Abstract
We assessed the ability of gene transfer to reverse vancomycin resistance in class A (VanA) glycopeptide-resistant Enterococcus faecalis. Recombinant shuttle vectors containing a vanH promoter-vanA antisense gene cassette fully restored vancomycin susceptibility through a combined transcriptional activator binding domain decoy and inducible vanA antisense RNA effect.
A growing prevalence of enterococci displaying several phenotypes of glycopeptide resistance has been described (14). Of these, class A glycopeptide resistance (VanA) is a commonly encountered phenotype. Class A resistance is genetically attributable to a transposon-based cluster consisting of seven genes that function in concert to allow for an alternative biosynthetic pathway for the production of cell wall precursors that less avidly bind vancomycin (Fig. 1) (1, 6, 19). Transcription from the vanH, -A, and -X genes within the vanA operon is under the control of the vanH promoter and is induced through the binding of the phosphorylated gene product of vanR (4).
FIG. 1.
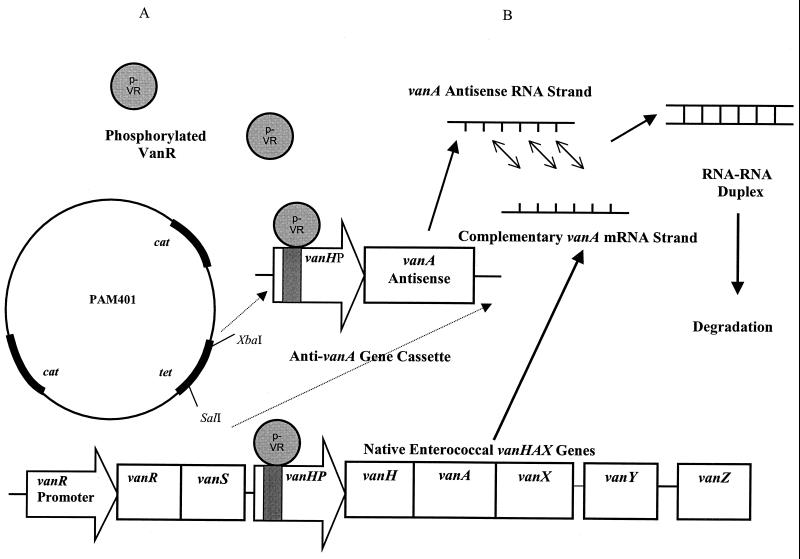
Proposed dual mechanism of action of the pAM401-vanH promoter-vanA antisense recombinant shuttle vector. The vancomycin susceptibility of a target VanA E. faecalis strain is enhanced through (A) the binding and sequestration of phosphorylated VanR from the native vanH promoter (vanHP) (shaded), thereby inhibiting the induction of the vanHAX genes, and (B) the phosphorylated-VanR-induced vanA antisense RNA effect.
In an attempt to inhibit pathogens that are resistant to conventional pharmacological antimicrobial agents, gene-based strategies have been studied, although primarily in eukaryotic systems (12, 13, 20). Here, we present a gene transfer model that employs a combination of transcriptional activation domain decoys and inducible antisense RNA to inhibit the expression of key vancomycin resistance determinants at the transcriptional and posttranscriptional levels. This resulted in the full restoration of vancomycin susceptibility in a glycopeptide-resistant enterococcus.
A vancomycin-resistant Enterococcus faecalis strain, designated A407, is a VanA phenotype clinical isolate. API-Rapid Strep strips (bioMerieux Vitex, Inc., Hazelwood, Mo.) were used for identification. The vanA genotype was confirmed by DNA probe as previously described (7). Vancomycin susceptibilities were determined by the National Committee for Clinical Laboratory Standards methods (15). Competent Escherichia coli DH5α (Life Technologies, Inc., Rockville, Md.) was used in the cloning of the recombinant vectors (18). The parent plasmid used in vector construction was pAM401 (American Type Culture Collection, Rockville, Md.). This multicopy shuttle vector contains both gram-negative (E. coli) and enterococcal (E. faecalis) elements necessary for replication in these two bacterial types as well as tetracycline and chloramphenicol resistance genes for selection (22). Plasmid yield in our experiments was >25 copies per bacterial cell. Plasmid pAMP1 (Life Technologies, Inc.) was employed for cloning of PCR-amplified fragments.
