POS and NEG cows as much as was possible. Initially, the estrous cycles of all cows were synchronized with two injections of Cloprostenol as indicated in Materials and methods. Between Days 1 and 7, the estrus detection was carried out as indicated in Supplementary 1 and on Day 7, ultrasound was performed on all cows. Thereafter, the treatment of G1, G2, and G3 was staggered by 1 day. Buserelin was administered and the cows received CIDRs for 6 days. Commencing on Day 13 and staggered by one day for each group, the CIDRs were withdrawn and the cows were given Cloprostenol to ensure the demise of any corpora lutea. One day later, the cows were blood sampled, transported to a local abattoir, and killed between 1500 and 1600 h.
Table 1.
Mean (± SEM) genetic indices, bodyweights, and prior PPAI of the fertility groups. BV were generated from 2017 New Zealand Animal Evaluation Limited, incorporating data from a number of generations (Supplementary S1). *P < 0.05, **P < 0.01, ***P < 0.001 compared with POS-CYC; #P < 0.05 compared with NEG-CYC. The difference between NEG-CYC and NEG-NONCYC for body condition score BV almost attained significance (P = 0.056)
| Parameter | POS-CYC (n = 8) |
NEG-CYC (n = 8) |
NEG-NonCYC (n = 8) |
|---|---|---|---|
| Fertility BV Bodyweight (kg) |
4.7 ± 0.27 456.9 ± 17.51 |
−5.3 ± 0.47 *** 465.7 ± 11.90 |
−4.4 ± 0.49 *** 472.5 ± 11.70 |
| Body condition score BV | 0.05 ± 0.02 | −0.06 ± 0.03* | −0.04 ± 0.02 |
| 2017 PPAI (days) | 47.5 ± 8.48 | 65.1 ± 9.54 | 101.0 ± 11.31**# |
The cows within each of the three groups were assigned to three replicates (n = 2–3) to allow logistic collection of tissues at slaughter over 3 days with regular slaughter times (Figure 1). The following details the protocol for Group 1 (G1), which are identical for the other two groups, staggered by 1 day. On Day 7 the cows received an intravaginal controlled internal drug releasing device (CIDR®) containing 1.38 g of progesterone (Pfizer Animal Health) and an i.m. injection of 8 μg of Buserelin, a synthetic GnRH analog (Receptal®, MSD Animal Health). At 1500 h on Day 14, the CIDR were removed and the cows received 500 μg Cloprostenol i.m. to ensure the luteolysis of any residual CL. On the day of slaughter blood samples were collected (vide infra), the cows were transported to an abbatoir and were killed between 1500 and 1600 h (Figure 1). The cows were humanely slaughtered by captive bolt directed into the hind-brain so as not to damage the hypothalamus and pituitary gland. Thus, slaughter and tissue collection were performed 23–25 h after CIDR withdrawal, during the induced early- to mid-follicular phase of the induced estrous cycle.
Sample collection
In order to confirm that the hormonal status of the cows was consistent with their being in the early stage of the follicular phase, three blood samples (32 mL) were collected prior to slaughter, from a coccygeal vessel into Lithium heparin Vacutainers® (Becton Dickinson) at 30 min intervals (1230, 1300 and 1330 h on the day of slaughter). The samples were centrifuged, and the plasma harvested and stored at −20°C for subsequent progesterone, LH, and FSH assay (vide infra).
Heads were skinned and a craniotomy performed using a band saw to enable removal of the brain and pituitary gland. The hypothalami, ME, and pituitary glands were dissected out. The ME were removed by cutting below the hypothalamus at the level where white matter joined the pink tissue of the pars tuberalis and was snap frozen on crushed dry ice. For the hypothalamus, cuts were made along the lateral fissures of the hypothalamus, on the base of the brain with a rostral cut made 5 mm anterior to the optic chiasma and a caudal cut immediately behind the mammillary bodies. Hypothalamic tissue was halved, with one-half being placed into RNAlater® (Ambion) and the other half into 10% formalin in saline. Notably, the walls of the third ventricle (3 V) were clearly visible on both halves of the hypothalamic tissues, verifying midline dissection. The tissues were kept at room temperature. The diaphragma sella was sliced off the top of the pituitary gland in situ and a v-section made to remove the posterior pituitary/intermediate lobe. One-half of the pars distalis of the pituitary was placed in RNA later and the other placed into 10% formalin (as above).
Quantitative polymerase chain reaction analysis
The hypothalamic halves and pituitary halves (pars distalis) that were snap frozen were placed on a cold stainless-steel plate and dissected into two regions. A POA block was taken from the posterior margin of the optic chiasm. The optic chiasm tissue was discarded, and the resulting rostral block was retained. An ARC block was taken from the rostral cut, extending caudally to include the mammillary bodies, with a horizontal cut made at a tangent to the base of the thalamus. The POA and ARC blocks and a block of pituitary tissue taken from the center of the anterior lobe of the gland were extracted for quantitative polymerase chain reaction (qPCR) using QIAzol Lysis Reagent (Qiagen Cat# 79306) and the standard protocol. Following RNA extraction, cDNA was prepared, and genomic DNA removed using QuantiTect Reverse Transcription Kit (Qiagen cat#205310). Real-time qPCR reactions for each sample were set up using 50 ng cDNA, in triplicate, using a QuantiNova SYBR Green Kit (Qiagen cat#208052) on the Rotor-Gene Q real-time PCR machine (95°C 2 min; combined extension and annealing 60°C 1 min for 40 cycles) for all primer pairs. In each case, purified DNA of known concentration was used as the assay standard. Optimization of each primer set involved the separation of qPCR products by agarose gel electrophoresis, followed by purification and sequencing to determine identity. The levels of expression of each mRNA and the estimated concentrations were determined relative to the standard preparation using Qiagen Rotor-Gene Q computer software. Similar amounts of RNA were used for each amplification and the ratio of each mRNA to the geometric mean of three reference genes (PPIA, GAPDH, and MDH1) was calculated to correct for differences in the total amount of RNA used between samples. Expression of the GnRH gene (GNRH1) (POA only) and Kiss1 expression (ARC and POA) and pituitary gene expression of the LHB and FSHB was measured using qPCR (Qiagen Rotor-Gene Q machine) with primer sequences as given in Table 2.
Table 2.
Primer sequences for genes of interest and reference genes
| Name | Accession number | Forward primer | Reverse primer |
|---|---|---|---|
| Kiss1 | NM_001306104 | GTGCAGCGGGAGAAGGAC | AGGCCGAAGGAGTTCCAGT |
| GNRH1 | XM_015093089 | CTGGAGGAAAGAGAAATGC | TTCAATCAGACTTTCCAGAG |
| LHB | NM_001009380 | GGCTACTGCCTCAGCATGAA | AAGGAGACCATTGGGTCCAC |
| FSHB | NM_001009798 | TATTGCTACACCCGGGACTT | TTTCACCGTCTCGTACACCA |
| PPIA | NM_001308578 | GCATACAGGTCCTGGCATCT | CATGCCCTCTTTCACTTTGC |
| GAPDH | NM_001190390 | TCAAGAAGGTGGTGAAGCAG | CCCAGCATCGAAGGTAGAA |
| MDH1 | AF233351 | CGTTGCAGAGCTGAAGGATT | GGTGCACTGAGAGATCAAGG |
In situ hybridization for Kiss1, GNRH1, TAC3, and PDYN
The hypothalami that had been fixed in 10% formalin were equilibrated to 15% sucrose, followed by 30% sucrose in 0.1 M phosphate buffer. The tissues were then cut by cryostat at 30 μm and stored in cryoprotectant solution (2% paraformaldehyde, 0.05 MPB, 30% ethylene glycol, 20% glycerol) at -20°C. Dissection of POA and ARC was as described above.
