ABSTRACT
The ongoing coronavirus disease 2019 (COVID-19) pandemic demonstrates the threat posed by novel coronaviruses to human health. Coronaviruses share a highly conserved cell entry mechanism mediated by the spike protein, the sole product of the S gene. The structural dynamics by which the spike protein orchestrates infection illuminate how antibodies neutralize virions and how S mutations contribute to viral fitness. Here, we review the process by which spike engages its proteinaceous receptor, angiotensin converting enzyme 2 (ACE2), and how host proteases prime and subsequently enable efficient membrane fusion between virions and target cells. We highlight mutations common among severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern and discuss implications for cell entry. Ultimately, we provide a model by which sarbecoviruses are activated for fusion competency and offer a framework for understanding the interplay between humoral immunity and the molecular evolution of the SARS-CoV-2 Spike. In particular, we emphasize the relevance of the Canyon Hypothesis (M. G. Rossmann, J Biol Chem 264:14587–14590, 1989) for understanding evolutionary trajectories of viral entry proteins during sustained intraspecies transmission of a novel viral pathogen.
KEYWORDS: SARS-CoV-2, adaptive mutations, coronavirus, COVID-19, evolution, genomics, glycoproteins, infectious disease, respiratory viruses, sarbecovirus, spike
INTRODUCTION
The ongoing coronavirus disease 2019 (COVID-19) pandemic is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel betacoronavirus in the subgenus Sarbecovirus. Since its emergence in late 2019 in Wuhan, China, the virus has caused at least 361 million confirmed infections and 5.62 million recorded deaths (1), underscoring the threat posed by coronaviruses to human and animal health (2, 3). Despite the four human coronaviruses, HKU1, 229E, NL63, and OC43, being traditionally associated with mild “common cold” illnesses (4), the 21st century has revealed the considerable threat posed by preemergent coronaviruses that circulate in various bat species.
In 2002, a sarbecovirus that ordinarily circulates in horseshoe bats (genus Rhinolophus) spilled into humans from masked palm civets (Paguma larvata) that are commonly sold in animal markets in Guangdong, China (5), causing an epidemic of viral pneumonia. By the time the epidemic was contained in 2003, the virus, now known as the original severe acute respiratory syndrome coronavirus (SARS-CoV; here SARS-CoV-1), had spread to 26 countries on five continents, infecting over 8,000 people and causing 774 deaths (6, 7). The case fatality rate of nearly 10% and apparent pandemic potential of SARS-CoV-1 provided an impetus to investigate wildlife for preemergent SARS-like coronaviruses and to revisit the pathogenic potential of the four human coronaviruses (hCoVs), which previously were assumed to cause only mild respiratory disease. It is now appreciated that seasonal hCoVs, such as OC43 and NL63, routinely cause outbreaks in long-term care facilities with high attack and mortality rates in the elderly and immunocompromised (8, 9), and a wide range of hCoVs have been identified as the causative agent in children hospitalized with pneumonia (10). Moreover, another betacoronavirus, the Middle East respiratory syndrome coronavirus (MERS-CoV), has spilled over into humans several times from camels. Since its identification in 2012, MERS has infected more than 2,500 people and caused at least 886 deaths, a case fatality rate of roughly 34% (11, 12).
Although the threat of coronaviruses to human health was not widely appreciated prior to the 2002–2003 SARS (SARS-CoV-1) epidemic, the threat these viruses pose to livestock has long been apparent. For example, the porcine epidemic diarrhea virus (PEDV) was first identified in Europe in the 1970s and has long been endemic to China (13, 14). However, in 2013, PEDV swept through the United States, causing a fatality rate of nearly 100% in piglets and nearly decimating the domestic pig population (15). The PEDV epizootic spurred development of an alphavirus RNA particle vaccine against porcine epidemic diarrhea virus (16, 17), which has been viewed as a precursor of contemporary mRNA vaccines (18). In 2016, the novel swine acute diarrhea syndrome coronavirus (SADS-CoV) killed ∼24,000 piglets across four farms in Guangdong province, China (19). Feline coronavirus (FCoV) is capable of causing disease in wild and domestic cats (20). Due to mutation and their ease of recombination (14, 21–24), coronaviruses are efficient explorers of host species and cell tropism. Therefore, coronaviruses are enduring and pervasive threats. Understanding their virology and evolution is essential to mitigating the ongoing SARS-CoV-2 pandemic as well as for establishing a toolkit to more effectively respond to future coronavirus epidemics.
CORONAVIRUS ENTRY
The mechanistic details of coronavirus entry have been covered elsewhere (3, 25–27). Our focus here will be on how the prefusion conformation of S engages with host cell receptors and undergoes proteolytic processing to reach a fusion-competent state. Like all class I viral membrane fusion proteins, S polypeptides assemble into homotrimers, with each subunit made up of two domains. The first subunit (S1) binds host receptors (e.g., ACE2 for SARS-CoV-1, SARS-CoV-2, and NL63) and in essence serves as a chaperone for the S2 subunit, which contains the spring-loaded machinery that executes membrane fusion. In the absence of S1, S2 rapidly and irreversibly transitions to its postfusion conformation. Many, but not all, coronaviruses encode a protease cleavage site at the S1/S2 boundary; this position, fittingly, is often referred to as S1/S2 (28). Cleavage at S1/S2 increases spike flexibility and has been suggested to promote a rapid mode of entry where fusion occurs at the cell surface (29). In the case of SARS-CoV-2 S, the S1/S2 motif enables preprocessing by the proprotein convertase furin (28, 30, 31). This feature, also known as the furin cleavage site (amino acids [aa] 681 to 685 in SARS-CoV-2 S), has been suggested to be crucial to the pandemic spread of SARS-CoV-2 (32) and has been an important locus of adaptive evolution in the SARS-CoV-2 S gene (33–35). Additionally, all coronaviruses have a second cleavage site within the S2 subunit called S2′ (28). Cleavage of this second site is strictly required for cell entry as it exposes the hydrophobic fusion peptide that anchors in the host cell membrane during the fusion process (36–38).
In coronaviruses such as SARS-CoV-2 that contain furin cleavage sites at the S1/S2 boundary, processing at this site is thought to occur primarily within the Golgi apparatus during maturation of newly synthesized S trimers. Such preprocessing at S1/S2 is thought to prime S for efficient membrane fusion at target cells, likely by promoting open conformations that facilitate interactions with receptors and enhancing cleavage at S2′ by cell surface proteases such as TMPRSS2 (30, 39). Without an S1/S2 furin cleavage motif, S1/S2 and S2′ sites both must be processed during target cell entry (Fig. 1A). This can be inefficient and restricts the virus to the slower endocytic entry route, in which the virus relies on pH-dependent cathepsins and may be more susceptible to restriction by certain innate immune factors, e.g., interferon-stimulated genes (40, 41) such as IFITMs, which rigidify cell membranes to prevent fusion (42, 43). Proprotein convertase (furin) preprocessing at the S1/S2 site leads to a more labile and fusogenic, but less stable, spike protein (32). The receptor binding domain (RBD) more readily binds ACE2 and the S2′ site can be efficiently cleaved by surface proteases like TMPRSS2 (44), facilitating direct entry and fast growth kinetics in respiratory epithelial cells (26, 28).
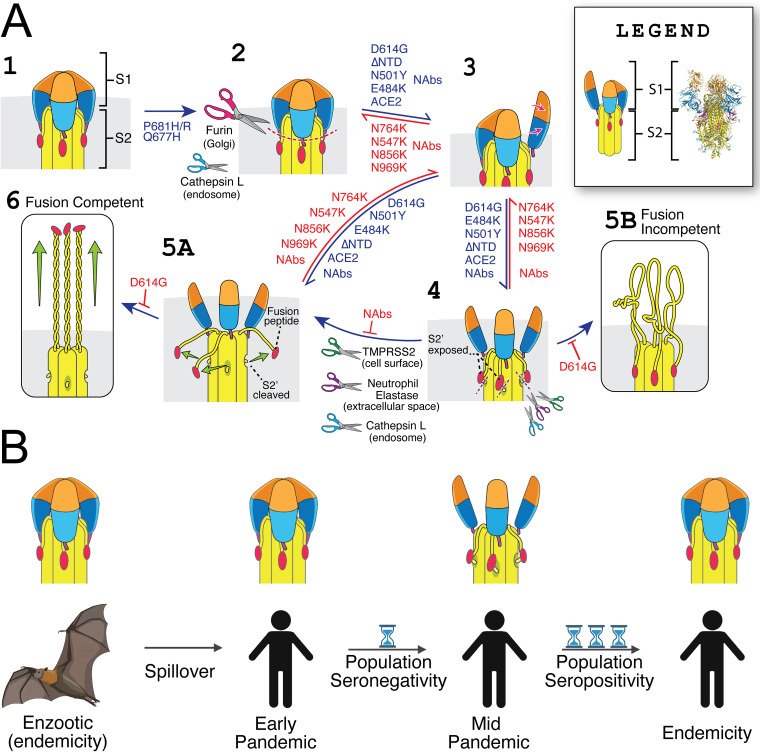
FIG 1.