A 459-bp fragment containing the vanH promoter was amplified using primers (Life Technologies) corresponding to bases 5563 to 5580 and 5986 to 6009 in the vanA resistance gene cluster (transposon Tn1546; GenBank accession no. M97297) (3, 5). The vanA gene (1,048 bp) was amplified using primers corresponding to bases 6967 to 6986 and 7996 to 8015 in the vanA gene cluster (transposon Tn1546; GenBank accession no. M97297) (3, 5).
PCR products, including the vanH promoter and vanA genes, were initially subcloned into pAMP1. Using XbaI and SalI, the promoter was then inserted into pAM401. The vanA gene in the sense and antisense orientations was then ligated downstream from the promoter after digestion with XhoI and SalI and ligation into the SalI site of the pAM401-vanH promoter construct.
As suggested by Arthur et al., the introduction of an exogenous vanH promoter could sequester phosphorylated VanR from the native vanH promoter, thereby interfering with the activation of vanH, -A, and -X expression (2). The phosphorylated-VanR binding domain within the vanH promoter has previously been attributed to an approximately 80-bp region that has the capacity to bind multiple phosphorylated-VanR molecules (11). Therefore, in order to differentiate these potential phosphorylated VanR binding domain decoy effects of the vanH promoter from the effect of the antisense vanA, shuttle vectors containing the entire vanH promoter, the phosphorylated-VanR binding domain portion of this promoter (primers corresponding to bases 5762 to 5782 and 5909 to 5928 in the vanA gene cluster), or a mutant version of the promoter with a deletion of this binding domain were constructed and transformed into E. faecalis A407.
Electrotransformation of the E. faecalis strains with pAM401, pAM401-vanH promoter, or pAM401-vanH promoter-vanA antisense was accomplished with a Bio-Rad Gene Pulser (8). The electroporation apparatus settings were 1.50 kV, 400 Ω, and 25 μF. Under these conditions, resultant time constants were typically in the 8- to 9-ms range. To confirm that MIC changes in transformants were not related to the loss of the vanA operon, the retention of the VanA gene cluster was confirmed by PCR amplification and sequencing of the relevant genes within the cluster. Antisense RNA expression was confirmed by reverse transcriptase PCR (RT-PCR). Briefly, bacterial RNA was prepared using the RNeasy protocol (Qiagen Inc., Valencia, Calif.) and incorporating the application of RNase-free DNase (Qiagen Inc.) directly on the QIAamp column to diminish DNA contamination. RT-PCR was performed using the Titan one-tube RT-PCR protocol (Roche Molecular Biochemicals, Indianapolis, Ind.). Primers corresponding to bases 6967 to 6986 of the VanA resistance gene cluster (5) and bases 2346 to 2328 of the sequence of the cloning vector pACYC184 (GenBank accession no. X06403) (17) were used to amplify 1,230 bp of bacterial vanA antisense RNA (including 182 bp from the vector pACYC184, a parent vector of pAM401). A PCR control was run in parallel with RT-PCR in order to evaluate the DNA contamination of each sample.
The vancomycin MIC for E. faecalis A407 transformed with the shuttle vector containing the vanH promoter was reduced from 512 to 64 μg/ml (Table 1). In contrast, in a control experiment, E. faecalis A407 transformed with the pAM401 vector alone maintained the baseline (MIC of 512 μg/ml) resistance phenotype (Table 1). The transformation with the phosphorylated VanR binding domain of the vanH promoter similarly resulted in a decrease in the vancomycin MIC to 64 μg/ml (Table 1). A control shuttle vector containing the mutant phosphorylated-VanR binding domain-deficient vanH promoter had no effect on vancomycin susceptibility (Table 1).
TABLE 1.
