ABSTRACT
Defective interfering particles (DIs) contain a considerably smaller genome than the parental virus but retain replication competency. As DIs can directly or indirectly alter propagation kinetics of the parental virus, they offer a novel approach to antiviral therapy, capitalizing on knowledge from natural infection. However, efforts to translate in vitro inhibition to in vivo screening models remain limited. We investigated the efficacy of virus-like particles containing DI genomes (therapeutic infectious particles [TIPs]) in the Syrian hamster model of lethal Nipah virus (NiV) disease. We found that coadministering a high dose of TIPs intraperitoneally with virus challenge improved clinical course and reduced lethality. To mimic natural exposure, we also evaluated lower-dose TIP delivery and virus challenge intranasally, finding equally efficacious reduction in disease severity and overall lethality. Eliminating TIP replicative capacity decreased efficacy, suggesting protection via direct inhibition. These data provide evidence that TIP-mediated treatment can confer protection against disease and lethal outcome in a robust animal NiV model, supporting further development of TIP treatment for NiV and other high-consequence pathogens.
KEYWORDS: Nipah virus, defective interfering particles, Syrian hamsters, animal model, henipavirus, treatment, antiviral, antiviral agents
INTRODUCTION
Nipah virus (NiV) is a highly pathogenic zoonotic paramyxovirus (family Paramyxoviridae, genus Henipavirus) capable of causing severe disease in humans (1), with case fatality rates ranging from 30% to 80% (2). First recognized during an outbreak of severe febrile encephalitic and respiratory illnesses in Malaysia and Singapore from 1998 to 1999 (3–5), NiV has caused nearly annual outbreaks in India and Bangladesh since 2001 (6–8). Transmission to humans occurs predominately via direct contact with the excreta of the reservoir species, the fruit bat (genus Pteropus) (9, 10), with pigs also recognized as an amplifying host and source of infection during the initial outbreak (3, 5, 11). Human-to-human transmission is frequently reported, most commonly between close family and caregivers of NiV-infected individuals (12, 13). Currently, there are no licensed vaccines or antiviral therapies available for NiV infection (https://www.who.int/activities/prioritizing-diseases-for-research-and-development-in-emergency-contexts).
Defective interfering particles (DIs) have been shown to modulate multiple aspects of viral pathogenesis, including inhibition of viral replication and immune stimulation (14, 15), and have been characterized in numerous RNA viruses, including influenza (16), Ebola (17), canine distemper (18), and Sendai (19) viruses. DI genomes are formed by errors during nascent viral genome synthesis, in which translocation of the viral polymerase during RNA replication results in truncated and functionally inoperative versions of the genome (20). While they have lost the ability to encode the complete suite of viral proteins, DIs retain their self-replicative capacity due to intact promoter motifs at genomic termini. Shorter than the parental viral genomes, DIs are replicated faster, resulting in increased sequestration of critical viral and host-cell components, thus, in turn, outcompeting replication of full-length genomes. While direct inhibition is believed to be the main mechanism of DI-mediated inhibition, DIs can also induce antiviral immune responses that further inhibit authentic viral replication (21–23).
We recently identified numerous naturally occurring NiV DI genomes consisting of two variants, (i) trailer copybacks, which have complementary trailer nucleotide sequences at each genomic terminus, resulting in either a partial or complete hairpin structure; and (ii) deletions, which have authentic leader and trailer sequences but are missing large (>90%) sections of the internal nucleotide sequences (24). Several of these DI genomes were shown to potently inhibit NiV replication when incorporated into virus-like particles and used to treat cells. While the in vitro inhibitory potential of DI particles has been well documented, in vivo efficacy to support their use as a treatment or therapeutic candidate has not been evaluated beyond a limited number of influenza studies (25–30).
Here, we investigate the use of DI-containing particles (termed therapeutic infectious particles, or TIPs) for treating NiV infection in the Syrian hamster model of disease (31–33). Infection in Syrian hamsters accurately recapitulates the two most common phenotypic clinical presentations of human NiV disease, respiratory distress and neurological complications (34, 35). After characterizing optimal challenge parameters for this model, we applied it to assess the in vivo efficacy of selected TIPs to combat NiV infection. We found that high-dose intraperitoneal delivery of TIPs resulted in dramatically improved clinical outcomes and reduced NiV-associated mortality in the lethal hamster model of NiV disease. Furthermore, intranasal delivery of TIPs given at a 2,000-fold lower dose also conferred protection. These data demonstrate that exploiting the natural inhibitory effects of DIs (21, 36, 37) can significantly reduce disease severity and support the continued development of this unique treatment approach to control NiV and other highly lethal viral infections.
RESULTS
NiV-infected Syrian hamsters recapitulate both respiratory and neurological disease phenotypes, providing a clinically relevant challenge model for TIP assessment.
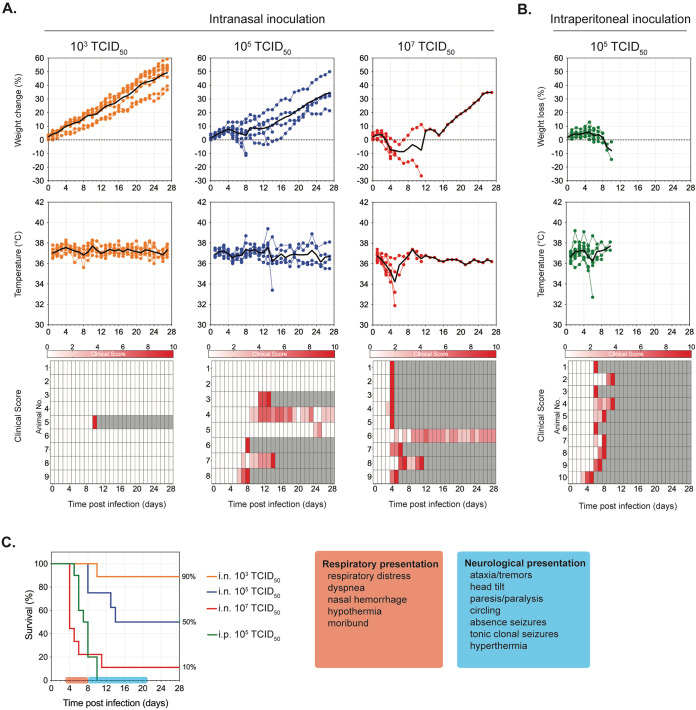
Prior to evaluating the potential of TIP treatment, we performed additional in-house characterization and clinical evaluation of the Syrian hamster model (31–33) using two inoculation routes, intranasal (i.n.) and intraperitoneal (i.p.), and a range of challenge doses. Besides monitoring body weights and temperatures, we developed a detailed clinical scoring system to better resolve NiV disease progression in hamsters. With i.n. inoculation, increased clinical severity was directly associated with increases in challenge dose (Fig. 1A); with i.p. inoculation lower doses achieved higher lethality than in i.n. inoculation (Fig. 1B).
FIG 1.
Optimal challenge dose determination in Nipah virus-infected Syrian hamsters. Hamsters were inoculated with Nipah virus (NiV) strain Malaysia via the intranasal (i.n.) route at 103 (orange, n = 9), 105 (blue, n = 8), or 107 (red, n = 9) TCID50 (A) or via the intraperitoneal (i.p.) route with 105 (green, n = 10) TCID50 (B). Graphs represent percent weight change from baseline (taken at −1 dpi), body temperatures, and daily clinical scores (scored from 0 to 10), with severity depicted by increasing intensity of red. Animals scoring ≥10 were humanely euthanized; any animals that succumbed to disease prior to euthanasia were allocated a score of 10. Gray boxes indicate the end of monitoring/scoring due to euthanasia/death. Individual animals are represented, with the black line indicating the mean value each day. (C) Combined survival curves for both i.n. and i.p. challenge routes indicating typical clinical signs observed at indicated times postinfection. Independently of inoculation dose and route, the initial disease presentation is respiratory distress beginning 3 to 6 dpi followed by a primarily neurological presentation at 8 to 20 dpi.