Structural and evolutionary dynamics of the SARS-CoV-2 Spike protein. (A) Model for how sarbecovirus Spike proteins achieve fusion competency and effects of common mutations. (Stage 1) An uncleaved S protein trimer. (Stage 2) Cleavage at the S1/S2 site can occur within the Golgi apparatus of an infected cell during the production of viral progeny but strictly speaking is not required for infectivity. This proteolytic processing at S1/S2 is made more efficient by a substitution such as P681H/R or Q677H. Alternatively, processing at S1/S2 could occur subsequent to egress of viral progeny, e.g., during endosomal entry. S1/S2 cleavage destabilizes the prefusion conformer, which promotes opening of RBD and the transition to stages 3 to 6. The D614G substitution, NTD loop deletions, and RBD mutations such as N501Y and E484K likewise increase RBD opening, which promotes binding to ACE2. When the three subunits of an S homotrimer simultaneously adopt the RBD open conformation, a state that is stabilized by receptor (ACE2) binding, the S2 subunit adopts increased flexibility, exposing the S2′ site for cleavage by host proteases (e.g., TMPRSS2, neutrophil elastase, and cathepsin L) (4). However, should S1 dissociate prior to S2′ cleavage (stage 5B), the S2 subunit prematurely transitions to its postfusion conformation, which is irreversible and tantamount to a noninfectious dead end. Alternatively, when S2′ cleavage occurs prior to dissociation of the S1 subunit (stage 5A), S is fully activated and competent to mediate fusion (stage 6). Therefore, stage 4 likely represents an unstable, transient state wherein S protomers can achieve fusion competency. Importantly, stage 6 must occur in close proximity to a target membrane (e.g., a host cell phospholipid bilayer) in order to achieve fusion. By decreasing the rate of S1 dissociation, the D614G substitution limits the occurrence of misfiring events (stage 5B), making fusion more efficient and offsetting stability costs of mutations that increase preprocessing at S1/S2 and/or enhance sampling of RBD open states. Neutralizing antibodies (NAbs) can impact viral entry in many different ways, depending on where they bind and how they affect S protein structure. The legend inlay indicates side-by-side PyMol rendering of a cryo-EM structure of S (PDB entry 6VYB) (30) and its cartoon interpretation; coloring is harmonized across domains. (B) The Canyon Hypothesis applied to zoonotic spillover. During circulation in populations with high rates of humoral immunity, viral entry proteins favor predominantly closed RBD configurations (112). Immediately after spillover into a population that lacks immunity, the newly emergent virus remains closely related to its ancestor and, hence, favors closed RBD configurations. During sustained transmission between seronegative individuals, large viral population sizes and wide transmission bottlenecks facilitate rapid emergence of variants that favor open RBD configurations to spread rapidly between hosts. Over time, the evolutionary entanglement between viral entry proteins and humoral immunity gradually leads to a return to closed RBDs as repeat exposures facilitate the affinity maturation of expansive antibody repertoires that are disproportionately costly to open RBD configurations. Panel B was generated using biorender.com.
STRUCTURAL DETERMINANTS OF VIRAL ENTRY
Due to the unidirectional conformational transition characteristic of class I viral fusion proteins, they have evolved sophisticated coincidence detection mechanisms to prevent misfiring. Influenza hemagglutinin, for example, combines proteolytic cleavage with endosomal pH reductions to destabilize the interface between its receptor engaging chaperone and fusogenic subunits (45). In contrast, SARS-CoV-2, and perhaps all coronavirus S proteins, may rely on 3 RBD simultaneously adopting the open or up conformation (3 RBD open or 3 RBD up). Cleavage at S1/S2, at least in some lineages, facilitates transition to the 3 RBD up state and may further destabilize the interface between the S1 (chaperone) and S2 (fusogen) subunits (e.g., Fig. 1A, stage 4). However, certain alphacoronaviruses lack an S1/S2 site entirely (28), and the SARS-CoV-2 spike protein can transition into the postfusion conformation in the complete absence of ACE2 and without proteolytic cleavage of either its S1/S2 or S2′ site (46). Despite this, S2′ cleavage usually occurs after S1/S2 cleavage and receptor binding (47).
Structural, computational, and biochemical evidence help clarify these seemingly conflicting observations. First, the 3 RBD open state was observed only with the inclusion of the D614G mutation (48), which simultaneously promoted opening while stabilizing the prefusion conformation (49). Molecular docking simulations between TMPRSS2 and a prefusion stabilized spike protein, in which only one of the 3 RBDs is oriented upward (e.g., Fig. 1A, stage 3), can align the S2′ loop with the protease active site (50), but R815 of the S2′ loop is not accessible because in this structure it is engaged in interactions with D820 and F823 (30). However, binding to ACE2 stabilizes Spike in the 3 RBD open conformation, causing R815 to become much more exposed (49) and therefore accessible to proteases. Moreover, D614G synergizes with N-terminal domain (NTD) loop deletions to enable ACE2-independent S2′ cleavage (51). Therefore, S1/S2 cleavage, RBD opening, S2′ processing, and the postfusion conformational transition are not steps that must occur in fixed linear order. Rather, these events are allosterically regulated through the stochastic mechanism of spike stability. The 3 RBD open conformation likely represents a transient state in which the S2′ loop is sufficiently disordered to be cleaved by host proteases. Independent of S2′ cleavage, this transient state is exited when the S1 subunit dissociates and the spike undergoes an irreversible transition to the postfusion conformer or when one or more RBDs fall back down into the closed conformation (Fig. 1A, stages 5A and 6). While the transition to the postfusion conformation can occur without S2′ cleavage (46), proteolytic processing is necessary for a fusion-competent conformational transition. Receptor binding appears to stabilize RBDs in the open conformation, promoting S2′ cleavage and the transition to the postfusion state independently.
Notably, TMPRSS2 is not the only protease capable of releasing the SARS-CoV-2 S fusion peptide. For instance, neutrophil elastase (NE) is also capable of cleaving the S2′ site (52), and an elevated neutrophil/lymphocyte ratio (NLR) during early illness clinically correlates with development of severe disease (53). Although elastase release might only modestly increase cell entry in the upper respiratory tract, due to its high TMPRSS2 expression (54), it may more markedly enhance infection of cells that express ACE2 but not TMPRSS2 (55). Therefore, elevated NE levels may be a common feature of severe COVID-19 due to an imbalanced immune response that enhances S2′ cleavage within extracellular spaces. This phenomenon might promote intrahost spread, particularly in tissues with low TMPRSS2 expression. Moreover, neuropilin-1 (56) and SR-B1 (57) improve the efficiency of ACE2-dependent entry, while soluble ACE2 enables endosomal entry in ACE2 and TMPRSS2-deficient cells through the renin-angiotensin system (58). One might even speculate that extracellular NE and soluble ACE2 could facilitate surface entry into ACE2- and TMPRSS2-deficient cells.
Thus, it would appear that the pre-Omicron variants selected during the COVID-19 pandemic combine S mutations that increase RBD opening and overall lability with stabilizing mutations that decrease S1 dissociation. This increases sampling of and tolerance for the 3 RBD open conformation in which the S2′ site can be cleaved, which is necessary to achieve a fusion-competent state before the S1 subunit dissociates and the S2 subunit is committed to its irreversible conformational transition. This facilitates early entry and rapid growth kinetics, promoting both intrahost replication and interhost transmission.
HOW D614G ENABLED FUSOGENICITY-ENHANCING SPIKE MUTATIONS
During the first 2 years of the pandemic, we have seen the steady emergence of highly fit SARS-CoV-2 variants. Each of these variants contains a constellation of mutations, and their ultimate phenotype is determined by the epistatic interactions between these mutations. Over the pandemic, the evolution of the spike protein has undergone a series of selective phases (59). While the ancestral SARS-CoV-2 virus likely had an alanine at the 372 position of its spike protein, during the early stages of the pandemic (likely before its detection and widespread sequencing) the virus acquired the A372T mutation (60). This mutation was an early adaptation to humans that increased ACE2 affinity (60). Once the virus was circulating and differentiating into clades, the virus underwent a second selective sweep. First lineage B and then lineage A acquired the D614G mutation. This mutation did not have immediately apparent phenotypic effects beyond modestly improved ACE2 affinity, greater thermal stability, and only slightly improved transmissibility (49, 61–66). Afterwards, a host of increasingly fit variants rapidly emerged and displaced each other (67–70) (Fig. 2) (reviewed in reference 71). As each variant’s overall fitness is a complex function of many variables, we will minimize discussion of particular lineages and instead prioritize the general principles governing their evolution.
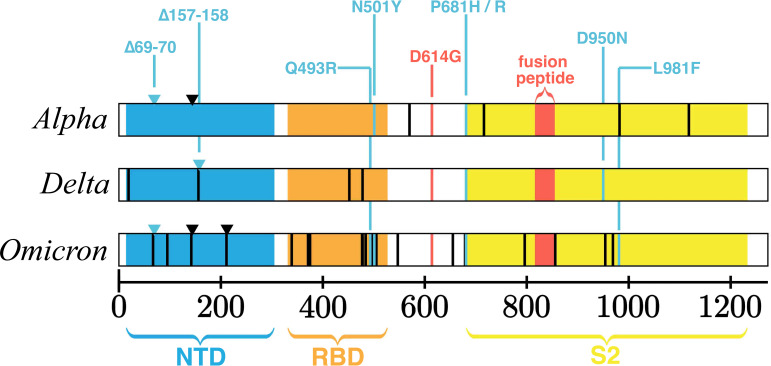
FIG 2.
SARS-CoV-2 S domain structure and characteristic mutations of variants of concern. The SARS-CoV-2 spike polypeptide labeled by its domains and annotated for amino acid substitutions and deletions with 70% or higher prevalence in GISAID data for three prominent variants of concern, Alpha (B.1.1.7 + Q.x), Delta (B.1.617.2 + AY.x), and Omicron (B.1.1.529 + BA.x), as tabulated by outbreak.info using 12 January 2022 data.
Although the combinatorial interactions are quite complex, the evolution of the SARS-CoV-2 spike protein highlights a relatively straightforward theme: tradeoffs between stability and fusogenicity (72, 73). The combinatorial interactions contribute to the exquisite tuning of spike conformational transitions to suit the environments encountered during infection and transmission. The D614G mutation will receive distinct attention due to its epistatic significance (59). Other mutations, while also important, will be discussed in groups. Most of our discussion will focus on how various mutations influence spike structural dynamics. These changes then have consequences for infectivity and transmission, processes involving viral particles in aggregate. For instance, a variant with a less fusogenic spike can be more transmissible despite having lower titers if it is unlikely to misfire before reaching a new host. Meanwhile, a variant with a more fusogenic spike can be more transmissible if it overcomes the drawbacks of instability through rapid growth kinetics, high titers, and mass action.