Agar dilution assay resultsa
| Strain | Vancomycin MIC (μg/ml) |
|---|---|
| E. faecalis A407-pAM401 | 512 |
| E. faecalis A407-pAM401-vanH promoter | 64 |
| E. faecalis A407–pAM401–phosphorylated-VanR binding domain of the vanH promoter | 64 |
| E. faecalis A407–pAM401–phosphorylated-VanR binding domain-deficient vanH promoter | 512 |
| E. faecalis A407-pAM401-vanH promoter-vanA antisense | 2 |
| E. faecalis A407-pAM401-vanH promoter-vanA sense | 256 |
Chloramphenicol at 10 μg/ml was added to the media.
In E. faecalis A407 electroporated with the pAM401-vanH promoter-vanA antisense, the vancomycin MIC was further reduced to a susceptible range, from 512 to 2 μg/ml (Table 1). In contrast the MIC for E. faecalis A407 transformed with the same construct but with the vanA gene in the sense orientation remained at or within 1 dilution of the baseline level. Synthesis of the vanH promoter-vanA antisense mRNA was confirmed by RT-PCR (data not shown).
Our study presents a model for reversing high-level vancomycin resistance with anti-drug resistance determinant gene transfer in enterococci. As a demonstration of the concept, antiresistance determinant genes were cloned into a shuttle vector plasmid and electrotransformed into the target vancomycin-resistant enterococci. However, several clinically applicable prokaryotic gene delivery modalities could be employed in future studies. These include enterococcal bacteriophage, conjugative plasmids-transposons, or modified or liposomally packaged oligonucleotides. Recombinant versions of these vectors could have potential use not only for the treatment of vancomycin-resistant enterococcus infections but additionally for altering the drug resistance phenotype of enterococci colonizing individual patients and contaminating endemic environments such as health care facilities and regional agricultural and livestock industries (16; A. E. van den Bogaard, L. B. Jensen, and E. E. Stobberingh, Letter, N. Engl. J. Med. 337:1558–1559, 1997; A. E. van den Bogaard, P. Mertens, N. H. London, and E. E. Stobberingh, Letter, J. Antimicrob. Chemother. 40:454–456, 1997).
The vanH promoter employed in the recombinant shuttle vectors is the same inducible enterococcal promoter that drives expression of the vanH, -A, and -X resistance determinants. When configured with a downstream vanA antisense gene, the resulting cassette inhibits class A glycopeptide resistance both by antisense RNA expression and by functioning as a phosphorylated-VanR transcriptional factor binding decoy (2), thereby attenuating up-regulation of native vanH, vanA, and vanX gene expression (Fig. 1). Reflective of such a dual mechanism, recombinant shuttle vectors containing the vanH promoter or the phosphorylated-VanR binding domain effected a partial restoration of vancomycin susceptibility. In contrast, no effect on resistance was noted with the introduction of a mutant vanH promoter with a deletion of this domain. Full restoration of vancomycin susceptibility resulted from the introduction of a vector containing both the vanH promoter and the vanA antisense gene.
Preliminary studies have examined the utility of gene transfer in the prokaryotic realm (9, 10, 21 ). One of the studies used antisense oligonucleotides targeting the multiple antibiotic resistance operon in Escherichia coli (21). Although the potential of gene transfer as an adjuvant for the treatment of multidrug resistant enterococcal infections has yet to be exploited, the present study provides preliminary support for such an approach.
Acknowledgments
The first two authors contributed equally to this work.
This research was funded in part by National Institute of Allergy and Infectious Diseases grant K08-AI01518. S.T. was supported by the Clinical Investigator Training Program of the Harvard-MIT Division of Health Sciences and Technology and Pfizer Inc.
