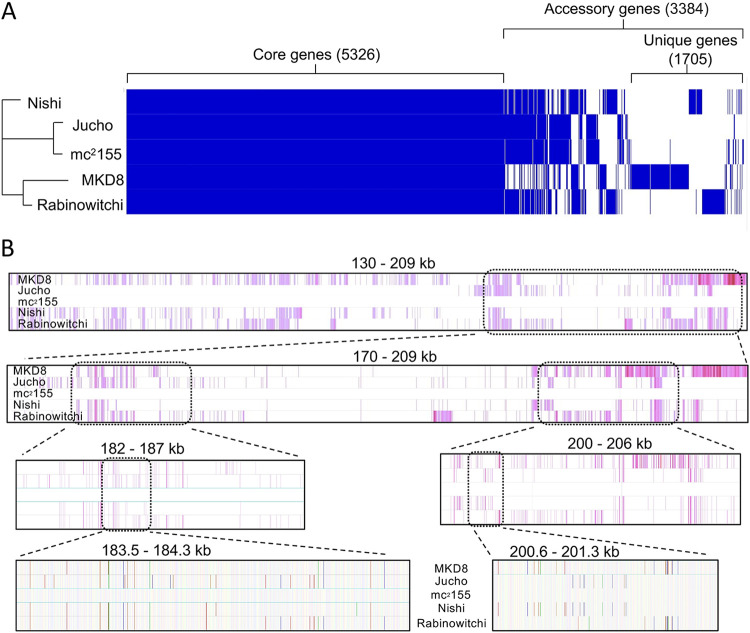
FIG 3.
Genome comparisons of the five independent environmental isolates of M. smegmatis. (A) Pangenome analysis identifies conserved core genes among the M. smegmatis isolates. Roary v.3.13.0 (42) was used to perform a pangenome analysis with default settings. Orthologs were required to share at least 95% global amino acid identity and are indicated by a vertical blue line in each genome along the x axis (the pangenome); the white (no blue) line indicates that an ortholog of >95% identity is absent in that genome. Clustered are conserved core genes (left), accessory genes (middle right), or lineage-specific genes (right) found in only one of the five strains. Results were visualized with Phandango v.1.3.0 (43) and Figtree v.1.4.4 (https://github.com/rambaut/Figtree). (B) A multiple alignment of the five mycobacterial genomes was performed with Parsnp and visualized using Gingr (37) to show collinearity and SNVs in a 79-kb region spanning nt coordinates 130000 to 209000 in the mc2155 genome. In the upper panels, vertical purple bars indicate SNVs present in each genome, using mc2155 as the reference sequence, which is depicted in the middle row as a white bar. Consecutive panels zoom in on the indicated regions. The bottom panel is at single-nucleotide resolution, and the colors correspond to GCAT (yellow, blue, green, and red, respectively). Bold colors are SNVs, and faintly colored vertical bars match the mc2155 sequence.