The onset of clinical signs and overall disease progression were broadly comparable and independent of challenge dose and route. Respiratory signs (increased respiratory rate, dyspnea, and, in more severe cases, nasal and oral hemorrhage) appeared 3 to 6 days postinfection (dpi) (Fig. 1C). Disease progression appeared more acute in i.p.-challenged animals, transitioning from an absence of clinical signs to severe disease in 12 to 14 h (versus up to 3 days in i.n.-challenged animals). In hamsters not developing or succumbing to respiratory disease, neurological signs (mild to severe, with either a sudden or gradual onset) generally become apparent from ∼8 dpi onwards. Mild to moderate clinical signs usually started as ataxia, limb weakness, head tilt progression from sporadic to persistent, and a generalized abnormality in behavior (very quiet, cautious movements, low creeping gait). These progressed to a more severe presentation that included limb paralysis, severe head tilt (>90°) associated with inability to stand on all four legs and a constant “corkscrewing” behavior, and short to constant tonic-clonal seizures associated with hyperthermia. Sudden onset (<24 h after appearing healthy) of these severe neurological signs was common. Some animals exhibiting mild to moderate clinical signs maintained this presentation until study end, and some recovered. The latest observed onset of neurological signs eventually requiring euthanasia (in a previously healthy hamster) was 13 dpi in a hamster challenged i.n. with 106 TCID50; the animal was euthanized at 16 dpi. Based on these data and previously generated data (38, 39), we chose challenge doses of 104 TCID50 i.p. or 106 TCID50 i.n. as optimal for evaluating efficacy in vivo. At these doses, we expected disease progression to be slow enough to allow manifestation of observable clinical signs and mortality rates to be sufficient to evaluate protective efficacy against lethal outcome.
TIP treatment provides robust protection against NiV disease in hamsters, and efficacy is enhanced when TIPs are active and able to directly inhibit viral replication and transcription.
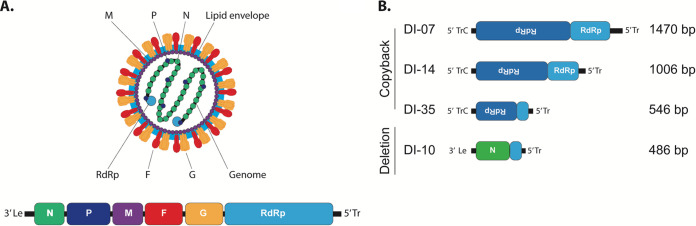
The NiV genome is a single-stranded negative-sense RNA containing 6 genes encoding 9 proteins (Fig. 2A). Four promising TIP candidates previously characterized to result in the greatest inhibition of NiV replication in vitro were selected for in vivo evaluation (24). Three of these were copyback variants DI-07 (1,470 nucleotides [nt] in length), DI-14 (1,006 nt), and DI-35 (546 nt), and one was deletion variant DI-10 (486 nt) (Fig. 2B). In vitro experiments indicated that a high ratio of TIP to virus (>2,500:1) was required for significant virus inhibition (24). Thus, to maximize the TIP/NiV treatment ratio in hamsters for initial screening, we chose the i.p. route of inoculation because larger delivery volumes can be used than with other routes. TIPs were given simultaneously with virus challenge at a DI/NiV ratio of ∼20,000:1 (2 mL total volume). Hamsters were challenged with a combination of NiV mixed with either: active TIPs (treated with 100 mJ/cm2 UV to destroy the full-length producer NiV genomes carried over from TIP production); inactive TIPs (treated with 2,400 mJ/cm2 UV to destroy both full-length NIV and TIP genomes); or Dulbecco’s modified Eagle’s medium (DMEM) only (control animals).
FIG 2.
Genome schematics and organization of NiV and DI genomes packaged as TIPs. (A) Virion and viral genome schematic of NiV. Viral genome is represented in the genomic polarity. 3′ Le, leader sequence; N, nucleoprotein; P, phosphoprotein; M, matrix protein; F, fusion protein; G, glycoprotein; RdRp, viral RNA-dependent RNA polymerase; 5′ Tr, trailer sequence. (B) Schematics of the DI genomes incorporated into TIPs.
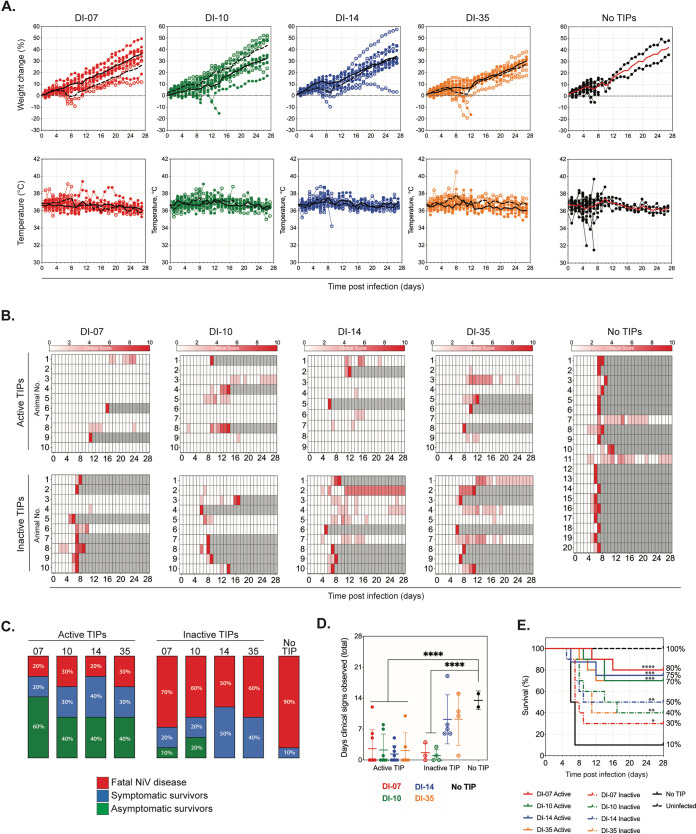
All mock-treated animals (no TIPs) exhibited weight loss between 3 to 7 dpi (Fig. 3A), as well as other clinical signs of infection (Fig. 3B); infection was lethal in 18 out of 20 (90%) hamsters. Both hamsters that survived until the end of challenge (28 dpi) exhibited persistent mild to moderate neurological signs of infection. In contrast, the majority of hamsters treated with active TIPs did not exhibit overt weight loss or temperature changes (Fig. 3A). Six of 10 (60%) of hamsters treated with active DI-07, 4 of 10 (40%) treated with active DI-10 and DI-35, and 3 of 9 (33%) treated with active DI-14 had no clinical signs of disease throughout the challenge period (Fig. 3C). Besides producing fewer clinical signs, disease in hamsters treated with active TIPs was significantly shorter than disease in mock-treated hamsters (Fig. 3D). On average, surviving active TIP-treated hamsters (n = 11) underwent 4.8 days of clinical signs, compared to 7.7 days for inactive TIP-treated survivors (n = 12) and 14 days for mock-treated animals (n = 2). Hamsters treated with inactive TIPs followed a clinical course closer to that of the mock-treated animals: the majority displayed weight loss between 3 to 7 dpi accompanied by other clinical signs of infection. When clinical signs were present in TIP-treated hamsters, they were more likely to appear in those treated with inactive TIPs (92.5%) than those treated with active TIPs (60.5%).
FIG 3.
Intraperitoneal NiV challenge and treatment with active or inactive TIPs. Hamsters were inoculated i.p. with 2 mL DMEM containing 104 TCID50 NiV strain Malaysia in combination with approximately 2 × 109 active TIPs (treated with 100 mJ/cm2 of UV), inactive TIPs (treated with 2,400 mJ/cm2 of UV), or no TIPs (mock treated). (A) Graphs represent percentage of weight change from baseline (taken at −1 dpi) and body temperatures. Individual animals are represented. DI-07, red; DI-10, green; DI-14, blue; DI-35, orange; no TIP (mock treated), black; closed circles represent hamsters treated with active TIPs, and open circles represent those treated with inactive TIPs. Lines indicate the mean value each day as follows: solid line for hamsters treated with active TIPs and dashed line for those treated with inactive TIPs. (B) Clinical signs (scored from 0 to 10), with severity depicted by increasing intensity of red. Animals scoring ≥10 were humanely euthanized; any animals that succumbed to disease prior to euthanasia were allocated a score of 10. Gray boxes indicate the end of monitoring/scoring due to euthanasia/death. (C) The proportion of hamsters from each group that succumbed (euthanized due to clinical signs or found dead) to NiV disease (red), those that survived to end of study at 28 dpi with having displayed at least 1 day of clinical signs (blue), or with no clinical signs (green). (D) Number of days that any hamster surviving to study end (28 dpi) exhibited clinical signs. Closed circles represent animals treated with active TIPs; open circles, those treated with inactive TIPs; and black circles, mock treated. Individual animals are represented, with lines indicating mean and standard deviation. Significance calculated by multiple t tests; ****, P ≤ 0.0001. (E) Survival of hamsters in each group. In groups containing treated hamsters, solid line represents active TIPs, and dashed line represents inactive TIPs. Significance calculated by log-rank (Mantel-Cox) test as follows: ****, P ≤ 0.0001; ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05.