Antisense riboprobes for ovine Kiss1, GNRH1, TAC3, and PDYN were synthesized with SP6 or T7 transcription kits (Ambion, Austin, TX, USA) as previously described (Li et al. [31] using corresponding DNA fragments as templates. In situ hybridization (ISH) was performed as described previously [32] using S35-labeled probes. For Kiss1, TAC3, and PDYN ISH, three sections representing rostral, middle and caudal area of ARC and for GNRH1, three sections representing rostral, middle and caudle area of POA were chosen for each cow. Sections were mounted on SuperFrost plus slides and air-dried overnight. Following 0.001% proteinase K treatment for 30 min at 37°C, sections were acetylated with 0.0025% acetic anhydride in 0.1 M TEA buffer for 10 min. After rinsing in 2 × SSC (sodium chloride and sodium citrate), the sections were dehydrated through ascending series of ethanol, delipidated in chloroform, rinsed in absolute ethanol and air-dried. The hybridization solutions containing S35 labeled antisense probe (5 × 106 cpm/mL) in a cocktail solution of 50% formamide, 5 × SSC, pH 7.0, 250 μg/mL herring sperm DNA, 100 μg/mL yeast tRNA, 5% dextran sulfate, 1× Denhardt’s solution, 0.1% Tween-20 was applied to sections and hybridized overnight at 53°C. After hybridization, sections were washed in decreasing concentrations of SSC, dehydrated, and coated with emulsion (Ilford Imaging, Melbourne). Exposure was for 1 week in dark at 4°C.
Image analysis was carried out under dark-field illumination with software designed to count the total number of cells and the number of silver grains per cell (ImagePro plus, Media Cybernetics, Inc., Silver Springs, MD). Cells were counted when silver grain density was greater than five times background. Data are expressed as the mean number of identifiable cells per section and the mean number of silver grains per cell (a semiquantitative index of mRNA expression/cell).
Immunohistochemistry to quantify LHβ and FSHβ cells in anterior pituitary tissue
Formalin-fixed tissues were initially treated as for hypothalamic tissues. Three free floating pituitary sections were washed in PBS and incubated with guinea pig anti-LH (Antibodies Australia, 1:2000) and mouse anti-FSH (Thermo Fisher Scientific, 1:1000) antibodies in Tris-buffered saline/0.3% triton X-100/5% normal goat serum. The LH and FSH immunoreactive (−ir) cells were revealed by Alexa 488 goat anti-guinea pig IgG and Alexa 546 goat anti-mouse IgG secondary antibody (Thermo Fisher Scientific).
The image analysis of fluorescent staining of immunohistochemistry for LH and FSH was processed under Zeiss fluorescent microscope. Cell numbers were counted from three microphotographs at 10× magnification (0.98 mm2) taken in areas of low, medium and high density from pituitary of each animal to obtain total cell numbers.
ME extraction for radioimmunoassay
Frozen ME were weighed and placed in a tube containing 1 mL of 2 N acetic acid and homogenized. The homogenate was centrifuged (14,000 g; 20 min; 4°C) after which the supernatant was collected and diluted 1:10 for Kisspeptin RIA and 1:1000 for GnRH RIA.
Peptide/hormone RIA
For all assays, the samples were assayed in duplicate. The GnRH was measured in ME extracts by RIA, using EL14 (courtesy of TM Nett, Colorado State University) as a primary antiserum GnRH for iodination (Auspep) and assay standard. The extracts were assayed as described previously [33], with all samples were included in a single assay; sensitivity 3.1 pg/mL and intra-assay coefficient of variation (CV) <10% between 29 and 101 pg/mL.
Kisspeptin was also measured in ME extracts by RIA, using a rabbit-anti-Kiss primary antiserum (#564) (RRID 2314708), courtesy of M. Beltramo (INRA, France) and ovine Kisspeptin-10 (Auspep) as the assay standard and for iodination. All samples were assayed in a single assay; sensitivity 64 pg/mL and intra-assay CV < 10% between 0.58 and 6.18 ng/mL.
The bovine LH RIA was performed using AFP-240580 rabbit anti-bovine primary antiserum and bovine LH (AFP-11118B) as standard (Dr A.F. Parlow; Lundquist Institute for Biomedical Innovation, Torrance, CA). The procedure was as described previously [34]. All samples were assayed in a single assay with a sensitivity of 0.07 ng/mL and intra-assay CV < 10% between 0.54 and 7.55 ng/mL.
The bovine FSH radioimmunoassay (RIA) was performed using bovine FSH primary antiserum raised in rabbit (AFP-7722291) (Dr A.F. Parlow) and bovine FSH (AFP-9294C) (Dr A.F. Parlow) as the assay standard. The samples were assayed in a single assay; sensitivity 0.08 ng/mL and intra-assay CV <10% between 1.18 and 9.05 ng/mL.
Concentrations of progesterone were determined using a commercial double-antibody RIA (ImmuChem, MP Biomedicals, CA). The manufacturer’s protocol was followed with the exception that all reagent volumes were halved. This modification was validated for repeatability, parallelism, and recovery. For repeatability, 14 assay reaction tubes for each of two control plasma samples resulted in CV% of 10.9 and 14.3 for standard concentrations of 1.45 and 0.24 ng/mL, respectively. For parallelism, two standard samples were diluted 100:0, 75:25, 50:50, and 25:75 with the kit diluent and parallelism was confirmed visually against the standard curve. For recovery, two plasma control samples were spiked with either 0.5, 2, or 5 ng/mL progesterone standards, with average recoveries being 89.9 and 92.0% for the two control samples. We also tested samples that had been previously assayed in a routinely used and validated kit (Coat-a-count, Siemens, Cat. # TKPG) and determined a good relationship (y = 1.26*X + 0.7; R2 = 0.94) between these two kits. The sensitivity of the assay was 0.3 ng/mL and intra-assay CV of 13.2%. Samples with a concentration less than sensitivity were assigned a value of 0.15 ng/mL before data analysis.
Statistical analysis
Data are presented as means ± standard error of mean (SEM). Following Bartlett’s test for heterogeneity of variance [35] and repeated measures analysis of variance (ANOVA) (SPSS) was used to ascertain statistical differences in means for gene expression and peptide levels. Progesterone data were log10 transformed then analyzed by repeated measures ANOVA (SAS/STAT 15.1; SAS Institute Inc., 2018). Differences were considered significant at P < 0.05.
