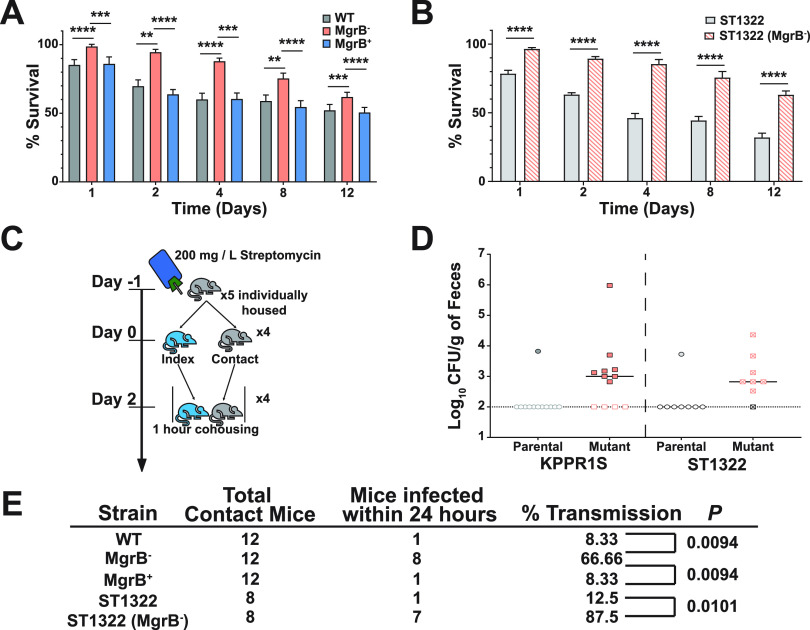
FIG 4.
MgrB inactivation affects survival outside the host and enhances host-to-host transmission. (A and B) Ex vivo solid surface survival of K. pneumoniae isolate KPPR1S and ST1322, their ΔmgrB isogenic mutants, and the chromosomal complement mgrB+. Bacterial strains were grown overnight and resuspended in PBS, their OD600 was adjusted to 4, and they were spotted onto nitrocellulose discs on 1% agarose pads in 6-well polystyrene plates. Discs were removed at appropriate time intervals, bacteria were resuspended in PBS, and viable bacterial counts were determined. Shown is the mean and SEM of six independent assays (in duplicate). Statistical analysis was carried out using Kruskal-Wallis tests with Dunn’s test of multiple comparisons at each time point (A) or using Mann-Whitney U tests at each time point (B). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (C) Schematic representation of the protocol for the transmission studies. Mice were placed on antibiotic water 24 hours before separating and infecting the index mouse and remained on antibiotic water for the duration of the study. Once the index mouse was robustly colonized, the individually housed contact mice were exposed to the index mouse for 1 hour. Fecal shedding was collected the following day to determine if transmission had occurred. (D) Shows fecal shedding of K. pneumoniae parental isolates (KPPR1S and ST1322) and their isogenic mutant (ΔmgrB) from contact mice after a single exposure; the bar indicates the median shedding, and the dotted line indicates limit of detection. (E) Summary of transmission data for the indicated strains after a single 1-hour exposure. For WT, MgrB−, and MgrB+, three groups of five mice (one index and four contact) were used, and for the ST1322 strains, two groups of five mice (one index and four contact) were used. The P value was calculated using Fisher’s exact test.