Initial studies of the S:D614G spike focused on potentially improved ACE2 affinity (66). However, cryogenic electron microscopy (cryo-EM) experiments revealed that its RBDs were more likely to hold the open conformation, because the S:D614G mutation disrupted an interaction between D614 in the C-terminal domain 2 (CTD2) of the S1 subunit and fusion peptide proximal region (FPPR) of the S2 subunit. Moreover, D614G enabled the first observation of sarbecovirus S trimers with 3 RBDs simultaneously in the open conformation (48). While this finding shed light on how D614G improved ACE2 affinity and made S more susceptible to neutralization, it did not explain why D614G particles were less likely to spontaneously transition to the postfusion conformation (51, 61). Additional cryo-EM studies demonstrated that D614G also caused structural rearrangements near the interface of the Spike protomers. The mutation rigidified the nearby 630 loop of CTD2, allowing it to wedge between the NTD and C-terminal domain 1 (CTD1). This rearrangement of the 630 loop likely strengthened engagement between the S1 and S2 subunits through additional hydrophobic interactions. By slowing overall kinetics, D614G simultaneously slowed firing and increased its tolerance of the 3 RBD open state critical for fusion-enabling S2′ cleavage (49). Thus, the overstabilized D614G spike protein was able to tolerate fusogenicity-enhancing mutations that otherwise would have impaired transmission due to instability-induced misfiring (51).
Arguably the most important mutational hot spot for its multilayered roles in S stability, fusogenicity, transmissibility, and the overall emergence of the COVID-19 pandemic is the S1/S2 furin cleavage site (FCS) (reviewed in reference 74). Because early SARS-CoV-2 isolates were inefficiently processed at S1/S2 (75), an improved FCS would result in viral particles whose spike proteins were more labile and whose RBDs sample the open state more frequently. Although other mutations near the S1/S2 site have been observed (76), the most successful lineages have converged on P681H or P681R (33, 34, 77, 78). Although mutations at the S:677 position may modestly affect furin processing (79), position 681 is the P5 site of the cleavage motif. As furin prefers a basic residue at this site (44), replacement with arginine confers the greatest increase in cleavage (78, 80). However, the transition to the postfusion conformation is irreversible, and multiple simultaneous open RBDs are inherently unstable (48). Thus, S1/S2 sites do not affect fitness in isolation, and there is probably significance to the observation that variants of concern (VOC) differ in their S1/S2 cleavage efficiencies (34).
The hypervariable region of the NTD is another hot spot in which many variants feature deletions and other mutations (81–83). While many studies have addressed their evasion of antibodies given their frequent emergence in immunocompromised patients (83–89), a recent study using SARS-CoV-2 virus-like particles sheds tremendous light on allosteric regulation and epistatic interactions within the spike protein (51). SARS-CoV-1 lacks the S1/S2 furin site of SARS-CoV-2 but possesses deletions in all 3 NTD loops, whereas SARS-CoV-2 variants of concern only possess deletions in 1 or 2 of these loops. By swapping the variable portions of the S1 NTD between SARS-CoV-1, SARS-CoV-2, and SARS-CoV-2 carrying S:D614G, the authors made several key observations. While the spike proteins of SARS-CoV-1 and SARS-CoV-2 showed similar fusogenicity to each other, deletions in the SARS-CoV-2 spike’s NTD loops significantly reduced its fusogenicity. However, the SARS-CoV-2 spike carrying D614G became more fusogenic with NTD loop deletions. Therefore, NTD loop deletions can increase S fusogenicity, but only in the context of the D614G substitution.
The next set of experiments involved centrifuging virus-like particles through sucrose solutions, followed by assessing fusogenicity. Postcentrifugation fusion assays showed that the SARS-CoV-2 D614G spike, which was formerly poorly fusogenic, was now the most fusogenic. However, when NTD deletions were included, the D614G SARS-CoV-2 spike became less fusogenic than the ancestral D614 SARS-CoV-2 spike, i.e., that encoded by hCoV-19/Wuhan/WIV04/2019 (WIV04). Thus, because mutations that increase fusogenicity often decrease stability, a more fusogenic spike protein often results in a less infectious viral particle through irreversible instability-induced misfiring (Fig. 1A).
Given this context, mutations in the RBD are not straightforward. Increased ACE2 binding can be through stronger interactions with the receptor or enhanced RBD opening (49). Similar to D614G and NTD deletions, RBD mutations have important consequences for spike stability. While mutations such as E484K (90–92), N501Y (90), L452R (67), and many others can promote ACE2 binding, they often partially mediate this through local destabilizing effects (91) that allosterically regulate RBD opening via interactions within the S1 subunit. Indeed, a structural remodeling of the RBD that establishes additional salt bridges with ACE2 is one of the distinguishing features of the Omicron variant (93, 94).
Furthermore, there are other convergent mutations within S as well as in other viral genes that likely impact fitness but have not yet been thoroughly characterized. For example, multiple SARS-CoV-2 variants carry profusogenic deletions in their cytoplasmic tails (95). Meanwhile, D950N, a signature mutation of the Delta (B.1.617.2 + AY.x) lineage, which is also found in Mu and a number of other variants (35), lies within the HR1 domain (96). Such convergence is a strong indicator of functional relevance and in this case remains largely unexplored. Further, although S protein stability and fusogenicity is an important axis, the S gene is not the only part of the genome that affects viral fitness (97). Mutations in the nucleocapsid gene often appear in variants of concern and can contribute to fitness (98, 99), as can those in the putative viroporin encoded by ORF3A (100), ORF8A (101), and NSP6 (102, 103), among other genes (104). Moreover, mutations in different genes are capable of epistatic interaction (59). Therefore, each variant exists as a complex and multifaceted configuration.
ARE HIGHLY OPEN RECEPTOR BINDING DOMAINS AN ADAPTATION TO ZOONOTIC SPILLOVER?
Despite the transmissibility benefits of a stable S protein that tolerates efficient proteolytic processing and predominantly open RBDs, spike proteins of endemic coronaviruses typically favor closed RBDs (105–108). Although SARS-CoV-2 spike variants with more open RBDs showed strong transmission advantages early during the pandemic, the same characteristics that promote receptor binding and rapid entry also increase susceptibility to neutralizing antibodies (48).
Along these lines, the emergence of the S:D614G mutation early during the pandemic set the stage for even more infectious SARS-CoV-2 variants. The D614G S exhibits decreased S1 dissociation after priming (S1/S2 cleavage), which in large part negates the otherwise high fitness costs of frequently sampling the 3 RBD open state necessary for S2′ cleavage. Consequently, the D614G change enabled selection for additional S mutations observed in many variants, with many exhibiting convergent evolution, such as N501Y, E484K, NTD deletions, and P681H/R. Hence, the emergence of D614G provided an epistatic shift in the fitness landscape by increasing the S protein’s tolerance for the 3 RBD open state (51). Although the ancestral Spike encoded by the December 2019 SARS-CoV-2 genome, i.e., WIV04, was sufficiently stable and fusogenic to enable respiratory transmission (32, 72, 109), more efficient proteolytic preprocessing at S1/S2 or a greater propensity toward open RBDs may have been deleterious in this context. In particular, we expect that alteration of proline 681 to either histidine or arginine, which is posited to be critical for the enhanced transmissibility of variant of concern lineages (33–35), would cause instability-induced misfiring in the context of the Dec 2019 WIV04 Spike. In other words, more efficient preprocessing by furin at S1/S2 may confer fitness advantages only when the S2 fusogen is stabilized by mutations found in VOCs (which occur primarily within the S1 subunit).
Moreover, the NTD is an antigenic supersite that is frequently targeted by neutralizing antibodies during primary infection. The structural remodeling induced by NTD loop deletions appears to simultaneously avoid binding of certain classes of NTD-directed antibodies (84, 86, 94) and promote RBD opening (51). Deep sequencing and mathematical modeling suggest that, due to a temporal mismatch between viral replication and antibody production, antigenic evolution can be nearly neutral within the host but highly adaptive at the level of circulating variants (110, 111). Therefore, it seems plausible that a similar process could have played out in the early stages of the SARS-CoV-2 pandemic, in which nearly neutral within-host antigenic evolution to escape NTD and RBD down-directed antibodies facilitated the emergence of variants with NTD loop deletions and a preference for open RBD states. Such variants were even more transmissible at the population level due to rapid growth kinetics in the absence of prior humoral immunity (Fig. 1B).
Overall, the Spike-mediated determinants of coronavirus transmissibility appear to involve a combination of S protein stability, proteolytic processing, propensity for sampling RBD open states necessary to interact with the host receptor (112), the strength of the interaction when such contact occurs, and additional factors, such as the fusion peptide’s capacity for membrane ordering (27, 37). Moreover, we hypothesize that the level of fitness conferred by these features is strongly impacted by the immune status of the host population. S proteins that are efficiently cleaved and frequently sample open RBD states show efficient receptor engagement and S2′ processing. Consequently, variants encoding S proteins with such features spread rapidly within and between seronegative individuals. However, antibodies present in recovered and vaccinated individuals compete with receptors and proteases for these epitopes and can even induce S1 dissociation and misfiring (46). Indeed, it has long been hypothesized that humoral immunity prevents selection of the viral entry protein configurations that would otherwise be the most transmissible in an antibody-deficient population (113).
CONCLUSIONS
Although the continued emergence of additional SARS-CoV-2 S variants is all but inevitable, it is difficult to predict precisely what changes will occur or to confidently assign time scales. Most changes in S prior to the emergence of the Omicron variant appear to have been driven by selection for improved transmission between immunologically naive hosts. However, this adaptive force should gradually weaken as reservoirs of seronegative individuals are depleted and the virus reaches a state where most transmission is driven by reinfection mediated by antigenic drift and naive infections are limited to young children.
Recent cryo-EM studies of the Omicron S protein suggest that while the Delta Spike predominantly occupies conformations with 1 or more RBDs open simultaneously, the Omicron Spike appears to prefer conformations with 0 or 1 open RBD (94, 114). Additionally, cryo-EM of the Omicron Spike with ACE2 or the S309 antibody occupied only the 1 or 2 open RBD states (93). This contrasts with the D614G Spike, which was also observed in the 3 RBD open state. The increased preference for the closed RBD state in Omicron is partially explained by the introduction of additional electrostatic contacts between the S1 and S2 subunits due to the mutations N764K, T547K, N856K, and N969K. These interactions support a structural basis for the Omicron variant’s increased preference for closed RBDs and decreased S1 shedding (93, 114). Furthermore, these mutations may compensate for the presence of NTD deletions that promote RBD opening and Spike destabilization. Intriguingly, the Omicron variant appears to exhibit less efficient S1/S2 furin cleavage despite the presence of two mutations, P681H and N679K, that individually increase preprocessing at S1/S2 (115, 116). However, a decrease in RBD opening and an increase in overall stability is consistent with impaired S1/S2 cleavage. Further, the L981F mutation appears to improve hydrophobic packing of the S2 subunit, which would likely also contribute to overall stability (93). In contrast, the mutations Q493R, G396S, and Q498R appear to introduce two new salt bridges and one additional hydrogen bond with ACE2 (93, 114). Curiously, the Omicron variant also appears to demonstrate reduced sensitivity to IFITMs (115), which may indicate changes in the function of its fusion peptide, perhaps involving the nearby N856K mutation. Thus, while the Omicron Spike is less likely to occupy the open RBD states necessary for ACE2 engagement, it may compensate by binding the receptor more strongly when such an interaction occurs and by having adapted to resist innate defenses. Such changes would be consistent with the broad pattern of host adaptation suggested by the presence of additional convergent mutations that were observed in prior variants (59, 103).