Results
One experiment was undertaken to determine whether changes in the steady-state expression of genes within the hypothalamo-pituitary axis were associated with high or low fertility in dairy cows. Cows of high or low fertility were prepared by synchronizing estrous cycles. Blood samples were taken to measure plasma LH, FSH and progesterone levels and the cows were slaughtered in the early follicular phase of the cycle. Brains and pituitary glands were collected for quantification of gene expression in particular genes of interest, as detailed in the Introduction. In particular, we focused on genes expressed by the kisspeptin cells of the hypothalamus, but also quantified gene expression for in GnRH cells, GnRH, and kisspeptin peptide levels in the ME and beta-subunit expression in the pituitary gland.
Plasma LH, FSH, and progesterone concentrations
No difference was observed in mean plasma LH concentrations of the three groups of cows (data not shown). FSH was undetectable (<0.1 ng/mL) in plasma. Plasma progesterone concentrations at the time of CIDR removal were 7.1 ± 0.52 ng/mL and were reduced (P < 0.001) to non-detectable levels over the period −2.5 to −1.5 h prior to slaughter, with no treatment replicate interaction. These data showed that hormone levels in these cows were consistent with their being in the early- to mid-follicular phase of an estrous cycle.
GNRH1 expression in the POA and GnRH peptide levels in the ME
The expression of GNRH1in the POA, measured by qPCR, was similar in the three groups as were GnRH peptide levels in the ME (Table 3). ISH provided a sample of GnRH cells that are scattered through the POA/septal region and the mediobasal hypothalamus. Because the sampling method displayed only a fraction of the total number of GnRH cells, expression is only given on a per cell basis (silver grains/cell). In order to accurately count the number of GnRH cells in the hypothalamus, it would have been necessary to examine serial sections across the entire region, which was beyond the scope of the study. An example of the visualization of cells (clusters of silver grains) is shown in Figure 2A. Mean (±SEM) GnRH1 gene expression/cell was similar in all groups (n = 8) (POS-CYC: 149 ± 5.9; NEG-CYC: 146.5 ± 12.0; NEG-NONCYC: 159.0 ± 11.9).
Table 3.
Mean (± SEM) GNRH1 and Kiss1 expression in the POA and GnRH and Kiss peptide levels in the ME of POS-CYC, NEG-CYC, and NEG-NONCYC cows (n = 8/group). Levels of expression are given relative to the housekeeping genes, as indicated in Materials and methods
| Group | POS-CYC | NEG-CYC | NEG-NONCYC |
|---|---|---|---|
| POA GNRH1 | 5.73 ± 0.67 | 6.47 ± 0.94 | 6.01 ± 1.29 |
| POA Kiss1 | 25.69 ± 5.06 | 22.06 ± 3.34 | 22.69 ± 3.34 |
| ME GnRH peptide (ng/mg) | 1.14 ± 0.10 | 1.05 ± 0.14 | 0.90 ± 0.13 |
| ME Kiss peptide (ng/mg) | 0.037 ± 0.037 | 0.044 ± 0.026 | 0.035 ± 0.042 |
Figure 2.
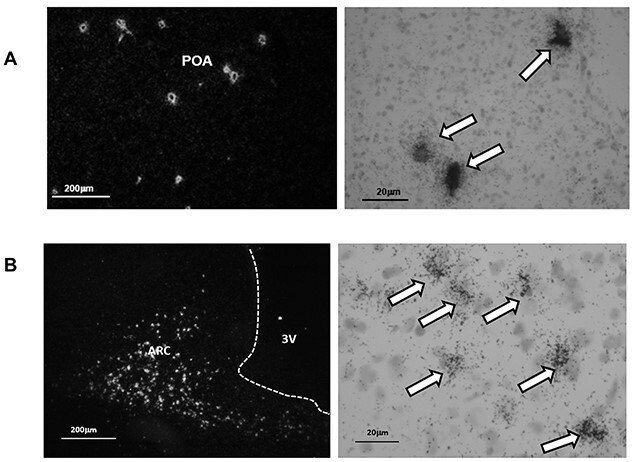
ISH displayed GNRH1 and Kiss1 cells with minimal background labeling. Panel A: Example of GNRH1 expression in single cells of the preoptic area visualized by clusters of silver grains. Panel B: Example of Kiss1 expression in the arcuate nucleus. The left-hand images are low power visualization of cells under dark field and the right-hand images show silver grains over cells under light field and higher power (arrows).
Figure 3.
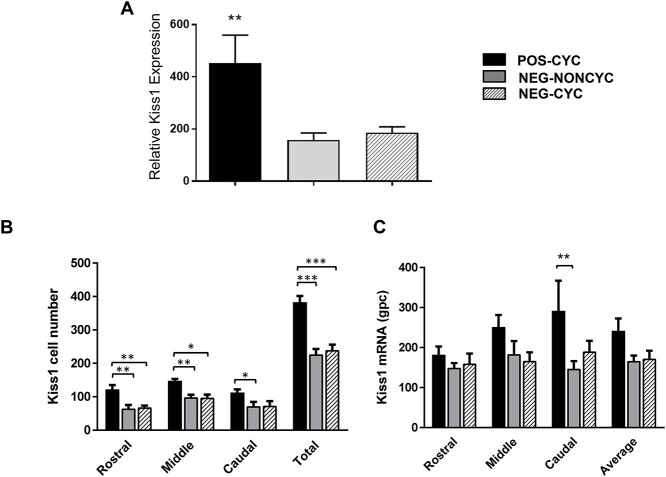
qPCR and ISH indicated that expression of Kiss1 in the arcuate nucleus of NEG cows was lower than that in POS cows. Mean ± SEM expression of Kiss1 in the arcuate nucleus of POS-CYC (n = 7), NEG-NONCYC (n = 8) and NEG-CYC cows (n = 6). Panel A shows quantification by qPCR, being relative to the geometric mean of housekeeping genes. Panels B (number of cells) and C (expression/cell silver grains/cell—gpc) show results from ISH. *P < 0.05, **P < 0.01, ***P < 0.001.
Kiss1 expression in the POA and Kisspeptin peptide levels in the ME
Kiss1 expression in the POA was assessed by qPCR only and was similar in the three groups (Table 3). Kiss1 expression was at least 2-fold higher (P < 0.01) in the ARC from POS-CYC cows than in the NEG cow groups (Figure 3A).
Two NEG-CYC cows and one POS-CYC cow were eliminated from the ISH analysis because of dissection error. The separation of the ME from the basal hypothalamus by gross dissection can sometimes be such that a portion of the latter is taken with the former. This results in loss of kisspeptin cells from the hypothalamic block, giving a false representation of the entire nucleus when analyzed by ISH. An example of the ISH signal is shown in Figure 2B, indicating a dense population of Kiss1 positive cells.
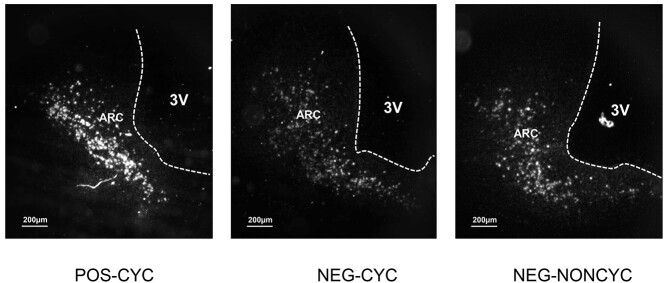
The number of cells expressing Kiss1 was higher in the POS-CYC group than in the NEG-CYC and NEG-NONCYC groups across the entire nucleus (P < 0.001; Figure 3B), with examples shown in Figure 4. Expression/cell was greater in the POS-CYC group in the caudal region of the nucleus (P < 0.01; Figure 2C), as displayed in Figure 4.