Overall, TIP treatment significantly (DI-07, P = 0.0001; DI-10, P = 0.0001; DI-14, P = 0.0009; DI-35, P = 0.0002) increased survival from 10% in mock-treated animals to 70 to 80% in active TIP-treated hamsters (Fig. 3E). Survival in hamsters treated with inactive TIPs was decreased compared to those treated with active TIPs, although these animals remained significantly protected from lethal disease (DI-07, P = 0.0367; DI-10, P = 0.0053; DI-14, P = 0.0094; DI-35, P = 0.0104) compared to mock-treated animals (30 to 50%). Corresponding to improved outcomes, levels of viral RNA in tissues were lower in survivors (both symptomatic and asymptomatic) than in animals that succumbed to infection (Fig. S2 in the supplemental material). Of the TIP-treated animals that succumbed to infection, reduction in RNA levels was more pronounced in hamsters treated with active TIPS than with inactive TIPs.
Determination of viral RNA levels in tissues taken from TIP-treated hamsters challenged intraperitoneally with NiV. RT-qPCR was performed based on primer and probe sets specific for NiV strain Malaysia (NiV-M) nucleoprotein (N) on select tissues taken from TIP-treated hamsters challenged i.p. with 104 TCID50 NiV. Hamsters were classified based on whether they succumbed (euthanized due to clinical signs or found dead) to NiV disease (fatal), survived to end of study at 28 dpi with least one day of clinical signs (symptomatic survivors), or survived with no clinical signs (asymptomatic survivors). Individual animals are represented; in groups containing treated hamsters, solid symbols represent hamsters treated with active TIPs, and open symbols represent animals treated with inactive TIPs. Download FIG S2, TIF file, 1.4 MB (1.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Low-dose TIPs can protect against severe outcomes when hamsters are treated and challenged intranasally.
Given the success of the simultaneous i.p. challenge and treatment experiment, we next evaluated TIP treatment using the i.n. route, which more closely mimics the natural exposure and transmission dynamics of NiV by monitoring clinical signs, outcome, and endpoint viral RNA in tissues (Fig. 4C; Fig. S3). Given the limitations regarding the maximum volume allowed for i.n. inoculation in hamsters, plus the requirement for a higher-challenge dose needed to see appreciable disease and mortality (106 TCID50), the maximum achievable TIP/NiV ratio for this experiment was only ∼100:1 (in 100 μL) compared to ∼20,000:1 achieved in the i.p. experiment (in 2 mL). Given the reduced protective effect we observed previously with inactive TIPs, hamsters were only challenged with a combination of NiV plus active TIPs.
FIG 4.
Intranasal NiV challenge and treatment with active TIPs. Hamsters were inoculated i.n. with 100 μL containing 106 TCID50 NiV strain Malaysia in combination with either 1 × 108 active (treated with 100 mJ/cm2 of UV) TIPs or no TIPs (mock treated). (A) Graphs represent the percent weight change from baseline (taken at −1 dpi) and body temperatures. Individual animals are represented. DI-07, red; DI-10, green; DI-14, blue; DI-35, orange; no TIP (mock treated), black; line indicates the mean value each day. (B) Clinical signs (scored from 0 to 10), with severity depicted by increasing intensity of red. Animals scoring ≥10 were humanely euthanized; any animals that succumbed to disease prior to euthanasia were allocated a score of 10. Gray boxes indicate the end of monitoring/scoring due to euthanasia/death. (C) Proportion of hamsters from each group that succumbed (euthanized due to clinical signs or found dead) to NiV disease (red) or survived to end of study at 28 dpi having displayed at least 1 day of clinical signs (blue) or no clinical signs (green). (D) Number of days that any hamster surviving to study end (28 dpi) exhibited clinical signs. Individual animals are represented, with lines indicating mean and standard deviation. Significance calculated by multiple t test; ****, P ≤ 0.0001. (E) Survival of hamsters in each group. In groups containing treated hamsters. Significance calculated by log-rank (Mantel-Cox) test as follows: ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05.
Determination of viral RNA levels in tissues taken from TIP-treated hamsters challenged intranasally with NiV. RT-qPCR was performed based on primer and probe sets specific for NiV strain Malaysia nucleoprotein (N) on select tissues taken from TIP-treated hamsters challenged i.n. with 106 TCID50 NiV strain Malaysia. Hamsters were classified based on whether they succumbed (euthanized due to clinical signs or found dead) to NiV disease (fatal), survived to end of study at 28 dpi having displayed at least one day of clinical signs (symptomatic survivors), or survived with no clinical signs (asymptomatic survivors). Individual animals are represented. Download FIG S3, TIF file, 1.2 MB (1.2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Even with a 200-fold reduction in TIP/NiV ratio compared to i.p. delivery, i.n. TIP treatment conferred high levels of protection from NiV infection. All mock-treated hamsters (n = 20) exhibited weight loss between 3 to 6 dpi, and temperature changes associated with hypoactivity (hypothermia) or severe seizures (hyperthermia) were observed in several animals (Fig. 4A); all animals displayed clinical signs at some point during the challenge period (Fig. 4B). In contrast, only 3 of 8 (37.5%) animals treated with DI-14 and 5 of 8 (62.5%) animals treated with DI-35 displayed any clinical signs throughout the challenge period (Fig. 4C). Again, the duration of observed clinical signs was significantly reduced in some TIP-treated hamsters (DI-14 and DI-35) compared to mock-treated animals, although the majority of DI-07- and DI-10-treated animals also had reduced clinical periods of observable disease (Fig. 4D). On average, surviving active TIP-treated hamsters (n = 14) underwent 6.1 days of clinical signs, while mock-treated animals underwent 13.4 days (n = 5). Intranasally treated hamsters had a significantly higher survival rate (63 to 80%; DI-07, P = 0.0008; DI-10, P = 0.0476; DI-14, P = 0.0085; DI-35, P = 0.0203) than mock-treated animals (25%) (Fig. 4E).
TIP treatment protects hamsters from death and clinical disease in a dose-dependent manner.
Previous data indicated that TIP-mediated inhibition of NiV in vitro was dependent on the TIP/NiV ratio (24). Here, we found that DI delivery in vivo also confers a significant protective effect at TIP/NiV ratio of 100:1 (for i.n. inoculation), even though no in vitro inhibition was observed at this ratio. To investigate the dose-dependent effect of TIP-mediated protection in vivo, we simultaneously administered TIPs and challenge virus i.p., as this route allowed a greater range of ratios to be investigated. We used DI-14, the best-performing candidate from previous in vivo experiments, to assess the efficacy of different TIP:NiV ratios from a maximum of 10,000:1 to a minimum of 123:1 in hamsters, alongside mock-treated (DMEM-only) control animals.
Clinical signs and mortality were greater in animals treated with lower TIP:NiV ratios than those treated with the original 20,000:1 ratio of TIP:NiV (Fig. 5A). Compared to mock-treated hamsters, significant differences in survival were conferred with treatment ratios of both 10,000:1 (P = 0.0030) and 3,333:1 (P = 0.0114), but not at the remaining lower ratios tested (Fig. 5B). While the two lower-delivery ratios (10,000:1 and 3,333:1) improved survival outcomes (per survival and/or mean time to death) compared to no treatment, they did not prevent clinical signs. Two of the three surviving animals in the highest ratio group tested (10,000:1) exhibited mild neurological signs over 1 to 3 days between 10 to 14 dpi but were free of clinical signs by study completion at 28 dpi (Fig. 5C). The remaining animal exhibited persistent mild to moderate neurological signs from 8 dpi until study end (20 days total). Similarly, the 1 survivor from the 3,333:1 ratio group exhibited mild to moderate neurological signs from 6 dpi until study end (19 days total). These data indicate that reduction in clinical disease and decreased mortality require high delivery ratios to allow optimal TIP-mediated protection.
FIG 5.
