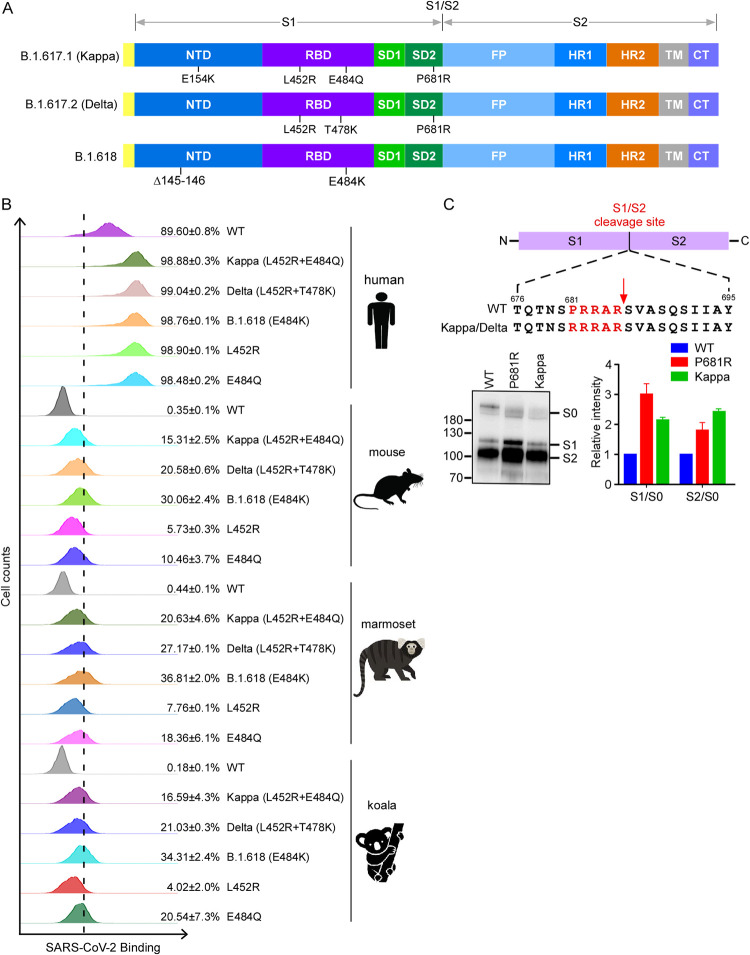
FIG 1.
Binding of variants’ spike proteins with ACE2 orthologs and spike protein processing. (A) Schematic overview of SARS-CoV-2 spike proteins of B.1.617.1 (Kappa), B.1.617.2 (Delta), and B.1.618, colored by domain. NTD, N-terminal domain; RBD, receptor binding domain; SD1, subdomain 1; SD2, subdomain 2; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane region; CT, cytoplasmic tail. (B) HeLa-ACE2 cells were incubated with recombinant RBD-His proteins bearing mutations of Kappa, Delta, and B.1.618 or an individual mutation. The binding of RBD-His with cells was analyzed by flow cytometry. Values are expressed as the percentage of cells positive for RBD-His among the ACE2-expressing cells (zsGreen1+ cells) and are shown as the means ± standard deviation (SD) from 3 biological replicates. This experiment was independently performed three times with similar results. (C) Immunoblot analysis of spike protein cleavage of pseudovirus of the Kappa and P681R variants using polyclonal antibodies against spike. Full-length spike (S0), S1, and S2 proteins are indicated. The ratio of S1 to S0 or S2 to S0 was quantitatively analyzed using ImageJ software. This experiment was repeated twice independently, and data were normalized to the WT (D614G) of the individual experiment.