Figure 4.
Examples of expression of Kiss1 in the ARC of POS-CYC, NEG-NONCYC, and NEG-CYC cows.
Kisspeptin peptide levels in the ME were similar in the three groups (Table 3).
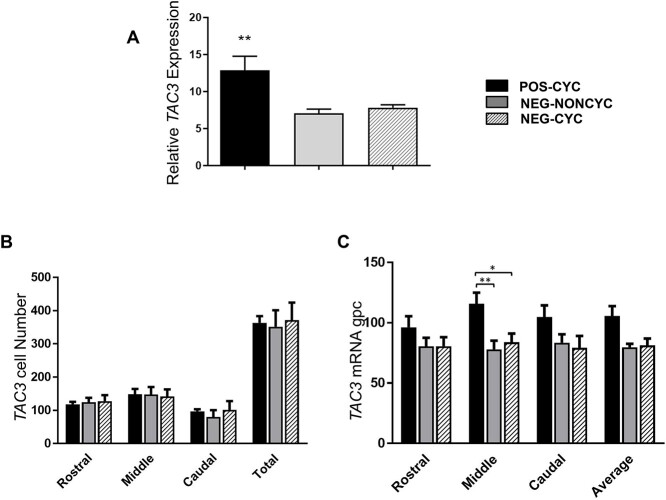
TAC3 and PDYN expression in the ARC
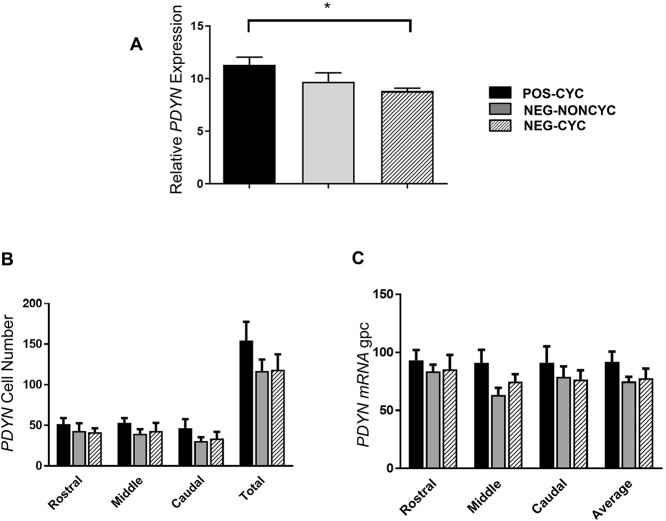
qPCR analysis showed that TAC3 expression in the ARC was 87% higher in POS-CYC cows than in NEG-NONCYC (P < 0.01) and NEG-CYC cows (P < 0.05) cows (Figure 5A). ISH analysis showed that TAC3 expression/cell was higher in the mid-ARC region of the POS-CYC cows than in the NEG FBV cows (Figure 5C), with no difference in the number of cells expressing TAC3 (Figure 5B). PDYN expression measured by qPCR was 27% higher (P < 0.05) in POS-CYC cows than in NEG-CYC cows (Figure 6A) but cell number and expression/cell for PDYN was similar in all groups when measured by ISH (Figure 6B and C).
Figure 5.
Mean ± SEM expression of TAC3 in the ARC of POS-CYC (n = 7), NEG-NONCYC (n = 8), and NEG-CYC cows (n = 6). Panel A shows quantification by qPCR, being relative to the geometric mean of housekeeping genes. Panels B (number of cells) and C (expression/cell—silver grains/cell—gpc) show results from ISH. *P < 0.05, **P < 0.01. TAC3 expression was higher in POS cows when measured by qPCR, with higher expression/cell detected by ISH in cells of the mid-region of the ARC.
Figure 6.
Mean ± SEM expression of PDYN in the arcuate nucleus of POS-CYC (n = 7), NEG-NONCYC (n = 8), and NEG-CYC cows (n = 6). Panel A shows quantification by qPCR, being relative to the geometric mean of housekeeping genes. Panels B (number of cells) and C (expression/cell silver grains/cell—gpc) show results from ISH. *P < 0.05. qPCR analysis showed that expression of PDYN was marginally higher (P < 0.05) in POS cows than in NEG-CYC cows, but this was not confirmed by ISH.
Gonadotropin subunit gene expression (ISH) in the anterior pituitary
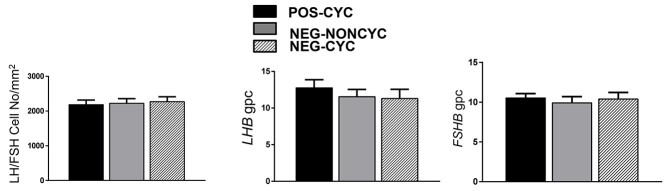
There was no difference in the number of LHB/FSHB expressing cells between the three groups (Figure 7), as was the level of expression of these genes (Figure 7).
Figure 7.
Mean ± SEM number of pituitary gonadotropes and levels of gene expression for LHB and FSHB in the anterior pituitary gland POS-CYC, NEG-NONCYC, and NEG-CYC cows (n = 8/group). There were no significant differences.
Discussion
The most significant finding in this study is the difference in ARC Kiss1 expression in cows of POS FBV and NEG FBV. Kisspeptin became prominent as a major element in the regulation of reproduction in 2003, with the publication of two papers indicating that inactivating mutations in the kisspeptin receptor gene (Kiss1R) caused pubertal failure in mice and humans [36, 37]. Kisspeptin cells of the ARC express estrogen and progesterone receptors and are believed to be the major intermediary of sex steroid regulation of GnRH secretion [17]. The estrous cycle is a finely tuned series of events that are dictated by the action of estrogen and progesterone on the brain and pituitary gland [10], so kisspeptin function is integral to this. In ewes, the kisspeptin cells of the ARC mediate negative feedback throughout the majority of the estrous cycle and also initiate positive feedback in the follicular phase, leading to the GnRH/LH surge and ovulation [11]. The lower level of expression of Kiss1 in cows of NEG FBV, whether displaying cycles or not in the previous breeding period, presumably compromises this cyclic function. This could be due to genetic variation in the 5′ regulatory region of the gene or differences in the 3′ untranslated region that is important for post-translational processing; testing this was beyond the scope of the present study. Other factors that may have contributed to the difference in Kiss1 expression in the ARC are discussed below. Alternatively, “upstream” regulators of kisspeptin cell activity in the ARC, such as Neurokinin (TAC3) or DYN, could be responsible for this difference. We believe that this is the first time non-binary variation in the level of expression of Kiss1 can be directly associated with genetic merit for FBVs in dairy cattle or in any other species.