Protection from NiV disease in hamsters treated with different TIP/NiV ratios. Hamsters were inoculated i.p. with 2 mL DMEM containing 104 TCID50 NiV strain Malaysia in combination with different concentrations of active TIPs DI-14 (treated with 100 mJ/cm2 of UV) or with no TIPs (mock treated). Final ratios of TIP/NiV were 10,000:1 (purple), 3,333:1 (green), 1,111:1 (orange), 370:1 (blue), 123:1 (cyan), and 0:1 (black). (A) Mean daily percent weight change from baseline (taken at −1 dpi) for each group. The weights of uninfected control hamsters are shown as daily means (dashed black line) and range (gray area). (B) Survival of hamsters in each group. Also shown with a dashed gray line are findings from a previous experiment (Fig. 2) in which hamsters were treated with active TIP DI-14 at 20,000:1 ratio for comparison. Significance calculated by log-rank (Mantel-Cox) test as follows: ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05. (C) Clinical signs (scored from 0 to 10), with severity depicted by increasing intensity of red. Animals scoring ≥10 were humanely euthanized; any animals that succumbed to disease prior to euthanasia were allocated a score of 10. Gray boxes indicate the end of monitoring/scoring due to euthanasia/death.
TIP treatment in hamsters results in strong but transient immunostimulatory response.
To investigate the immunostimulatory properties of TIP treatment in the hamster model and to assess its potential contribution to protective efficacy, groups of 5 hamsters were treated i.n. with (i) active TIP only (either DI-07 or DI-14), (ii) a combination of active TIP (either DI-07 or DI-14) and 106 TCID50 NiV, or (iii) 106 TCID50 NiV alone. Hamsters were euthanized at 1 and 4 dpi, and RNA extracted from lung tissues was examined for differentially expressed immune genes in interferon (IFN; sensing, induction, and signaling) and cytokine/chemokine expression pathways.
As expected, due to well-characterized IFN antagonism by NiV (40, 41), upregulation of IFN or interferon-stimulated genes (ISGs) was not observed in infected untreated hamsters (NiV alone). In TIP-treated hamsters, type I and type II interferon (IFNA-23-like, IFNA-13-like, IFNA-9-like, IFNE, IFNG, and IFNK) were strongly upregulated at 1 dpi (Fig. 6). However, this was not seen in TIP-treated infected hamsters (TIP plus NiV). Since TIP propagation initially relies on viral replication, some level of cell-to-cell spread of virus is likely to occur early in infection. This spread may confer immunosuppressive activity, counteracting the TIP-induced upregulation seen in tissues from animals receiving TIP alone. Pattern recognition receptor (PRR) genes involved in the antiviral response (DDX58, DHX58, and IFIH1) and those associated with IFN-mediated effector pathways (MX1 and OAS2) were slightly upregulated (2- to 3-fold) in DI-14 treated, but not DI-10 treated, hamsters. This may be due to the structural differences between the two DI variants (copyback versus deletion) and may explain the small difference in treatment efficacy noted between the two. However, these effects were transient, with changes observed at 1 dpi largely absent at 4 dpi. In hamsters treated with TIP plus NiV, any differentially expressed (DE) genes were largely similar to those observed in the NiV-alone control animals. A similar pattern was true for genes involved in cytokine and chemokine expression. At 1 dpi, in hamsters treated only with active TIP, several genes involved in inflammatory pathways and response to infection (CCL1, CSF3, IL-10, IL-12A, IL-17A, and IL-2) were strongly upregulated, which was not seen in TIP plus NIV and NiV-only groups (Fig. 6). Again, these effects were transient and absent by 4 dpi, and at both time points, the DE gene pattern was largely similar in both TIP plus NiV and the NiV-alone control animals.
FIG 6.
Early interferon signaling and cytokine/chemokine expression in hamsters treated with active TIPs. Hamsters inoculated i.n. with 100 μL DMEM containing 106 TCID50 NiV strain Malaysia in combination with either 1 × 108 active TIPs (treated with 100 mJ/cm2 of UV) or no TIPs (mock treated) were euthanized at 1 or 4 dpi (n = 5 each group). Lung tissues were collected for mRNA analysis. Heatmaps represent fold changes in transcript levels (compared to mock-treated hamsters at each day) of mRNA associated with genes from interferon induction and signaling or cytokine and chemokine signaling pathways. Intensity of color corresponds to magnitude of fold change over baseline.
DISCUSSION
Here, we demonstrate that TIPs can provide robust protection in a lethal hamster model of NiV disease. A single-dose TIP treatment reduced mortality, and TIP-treated hamsters were less likely to exhibit clinical signs of infection. If present, clinical signs in TIP-treated animals were shorter in duration than signs in mock-treated animals. These results are encouraging compared to other reported NiV treatment options. While some treatments, including the antiviral compounds T-705 (42) and remdesivir (43) and the neutralizing antibodies m102.4 (44, 45) and h5b3.1 (46), have been shown to protect against disease and decrease mortality when given pre- and/or postexposure, they all require multiple doses to achieve protection. Here, we assess single-dose concurrent delivery of TIPs with virus challenge. Additional characterization of TIPs given prophylactically and postexposure will be key to determining optimal timing for TIP treatment alone or in conjunction with other interventions and will provide data to help develop putative treatment plans for patient care.
To date, few reports have investigated inhibitory effects of DIs in vitro and in vivo. Inoculation with a high dose of highly purified, uncharacterized vesicular stomatitis virus (VSV) DI particles protected against lethal VSV challenge in mice (47, 48). Similarly, Semliki Forest virus (SFV) preparations high in uncharacterized DI genomes protected against lethal SFV challenge in mice (49), and further studies using a single cloned characterized DI candidate also protected mice against SFV challenge (50). The majority of work investigating DI treatment in vivo centers on influenza A virus (IAV). IAV DI244 (deletion in genome segment PB2), given either prophylactically or postexposure, increased survival and reduced clinical signs in IAV-infected mice and ferrets (25, 27–29), and DIG-3 (deletions in genome segments PB2, PB1, and polymerase acidic [PA]) protected against IAV challenge in mice (26). In addition, DI244 protected against heterologous influenza B virus challenge in mice (30). Due to differences in DI genome structures, DI quantification methods, experimental processes, challenge doses, and viral pathogenesis, comparisons between these data and ours are difficult. However, these promising findings for influenza and our findings regarding NiV support broader development and application of DIs for other diverse pathogens.
Hamsters were protected from NiV challenge using both a high-TIP/NiV-ratio i.p. dose (∼20,000:1) and a low-TIP/NiV-ratio i.n. dose (∼100:1). Dimmock et al. achieved protection using a ratio of DI244:IAV between 100,000:1 to 1,000,000:1 (51), but their studies did not evaluate lower ratios. Our data demonstrate that lower doses of DIs can provide protective effects and that efficacious ratios determined in vitro may underestimate the treatment potential of lower doses. However, our data also show that protective ratios are associated with route of delivery and infection. For example, in hamsters, NiV disease progresses more rapidly, and time to death is shorter with i.p. challenge than i.n. challenge (31, 52). Intranasal challenge may therefore allow TIPs more time to exert their protective effects. Alternatively, i.n. administration may allow more targeted delivery of TIPs. Respiratory disease in NiV-infected hamsters is characterized by extensive viral replication in the respiratory tract and lungs (32, 33, 53). Direct delivery of TIPs to the cells where initial NIV infection occurs may thus facilitate protection at lower-ratio doses.
Mechanisms of TIP-mediated viral inhibition can be broadly divided into two categories, direct and indirect. Direct inhibition is a result of DI genomes interfering with replication of viral genomes, whereas indirect inhibition may be due to nonspecific immunostimulatory properties of the TIPs (54–58). Several inherent factors in the DI structure have been shown to influence their direct inhibitory potential, including DI length (17, 24, 59, 60), relative strengths of the replication promoter elements incorporated into the DI (61–63), and efficiency of DI genome packaging (64, 65). However, these structural features can also influence mechanisms related to indirect interference. For example, double-stranded hairpin loops formed by nonencapsidated copyback genomes are readily recognized by cytosolic innate immune pattern recognition receptors like RIG-I, MDA5, and LGP2 (66–69).