Further, if the S2′ loop is only accessible for cleavage when the S2 subunit is destabilized, such as in the 3 RBD open state (Fig. 1A), then Omicron’s poor sampling of this conformation would predict a preference for endosomal entry, consistent with its reported phenotype. Moreover, the endosomal pathway may be less reliant on RBD opening and ACE2 binding-mediated destabilization of the S2 subunit. As endosomes mature, they experience an influx of calcium and acidify (117, 118). Low pH does not appear to destabilize coronavirus spike proteins (119) as it does the influenza hemagglutinin (HA) protein (45). However, divalent cations can also destabilize HA (120) and have been long established to influence protein folding and biological membrane curvature by modulating phenomena such as salt bridges, cation-pi interactions, and the hydrophobic effect (121, 122). Lastly, coronavirus fusion peptides require calcium ions to efficiently order membranes (36, 37).
The overall pattern of mutation, structural remodeling, and reduction in binding of antibodies generated against prior Spike structural configurations is consistent with antigenic drift. While most antibodies only recognize RBDs in the up or down state, repeat exposure, either through vaccine booster or breakthrough infection, triggers a memory response and further affinity maturation. Successive rounds of affinity maturation appear to promote the generation and maintenance of broadly neutralizing antibodies, including those capable of recognizing Spike proteins in both the RBD up and RBD down conformations (123, 124).
Moreover, the antigenicity of stabilizing elements such as the SARS-CoV-2 NTD, and the tendency of primary immune responses to generate a relatively limited repertoire of antibodies, may help explain the selection for open RBDs early in the pandemic as well as the subsequent shift in the selective landscape that led to the Omicron variant’s emergence and rapid sweep (125). The Canyon Hypothesis predicts that animal viruses will encode entry proteins that favor closed RBDs at the time of spillover because such viruses have adapted to host humoral immunity, which selects against highly exposed RBDs (113). A particularly intriguing implication when applying this hypothesis to pandemic viruses is that spillover may temporarily free viral entry proteins from a trade-off between infectivity and immune evasion. Moreover, immune responses targeting closed RBD conformations in the early stages of a pandemic may even select for increasingly open entry proteins that enhance transmissibility in seronegative populations (Fig. 1B). Viruses encoding open RBD configurations may spread rapidly due to fast entry kinetics and broad cell tropism but will likely be disfavored over long periods of time due to their inherent instability and susceptibility to neutralization or misfiring.
These principles are also relevant to other preemergent coronaviruses. For example, while MERS has furin recognition motifs at both its S1/S2 and S2′ sites, only S1/S2 is preprocessed. The rapid mode of direct entry at the cell surface is largely restricted to cells that express TMPRSS2 (126). Similarly, introducing a furin motif at the S2′ position of PEDV was unable to mediate S2′ preprocessing or surface entry (127). Artificial overexpression of furin, however, has been observed to enable cell surface entry by MERS in TMPRSS2-deficient cells, albeit inefficiently (128). Given that the MERS S protein has not been observed in the 3 RBD open conformation and has a highly ordered S2′ loop in its 1- and 2-RBD open states (129), its inefficient S2′ preprocessing may be due to infrequent sampling of the fully open state as well as its inherent instability. However, if MERS were to achieve sustained human-human transmission, then improved sampling and tolerance of RBD open states, which the Canyon Hypothesis predicts would be advantageous in the absence of prior humoral immunity, may also improve cleavage at its otherwise cryptic S2′ furin motif. This development would be concerning, as such a configuration may show expanded cell tropism, faster growth, and enhanced cell-to-cell spread. Moreover, acquisition of this feature has been observed during serial in ovo passage of avian infectious bronchitis virus, a gammacoronavirus (130).
At least one report demonstrates the expanded tropism possible if the S2′ site is processed before reaching the target cell (131). In this case, a SARS-CoV-2 isolate developed several S mutations during serial passage in cultured Vero E6 cells, including a 9-amino-acid NTD deletion as well as E484D, D614G, Q954H, and P812R, the last of which introduces a furin motif at the S2′ site. The authors observed that purified viral particles were competent to initiate rapid growth with extensive syncytia, even in A549 cells, which are deficient for ACE2 and TMPRSS2 (132). This phenotype suggests furin-mediated preprocessing at both the S1/S2 and the S2′ positions, similar to what is seen for the respiratory syncytial virus fusion protein (F) (133, 134), and may further indicate that furin- or TMPRSS2-mediated S2′ cleavage requires frequent RBD opening.
In conclusion, the SARS-CoV-2 Spike protein appears to have followed a general trend of RBD opening and increasing fusogenicity in the early pandemic, with what may now be the beginning of a gradual return to closed RBDs. Population immunity will likely be established as more and more individuals experience vaccination and serial exposures to divergent Spike variants through infection. Iterative rounds of affinity maturation appear to facilitate establishment of expansive antibody repertoires, including broadly neutralizing antibodies capable of recognizing diverse RBDs in both the up (open) and down (closed) conformations (123, 135). These antibodies impose disproportionate fitness costs on open RBDs, leading to an eventual long-term preference for Spike proteins that are stabilized in RBD down (or closed) states (108).
Given the antigenic evolution of seasonal human coronaviruses (136) and the intensity of the Omicron wave, we expect future variants to emerge with reduced sensitivity to the suite of neutralizing antibodies it most commonly elicits. Although Omicron appears to favor the RBD closed state, this does not guarantee that all subsequent variants will follow suit. Fitness landscapes are complex, and evolution is strongly influenced by viral population size (137, 138). Nonetheless, the general trend of RBD opening early in a pandemic, followed by their gradual closing, may help explain the observation that acute respiratory virus pandemics involve escalating waves in the first few years, followed by several years of elevated levels of illness (139–141), which presumably occur in cyclic fashion as population-level immunity is gradually established (Fig. 1B). The complex interactions between viruses, their entry proteins, and the immune systems of the hosts they infect exemplify the stochastic, yet path-dependent, nature of evolution (142–144). These dynamics, coupled with the inherently global nature of pandemics, underscore the importance of swift vaccination in response to emerging viral diseases, which in turn entails equitable and decentralized approaches for viral genomic surveillance and vaccine production. Vaccination, unlike infection-acquired immunity, affords a route to population immunity without viral replication or antigenic evolution (145, 146).
ACKNOWLEDGMENTS
We are grateful to Sally Griffith-Oh for generously providing expert graphic design assistance with Fig. 1. We also thank Dylan H. Morris (University of California, Los Angeles, Los Angeles, CA), Tom Gallagher (Loyola University Chicago, Maywood, IL), and Jasnah Kholin (@wanderer_jasnah), an anonymous coronavirus researcher in Hong Kong, for their helpful comments and discussion. We also apologize to the many colleagues whose important contributions we inadvertently failed to cite.
We gratefully acknowledge the funding agencies that supported this work. J.C.K. and K.A.W were supported by the National Science Foundation, award no. 1845890. J.P.K. was supported by the National Institutes of Health, grants no. P20GM121307-04S1 and P20GM134974; The Rockefeller Foundation, grant no. 2021 HTH 010; and a COVID-19 Fast Grant (no. 2239) from Emergent Ventures at the Mercatus Center at George Mason University.
Footnotes
[This article was published on 8 March 2022 with errors in Acknowledgments. The Acknowledgments were updated in the current version, posted on 4 April 2022.]
Contributor Information
Kyle A. Wolf, Email: kawolf2@wisc.edu.
Jeremy P. Kamil, Email: jeremy.kamil@lsuhs.edu.