Variants in Kiss1 have also been reported in humans with idiopathic hypothalamic hypogonadism (IHH) [38–40], but these are rare. In addition, activating variants in Kiss1 and Kiss1R genes have been observed in cases of central precocious puberty [41]. Inactivating mutations also occur in the GNRH1 [42, 43] and the GnRH receptor gene (GNRHR) [44] the latter accounting for up to 40% of familial cases of IHH in humans [45]. We do not know of any cases of such mutations occurring naturally in ungulates to cause infertility, but the present results indicate they may occur as a result of genetic selection for POS or NEG fertility. Importantly, however, conditional knockout of Kiss1 in ARC neurons in mice (Pdyn-Cre/Kiss1fl/fl) by markedly reduces pulsatile LH secretion and causes hypogonadism and persistent diestrus [20]). This transgenic model, generated by embryonic manipulation reduces Kiss1 expression by 90%, so it much more severe than that observed in NEG cows. On the other hand, work in humans suggests that lack of kisspeptin function may be the cause of infertility in certain circumstances. Thus, kisspeptin treatment stimulates LH pulsatility and the secretion of estrogen and inhibinB from the ovaries of hyperprolactinemic, hypogonadotropic amenorrheic women [46], suggesting that the cause of infertility may be due to lack of kisspeptin signaling. Other work in humans shows that continuous intravenous infusion of kisspeptin may increase pulsatile LH secretion in women with hypothalamic amenorrhea (HA) [47], which the authors inferred was due to action on the ME to increase GnRH secretion. Notably, there has been no follow-up study to indicate whether kisspeptin infusion restores fertility in women with HA [48]. In anestrous ewes, in which ARC expression of kisspeptin is lower than in ewes during the breeding season [14], continuous intravenous infusion elicits a rise in pulsatile LH secretion that culminates in a preovulatory surge and ovulation [49]. As far as we are aware, the association between relatively low kisspeptin expression in the ARC, rather than the profound reduction seen in transgenic mouse models, and lack of ovarian cyclicity in seasonally anestrous ewes is the only other instance, apart from the present work, in which these two parameters may be associated.
In sheep, kisspeptin cells also produce DYN and NKB [17], the former acting as a negative regulator of kisspeptin secretion and the latter being a stimulator [50, 51]. Furthermore, loss-of-function mutations in TAC3 or the NKB receptor gene (TACR3) cause hypothalamic hypogonadism in humans [52]. In the present study, we examined TAC3 and PDYN expression throughout the ARC because there are cells apart from kisspeptin cells that produce these peptides. TAC3 expression was higher in POS-CYC cows than in the NEG-CYC and NEG-NONCYC cows, as determined by qPCR. This difference was in the mid-region of the ARC, as determined by ISH, so it is most likely due to a difference in NKB cells that are not kisspeptin cells (which are predominantly in the caudal region). In the ewe, virtually all NKB cells express estrogen receptors [53]. Of particular relevance to the lactating dairy cow, one recent study [28] showed that Senktide, a TAC3R agonist, can advance the timing of first ovulation post-partum. Accordingly, higher TAC3 expression in POS-CYC cows, compared with NEG cows, may be related to the higher expression of TAC3 in the former, which may, in turn, drive higher expression of Kiss1. Further work to confirm the role of NKB/TAC3 in the regulation of reproduction in the dairy cow would be warranted. Although we found higher expression of PDYN in HIGH-CYC cows using qPCR, this was not confirmed by ISH studies, so the role of DYN as a negative regulator of Kiss1 cells (and GnRH secretion) in post-partum cows seems unlikely.
It is salient that the difference in expression of the Kiss1 was seen in the ARC, and not the POA. In addition to the role of NKB and DYN in the regulation of Kiss cells, a high percentage of kisspeptin cells also receive input from gamma amino-butyric acid and glutamatergic cells in sheep at least [19], so the function of these afferent cells could also be relevant. Further work is required to ascertain if such input to kisspeptin cells is of relevance to fertility in post-partum dairy cows.
The difference in ARC expression of Kiss1 and TAC3 between cows of divergent FBV is remarkable given that we did not observe a difference in expression of GNRH1 or Kiss1 in the POA. This is consistent with the importance of the ARC Kisspeptin cells in feedback regulation of GnRH secretion in ruminants as indicated above. Other work in sheep indicates that the kisspeptin cells of the POA facilitate the positive feedback effect of estrogen that causes ovulation [16, 54]. The level of GnRH and kisspeptin peptide in the ME was similar in cows of POS and NEG FBV. The origin of the Kiss peptide in the ME is the ARC cells [55]. Firstly, this indicates that the production of GnRH is not compromised in NEG FBV cows. Secondly, the equivalent amounts of kisspeptin in the ME of POS and NEG FBV cows suggests that the function of the ARC Kisspeptin cells rather than the absolute ability to produce kisspeptin may be the cause of low fertility in the NEG cows. Whether this is an inherent lesion in the ARC Kisspeptin cells or it is a function of lower NKB input remains to be determined.
We did not show any difference between NEG and POS cows in the level of GNRH1 expression in cells. This is consistent with a number of studies showing that levels of GnRH in the brain do not change with reproductive state of adult females (or males) [56]. In general, changes in reproductive status with season are not generally accompanied by changes in GnRH synthesis, whereas the change in the secretion of GnRH does so. This reflects the fact that secretion is controlled by a range of neuronal inputs to GnRH cells, whereas GnRH stores are maintained [10, 11, 54]. This is not withstanding changes in the synthesis of GnRH that may occur in the prepubertal period or with aging [56]. Thus, barring genetic mutations, it is the pattern of GnRH secretion rather than the synthesis of GnRH that determines reproductive status and kisspeptin controls GnRH secretion at the level of the ME [19]. We did not quantify the numbers of GnRH cells in the brains of the POS and NEG cows, but this would have required examination of serial sections across the entire POA and hypothalamus, a significant task that was beyond the scope of the study. Nevertheless, the equivalent levels of GnRH in the ME of the two groups, suggest that GnRH production is not a limiting factor in NEG cows.
Our current model for control of pulsatile GnRH secretion is that it is regulated by kisspeptin cells that project from the ARC to the ME in sheep [19] and the bovine [13]. Although we did not show any difference in the levels of LH in the two groups of cows, this might be expected because of the limited number of samples taken. Samples taken shortly before slaughter were intended to demonstrate that the cows were in early/mid-follicular phase, displaying low progesterone and gonadotropin levels, rather than in a preovulatory state where surge levels of LH would be expected. In order to ascertain whether there are differences in the secretion of GnRH, LH, and FSH, much more extensive sampling protocols would be required and this was not an objective of the present study. It would have also been desirable to measure plasma estrogen levels, although this would have been relevant only if we had conducted serial sampling at short intervals over a longer period to determine values following LH pulses. It is possible that the difference in expression of Kiss1 and TAC3 is due to differences in estrogen status, which will require further studies of the endocrine status of POS and NEG cows. Because pulses of estrogen secretion follow pulses of LH [57], accurate measurement of the former would necessitate serial sampling and to undertake such an exercise would have imposed a significant further experimental manipulation. We adopted the protocol as shown in Figure 1 to cause minimal disruption to the cows.