Our data suggest that while both direct and indirect mechanisms play a role, direct interference is the predominant mechanism for protection by NiV TIPs. When the RNA component of TIPs was destroyed by UV irradiation, eliminating direct interference capacity (inactive TIPs), we saw a significant reduction in the protective effect of TIP treatment. Furthermore, of the candidates evaluated, copyback DIs conferred the highest levels of protection. Copyback DIs have two copies of the 5′-trailer sequence, which contains the stronger of the two cis-acting replication promoter elements found in the NiV genome. Therefore, copyback DI genomes are generated at a higher rate than deletion Dis. They may thus exert greater direct inhibitory pressure on full-length viral genome replication and may also result in comparably higher, albeit still low, nonspecific immunostimulatory effects. Interestingly, the shortest copyback (DI-35) performed more poorly than the slightly longer copybacks (DI-07 and DI-14). The shorter length of DI-35 may have resulted in the removal of one or more elements involved in efficient packaging, suggesting that the optimal length of DI genomes is 1.0 to 1.5 kb.
Destruction of the RNA component of TIPs did not completely abolish their inhibitory potential. Inactive TIPs are structurally similar to classical VLPs, and protection from respiratory diseases via VLP vaccination is reported for respiratory syncytial virus (RSV) (70, 71), influenza (72), and SARS1 (73). The low-level protection we observed by TIPs in this “inactive” state was likely due to indirect stimulation of the innate immune system. “Inactive” TIPs are likely not completely inert and may still produce pathogen-associated molecular patterns (PAMPs) that can activate some level of innate immune response; this is supported by the observed upregulation of several pathways for IFN production and cytokine signaling in animals treated with TIPs alone. While fast-acting and strong for many genes, the immunostimulatory effects of the TIPs in our study were predominantly transient in effect, with a return to basal levels by 4 dpi. TIPs are presumed to be inert in the absence of parental virus, however, low-level replication activity may occur due to co-packaging of viral polymerase with DI genome. This activity may have led to the transient nature of the immunostimulation. As nucleotide sequences and motifs can be manipulated to enhance and prolong immunostimulatory effects (74, 75), DI genomes may be further optimized to increase the contribution of indirect inhibition, thereby improving the overall protective efficacy of TIPs.
Here, we provide support for continued studies of the clinical utility of TIP treatment. NiV is a highly lethal disease circulating in areas with exceptionally dense human populations. Developing a therapeutic candidate that is easily administered and achieves high levels of protection is both indicated and required to prevent human disease and to provide intervention options in outbreak response. Here, we provide the first evidence that TIPs can confer protection against disease and lethal outcome in a robust animal model of NiV, supporting further investigations of both treatment and therapeutic use of DI for NiV and other high-consequence pathogens.
MATERIALS AND METHODS
Biosafety.
All work with infectious virus or infected animals was conducted in a biosafety level 4 (BSL-4) laboratory at the Centers for Disease Control and Prevention (CDC) following established BSL-4 standard operating procedures approved by the Institutional Biosafety Committee. All recombinant virus work was approved by the Centers for Disease Control and Prevention Institutional Biosafety Committee.
Cells and viruses.
Vero-E6 and BSR-T7/5 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% (vol/vol) fetal calf serum, nonessential amino acids, 1 mM sodium pyruvate, 2 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. HSAEC-KT1 cells (ATCC, USA) were cultured in airway epithelial cell basal medium paired with the bronchial epithelial cell growth kit medium (both ATCC, USA). NiV strain Malaysia (NiV-M; GenBank accession no. AF212302) was originally obtained from a clinical isolate, passaged once on Vero-E6 cells for isolation from clinical sample, and further amplified on Vero-E6 cells. Viral titers were calculated as 50% tissue culture infective dose (TCID50) in Vero-E6 cells (76). All viral stocks were verified by next-generation sequencing and confirmed to be mycoplasma free.
TIP production.
TIPs were produced as previously described (Fig. S1 in the supplemental material) (24). To generate these virus-like particles containing the DI genomes, a modified NiV reverse genetics rescue system (40) was developed in which a plasmid transcribing the DI genome plus support plasmids expressing the proteins required for transcription and genome replication were transfected into BSR-T7/5 cells. After 2 days, the transfected cells were overlaid with Vero-E6 cells and infected with a recombinant NiV-M-expressing ZsG fluorescent protein (rNiV-M/ZsG) in order to supply the necessary proteins for assembly and release of the virus-like particles. To remove the infectious rNiV-M/ZsG from the final product, a precise dose of UV radiation was used (100 mJ/cm2 of UV radiation [CX-2000 Crosslinker; UVP] in a 6-well plate [1 mL per well]), which has been previously shown to result in breakage of full-length RNA genomes (uncoupling the leader and trailer cis-acting elements and therefore removing the ability to replicate full-length genome) while leaving the shorter DI genomes intact (24). The resulting products were termed active TIPs. The TIPs were quantified using a DI genome-specific digital droplet quantitative reverse transcription-PCR (RT-qPCR) assay; the above method produced an average of approximately 1 × 109 DI genome copies/mL. To create stocks depleted of both the producer virus and DI genome (termed inactive TIPs), stocks were treated with 10 consecutive treatments of 240 mJ/cm2 of UV radiation (2,400 mJ/cm2 total).
Generation of TIPs for in vivo studies. All plasmids required for genome replication and encapsidation processes were transfected into BSR-T7/5 cells, followed 2 days later by addition of Vero-E6 cells and infectious rNiV-M/ZsG (recombinant Nipah virus strain Malaysia [NiV-M] capable of expressing the reporter protein ZsGreen1), which supplies all required proteins for TIP assembly and release. Supernatants containing DIs and nascent NiV virions are treated with UV to remove infectious rNiV-M/ZsG. Download FIG S1, TIF file, 0.6 MB (589.3KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Digital droplet quantitative reverse transcription-PCR.
A 20× mix of DI-specific or NiV-specific primers and probes (18 μM each primer and 5 μM probe) were used in conjunction with 1-Step RT-digital droplet PCR (ddPCR) advanced kit for probes and droplet generation oil for probes (both from Bio-Rad). Duplex RT-qPCR assays were set up and run per manufacturers’ conditions, with droplets generated using QX200 droplet generator (Bio-Rad). Results were analyzed using QX200 droplet digital PCR system, and quantification data were generated using QuantaSoft software (both from Bio-Rad).
Hamster studies.
All animal experiments were approved by the CDC Institutional Animal Care and Use Committee and performed in an AAALAC International-approved facility. Data are based on four independent hamster studies using 6-week-old HsdHan:Aura Syrian hamsters (Envigo; catalog no. 8903F or 8903M). The DI dose-effect study used experimental groups of 8 to 10 hamsters (half female and half male). All other hamster studies used experimental groups of 5 to 10 female hamsters. Hamsters were inoculated either intranasally (i.n.; 100 μL divided bilaterally) or intraperitoneally (i.p.; 2 mL) with Dulbecco’s modified Eagle’s medium (DMEM) alone, NiV strain Malaysia (NiV-M) in DMEM, or a mixture of NiV-M and TIP in DMEM. Hamsters were kept in a climate-controlled laboratory with a 12-h day/night cycle, provided Teklad global 18% protein rodent diet (Envigo) and water ad libitum, and group housed on corn cob bedding (Bed-o'Cobs, ¼ in.; Anderson Lab Bedding, Maumee, OH, USA) with cotton nestlets and crinkle paper in an isolator caging system (Thoren Caging, Inc., Hazleton, PA, USA) with a HEPA-filtered inlet and exhaust air supply. Microchip transponders (BMDS; IPTT-300) were placed subcutaneously in the interscapular region at −4 dpi for individual identification and to assess body temperature. Baseline weights were taken −1 dpi, and hamsters’ weight change, body temperatures, and clinical signs were assessed daily. Animals were scored by the following criteria: quiet, dull responsive, hunched back/ruffled coat, hypoactivity, mild neurological signs, each 2 points; abnormal breathing (i.e., increased respiratory rate, dyspnea), hypothermia (<34°C), moderate neurological signs, each 5 points; and paralysis, frank hemorrhage, moribund, weight loss >25% of baseline (measured at −1 dpi), severe neurological signs, each 10 points. Neurological signs were classified as (i) mild, with abnormal gait or movement and/or mild head tilt (∼0 to 30° from vertical or sporadic); (ii) moderate, with tremors, ataxia, circling, absence seizures, and/or moderate head tilt (∼30 to 90° from vertical and persistent) while retaining the ability to walk, feed, and drink; or (iii) severe, with limb paralysis, tonic clonal seizure, inability to return to normal posture, and/or severe head tilt (>90° from vertical). Hamsters were euthanized with isoflurane vapor when they met euthanasia criteria (score ≥ 10) or at the completion of the study (28 dpi). Survival data from the 10 hamsters challenged with 105 TCID50 i.p. in Fig. 1B were previously reported unvaccinated control animals (38).