Vinayaka R. Prasad, Albert Einstein College of Medicine
REFERENCES
- 1.Dong E, Du H, Gardner L. 2020. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menachery VD, Yount BL, Jr, Debbink K, Agnihothram S, Gralinski LE, Plante JA, Graham RL, Scobey T, Ge X-Y, Donaldson EF, Randell SH, Lanzavecchia A, Marasco WA, Shi Z-L, Baric RS. 2016. Corrigendum: a SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 22:446. doi: 10.1038/nm0416-446d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li F. 2016. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman VM, Muth D, Niemeyer D, Drosten C. 2018. Hosts and sources of endemic human coronaviruses. Adv Virus Res 100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu B, Zeng L-P, Yang X-L, Ge X-Y, Zhang W, Li B, Xie J-Z, Shen X-R, Zhang Y-Z, Wang N, Luo D-S, Zheng X-S, Wang M-N, Daszak P, Wang L-F, Cui J, Shi Z-L. 2017. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog 13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherry JD, Krogstad P. 2004. SARS: the first pandemic of the 21st century. Pediatr Res 56:1–5. doi: 10.1203/01.PDR.0000129184.87042.FC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peiris JSM, Yuen KY, Osterhaus ADME, Stöhr K. 2003. The severe acute respiratory syndrome. N Engl J Med 349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 8.Birch CJ, Clothier HJ, Seccull A, Tran T, Catton MC, Lambert SB, Druce JD. 2005. Human coronavirus OC43 causes influenza-like illness in residents and staff of aged-care facilities in Melbourne, Australia. Epidemiol Infect 133:273–277. doi: 10.1017/s0950268804003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hand J, Rose EB, Salinas A, Lu X, Sakthivel SK, Schneider E, Watson JT. 2018. Severe respiratory illness outbreak associated with human coronavirus NL63 in a long-term care facility. Emerg Infect Dis 24:1964–1966. doi: 10.3201/eid2410.180862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng Z-Q, Chen D-H, Tan W-P, Qiu S-Y, Xu D, Liang H-X, Chen M-X, Li X, Lin Z-S, Liu W-K, Zhou R. 2018. Epidemiology and clinical characteristics of human coronaviruses OC43, 229E, NL63, and HKU1: a study of hospitalized children with acute respiratory tract infection in Guangzhou, China. Eur J Clin Microbiol Infect Dis 37:363–369. doi: 10.1007/s10096-017-3144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RAM, Galiano M, Gorbalenya AE, Memish ZA, Perlman S, Poon LLM, Snijder EJ, Stephens GM, Woo PCY, Zaki AM, Zambon M, Ziebuhr J. 2013. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol 87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramadan N, Shaib H. 2019. Middle East respiratory syndrome coronavirus (MERS-CoV): a review. Germs 9:35–42. doi: 10.18683/germs.2019.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song D, Park B. 2012. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes 44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y-W, Dickerman AW, Piñeyro P, Li L, Fang L, Kiehne R, Opriessnig T, Meng X-J. 2013. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 4:e00737-13. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung K, Saif LJ. 2015. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet J 204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerdts V, Zakhartchouk A. 2017. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet Microbiol 206:45–51. doi: 10.1016/j.vetmic.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mogler MA, Gander J, Harris DLH. 2014. Development of an alphavirus RNA particle vaccine against porcine epidemic diarrhea virus. Ann Proc Am Assoc Swine Vet 2014:63–64. [Google Scholar]
- 18.Pardi N, Hogan MJ, Porter FW, Weissman D. 2018. mRNA vaccines–a new era in vaccinology. Nat Rev Drug Discov 17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou P, Fan H, Lan T, Yang X-L, Shi W-F, Zhang W, Zhu Y, Zhang Y-W, Xie Q-M, Mani S, Zheng X-S, Li B, Li J-M, Guo H, Pei G-Q, An X-P, Chen J-W, Zhou L, Mai K-J, Wu Z-X, Li D, Anderson DE, Zhang L-B, Li S-Y, Mi Z-Q, He T-T, Cong F, Guo P-J, Huang R, Luo Y, Liu X-L, Chen J, Huang Y, Sun Q, Zhang X-L-L, Wang Y-Y, Xing S-Z, Chen Y-S, Sun Y, Li J, Daszak P, Wang L-F, Shi Z-L, Tong Y-G, Ma J-Y. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaimes JA, Millet JK, Stout AE, André NM, Whittaker GR. 2020. A tale of two viruses: the distinct spike glycoproteins of feline coronaviruses. Viruses 12:83. doi: 10.3390/v12010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein SA, Brown J, Pedersen BS, Quinlan AR, Elde NC. 2021. Extensive recombination-driven coronavirus diversification expands the pool of potential pandemic pathogens. bioRxiv doi: 10.1101/2021.02.03.429646. [DOI] [PMC free article] [PubMed]
- 22.Graham RL, Baric RS. 2010. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol 84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F. 2013. Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res 100:246–254. doi: 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, Wong S-K, Li F, Kuhn JH, Huang I-C, Choe H, Farzan M. 2006. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J Virol 80:4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrison SC. 2015. Viral membrane fusion. Virology 479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. 2020. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res 178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whittaker GR, Daniel S, Millet JK. 2021. Coronavirus entry: how we arrived at SARS-CoV-2. Curr Opin Virol 47:113–120. doi: 10.1016/j.coviro.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millet JK, Whittaker GR. 2015. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res 202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heald-Sargent T, Gallagher T. 2012. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses 4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boni MF, Lemey P, Jiang X, Lam TT-Y, Perry BW, Castoe TA, Rambaut A, Robertson DL. 2020. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol 5:1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- 32.Johnson BA, Xie X, Bailey AL, Kalveram B, Lokugamage KG, Muruato A, Zou J, Zhang X, Juelich T, Smith JK, Zhang L, Bopp N, Schindewolf C, Vu M, Vanderheiden A, Winkler ES, Swetnam D, Plante JA, Aguilar P, Plante KS, Popov V, Lee B, Weaver SC, Suthar MS, Routh AL, Ren P, Ku Z, An Z, Debbink K, Diamond MS, Shi P-Y, Freiberg AN, Menachery VD. 2021. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 591:293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lubinski B, Tang T, Daniel S, Jaimes JA, Whittaker GR. 2021. Functional evaluation of proteolytic activation for the SARS-CoV-2 variant B.1.1.7: role of the P681H mutation. bioRxiv doi: 10.1101/2021.04.06.438731. [DOI] [PMC free article] [PubMed]
- 34.Liu Y, Liu J, Johnson BA, Xia H, Ku Z, Schindewolf C, Widen SG, An Z, Weaver SC, Menachery VD, Xie X, Shi P-Y. 2021. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv doi: 10.1101/2021.08.12.456173. [DOI] [PMC free article] [PubMed]
- 35.Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, Rakshit P, Singh S, Abraham P, Panda S, NIC Team . 2021. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms 9:1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai AL, Millet JK, Daniel S, Freed JH, Whittaker GR. 2017. The SARS-CoV fusion peptide forms an extended bipartite fusion platform that perturbs membrane order in a calcium-dependent manner. J Mol Biol 429:3875–3892. doi: 10.1016/j.jmb.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai AL, Freed JH. 2021. SARS-CoV-2 fusion peptide has a greater membrane perturbating effect than SARS-CoV with highly specific dependence on Ca2. J Mol Biol 433:166946. doi: 10.1016/j.jmb.2021.166946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madu IG, Roth SL, Belouzard S, Whittaker GR. 2009. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J Virol 83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park J-E, Li K, Barlan A, Fehr AR, Perlman S, McCray PB, Jr, Gallagher T. 2016. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci USA 113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaender S, Mar KB, Michailidis E, Kratzel A, Boys IN, V'kovski P, Fan W, Kelly JN, Hirt D, Ebert N, Stalder H, Kleine-Weber H, Hoffmann M, Hoffmann H-H, Saeed M, Dijkman R, Steinmann E, Wight-Carter M, McDougal MB, Hanners NW, Pöhlmann S, Gallagher T, Todt D, Zimmer G, Rice CM, Schoggins JW, Thiel V. 2020. LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat Microbiol 5:1330–1339. doi: 10.1038/s41564-020-0769-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shalamova L, Felgenhauer U, Schaubmar AR, Büttner K, Widera M, Ciesek S, Weber F. 2022. Omicron variant of SARS-CoV-2 exhibits an increased resilience to the antiviral type I interferon response. bioRxiv doi: 10.1101/2022.01.20.476754. [DOI] [PMC free article] [PubMed]