This study did not extend to the quantification of the expression levels of kisspeptin receptors by GnRH cells or the expression of the GnRH receptor on gonadotropes and both could be compromised in NEG FBV cows. What is clear from the present study, however, is that production of gonadotropins by the pituitary gonadotropes is similar in cows of POS and NEG FBV, indicating that a pituitary factor is unlikely to be the cause of the difference in fertility. It seems more likely that lowered function of kisspeptin cells in the ARC of NEG FBV cows would lead to reduced GnRH secretion, with consequent lowering of the pulsatile secretion of LH and FSH. Serial blood sampling of such cows would be instructive in this regard. The condition may be compared to that of seasonally anestrous ewes, in which pulsatile GnRH/LH secretion is lower than during the breeding season, when ewes are displaying regular estrous cycles [58]. The issue of whether there might be a difference in the pulsatile secretion of FSH is somewhat more difficult to predict, since the pattern of FSH secretion, in the ewe at least, is not tightly linked to that of GnRH/LH, displaying an entropic nature with some pulses linked to those of LH and others not so [59].
It could be argued that the difference seen in ARC gene expression in POS and NEG cows relates to a particular trait taken into account for the calculation of FBV, but this would require identification of cows that segregate specifically for a single trait; this would require a very significantly expanded program of genetic selection that is well beyond the scope of the present work. Furthermore, in focusing on any specific trait used for FBV calculation, it is most likely that other traits would segregate with the same, making it virtually impossible to isolate each trait as a single non-linked entity. In essence, we are interested in identifying factors that may relate to the differing levels of fertility and provide an indication of the possible utility of remediation of infertility. The present data suggest that, as for anestrous ewes, therapeutic enhancement of kisspeptin status, whether by TAC3 agonist or a kisspeptin, may be efficacious.
In conclusion, we have demonstrated reduced expression of Kiss1 and TAC3 in the ARC of NEG FBV cows, strongly suggesting that this is a causative factor in their lowered fertility. There were no differences between NEG-CYC and NEG-NONCYC cows, suggesting that the compromise in fertility in the latter could also have other causes. Nevertheless, given the importance of kisspeptin in the control of GnRH secretion, the present results indicate at least one cause of lowered fertility. Such an observation is novel since we know of no other case where reduced steady-state expression of a key gene is directly associated with fertility. The production of GnRH and the gonadotropins is not different in POS and NEG FBV cows, so the function of kisspeptin and NKB cells in the ARC is the singular neuroendocrine lesion identified. This suggests that fertility might be improved in such cows by treatment with a TAC3R receptor agonist, such as Senktide [28] or with a kisspeptin agonist [60]. This may provide an alternative to the progesterone-Ovsynch protocol, which is considered expensive [61]. Finally, in depth genetic analysis of the TAC3 and Kiss1 genes (and/or other genes regulating kisspeptin cells) may identify a genetic cause for the low expression of these genes in NEG FBV cows.
Supplementary Material
Acknowledgments
We thank the staff of Greenlea Meats Ltd (Morrinsville, New Zealand) for the generous provision of space and their assistance with the collection of brain and pituitary tissues. We thank Prof TM Nett for provision of the GnRH antibody and Dr M Beltramo for the Kiss antibody. We thank the DairyNZ technicians for animal handling and tissue collection and Ms Alix Rao for laboratory analysis. We thank Ms Mandy Curd for assistance in manuscript preparation.
Footnotes
† Grant Support: This work was funded by dairy farmers through DairyNZ Incorporated with matched co-funding from the Ministry of Business, Innovation and Employment (DRCX1302). Other support and resources were kindly provided by LIC, CRV Ambreed, and AgResearch.
Contributor Information
Iain J Clarke, Neuroscience Program, Monash Biomedicine Discovery Institute and Department of Physiology, Monash University, Melbourne, Victoria, Australia.
Charlotte B Reed, DairyNZ, Private Bag 3221, Hamilton 3240, New Zealand.
Chris R Burke, DairyNZ, Private Bag 3221, Hamilton 3240, New Zealand.
Qun Li, Neuroscience Program, Monash Biomedicine Discovery Institute and Department of Physiology, Monash University, Melbourne, Victoria, Australia.
Susanne Meier, DairyNZ, Private Bag 3221, Hamilton 3240, New Zealand.
Author contributions
IJC: 30%
CBR: 20%
CRB: 10%
QL: 30%
SM: 10%
This paper demonstrates that the expression of Kiss1 in the hypothalamus of dairy cows of relatively high fertility is twice that of cows with relatively low fertility and is directly relevant to productivity in the dairy industry.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Conflict of interest
The authors have declared that no conflict of interest exists.
References
- 1. Roche JR, Burke CR, Crookenden MA, Heiser A, Loor JL, Meier S, Mitchell MD, Phyn CVC, Turner S-A. Fertility and the transition dairy cow. Reprod Fertil Dev 2018; 30:85–100. [DOI] [PubMed] [Google Scholar]
- 2. Roche JR, Burke CR, Meier S, Walker CG. Nutrition x reproduction interaction in pasture-based systems: Is nutrition a factor in reproductive failure? Animal Product Sci 2011; 51:1045–10-9-66. [Google Scholar]
- 3. Weller JI, Ezra E, Ron M. Invited review: a perspective on the future of genomic selection in dairy cattle. J Dairy Sci 2017; 100:8633–8644. [DOI] [PubMed] [Google Scholar]
- 4. Royal MD, Darwash AO, Flint APF, Webb R, Woolliams JA, Lamming GE. Declining fertility in dairy cattle: changes in traditional and endocrine parameters of fertility. Animal Sci 2000; 70:487–501. [Google Scholar]
- 5. Berry PW, E., Pryce JE. Genetics and genomics of reproductive performance in dairy and beef cattle. Animal 2014; 8:105–121. [DOI] [PubMed] [Google Scholar]
- 6. Dennis NA, Stachowicz K, Visser B, Hely FS, Berg DK, Friggens NC, Amer PR, Meier S, Burke CR. Combining genetic and physiological data to identify predictors of lifetime reproductive success and the effect of selection on these predictors on underlying fertility traits. J Dairy Sci 2018; 101:3176–3192. [DOI] [PubMed] [Google Scholar]
- 7. Crowe MA. Resumption of ovarian cyclicity in post-partum beef and dairy cows. Reprod Domest Anim 2008; 43:20–28. [DOI] [PubMed] [Google Scholar]
- 8. Ayres H, Ferreira RM, Cunha AP, Araujo RR, Wiltbank MC. Double-Ovsynch in high-producing dairy cows: effects on progesterone concentrations and ovulation to GnRH treatments. Theriogenology 2013; 79:159–164. [DOI] [PubMed] [Google Scholar]
