Abstract
Resistance mechanism relatedness was studied in 18 clinical, European vanA vancomycin-resistant enterococci. Molecular analysis revealed 10 Tn1546-like elements, suggesting two evolutionary lineages. Lineage I dominated the European mainland, and lineage II dominated the United Kingdom and Israel. Geographic clustering reflected different types of meat consumption between countries, since each lineage is associated with colonization of different animals.
Over the past decade, vancomycin-resistant enterococci (VRE) have emerged worldwide (7, 10, 15). Prevalences vary between the United States and Europe, and a different epidemiology for these continents has been postulated (7, 15). Six vancomycin resistance types in enterococci have been described: VanA, VanB, VanC, VanD, VanE, and VanG (6, 17). VanF glycopeptide resistance has been described but has not yet been seen in enterococci (19). VanA type resistance, characterized by high-level inducible vancomycin resistance (MICs of 64 to >1,024 mg/liter) and teicoplanin resistance (MICs of 16 to >512 mg/liter), is most frequently encountered. VanA resistance results from VanA transposon Tn1546 acquisition. Detailed molecular analysis of Tn1546-like elements in enterococci isolated from human and animal sources has revealed the presence of different Tn1546 subtypes. These differences include point mutations, insertions of insertion sequence (IS) elements, and deletions (2–4, 8, 9, 11, 12, 14, 18, 21–25, 27–29; A. L. da Costa Darini, M.-F. I. Palepou, D. James, and N. Woodford, Letter, Antimicrob. Agents Chemother. 43:995–996, 1999; L. B. Jensen, Letter, Antimicrob. Agents Chemother. 42:2463–2464, 1998; L. B. Jensen, A. M. Hammerum, R. L. Poulsen, and H. Westh, Letter, Antimicrob. Agents Chemother. 43:724–725, 1999; G. S. Simonsen, K. H. Dahl, M. R. Mikalsen, O. Ølsvik, and A. Sundsfjord, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother, abstr. C-82, p. 92, 1998). Although genetic diversity of Tn1546 like elements has been described in great detail by several authors, the different Tn1546 subtypes, which were found in different European studies, are difficult to compare, since various molecular techniques were used (11, 12, 18, 21, 24, 27, 29; Jensen, Letter).
The aim of this study was to investigate the genetic relationships between the vanA transposons of VRE from different European countries.
Early in 1997, 4,208 clinical enterococcal strains were collected by 49 centers representing 27 European countries (20; M. A. Schouten, J. A. A. Hoogkamp-Korstanje, C. J. M. Bartels, H. J. G. R. Roelofs-Willemse, Y. J. M. Peters, and A. Voss, Abstr. Eighth Annu. Meet. Soc. Healthcare Epidemiol. Am., abstr. 18, p. 28, 1998). Fifty-one strains exhibiting vancomycin resistance were characterized for the presence of the vanA, vanB, and vanC genes by means of a multiplex PCR as described by Dutka-Malen et al. (5). In 18 isolates, the vanA gene was detected, while vanB and vanC genes were detected in 5 and 28 isolates, respectively. This means that the overall prevalence of VRE among clinical European isolates is still very low (Schouten et al., Abstr. Eighth Annu. Meet. Soc. Healthcare Epidemiol. Am., 1998).
Detailed molecular characterization of Tn1546-like elements in the 18 VanA VRE was performed by a combination of restriction fragment length polymorphism (RFLP) analysis and DNA sequencing of internal PCR fragments of VanA transposons as described previously (27). All VRE were analyzed for the presence of point mutations at positions 1226, 4847, 7658, 8234, and 9692; insertions of IS1216V in the vanX-vanY intergenic region; and left-end deletions (26, 27). The exact integration site and orientation of the IS1542 insertion in the orf2-vanR intergenic region were determined by amplifying and sequencing a DNA fragment with the primer combination IS1216V.E (5′-AGCTTAAATCATAGATACCGTAAGG)-Tn1546 4511.R (5′-TCGGAGCTAACCACATTC). Strain origin, species, Tn1546 type, and epidemiological data concerning patients from whom strains were isolated are shown in Table 1.
TABLE 1.