- 42.Li K, Markosyan RM, Zheng Y-M, Golfetto O, Bungart B, Li M, Ding S, He Y, Liang C, Lee JC, Gratton E, Cohen FS, Liu S-L. 2013. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog 9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, Planchais C, Porrot F, Guivel-Benhassine F, Van der Werf S, Casartelli N, Mouquet H, Bruel T, Schwartz O. 2020. Syncytia formation by SARS-CoV-2-infected cells. EMBO J 39:e106267. doi: 10.15252/embj.2020106267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blijleven JS, Boonstra S, Onck PR, van der Giessen E, van Oijen AM. 2016. Mechanisms of influenza viral membrane fusion. Semin Cell Dev Biol 60:78–88. doi: 10.1016/j.semcdb.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Walls AC, Xiong X, Park Y-J, Tortorici MA, Snijder J, Quispe J, Cameroni E, Gopal R, Dai M, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D. 2019. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176:1026–1039. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belouzard S, Chu VC, Whittaker GR. 2009. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci USA 106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, Baum A, Diehl WE, Dauphin A, Carbone C, Veinotte K, Egri SB, Schaffner SF, Lemieux JE, Munro JB, Rafique A, Barve A, Sabeti PC, Kyratsous CA, Dudkina NV, Shen K, Luban J. 2020. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 183:739–751. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Cai Y, Xiao T, Lu J, Peng H, Sterling SM, Walsh RM, Jr, Rits-Volloch S, Zhu H, Woosley AN, Yang W, Sliz P, Chen B. 2021. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science 372:525–530. doi: 10.1126/science.abf2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hussain M, Jabeen N, Amanullah A, Baig AA, Aziz B, Shabbir S, Raza F, Uddin N. 2020. Molecular docking between human TMPRSS2 and SARS-CoV-2 spike protein: conformation and intermolecular interactions. AIMS Microbiol 6:350–360. doi: 10.3934/microbiol.2020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qing E, Kicmal T, Kumar B, Hawkins GM, Timm E, Perlman S, Gallagher T. 2021. Dynamics of SARS-CoV-2 spike proteins in cell entry: control elements in the amino-terminal domains. mBio 12:e0159021. doi: 10.1128/mBio.01590-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevens CS, Oguntuyo KY, Lee B. 2021. Proteases and variants: context matters for SARS-CoV-2 entry assays. Curr Opin Virol 50:49–58. doi: 10.1016/j.coviro.2021.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Liu C, Mao Z, Xiao M, Wang L, Qi S, Zhou F. 2020. Predictive values of neutrophil-to-lymphocyte ratio on disease severity and mortality in COVID-19 patients: a systematic review and meta-analysis. Crit Care 24:647. doi: 10.1186/s13054-020-03374-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh M, Bansal V, Feschotte C. 2020. A single-cell RNA expression map of human coronavirus entry factors. Cell Rep 32:108175. doi: 10.1016/j.celrep.2020.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahmadian E, Hosseiniyan Khatibi SM, Razi Soofiyani S, Abediazar S, Shoja MM, Ardalan M, Zununi Vahed S. 2021. Covid-19 and kidney injury: pathophysiology and molecular mechanisms. Rev Med Virol 31:e2176. doi: 10.1002/rmv.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, van der Meer F, Kallio K, Kaya T, Anastasina M, Smura T, Levanov L, Szirovicza L, Tobi A, Kallio-Kokko H, Österlund P, Joensuu M, Meunier FA, Butcher SJ, Winkler MS, Mollenhauer B, Helenius A, Gokce O, Teesalu T, Hepojoki J, Vapalahti O, Stadelmann C, Balistreri G, Simons M. 2020. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei C, Wan L, Yan Q, Wang X, Zhang J, Yang X, Zhang Y, Fan C, Li D, Deng Y, Sun J, Gong J, Yang X, Wang Y, Wang X, Li J, Yang H, Li H, Zhang Z, Wang R, Du P, Zong Y, Yin F, Zhang W, Wang N, Peng Y, Lin H, Feng J, Qin C, Chen W, Gao Q, Zhang R, Cao Y, Zhong H. 2020. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat Metab 2:1391–1400. doi: 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]
- 58.Yeung ML, Teng JLL, Jia L, Zhang C, Huang C, Cai J-P, Zhou R, Chan K-H, Zhao H, Zhu L, Siu K-L, Fung S-Y, Yung S, Chan TM, To KK-W, Chan JF-W, Cai Z, Lau SKP, Chen Z, Jin D-Y, Woo PCY, Yuen K-Y. 2021. Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system. Cell 184:2212–2228. doi: 10.1016/j.cell.2021.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rochman ND, Wolf YI, Faure G, Mutz P, Zhang F, Koonin EV. 2021. Ongoing global and regional adaptive evolution of SARS-CoV-2. Proc Natl Acad Sci USA 118:e2104241118. doi: 10.1073/pnas.2104241118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang L, He G, Sharp AK, Wang X, Brown AM, Michalak P, Weger-Lucarelli J. 2021. A selective sweep in the Spike gene has driven SARS-CoV-2 human adaptation. Cell 184:4392–4400.e4. doi: 10.1016/j.cell.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang S-Y, Kung Y-A, Huang P-N, Chang S-Y, Gong Y-N, Han Y-J, Chiang H-J, Liu K-T, Lee K-M, Chang C-Y, Chang C-C, Huang C-G, Shih S-R. 2021. Stability of SARS-CoV-2 Spike G614 variant surpasses that of the D614 variant after cold storage. mSphere 6:e00104-21. doi: 10.1128/mSphere.00104-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, Zhang X, Muruato AE, Zou J, Fontes-Garfias CR, Mirchandani D, Scharton D, Bilello JP, Ku Z, An Z, Kalveram B, Freiberg AN, Menachery VD, Xie X, Plante KS, Weaver SC, Shi P-Y. 2021. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou B, Thao TTN, Hoffmann D, Taddeo A, Ebert N, Labroussaa F, Pohlmann A, King J, Steiner S, Kelly JN, Portmann J, Halwe NJ, Ulrich L, Trüeb BS, Fan X, Hoffmann B, Wang L, Thomann L, Lin X, Stalder H, Pozzi B, de Brot S, Jiang N, Cui D, Hossain J, Wilson MM, Keller MW, Stark TJ, Barnes JR, Dijkman R, Jores J, Benarafa C, Wentworth DE, Thiel V, Beer M. 2021. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 592:122–127. doi: 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- 64.Laporte M, Raeymaekers V, Van Berwaer R, Vandeput J, Marchand-Casas I, Thibaut H-J, Van Looveren D, Martens K, Hoffmann M, Maes P, Pöhlmann S, Naesens L, Stevaert A. 2021. The SARS-CoV-2 and other human coronavirus spike proteins are fine-tuned towards temperature and proteases of the human airways. PLoS Pathog 17:e1009500. doi: 10.1371/journal.ppat.1009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O'Toole Á, Southgate J, Johnson R, Jackson B, Nascimento FF, Rey SM, Nicholls SM, Colquhoun RM, da Silva Filipe A, Shepherd J, Pascall DJ, Shah R, Jesudason N, Li K, Jarrett R, Pacchiarini N, Bull M, Geidelberg L, Siveroni I, Goodfellow I, Loman NJ, Pybus OG, Robertson DL, Thomson EC, Rambaut A, Connor TR, COG-UK Consortium . 2021. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 184:64–75. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Sheffield COVID-19 Genomics Group . 2020. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, Sotomayor-González A, Glasner DR, Reyes KR, Gliwa AS, Reddy NP, Sanchez San Martin C, Federman S, Cheng J, Balcerek J, Taylor J, Streithorst JA, Miller S, Sreekumar B, Chen P-Y, Schulze-Gahmen U, Taha TY, Hayashi JM, Simoneau CR, Kumar GR, McMahon S, Lidsky PV, Xiao Y, Hemarajata P, Green NM, Espinosa A, Kath C, Haw M, Bell J, Hacker JK, Hanson C, Wadford DA, Anaya C, Ferguson D, Frankino PA, Shivram H, Lareau LF, Wyman SK, Ott M, Andino R, Chiu CY. 2021. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell 184:3426–3437. doi: 10.1016/j.cell.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramanathan M, Ferguson ID, Miao W, Khavari PA. 2021. SARS-CoV-2 B.1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity. Lancet Infect Dis doi: 10.1016/S1473-3099(21)00262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin DP, Weaver S, Tegally H, San EJ, Shank SD, Wilkinson E, Giandhari J, Naidoo S, Pillay Y, Singh L, Lessells RJ, Gupta RK, Wertheim JO, Nekturenko A, Murrell B, Harkins GW, Lemey P, MacLean O, Robertson DL, de Oliveira T, Kosakovsky Pond SL, NGS-SA Consortium, COVID-19 Genomics UK (COG-UK) Consortium . 2021. The emergence and ongoing convergent evolution of the N501Y lineages coincides with a major global shift in the SARS-CoV-2 selective landscape. medRxiv doi: 10.1101/2021.02.23.21252268. [DOI] [Google Scholar]
- 70.Naveca FG, Nascimento V, de Souza VC, Corado A, de L, Nascimento F, Silva G, Costa Á, Duarte D, Pessoa K, Mejía M, Brandão MJ, Jesus M, Gonçalves L, da Costa CF, Sampaio V, Barros D, Silva M, Mattos T, Pontes G, Abdalla L, Santos JH, Arantes I, Dezordi FZ, Siqueira MM, Wallau GL, Resende PC, Delatorre E, Gräf T, Bello G. 2021. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat Med 27:1230–1238. doi: 10.1038/s41591-021-01378-7. [DOI] [PubMed] [Google Scholar]
- 71.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Peacock SJ, Robertson DL, COVID-19 Genomics UK (COG-UK) Consortium . 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berger I, Schaffitzel C. 2020. The SARS-CoV-2 spike protein: balancing stability and infectivity. Cell Res 30:1059–1060. doi: 10.1038/s41422-020-00430-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gong LI, Suchard MA, Bloom JD. 2013. Stability-mediated epistasis constrains the evolution of an influenza protein. Elife 2:e00631. doi: 10.7554/eLife.00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holmes EC, Goldstein SA, Rasmussen AL, Robertson DL, Crits-Christoph A, Wertheim JO, Anthony SJ, Barclay WS, Boni MF, Doherty PC, Farrar J, Geoghegan JL, Jiang X, Leibowitz JL, Neil SJD, Skern T, Weiss SR, Worobey M, Andersen KG, Garry RF, Rambaut A. 2021. The origins of SARS-CoV-2: a critical review. Cell 184:4848–4856. doi: 10.1016/j.cell.2021.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Whittaker GR. 2021. SARS-CoV-2 spike and its adaptable furin cleavage site. Lancet Microbe doi: 10.1016/S2666-5247(21)00174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hodcroft EB, Domman DB, Snyder DJ, Oguntuyo KY, Van Diest M, Densmore KH, Schwalm KC, Femling J, Carroll JL, Scott RS, Whyte MM, Edwards MW, Hull NC, Kevil CG, Vanchiere JA, Lee B, Dinwiddie DL, Cooper VS, Kamil JP. 2021. Emergence in late 2020 of multiple lineages of SARS-CoV-2 Spike protein variants affecting amino acid position 677. medRxiv doi: 10.1101/2021.02.12.21251658. [DOI] [Google Scholar]
- 77.Tang T, Jaimes JA, Bidon MK, Straus MR, Daniel S, Whittaker GR. 2021. Proteolytic activation of SARS-CoV-2 Spike at the S1/S2 boundary: potential role of proteases beyond furin. ACS Infect Dis 7:264–272. doi: 10.1021/acsinfecdis.0c00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lubinski B, Frazier LE, Phan MVT, Bugembe DL, Tang T, Daniel S, Cotten M, Jaimes JA, Whittaker GR. 2021. Spike protein cleavage-activation mediated by the SARS-CoV-2 P681R mutation: a case-study from its first appearance in variant of interest (VOI) A.23.1 identified in Uganda. bioRxiv doi: 10.1101/2021.06.30.450632. [DOI] [PMC free article] [PubMed]