- 9. Nowicki A, Baranski W, Baryczka A, Janowski T. OvSynch protocol and its modifications in the reproduction management of dairy cattle herds - an update. J Vet Res 2017; 61:329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clarke IJ, Campbell R, Smith JT, Wray S. Neuroendocrine control of reproduction. In: Fink G, Pfaff DW, Levine JE (eds.), Handbook of Neuroendocrinology, vol. 9. London, Waltham, San Diego: Academic press, Elsevier. Chapter 9, 197-235; 2011: 197–235. [Google Scholar]
- 11. Clarke IJ. Control of GnRH secretion: one step back. Front Neuroendocrinol 2011; 32:367–375. [DOI] [PubMed] [Google Scholar]
- 12. Hassaneen A, Naniwa Y, Suetomi Y, Matsuyama S, Kimura K, Ieda N, Inoue N, Uenoyama Y, Tsukamura H, Maeda KI, Matsuda F, Ohkura S. Immunohistochemical characterization of the arcuate kisspeptin/neurokinin B/dynorphin (KNDy) and preoptic kisspeptin neuronal populations in the hypothalamus during the estrous cycle in heifers. J Reprod Dev 2016; 62:471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tanco VM, Whitlock BK, Jones MA, Wilborn RR, Brandebourg TD, Foradori CD. Distribution and regulation of gonadotropin-releasing hormone, kisspeptin, RF-amide related peptide-3, and dynorphin in the bovine hypothalamus. PeerJ 2016; 4:e1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith JT, Clay CM, Caraty A, Clarke IJ. KiSS-1 messenger ribonucleic acid expression in the hypothalamus of the ewe is regulated by sex steroids and season. Endocrinology 2007; 148:1150–1157. [DOI] [PubMed] [Google Scholar]
- 15. Smith JT, Li Q, Pereira A, Clarke IJ. Kisspeptin neurons in the ovine arcuate nucleus and preoptic area are involved in the preovulatory luteinizing hormone surge. Endocrinology 2009; 150:5530–5538. [DOI] [PubMed] [Google Scholar]
- 16. Hoffman GE, Le WW, Franceschini I, Caraty A, Advis JP. Expression of fos and in vivo median eminence release of LHRH identifies an active role for preoptic area kisspeptin neurons in synchronized surges of LH and LHRH in the ewe. Endocrinology 2011; 152:214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goodman RL, Lehman MN, Smith JT, Coolen LM, de Oliveira CV, Jafarzadehshirazi MR, Pereira A, Iqbal J, Caraty A, Ciofi P, Clarke IJ. Kisspeptin neurons in the arcuate nucleus of the ewe express both dynorphin a and neurokinin B. Endocrinology 2007; 148:5752–5760. [DOI] [PubMed] [Google Scholar]
- 18. Goodman RL, Hileman SM, Nestor CC, Porter KL, Connors JM, Hardy SL, Millar RP, Cernea M, Coolen LM, Lehman MN. Kisspeptin, neurokinin B, and dynorphin act in the arcuate nucleus to control activity of the GnRH pulse generator in ewes. Endocrinology 2013; 154:4259–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ezzat A, Pereira A, Clarke IJ. Kisspeptin is a component of the pulse generator for GnRH secretion in female sheep but not the pulse generator. Endocrinology 2015; 156:1828–1837. [DOI] [PubMed] [Google Scholar]
- 20. Nandankar N, Negron AL, Wolfe A, Levine JE, Radovick S. Deficiency of arcuate nucleus kisspeptin results in postpubertal central hypogonadism. Am J Physiol Endocrinol Metab 2021; 321:E264–E280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wahab F, Quinton R, Seminara SB. The kisspeptin signaling pathway and its role in human isolated GnRH deficiency. Mol Cell Endocrinol 2011; 346:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uenoyama Y, Inoue N, Maeda KI, Tsukamura H. The roles of kisspeptin in the mechanism underlying reproductive functions in mammals. J Reprod Dev 2018; 64:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harris BL, Pryce JE, Xu ZZ, Montgomerie WA. Development of new fertility breeding values for the dairy industry. Proceedings of the New Zealand Society of Animal Production 2006; 66:107–112. [Google Scholar]
- 24. Meier S, Fisher B, Eketone K, McNaughton LR, Amer PR, Beatson P, Bryant JR, Dodds KG, Spelman R, Roche JR, Burke CR. Calf and heifer development and the onset of puberty in dairy cows with divergent genetic merit for fertility. Proc. NZ Soc. Anim. Prod. 2017; 77:205–210. [Google Scholar]
- 25. Burke CR, Meier S, Phyn CV, Stephen M, Bryant JR, Amer PR, Roche JR. Identifying new traits to improve genetic merit for cow fertility. Proc Soc Dairy Cattle Veterinarians of NZVA 2019; 103–106. [Google Scholar]
- 26. Meier S, McNaughton LR, Handcock R, Amer PR, Beatson PR, Bryant JR, Dodds KG, Spelman R, Roche JR, Burke CR. Heifers with positive genetic merit for fertility traits reach puberty earlier and have a greater pregnancy rate than heifers with negative genetic merit for fertility traits. J Dairy Sci 2021; 104:3707–3721. [DOI] [PubMed] [Google Scholar]
- 27. Giordano J. Increased fertility in lactating dairy cows resynchronized with double-Ovsynch compared with Ovsynch initiated 32 d after timed artificial insemination. J Dairy Sci 2012; 95:639–653. [DOI] [PubMed] [Google Scholar]
- 28. Nakamura S, Wakabayashi Y, Yamamura T, Ohkura S, Matsuyama S. A neurokinin 3 receptor-selective agonist accelerates pulsatile luteinizing hormone secretion in lactating cattle. Biol Reprod 2017; 97:81–90. [DOI] [PubMed] [Google Scholar]
- 29. Lehman MN, Coolen LM, Goodman RL. Minireview: kisspeptin/neurokinin B/dynorphin (KNDy) cells of the arcuate nucleus: a central node in the control of gonadotropin-releasing hormone secretion. Endocrinology 2010; 151:3479–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wakabayashi Y, Nakada T, Murata K, Ohkura S, Mogi K, Navarro VM, Clifton DK, Mori Y, Tsukamura H, Maeda K, Steiner RA, Okamura H. Neurokinin B and dynorphin a in kisspeptin neurons of the arcuate nucleus participate in generation of periodic oscillation of neural activity driving pulsatile gonadotropin-releasing hormone secretion in the goat. J Neurosci 2010; 30:3124–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Q, Smith JT, Henry BA, Rao A, Pereira A, Clarke IJ. Expression of genes for kisspeptin (KISS1), neurokinin B (TAC3), Prodynorphin (PDYN) and gonadotropin inhibitory hormone (RFRP) across natural puberty in sheep. Physiological Reports 2020; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simmons DMAJ, Swanson LW. A complete protocol for in situ hybridization of messenger RNAs in brain and other tissues with radio-labelled single-stranded RNA probes. J Histotechnol 1989; 12:169–181. [Google Scholar]
- 33. Jonas HA, Burger HG, Cumming IA, Findlay JK, de Kretser DM. Radioimmunoassay for luteinizing hormone-releasing hormone (LHRH): its application to the measurement of LHRH in ovine and human plasma. Endocrinology 1975; 96:384–393. [DOI] [PubMed] [Google Scholar]
- 34. Otto JR, Malau-Aduli BS, Rao A, Clarke IJ, Malau-Aduli AEO. Effect of incremental levels of crude degummed canola oil on milk progesterone, plasma luteinizing and follicle stimulating hormones of primiparous Holstein-Friesian cows in a pasture-based system. Int J Veterin Sci Med 2014; 2:122–129. [Google Scholar]