REFERENCES
- 1.Arthur M, Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993;37:1563–1571. doi: 10.1128/aac.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Depardieu F, Courvalin P. Regulated interactions between partner and non-partner sensors and response regulators that control glycopeptide resistance gene expression in enterococci. Microbiology. 1999;145:1849–1858. doi: 10.1099/13500872-145-8-1849. [DOI] [PubMed] [Google Scholar]
- 3.Arthur M, Depardieu F, Molinas C, Reynolds P, Courvalin P. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene. 1995;154:87–92. doi: 10.1016/0378-1119(94)00851-i. [DOI] [PubMed] [Google Scholar]
- 4.Arthur M, Molinas C, Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992;174:2582–2591. doi: 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bugg T D, Dutka-Malen S, Arthur M, Courvalin P, Walsh C T. Identification of vancomycin resistance protein VanA as a d-alanine:d-alanine ligase of altered substrate specificity. Biochemistry. 1991;30:2017–2021. doi: 10.1021/bi00222a002. [DOI] [PubMed] [Google Scholar]
- 7.Eliopoulos G M, Wennersten C B, Gold H S, Schulin T, Souli M, Farris M G, Cerwinka S, Nadler H L, Dowzicky M, Talbot G H, Moellering R C., Jr Characterization of vancomycin-resistant Enterococcus faecium isolates from the United States and their susceptibility in vitro to dalfopristin-quinupristin. Antimicrob Agents Chemother. 1998;42:1088–1092. doi: 10.1128/aac.42.5.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friesenegger A, Fiedler S, Devriese L A, Wirth R. Genetic transformation of various species of Enterococcus by electroporation. FEMS Microbiol Lett. 1991;63:323–327. doi: 10.1016/0378-1097(91)90106-k. [DOI] [PubMed] [Google Scholar]
- 9.Good L, Nielsen P E. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat Biotechnol. 1998;16:355–358. doi: 10.1038/nbt0498-355. [DOI] [PubMed] [Google Scholar]
- 10.Guerrier-Takada C, Salavati R, Altman S. Phenotypic conversion of drug-resistant bacteria to drug sensitivity. Proc Natl Acad Sci USA. 1997;94:8468–8472. doi: 10.1073/pnas.94.16.8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holman T R, Wu Z, Wanner B L, Walsh C T. Identification of the DNA-binding site for the phosphorylated VanR protein required for vancomycin resistance in Enterococcus faecium. Biochemistry. 1994;33:4625–4631. doi: 10.1021/bi00181a024. [DOI] [PubMed] [Google Scholar]
- 12.Inouye R, Gillis J, Hammer S M. Combination genetic-pharmacological therapy for the treatment of drug-resistant and susceptible HIV-1. Antivir Ther. 1999;4(Suppl. 1):121. [Google Scholar]
- 13.Mercola D, Cohen J S. Antisense approaches to cancer gene therapy. Cancer Gene Ther. 1995;2:47–59. [PubMed] [Google Scholar]
- 14.Murray B E. Vancomycin-resistant enterococcal infections. N Engl J Med. 2000;342:710–721. doi: 10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 16.Robredo B, Singh K V, Baquero F, Murray B E, Torres C. Vancomycin-resistant enterococci isolated from animals and food. Int J Food Microbiol. 2000;54:197–204. doi: 10.1016/s0168-1605(99)00195-6. [DOI] [PubMed] [Google Scholar]
- 17.Rose R E. The nucleotide sequence of pACYC184. Nucleic Acids Res. 1988;16:355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Walsh C T. Enzymes in the d-alanine branch of bacterial cell wall peptidoglycan assembly. J Biol Chem. 1989;264:2393–2396. [PubMed] [Google Scholar]
- 20.Weiss B, Davidkova G, Zhou L W. Antisense RNA gene therapy for studying and modulating biological processes. Cell Mol Life Sci. 1999;55:334–358. doi: 10.1007/s000180050296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White D G, Maneewannakul K, von Hofe E, Zillman M, Eisenberg W, Field A K, Levy S B. Inhibition of the multiple antibiotic resistance (mar) operon in Escherichia coli by antisense DNA analogs. Antimicrob Agents Chemother. 1997;41:2699–2704. doi: 10.1128/aac.41.12.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wirth R, An F Y, Clewell D B. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli- S. faecalis shuttle vector. J Bacteriol. 1986;165:831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]