Quantitative RT-PCR.
Viral RNA was extracted from homogenized tissue samples and whole blood using the MagMAX-96 total RNA isolation kit (Thermo Fisher Scientific) on a 96-well ABI MagMAX extraction platform with a DNase I treatment step according to the manufacturer’s instructions. RNA was detected using a RT-qPCR assay targeting the NiV NP gene sequence (NiV forward, 5′-CTGGTCTCTGCAGTTATCACCATCGA-3′; NiV reverse, 5′-ACGTACTTAGCCCATCTTCTAGTTTCA-3′; and NiV probe, 5′-FAM-CAGCTCCCGACACTGCCGAGGAT-BHQ1-3′; all from IDT), with levels normalized to 18S RNA values using a commercial endogenous control assay (Thermo Fisher). Genome copy numbers were determined using standards prepared from in vitro-transcribed NP RNA.
Statistics and graphing.
Survival statistics were calculated using the log-rank (Mantel-Cox) test. Significance in duration of observable clinical signs was calculated using multiple t tests. All graphs and statistical analyses were created in GraphPad Prism (v9.3.0).
ACKNOWLEDGMENTS
We thank Tatyana Klimova for assistance with editing the manuscript.
This work was partially supported by an appointment to the Research Participation Program at the Centers for Disease Control and Prevention (CDC) administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and CDC (S.R.W.), the DARPA INTERfering and Co-Evolving Prevention and Therapy (INTERCEPT) program (DARPA-BAA-16-35) managed by Jim Gimlett, Brad Ringiesen, and Seth Cohen, and by CDC Emerging Infectious Disease Research Core Funds.
We have no reported conflicts of interest.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Contributor Information
Christina F. Spiropoulou, Email: ccs8@cdc.gov.
Dimitrios Paraskevis, Medical School, National and Kapodistrian University of Athens.
REFERENCES
- 1.Amarasinghe GK, Bào Y, Basler CF, Bavari S, Beer M, Bejerman N, Blasdell KR, Bochnowski A, Briese T, Bukreyev A, Calisher CH, Chandran K, Collins PL, Dietzgen RG, Dolnik O, Dürrwald R, Dye JM, Easton AJ, Ebihara H, Fang Q, Formenty P, Fouchier RAM, Ghedin E, Harding RM, Hewson R, Higgins CM, Hong J, Horie M, James AP, Jiāng D, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lee B, Leroy EM, Li M, Maisner A, Mühlberger E, Netesov SV, Nowotny N, Patterson JL, Payne SL, Paweska JT, Pearson MN, Randall RE, Revill PA, Rima BK, Rota P, Rubbenstroth D, et al. 2017. Taxonomy of the order Mononegavirales: update 2017. Arch Virol 162:2493–2504. doi: 10.1007/s00705-017-3311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lo MK, Rota PA. 2008. The emergence of Nipah virus, a highly pathogenic paramyxovirus. J Clin Virol 43:396–400. doi: 10.1016/j.jcv.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Chua KB, Goh KJ, Wong KT, Kamarulzaman A, Tan PS, Ksiazek TG, Zaki SR, Paul G, Lam SK, Tan CT. 1999. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet (London, England) 354:1257–1259. doi: 10.1016/S0140-6736(99)04299-3. [DOI] [PubMed] [Google Scholar]
- 4.Chua KB, Lam SK, Goh KJ, Hooi PS, Ksiazek TG, Kamarulzaman A, Olson J, Tan CT. 2001. The presence of Nipah virus in respiratory secretions and urine of patients during an outbreak of Nipah virus encephalitis in Malaysia. J Infect 42:40–43. doi: 10.1053/jinf.2000.0782. [DOI] [PubMed] [Google Scholar]
- 5.Paton NI, Leo YS, Zaki SR, Auchus AP, Lee KE, Ling AE, Chew SK, Ang B, Rollin PE, Umapathi T, Sng I, Lee CC, Lim E, Ksiazek TG. 1999. Outbreak of Nipah-virus infection among abattoir workers in Singapore. Lancet (London, England) 354:1253–1256. doi: 10.1016/S0140-6736(99)04379-2. [DOI] [PubMed] [Google Scholar]
- 6.Hsu VP, Hossain MJ, Parashar UD, Ali MM, Ksiazek TG, Kuzmin I, Niezgoda M, Rupprecht C, Bresee J, Breiman RF. 2004. Nipah virus encephalitis reemergence, Bangladesh. Emerg Infect Dis 10:2082–2087. doi: 10.3201/eid1012.040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chadha MS, Comer JA, Lowe L, Rota PA, Rollin PE, Bellini WJ, Ksiazek TG, Mishra A. 2006. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis 12:235–240. doi: 10.3201/eid1202.051247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luby SP. 2013. The pandemic potential of Nipah virus. Antiviral Res 100:38–43. doi: 10.1016/j.antiviral.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Chua KB, Koh CL, Hooi PS, Wee KF, Khong JH, Chua BH, Chan YP, Lim ME, Lam SK. 2002. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect 4:145–151. doi: 10.1016/s1286-4579(01)01522-2. [DOI] [PubMed] [Google Scholar]
- 10.Halpin K, Hyatt AD, Fogarty R, Middleton D, Bingham J, Epstein JH, Rahman SA, Hughes T, Smith C, Field HE, Daszak P, Henipavirus Ecology Research Group . 2011. Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am J Trop Med Hyg 85:946–951. doi: 10.4269/ajtmh.2011.10-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chew MH, Arguin PM, Shay DK, Goh KT, Rollin PE, Shieh WJ, Zaki SR, Rota PA, Ling AE, Ksiazek TG, Chew SK, Anderson LJ. 2000. Risk factors for Nipah virus infection among abattoir workers in Singapore. J Infect Dis 181:1760–1763. doi: 10.1086/315443. [DOI] [PubMed] [Google Scholar]
- 12.Gurley ES, Montgomery JM, Hossain MJ, Bell M, Azad AK, Islam MR, Molla MAR, Carroll DS, Ksiazek TG, Rota PA, Lowe L, Comer JA, Rollin P, Czub M, Grolla A, Feldmann H, Luby SP, Woodward JL, Breiman RF. 2007. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis 13:1031–1037. doi: 10.3201/eid1307.061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikolay B, Salje H, Hossain MJ, Khan AKMD, Sazzad HMS, Rahman M, Daszak P, Ströher U, Pulliam JRC, Kilpatrick AM, Nichol ST, Klena JD, Sultana S, Afroj S, Luby SP, Cauchemez S, Gurley ES. 2019. Transmission of Nipah virus - 14 years of investigations in Bangladesh. N Engl J Med 380:1804–1814. doi: 10.1056/NEJMoa1805376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roux L, Simon AE, Holland JJ. 1991. Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv Virus Res 40:181–211. doi: 10.1016/s0065-3527(08)60279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vignuzzi M, López CB. 2019. Defective viral genomes are key drivers of the virus–host interaction. Nat Microbiol 4:1075–1087. doi: 10.1038/s41564-019-0465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alnaji FG, Brooke CB. 2020. Influenza virus DI particles: defective interfering or delightfully interesting? PLoS Pathog 16:e1008436. doi: 10.1371/journal.ppat.1008436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calain P, Monroe MC, Nichol ST. 1999. Ebola virus defective interfering particles and persistent infection. Virology 262:114–128. doi: 10.1006/viro.1999.9915. [DOI] [PubMed] [Google Scholar]
- 18.Tilston-Lunel NL, Welch SR, Nambulli S, de Vries RD, Ho GW, Wentworth DE, Shabman R, Nichol ST, Spiropoulou CF, de Swart RL, Rennick LJ, Duprex WP. 2021. Sustained replication of synthetic canine distemper virus defective genomes in vitro and in vivo. mSphere 6:e0053721. doi: 10.1128/mSphere.00537-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strahle L, Garcin D, Kolakofsky D. 2006. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology 351:101–111. doi: 10.1016/j.virol.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 20.Ziegler CM, Botten JW. 2020. Defective interfering particles of negative-strand RNA viruses. Trends Microbiol 28:554–565. doi: 10.1016/j.tim.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Lyu T, Zhou R, He X, Ye K, Xie Q, Zhu L, Chen T, Shen C, Wu Q, Zhang B, Zhao W. 2019. The antiviral and antitumor effects of defective interfering particles/genomes and their mechanisms. Front Microbiol 10:1852. doi: 10.3389/fmicb.2019.01852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Y, Jain D, Koziol-White CJ, Genoyer E, Gilbert M, Tapia K, Panettieri RA, Hodinka RL, López CB. 2015. Immunostimulatory defective viral genomes from respiratory syncytial virus promote a strong innate antiviral response during infection in mice and humans. PLoS Pathog 11:e1005122. doi: 10.1371/journal.ppat.1005122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yount JS, Gitlin L, Moran TM, Lopez CB, López CB. 2008. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to sendai virus defective interfering particles. J Immunol 180:4910–4918. doi: 10.4049/jimmunol.180.7.4910. [DOI] [PubMed] [Google Scholar]