- 79.Zeng C, Evans JP, Faraone JN, Qu P, Zheng Y-M, Saif L, Oltz EM, Lozanski G, Gumina RJ, Liu S-L. 2021. Neutralization of SARS-CoV-2 variants of concern harboring Q677H. mBio 12:e0251021. doi: 10.1128/mBio.02510-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jaimes JA, Millet JK, Whittaker GR. 2020. Proteolytic cleavage of the SARS-CoV-2 Spike protein and the role of the novel S1/S2 site. iScience 23:101212. doi: 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCallum M, De Marco A, Lempp FA, Alejandra Tortorici M, Pinto D, Walls AC, Beltramello M, Chen A, Liu Z, Zatta F, Zepeda S, di Iulio J, Bowen JE, Montiel-Ruiz M, Zhou J, Rosen LE, Bianchi S, Guarino B, Fregni CS, Abdelnabi R, Foo S-YC, Rothlauf PW, Bloyet L-M, Benigni F, Cameroni E, Neyts J, Riva A, Snell G, Telenti A, Whelan SPJ, Virgin HW, Corti D, Pizzuto MS, Veesler D. 2021. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 184:2332–2347. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tortorici MA, Veesler D. 2019. Structural insights into coronavirus entry. Adv Virus Res 105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McCarthy KR, Rennick LJ, Nambulli S, Robinson-McCarthy LR, Bain WG, Haidar G, Duprex WP. 2021. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang G-Y, Ho DD, Sheng Z, Kwong PD, Shapiro L. 2021. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe 29:819–833. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Voss WN, Hou YJ, Johnson NV, Delidakis G, Kim JE, Javanmardi K, Horton AP, Bartzoka F, Paresi CJ, Tanno Y, Chou C-W, Abbasi SA, Pickens W, George K, Boutz DR, Towers DM, McDaniel JR, Billick D, Goike J, Rowe L, Batra D, Pohl J, Lee J, Gangappa S, Sambhara S, Gadush M, Wang N, Person MD, Iverson BL, Gollihar JD, Dye JM, Herbert AS, Finkelstein IJ, Baric RS, McLellan JS, Georgiou G, Lavinder JJ, Ippolito GC. 2021. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes. Science 372:1108–1112. doi: 10.1126/science.abg5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Planas D, Bruel T, Grzelak L, Guivel-Benhassine F, Staropoli I, Porrot F, Planchais C, Buchrieser J, Rajah MM, Bishop E, Albert M, Donati F, Prot M, Behillil S, Enouf V, Maquart M, Smati-Lafarge M, Varon E, Schortgen F, Yahyaoui L, Gonzalez M, De Sèze J, Péré H, Veyer D, Sève A, Simon-Lorière E, Fafi-Kremer S, Stefic K, Mouquet H, Hocqueloux L, van der Werf S, Prazuck T, Schwartz O. 2021. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med 27:917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 87.Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, Solomon IH, Kuo H-H, Boucau J, Bowman K, Adhikari UD, Winkler ML, Mueller AA, Hsu TY-T, Desjardins M, Baden LR, Chan BT, Walker BD, Lichterfeld M, Brigl M, Kwon DS, Kanjilal S, Richardson ET, Jonsson AH, Alter G, Barczak AK, Hanage WP, Yu XG, Gaiha GD, Seaman MS, Cernadas M, Li JZ. 2020. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aydillo T, Gonzalez-Reiche AS, Aslam S, van de Guchte A, Khan Z, Obla A, Dutta J, van Bakel H, Aberg J, García-Sastre A, Shah G, Hohl T, Papanicolaou G, Perales M-A, Sepkowitz K, Babady NE, Kamboj M. 2020. Shedding of viable SARS-CoV-2 after immunosuppressive therapy for cancer. N Engl J Med 383:2586–2588. doi: 10.1056/NEJMc2031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hensley MK, Bain WG, Jacobs J, Nambulli S, Parikh U, Cillo A, Staines B, Heaps A, Sobolewski MD, Rennick LJ, Macatangay BJC, Klamar-Blain C, Kitsios GD, Methé B, Somasundaram A, Bruno TC, Cardello C, Shan F, Workman C, Ray P, Ray A, Lee J, Sethi R, Schwarzmann WE, Ladinsky MS, Bjorkman PJ, Vignali DA, Duprex WP, Agha ME, Mellors JW, McCormick KD, Morris A, Haidar G. 2021. Intractable coronavirus disease 2019 (COVID-19) and prolonged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication in a chimeric antigen receptor-modified T-cell therapy recipient: a case study. Clin Infect Dis 73:e815–e821. doi: 10.1093/cid/ciab072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Khan A, Zia T, Suleman M, Khan T, Ali SS, Abbasi AA, Mohammad A, Wei D-Q. 2021. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J Cell Physiol 236:7045–7057. doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gobeil SM-C, Janowska K, McDowell S, Mansouri K, Parks R, Stalls V, Kopp MF, Manne K, Li D, Wiehe K, Saunders KO, Edwards RJ, Korber B, Haynes BF, Henderson R, Acharya P. 2021. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science 373:eabi6226. doi: 10.1126/science.abi6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Laiton-Donato K, Franco-Muñoz C, Álvarez-Díaz DA, Ruiz-Moreno HA, Usme-Ciro JA, Prada DA, Reales-González J, Corchuelo S, Herrera-Sepúlveda MT, Naizaque J, Santamaría G, Rivera J, Rojas P, Ortiz JH, Cardona A, Malo D, Prieto-Alvarado F, Gómez FR, Wiesner M, Martínez MLO, Mercado-Reyes M. 2021. Characterization of the emerging B.1.621 variant of interest of SARS-CoV-2. Infect Genet Evol 95:105038. doi: 10.1016/j.meegid.2021.105038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McCallum M, Czudnochowski N, Rosen LE, Zepeda SK, Bowen JE, Walls AC, Hauser K, Joshi A, Stewart C, Dillen JR, Powell AE, Croll TI, Nix J, Virgin HW, Corti D, Snell G, Veesler D. 2022. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science doi: 10.1126/science.abn8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mannar D, Saville JW, Zhu X, Srivastava SS, Berezuk AM, Tuttle KS, Marquez AC, Sekirov I, Subramaniam S. 2022. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein–ACE2 complex. Science doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu J, Li Z, He X, Gebre MS, Bondzie EA, Wan H, Jacob-Dolan C, Martinez DR, Nkolola JP, Baric RS, Barouch DH. 2021. Deletion of the SARS-CoV-2 spike cytoplasmic tail increases infectivity in pseudovirus neutralization assays. J Virol 95:e00044-21. doi: 10.1128/JVI.00044-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xia S, Liu M, Wang C, Xu W, Lan Q, Feng S, Qi F, Bao L, Du L, Liu S, Qin C, Sun F, Shi Z, Zhu Y, Jiang S, Lu L. 2020. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res 30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Obermeyer F, Schaffner SF, Jankowiak M, Barkas N, Pyle JD, Park DJ, MacInnis BL, Luban J, Sabeti PC, Lemieux JE. 2021. Analysis of 2.1 million SARS-CoV-2 genomes identifies mutations associated with transmissibility. medRxiv doi: 10.1101/2021.09.07.21263228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Johnson BA, Zhou Y, Lokugamage KG, Vu MN, Bopp N, Crocquet-Valdes PA, Schindewolf C, Liu Y, Scharton D, Plante JA, Xie X, Aguilar P, Weaver SC, Shi P-Y, Walker DH, Routh AL, Plante KS, Menachery VD. 2021. Nucleocapsid mutations in SARS-CoV-2 augment replication and pathogenesis. bioRxiv doi: 10.1101/2021.10.14.464390. [DOI] [PMC free article] [PubMed]
- 99.Wu H, Xing N, Meng K, Fu B, Xue W, Dong P, Tang W, Xiao Y, Liu G, Luo H, Zhu W, Lin X, Meng G, Zhu Z. 2021. Nucleocapsid mutations R203K/G204R increase the infectivity, fitness, and virulence of SARS-CoV-2. Cell Host Microbe 29:1788–1801. doi: 10.1016/j.chom.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y, Sun H, Pei R, Mao B, Zhao Z, Li H, Lin Y, Lu K. 2021. The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov 7:31. doi: 10.1038/s41421-021-00268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zinzula L. 2021. Lost in deletion: the enigmatic ORF8 protein of SARS-CoV-2. Biochem Biophys Res Commun 538:116–124. doi: 10.1016/j.bbrc.2020.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bugembe DL, Phan MVT, Ssewanyana I, Semanda P, Nansumba H, Dhaala B, Nabadda S, O'Toole ÁN, Rambaut A, Kaleebu P, Cotten M. 2021. Emergence and spread of a SARS-CoV-2 lineage A variant (A.23.1) with altered spike protein in Uganda. Nat Microbiol 6:1094–1101. doi: 10.1038/s41564-021-00933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Garvin MR, Prates ET, Romero J, Cliff A, Gazolla JGF, Pickholz M, Pavicic M, Jacobson D. 2021. Rapid expansion of SARS-CoV-2 variants of concern is a result of adaptive epistasis. bioRxiv doi: 10.1101/2021.08.03.454981. [DOI]
- 104.Plante JA, Mitchell BM, Plante KS, Debbink K, Weaver SC, Menachery VD. 2021. The variant gambit: COVID-19’s next move. Cell Host Microbe 29:508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, Cottrell CA, Becker MM, Wang L, Shi W, Kong W-P, Andres EL, Kettenbach AN, Denison MR, Chappell JD, Graham BS, Ward AB, McLellan JS. 2017. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci USA 114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang K, Li S, Pintilie G, Chmielewski D, Schmid MF, Simmons G, Jin J, Chiu W. 2020. A 3.4-Å cryo-EM structure of the human coronavirus spike trimer computationally derived from vitrified NL63 virus particles. bioRxiv doi: 10.1101/2020.08.11.245696. [DOI] [PMC free article] [PubMed]
