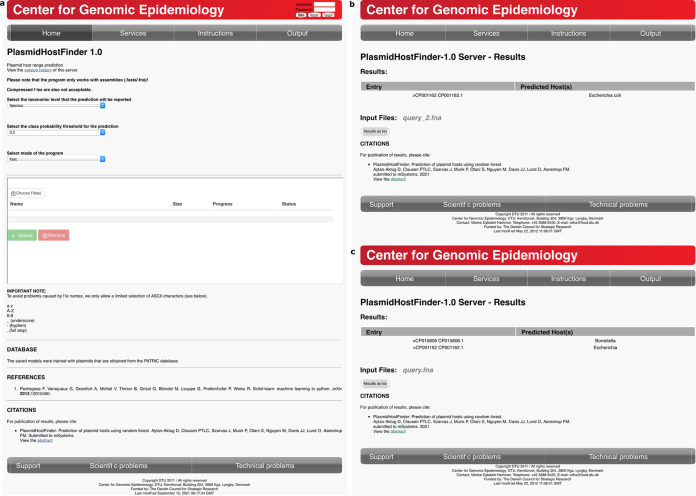
FIG 9.
Web server for PlasmidHostFinder. (a) The interface of the PlamidHostFinder web server is shown. The web server runs online after uploading a query file in FASTA format and selecting appropriate parameters for the taxonomic level (species to order), class probability threshold (0.1 to 0.9), and mode of the program (fast or slow). (b) An example of output of the PlasmidHostFinder web server is shown. In this example, the query_2.fna file was run using the following parameters: species level, 0.5 class probability threshold, and fast mode. The tool has two kinds of output files: HTML table and downloadable TSV file. While the HTML table includes only the detected plasmid host with at least 0.5 host probability, the TSV file includes the probabilities of all possible hosts at the species level. (c) Another example of the PlasmidHostFinder output. In this example, the query.fna file was run using the following parameters: genus level, 0.5 class probability threshold, and fast mode. Differently, this query file includes two contigs in the same file. Thus, the predicted hosts and all of the host probabilities were reported per contig in the HTML table and the TSV file, respectively.