- 24.Welch SR, Tilston NL, Lo MK, Whitmer SLM, Harmon JR, Scholte FEM, Spengler JR, Duprex WP, Nichol ST, Spiropoulou CF. 2020. Inhibition of Nipah virus by defective interfering particles. J Infect Dis 221:S460–S470. doi: 10.1093/infdis/jiz564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimmock NJ, Rainsford EW, Scott PD, Marriott AC. 2008. Influenza virus protecting RNA: an effective prophylactic and therapeutic antiviral. J Virol 82:8570–8578. doi: 10.1128/JVI.00743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao H, To KKW, Chu H, Ding Q, Zhao X, Li C, Shuai H, Yuan S, Zhou J, Kok K-H, Jiang S, Yuen K-Y. 2018. Dual-functional peptide with defective interfering genes effectively protects mice against avian and seasonal influenza. Nat Commun 9:2358. doi: 10.1038/s41467-018-04792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimmock NJ, Dove BK, Scott PD, Meng B, Taylor I, Cheung L, Hallis B, Marriott AC, Carroll MW, Easton AJ. 2012. Cloned defective interfering influenza virus protects ferrets from pandemic 2009 influenza A virus and allows protective immunity to be established. PLoS One 7:e49394. doi: 10.1371/journal.pone.0049394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimmock NJ, Dove BK, Meng B, Scott PD, Taylor I, Cheung L, Hallis B, Marriott AC, Carroll MW, Easton AJ. 2012. Comparison of the protection of ferrets against pandemic 2009 influenza A virus (H1N1) by 244 DI influenza virus and oseltamivir. Antiviral Res 96:376–385. doi: 10.1016/j.antiviral.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott PD, Meng B, Marriott AC, Easton AJ, Dimmock NJ. 2011. Defective interfering influenza virus confers only short-lived protection against influenza virus disease: evidence for a role for adaptive immunity in DI virus-mediated protection in vivo. Vaccine 29:6584–6591. doi: 10.1016/j.vaccine.2011.06.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott PD, Meng B, Marriott AC, Easton AJ, Dimmock NJ. 2011. Defective interfering influenza A virus protects in vivo against disease caused by a heterologous influenza B virus. J Gen Virol 92:2122–2132. doi: 10.1099/vir.0.034132-0. [DOI] [PubMed] [Google Scholar]
- 31.Wong KT, Grosjean I, Brisson C, Blanquier B, Fevre-Montange M, Bernard A, Loth P, Georges-Courbot M-CC, Chevallier M, Akaoka H, Marianneau P, Lam SK, Wild TF, Deubel V. 2003. A Golden hamster model for human acute Nipah virus infection. Am J Pathol 163:2127–2137. doi: 10.1016/S0002-9440(10)63569-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rockx B, Brining D, Kramer J, Callison J, Ebihara H, Mansfield K, Feldmann H. 2011. Clinical outcome of henipavirus infection in hamsters is determined by the route and dose of infection. J Virol 85:7658–7671. doi: 10.1128/JVI.00473-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Wit E, Bushmaker T, Scott D, Feldmann H, Munster VJ. 2011. Nipah virus transmission in a hamster model. PLoS Negl Trop Dis 5:e1432. doi: 10.1371/journal.pntd.0001432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rockx B. 2014. Recent developments in experimental animal models of henipavirus infection. Pathog Dis 71:199–206. doi: 10.1111/2049-632X.12149. [DOI] [PubMed] [Google Scholar]
- 35.Geisbert TW, Feldmann H, Broder CC. 2012. Animal challenge models of henipavirus infection and pathogenesis. Curr Top Microbiol Immunol 359:153–177. doi: 10.1007/82_2012_208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manzoni TB, López CB. 2018. Defective (interfering) viral genomes re-explored: impact on antiviral immunity and virus persistence. Future Virol 13:493–503. doi: 10.2217/fvl-2018-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genoyer E, López CB. 2019. The impact of defective viruses on infection and immunity. Annu Rev Virol 6:547–566. doi: 10.1146/annurev-virology-092818-015652. [DOI] [PubMed] [Google Scholar]
- 38.Lo MK, Spengler JR, Welch SR, Harmon JR, Coleman-McCray JD, Scholte FEM, Shrivastava-Ranjan P, Montgomery JM, Nichol ST, Weissman D, Spiropoulou CF. 2020. Evaluation of a single-dose nucleoside-modified messenger RNA vaccine encoding Hendra virus-soluble glycoprotein against lethal Nipah virus challenge in Syrian hamsters. J Infect Dis 221:S493–S498. doi: 10.1093/infdis/jiz553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lo MK, Spengler JR, Krumpe LRH, Welch SR, Chattopadhyay A, Harmon JR, Coleman-McCray JD, Scholte FEM, Hotard AL, Fuqua JL, Rose JK, Nichol ST, Palmer KE, O'Keefe BR, Spiropoulou CF. 2020. Griffithsin inhibits Nipah virus entry and fusion and can protect Syrian golden hamsters from lethal Nipah virus challenge. J Infect Dis 221:S480–S492. doi: 10.1093/infdis/jiz630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo MK, Peeples ME, Bellini WJ, Nichol ST, Rota P.a, Spiropoulou CF. 2012. Distinct and overlapping roles of Nipah virus P gene products in modulating the human endothelial cell antiviral response. PLoS One 7:e47790. doi: 10.1371/journal.pone.0047790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glennon NB, Jabado O, Lo MK, Shaw ML. 2015. Transcriptome profiling of the virus-induced innate immune response in Pteropus vampyrus and its attenuation by Nipah virus interferon antagonist functions. J Virol 89:7550–7566. doi: 10.1128/JVI.00302-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dawes BE, Kalveram B, Ikegami T, Juelich T, Smith JK, Zhang L, Park A, Lee B, Komeno T, Furuta Y, Freiberg AN. 2018. Favipiravir (T-705) protects against Nipah virus infection in the hamster model. Sci Rep 8:7604. doi: 10.1038/s41598-018-25780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo MK, Feldmann F, Gary JM, Jordan R, Bannister R, Cronin J, Patel NR, Klena JD, Nichol ST, Cihlar T, Zaki SR, Feldmann H, Spiropoulou CF, de Wit E. 2019. Remdesivir (GS-5734) protects African green monkeys from Nipah virus challenge. Sci Transl Med 11:eaau9242. doi: 10.1126/scitranslmed.aau9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bossart KN, Zhu Z, Middleton D, Klippel J, Crameri G, Bingham J, McEachern JA, Green D, Hancock TJ, Chan YP, Hickey AC, Dimitrov DS, Wang LF, Broder CC. 2009. A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute Nipah virus infection. PLoS Pathog 5:e1000642. doi: 10.1371/journal.ppat.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bossart KN, Geisbert TW, Feldmann H, Zhu Z, Feldmann F, Geisbert JB, Yan L, Feng Y-R, Brining D, Scott D, Wang Y, Dimitrov AS, Callison J, Chan Y-P, Hickey AC, Dimitrov DS, Broder CC, Rockx B. 2011. A neutralizing human monoclonal antibody protects African green monkeys from Hendra virus challenge. Sci Transl Med 3:105ra103. doi: 10.1126/scitranslmed.3002901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mire CE, Chan Y-P, Borisevich V, Cross RW, Yan L, Agans KN, Dang HV, Veesler D, Fenton KA, Geisbert TW, Broder CC. 2020. A cross-reactive humanized monoclonal antibody targeting fusion glycoprotein function protects ferrets against lethal Nipah virus and Hendra virus infection. J Infect Dis 221:S471–S479. doi: 10.1093/infdis/jiz515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holland JJ, Doyle M. 1973. Attempts to detect homologous autointerference in vivo with influenza virus and vesicular stomatitis virus. Infect Immun 7:526–531. doi: 10.1128/iai.7.4.526-531.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doyle M, Holland JJ. 1973. Prophylaxis and immunization in mice by use of virus-free defective T particles to protect against intracerebral infection by vesicular stomatitis virus. Proc Natl Acad Sci USA 70:2105–2108. doi: 10.1073/pnas.70.7.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dimmock NJ, Kennedy SI. 1978. Prevention of death in Semliki Forest virus-infected mice by administration of defective-interfering Semliki Forest virus. J Gen Virol 39:231–242. doi: 10.1099/0022-1317-39-2-231. [DOI] [PubMed] [Google Scholar]
- 50.Thomson M, White CL, Dimmock NJ. 1998. The genomic sequence of defective interfering Semliki Forest virus (SFV) determines its ability to be replicated in mouse brain and to protect against a lethal SFV infection in vivo. Virology 241:215–223. doi: 10.1006/viro.1997.8975. [DOI] [PubMed] [Google Scholar]