- 35. Sokal RR. F.J R. Biometry. WH Freeman and Coy: Baltimore; 1969. [Google Scholar]
- 36. de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci U S A 2003; 100:10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J et al. The GPR54 gene as a regulator of puberty. N Engl J Med 2003; 349:1614–1627. [DOI] [PubMed] [Google Scholar]
- 38. Gajdos ZK, Butler JL, Henderson KD, He C, Supelak PJ, Egyud M, Price A, Reich D, Clayton PE, Le Marchand L, Hunter DJ, Henderson BE et al. Association studies of common variants in 10 hypogonadotropic hypogonadism genes with age at menarche. J Clin Endocrinol Metab 2008; 93:4290–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chan YM, Broder-Fingert S, Paraschos S, Lapatto R, Au M, Hughes V, Bianco SD, Min L, Plummer L, Cerrato F, De Guillebon A, Wu IH et al. GnRH-deficient phenotypes in humans and mice with heterozygous variants in KISS1/Kiss1. J Clin Endocrinol Metab 2011; 96:E1771–E1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Topaloglu AK, Tello JA, Kotan LD, Ozbek MN, Yilmaz MB, Erdogan S, Gurbuz F, Temiz F, Millar RP, Yuksel B. Inactivating KISS1 mutation and hypogonadotropic hypogonadism. N Engl J Med 2012; 366:629–635. [DOI] [PubMed] [Google Scholar]
- 41. Teles MG, Bianco SD, Brito VN, Trarbach EB, Kuohung W, Xu S, Seminara SB, Mendonca BB, Kaiser UB, Latronico AC. A GPR54-activating mutation in a patient with central precocious puberty. N Engl J Med 2008; 358:709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bouligand J, Ghervan C, Tello JA, Brailly-Tabard S, Salenave S, Chanson P, Lombes M, Millar RP, Guiochon-Mantel A, Young J. Isolated familial hypogonadotropic hypogonadism and a GNRH1 mutation. N Engl J Med 2009; 360:2742–2748. [DOI] [PubMed] [Google Scholar]
- 43. Chan YM, de Guillebon A, Lang-Muritano M, Plummer L, Cerrato F, Tsiaras S, Gaspert A, Lavoie HB, Wu CH, Crowley WF Jr, Amory JK, Pitteloud N et al. GNRH1 mutations in patients with idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci U S A 2009; 106:11703–11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Silveira LF, Trarbach EB, Latronico AC. Genetics basis for GnRH-dependent pubertal disorders in humans. Mol Cell Endocrinol 2010; 324:30–38. [DOI] [PubMed] [Google Scholar]
- 45. Beranova M, Oliveira LM, Bedecarrats GY, Schipani E, Vallejo M, Ammini AC, Quintos JB, Hall JE, Martin KA, Hayes FJ, Pitteloud N, Kaiser UB et al. Prevalence, phenotypic spectrum, and modes of inheritance of gonadotropin-releasing hormone receptor mutations in idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab 2001; 86:1580–1588. [DOI] [PubMed] [Google Scholar]
- 46. Millar RP, Sonigo C, Anderson RA, George J, Maione L, Brailly-Tabard S, Chanson P, Binart N, Young J. Hypothalamic-pituitary-ovarian Axis reactivation by kisspeptin-10 in hyperprolactinemic women with chronic amenorrhea. J Endocr Soc 2017; 1:1362–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jayasena CN, Abbara A, Veldhuis JD, Comninos AN, Ratnasabapathy R, De Silva A, Nijher GM, Ganiyu-Dada Z, Mehta A, Todd C, Ghatei MA, Bloom SR et al. Increasing LH pulsatility in women with hypothalamic amenorrhoea using intravenous infusion of kisspeptin-54. J Clin Endocrinol Metab 2014; 99:E953–E961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roberts RE, Farahani L, Webber L, Jayasena C. Current understanding of hypothalamic amenorrhoea. Ther Adv Endocrinol Metab 2020; 11:2042018820945854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Caraty A, Smith JT, Lomet D, Ben Said S, Morrissey A, Cognie J, Doughton B, Baril G, Briant C, Clarke IJ. Kisspeptin synchronizes preovulatory surges in cyclical ewes and causes ovulation in seasonally acyclic ewes. Endocrinology 2007; 148:5258–5267. [DOI] [PubMed] [Google Scholar]
- 50. Goodman RL, Coolen LM, Anderson GM, Hardy SL, Valent M, Connors JM, Fitzgerald ME, Lehman MN. Evidence that dynorphin plays a major role in mediating progesterone negative feedback on gonadotropin-releasing hormone neurons in sheep. Endocrinology 2004; 145:2959–2967. [DOI] [PubMed] [Google Scholar]
- 51. Grachev P, Porter KL, Coolen LM, McCosh RB, Connors JM, Hileman SM, Lehman MN, Goodman RL. Surge-like luteinising hormone secretion induced by retrochiasmatic area NK3R activation is mediated primarily by arcuate kisspeptin neurones in the ewe. J Neuroendocrinol 2016; 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rance NE, Krajewski SJ, Smith MA, Cholanian M, Dacks PA. Neurokinin B and the hypothalamic regulation of reproduction. Brain Res 2010; 1364:116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Goubillon ML, Forsdike RA, Robinson JE, Ciofi P, Caraty A, Herbison AE. Identification of neurokinin B-expressing neurons as an highly estrogen-receptive, sexually dimorphic cell group in the ovine arcuate nucleus. Endocrinology 2000; 141:4218–4225. [DOI] [PubMed] [Google Scholar]
- 54. Clarke IJ. Generation of the gonadotropin releasing hormone surge and the luteinising hormone surge, by the positive feedback effect of estrogen. In: Herbison AE, Plant TM (eds.), The GnRH Neuron and its Control. Wiley-Blackwell, Hoboken, New Jersey. 2017.
- 55. Smith JT, Li Q, Yap KS, Shahab M, Roseweir AK, Millar RP, Clarke IJ. Kisspeptin is essential for the full preovulatory LH surge and stimulates GnRH release from the isolated ovine median eminence. Endocrinology 2011; 152:1001–1012. [DOI] [PubMed] [Google Scholar]
- 56. Clarke IJ, Pompolo S. Synthesis and secretion of GnRH. Anim Reprod Sci 2005; 88:29–55. [DOI] [PubMed] [Google Scholar]
- 57. Baird DT. Pulsatile secretion of LH and ovarian estradiol during the follicular phase of the sheep estrous cycle. Biol Reprod 1978; 18:359–364. [DOI] [PubMed] [Google Scholar]
- 58. Barrell GK, Moenter SM, Caraty A, Karsch FJ. Seasonal changes of gonadotropin-releasing hormone secretion in the ewe. Biol Reprod 1992; 46:1130–1135. [DOI] [PubMed] [Google Scholar]
- 59. Clarke IJ, Moore L, Veldhuis J. Intensive direct cavernous sinus sampling identifies high-frequency, nearly random patterns of FSH secretion in ovariectomized ewes: combined appraisal by RIA and bioassay. Endocrinology 2002; 143:117–129. [DOI] [PubMed] [Google Scholar]
- 60. Leonardi CEP, Dias FCF, Adams GP, Araujo ER, Singh J. Kisspeptin induces ovulation in heifers under low plasma progesterone concentrations. Theriogenology 2020; 141:26–34. [DOI] [PubMed] [Google Scholar]
- 61. Rojas Canadas E, Gobikrushanth M, Fernandez P, Kenneally J, Lonergan P, Butler ST. Evaluation of alternative strategies to treat anoestrous dairy cows and implications for reproductive performance in pasture-based seasonal calving herds: a pilot study. Theriogenology 2019; 127:130–136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.