- 107.Song X, Shi Y, Ding W, Niu T, Sun L, Tan Y, Chen Y, Shi J, Xiong Q, Huang X, Xiao S, Zhu Y, Cheng C, Fu ZF, Liu Z-J, Peng G. 2021. Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes. Nat Commun 12:141. doi: 10.1038/s41467-020-20401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zimmerman MI, Porter JR, Ward MD, Singh S, Vithani N, Meller A, Mallimadugula UL, Kuhn CE, Borowsky JH, Wiewiora RP, Hurley MFD, Harbison AM, Fogarty CA, Coffland JE, Fadda E, Voelz VA, Chodera JD, Bowman GR. 2021. SARS-CoV-2 simulations go exascale to predict dramatic spike opening and cryptic pockets across the proteome. Nat Chem 13:651–659. doi: 10.1038/s41557-021-00707-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xie X, Liu Y, Liu J, Zhang X, Zou J, Fontes-Garfias CR, Xia H, Swanson KA, Cutler M, Cooper D, Menachery VD, Weaver SC, Dormitzer PR, Shi P-Y. 2021. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med 27:620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 110.Morris DH, Petrova VN, Rossine FW, Parker E, Grenfell BT, Neher RA, Levin SA, Russell CA. 2020. Asynchrony between virus diversity and antibody selection limits influenza virus evolution. Elife 9:e62105. doi: 10.7554/eLife.62105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xue KS, Moncla LH, Bedford T, Bloom JD. 2018. Within-host evolution of human influenza virus. Trends Microbiol 26:781–793. doi: 10.1016/j.tim.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang Z, Han Y, Ding S, Shi W, Zhou T, Finzi A, Kwong PD, Mothes W, Lu M. SARS-CoV-2 Variants Increase Kinetic Stability of Open Spike Conformations as an Evolutionary Strategy. mBio. 2022 Feb 15:e0322721. doi: 10.1128/mbio.03227-21. Epub ahead of print. PMID: 35164561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rossmann MG. 1989. The canyon hypothesis. Hiding the host cell receptor attachment site on a viral surface from immune surveillance. J Biol Chem 264:14587–14590. doi: 10.1016/S0021-9258(18)63732-9. [DOI] [PubMed] [Google Scholar]
- 114.Ni D, Lau K, Turelli P, Raclot C, Beckert B, Nazarov S, Pojer F, Myasnikov A, Stahlberg H, Trono D. 2021. Structural analysis of the Spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion. bioRxiv doi: 10.1101/2021.12.27.474250. [DOI]
- 115.Peacock TP, Brown JC, Zhou J, Thakur N, Newman J, Kugathasan R, Sukhova K, Kaforou M, Bailey D, Barclay WS. 2022. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv doi: 10.1101/2021.12.31.474653. [DOI]
- 116.Zeng C, Evans JP, Qu P, Faraone J, Zheng Y-M, Carlin C, Bednash JS, Zhou T, Lozanski G, Mallampalli R, Saif LJ, Oltz EM, Mohler P, Xu K, Gumina RJ, Liu S-L. 2021. Neutralization and stability of SARS-CoV-2 Omicron variant. bioRxiv doi: 10.1101/2021.12.16.472934. [DOI]
- 117.Huotari J, Helenius A. 2011. Endosome maturation. EMBO J 30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Albrecht T, Zhao Y, Nguyen TH, Campbell RE, Johnson JD. 2015. Fluorescent biosensors illuminate calcium levels within defined beta-cell endosome subpopulations. Cell Calcium 57:263–274. doi: 10.1016/j.ceca.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 119.Cui Z, Liu P, Wang N, Wang L, Fan K, Zhu Q, Wang K, Chen R, Feng R, Jia Z, Yang M, Zhu B, Fu W, Chu T, Feng L, Wang Y, Pei X, Yang P, Xie XS, Cao L, Cao YR, Wang X. 2022. Structural and functional characterizations of altered infectivity and immune evasion of SARS-CoV-2 Omicron variant. bioRxiv doi: 10.1101/2021.12.29.474402. [DOI] [PMC free article] [PubMed]
- 120.Seok JH, Kim H, Lee DB, An JS, Kim EJ, Lee J-H, Chung MS, Kim KH. 2020. Divalent cation-induced conformational changes of influenza virus hemagglutinin. Sci Rep 10:15457. doi: 10.1038/s41598-020-72368-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hofmeister F. 1888. Zur lehre von der wirkung der salze. Arch Exp Pathol Pharmakol 24:247–260. doi: 10.1007/BF01918191. [DOI] [Google Scholar]
- 122.Vlachy N, Jagoda-Cwiklik B, Vácha R, Touraud D, Jungwirth P, Kunz W. 2009. Hofmeister series and specific interactions of charged headgroups with aqueous ions. Adv Colloid Interface Sci 146:42–47. doi: 10.1016/j.cis.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 123.Sheward DJ, Pushparaj P, Das H, Kim C, Kim S, Hanke L, Dyrdak R, McInerney G, Albert J, Murrell B, Karlsson Hedestam GB, Martin Hällberg B. 2022. Structural basis of Omicron neutralization by affinity-matured public antibodies. bioRxiv doi: 10.1101/2022.01.03.474825. [DOI] [PMC free article] [PubMed]
- 124.Park Y-J, De Marco A, Starr TN, Liu Z, Pinto D, Walls AC, Zatta F, Zepeda SK, Bowen JE, Sprouse KR, Joshi A, Giurdanella M, Guarino B, Noack J, Abdelnabi R, Foo S-YC, Rosen LE, Lempp FA, Benigni F, Snell G, Neyts J, Whelan SPJ, Virgin HW, Bloom JD, Corti D, Pizzuto MS, Veesler D. 2022. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science doi: 10.1126/science.abm8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Viana R, Moyo S, Amoako DG, Tegally H, Scheepers C, Althaus CL, Anyaneji UJ, Bester PA, Boni MF, Chand M, Choga WT, Colquhoun R, Davids M, Deforche K, Doolabh D, Du Plessis L, Engelbrecht S, Everatt J, Giandhari J, Giovanetti M, Hardie D, Hill V, Hsiao N-Y, Iranzadeh A, Ismail A, Joseph C, Joseph R, Koopile L, Kosakovsky Pond SL, Kraemer MUG, Kuate-Lere L, Laguda-Akingba O, Lesetedi-Mafoko O, Lessells RJ, Lockman S, Lucaci AG, Maharaj A, Mahlangu B, Maponga T, Mahlakwane K, Makatini Z, Marais G, Maruapula D, Masupu K, Matshaba M, Mayaphi S, Mbhele N, Mbulawa MB, Mendes A, Mlisana K, et al. 2022. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature doi: 10.1038/d41586-021-03832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matsuyama S, Shirato K, Kawase M, Terada Y, Kawachi K, Fukushi S, Kamitani W. 2018. Middle East respiratory syndrome coronavirus Spike protein is not activated directly by cellular furin during viral entry into target cells. J Virol 92:e00683-18. doi: 10.1128/JVI.00683-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li W, Wicht O, van Kuppeveld FJM, He Q, Rottier PJM, Bosch B-J. 2015. A single point mutation creating a furin cleavage site in the Spike protein renders porcine epidemic diarrhea coronavirus trypsin independent for cell entry and fusion. J Virol 89:8077–8081. doi: 10.1128/JVI.00356-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Millet JK, Whittaker GR. 2014. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci USA 111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yuan Y, Cao D, Zhang Y, Ma J, Qi J, Wang Q, Lu G, Wu Y, Yan J, Shi Y, Zhang X, Gao GF. 2017. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun 8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bickerton E, Maier HJ, Stevenson-Leggett P, Armesto M, Britton P. 2018. The S2 subunit of infectious bronchitis virus Beaudette is a determinant of cellular tropism. J Virol 92:e01044-18. doi: 10.1128/JVI.01044-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Offersgaard A, Duarte Hernandez CR, Pihl AF, Costa R, Venkatesan NP, Lin X, Van Pham L, Feng S, Fahnøe U, Scheel TKH, Ramirez S, Reichl U, Bukh J, Genzel Y, Gottwein JM. 2021. SARS-CoV-2 production in a scalable high cell density bioreactor. Vaccines (Basel) 9:706. doi: 10.3390/vaccines9070706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ramirez S, Fernandez-Antunez C, Galli A, Underwood A, Pham LV, Ryberg LA, Feng S, Pedersen MS, Mikkelsen LS, Belouzard S, Dubuisson J, Sølund C, Weis N, Gottwein JM, Fahnøe U, Bukh J. 2021. Overcoming culture restriction for SARS-CoV-2 in human cells facilitates the screening of compounds inhibiting viral replication. Antimicrob Agents Chemother 65:e0009721. doi: 10.1128/AAC.00097-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McLellan JS, Ray WC, Peeples ME. 2013. Structure and function of respiratory syncytial virus surface glycoproteins. Curr Top Microbiol Immunol 372:83–104. doi: 10.1007/978-3-642-38919-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zimmer G, Budz L, Herrler G. 2001. Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J Biol Chem 276:31642–31650. doi: 10.1074/jbc.M102633200. [DOI] [PubMed] [Google Scholar]
- 135.Perugino CA, Liu H, Feldman J, Hauser BM, Jacob-Dolan C, Nathan A, Zhou Z, Kaseke C, Tano-Menka R, Getz MA, Senjobe F, Berrios C, Ofoman O, Lemieux JE, Goldberg MB, Nundel K, Moormann A, Marshak-Rothstein A, Iafrate JA, Gaiha G, Charles R, Balazs AB, Naranbhai V, Schmidt AG, Pillai S. 2022. Preferential expansion upon boosting of cross-reactive “pre-existing” switched memory B cells that recognize the SARS-CoV-2 Omicron variant Spike protein. medRxiv doi: 10.1101/2021.12.30.21268554. [DOI] [Google Scholar]
- 136.Eguia RT, Crawford KHD, Stevens-Ayers T, Kelnhofer-Millevolte L, Greninger AL, Englund JA, Boeckh MJ, Bloom JD. 2021. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog 17:e1009453. doi: 10.1371/journal.ppat.1009453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Marchi J, Lässig M, Walczak AM, Mora T. 2021. Antigenic waves of virus-immune coevolution. Proc Natl Acad Sci USA 118:e2103398118. doi: 10.1073/pnas.2103398118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.McCrone JT, Lauring AS. 2018. Genetic bottlenecks in intraspecies virus transmission. Curr Opin Virol 28:20–25. doi: 10.1016/j.coviro.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Robertson JD, Koehler G. 1918. Preliminary report on the influenza epidemic in Chicago. Am J Public Health (N Y) 8:849–856. doi: 10.2105/ajph.8.11.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Evans WA, Heckard MO. 1918. The 1890 epidemic of influenza in Chicago and its influence on mortality, 1890 to 1893 inclusive. Am J Public Health (N Y) 8:845–848. doi: 10.2105/ajph.8.11.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Radin MJ. 1920. Chronic lung disease following the influenza pandemic of 1918–1919. The American Journal of the Medical Sciences (1827–1924); Philadelphia 160 (2) (Aug 1920): 233. [Google Scholar]
- 142.Koonin EV. 2016. Splendor and misery of adaptation, or the importance of neutral null for understanding evolution. BMC Biol 14:114. doi: 10.1186/s12915-016-0338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lynch M. 2007. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci USA 104(Suppl 1):8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dobzhansky T. 1973. Nothing in biology makes sense except in the light of evolution. Am Biol Teach 35:125–129. doi: 10.2307/4444260. [DOI] [Google Scholar]
- 145.Rasmussen AL. 2020. Vaccination is the only acceptable path to herd immunity. Medicine (N Y) 1:21–23. doi: 10.1016/j.medj.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cobey S, Larremore DB, Grad YH, Lipsitch M. 2021. Concerns about SARS-CoV-2 evolution should not hold back efforts to expand vaccination. Nat Rev Immunol 21:330–335. doi: 10.1038/s41577-021-00544-9. [DOI] [PMC free article] [PubMed] [Google Scholar]