- 51.Dimmock NJ, Easton AJ. 2014. Defective interfering influenza virus RNAs: time to reevaluate their clinical potential as broad-spectrum antivirals? J Virol 88:5217–5227. doi: 10.1128/JVI.03193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeBuysscher BL, de Wit E, Munster VJ, Scott D, Feldmann H, Prescott J. 2013. Comparison of the pathogenicity of Nipah virus isolates from Bangladesh and Malaysia in the Syrian hamster. PLoS Negl Trop Dis 7:e2024. doi: 10.1371/journal.pntd.0002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baseler L, de Wit E, Scott DP, Munster VJ, Feldmann H. 2015. Syrian hamsters (Mesocricetus auratus) oronasally inoculated with a Nipah virus isolate from Bangladesh or Malaysia develop similar respiratory tract lesions. Vet Pathol 52:38–45. doi: 10.1177/0300985814556189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sekellick MJ, Marcus PI. 1978. Persistent infection. I Interferon-inducing defective-interfering particles as mediators of cell sparing: possible role in persistent infection by vesicular stomatitis virus. Virology 85:175–186. doi: 10.1016/0042-6822(78)90422-1. [DOI] [PubMed] [Google Scholar]
- 55.Marcus PI, Sekellick MJ. 1977. Defective interfering particles with covalently linked [±]RNA induce interferon. Nature 266:815–819. doi: 10.1038/266815a0. [DOI] [PubMed] [Google Scholar]
- 56.Salinas Y, Roux L. 2005. Replication and packaging properties of short Paramyxovirus defective RNAs. Virus Res 109:125–132. doi: 10.1016/j.virusres.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 57.Ho T-H, Kew C, Lui P-Y, Chan C-P, Satoh T, Akira S, Jin D-Y, Kok K-H. 2016. PACT- and RIG-I-dependent activation of type I interferon production by a defective interfering RNA derived from measles virus vaccine. J Virol 90:1557–1568. doi: 10.1128/JVI.02161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strahle L, Marq J-B, Brini A, Hausmann S, Kolakofsky D, Garcin D. 2007. Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J Virol 81:12227–12237. doi: 10.1128/JVI.01300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang AS, Baltimore D. 1970. Defective viral particles and viral disease processes. Nature 226:325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- 60.Lazzarini RA, Keene JD, Schubert M. 1981. The origins of defective interfering particles of the negative-strand RNA viruses. Cell 26:145–154. doi: 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- 61.Rao DD, Huang AS. 1982. Interference among defective interfering particles of vesicular stomatitis virus. J Virol 41:210–221. doi: 10.1128/JVI.41.1.210-221.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pattnaik AK, Ball LA, Legrone A, Wertz GW. 1995. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology 206:760–764. doi: 10.1016/s0042-6822(95)80005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li T, Pattnaik AK. 1997. Replication Signals in the genome of vesicular stomatitis virus and its defective interfering particles: identification of a sequence element that enhances DI RNA replication. Virology 232:248–259. doi: 10.1006/viro.1997.8571. [DOI] [PubMed] [Google Scholar]
- 64.Duhaut SD, McCauley JW. 1996. Defective RNAs inhibit the assembly of influenza virus genome segments in a segment-specific manner. Virology 216:326–337. doi: 10.1006/viro.1996.0068. [DOI] [PubMed] [Google Scholar]
- 65.Odagiri T, Tashiro M. 1997. Segment-specific noncoding sequences of the influenza virus genome RNA are involved in the specific competition between defective interfering RNA and its progenitor RNA segment at the virion assembly step. J Virol 71:2138–2145. doi: 10.1128/JVI.71.3.2138-2145.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science 314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 67.Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, Iskarpatyoti JA, Barchet W, Ludwig J, Dermody TS, Hartmann G, Reis e Sousa C. 2014. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5’-diphosphates. Nature 514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pichlmair A, Schulz O, Tan C-P, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol 83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blanco JCG, Pletneva LM, McGinnes-Cullen L, Otoa RO, Patel MC, Fernando LR, Boukhvalova MS, Morrison TG. 2018. Efficacy of a respiratory syncytial virus vaccine candidate in a maternal immunization model. Nat Commun 9:1904. doi: 10.1038/s41467-018-04216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim A-R, Lee D-H, Lee S-H, Rubino I, Choi H-J, Quan F-S. 2018. Protection induced by virus-like particle vaccine containing tandem repeat gene of respiratory syncytial virus G protein. PLoS One 13:e0191277. doi: 10.1371/journal.pone.0191277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quan F-S, Lee Y-T, Kim K-H, Kim M-C, Kang S-M. 2016. Progress in developing virus-like particle influenza vaccines. Expert Rev Vaccines 15:1281–1293. doi: 10.1080/14760584.2016.1175942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu B, Huang Y, Huang L, Li B, Zheng Z, Chen Z, Chen J, Hu Q, Wang H. 2010. Effect of mucosal and systemic immunization with virus-like particles of severe acute respiratory syndrome coronavirus in mice. Immunology 130:254–261. doi: 10.1111/j.1365-2567.2010.03231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chiang C, Beljanski V, Yin K, Olagnier D, Ben Yebdri F, Steel C, Goulet M-L, DeFilippis VR, Streblow DN, Haddad EK, Trautmann L, Ross T, Lin R, Hiscott J. 2015. Sequence-specific modifications enhance the broad-spectrum antiviral response activated by RIG-I agonists. J Virol 89:8011–8025. doi: 10.1128/JVI.00845-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Uzri D, Gehrke L. 2009. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol 83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reed LJ, Muench H. 1938. A simple method for estimating fifty percent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Determination of viral RNA levels in tissues taken from TIP-treated hamsters challenged intraperitoneally with NiV. RT-qPCR was performed based on primer and probe sets specific for NiV strain Malaysia (NiV-M) nucleoprotein (N) on select tissues taken from TIP-treated hamsters challenged i.p. with 104 TCID50 NiV. Hamsters were classified based on whether they succumbed (euthanized due to clinical signs or found dead) to NiV disease (fatal), survived to end of study at 28 dpi with least one day of clinical signs (symptomatic survivors), or survived with no clinical signs (asymptomatic survivors). Individual animals are represented; in groups containing treated hamsters, solid symbols represent hamsters treated with active TIPs, and open symbols represent animals treated with inactive TIPs. Download FIG S2, TIF file, 1.4 MB (1.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Determination of viral RNA levels in tissues taken from TIP-treated hamsters challenged intranasally with NiV. RT-qPCR was performed based on primer and probe sets specific for NiV strain Malaysia nucleoprotein (N) on select tissues taken from TIP-treated hamsters challenged i.n. with 106 TCID50 NiV strain Malaysia. Hamsters were classified based on whether they succumbed (euthanized due to clinical signs or found dead) to NiV disease (fatal), survived to end of study at 28 dpi having displayed at least one day of clinical signs (symptomatic survivors), or survived with no clinical signs (asymptomatic survivors). Individual animals are represented. Download FIG S3, TIF file, 1.2 MB (1.2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Generation of TIPs for in vivo studies. All plasmids required for genome replication and encapsidation processes were transfected into BSR-T7/5 cells, followed 2 days later by addition of Vero-E6 cells and infectious rNiV-M/ZsG (recombinant Nipah virus strain Malaysia [NiV-M] capable of expressing the reporter protein ZsGreen1), which supplies all required proteins for TIP assembly and release. Supernatants containing DIs and nascent NiV virions are treated with UV to remove infectious rNiV-M/ZsG. Download FIG S1, TIF file, 0.6 MB (589.3KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.