Strain origin, species, Tn1546 type, and patient data
| Strain no. | Tn1546 type | Country | City | Species | Sexa | Age groupb | Departmentc | Isolation sited |
|---|---|---|---|---|---|---|---|---|
| 4 | A1 | Germany | Aachen | E. faecium | M | A | Urology | Urogenital tract |
| 2 | A1 | Czech Republic | Prague | E. faecium | F | G | Internal medicine | Urogenital tract |
| 11 | A1 | Italy | Torino | E. faecium | F | G | Other | Urogenital tract |
| 12 | A1 | Italy | Torino | E. faecium | F | A | OPD | Digestive tract |
| 13 | A1 | Italy | Catania | E. faecium | F | A | Hematology | Skin |
| 14 | A1 | Slovak Republic | Bratislava | E. faecium | F | N | Intensive care | Digestive tract |
| 5 | A5 | Germany | Frankfurt am Main | E. faecium | M | A | Hematology | Digestive tract |
| 6 | A5 | Germany | Frankfurt am Main | E. faecium | M | A | OPD | Skin |
| 3 | A6 | France | Paris | E. faecium | M | C | Pediatrics | Digestive tract |
| 18 | B3 | United Kingdom | London | E. faecium | F | C | Other | Blood |
| 1 | B4 | Belgium | Brussels | E. faecalis | M | A | OPD | Respiratory tract |
| 16 | C | United Kingdom | Manchester | E. faecium | M | A | General practitioner | Other |
| 10 | E2 | Israel | Beer-Sheva | E. faecium | F | A | Internal medicine | Skin |
| 9 | E4 | Israel | Jerusalem | E. faecium | F | G | Other | Urogenital tract |
| 17 | E4 | United Kingdom | London | E. faecium | M | C | Intensive care | Other |
| 15 | E12 | United Kingdom | Manchester | E. faecium | F | G | Other | Other |
| 7 | E13 | Israel | Jerusalem | E. faecalis | M | A | Surgery | Skin |
| 8 | E13 | Israel | Jerusalem | E. faecium | F | G | Surgery | Blood |
M, male; F, female.
N, less than 1 year old; C, between 1 and 16 years old; A, between 16 and 65 years old; G, 65 years and older.
OPD, outpatient department; Other, not further specified.
Other, not further specified.
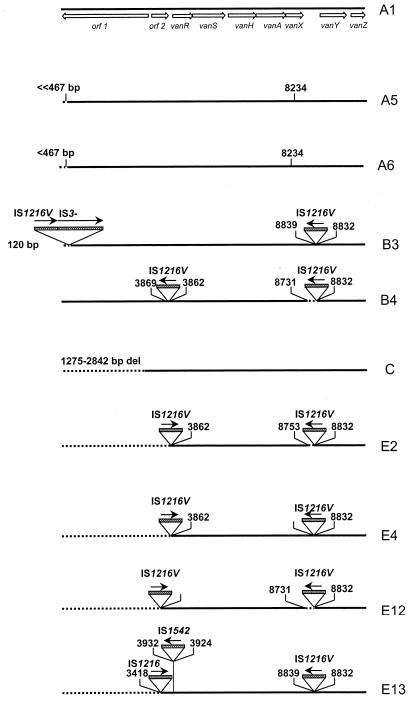
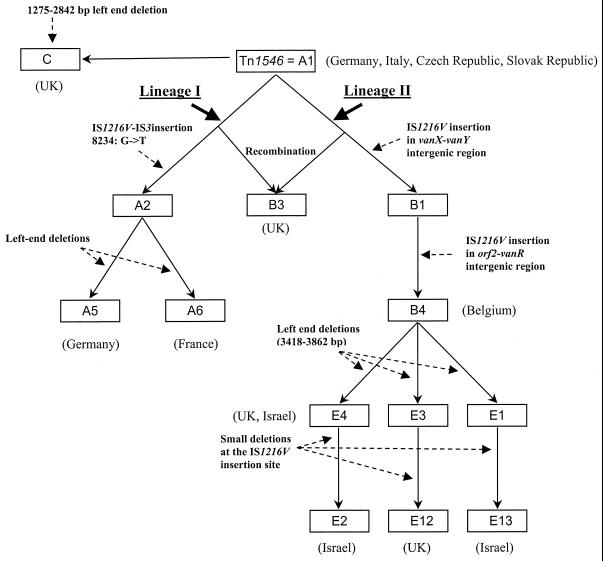
Among 18 VanA VRE, 10 different Tn1546-like elements could be distinguished (Fig. 1). A scheme was constructed that describes the hypothetical evolutionary relationship between Tn1546 variants found in this study and those found in earlier studies (Fig. 2). This scheme is comparable to the scheme described previously (27). Six isolates from Italy, regardless of the center of origin, Czech Republic, and the Slovak Republic contained type A1 transposon (Table 1), which is identical to transposon Tn1546, as described by Arthur et al. (1). This confirms earlier findings that VanA transposons indistinguishable from Tn1546 are frequently encountered in Europe (12, 18, 21, 27, 29; Jensen, Letter; Simonsen et al., 38th ICAAC). From type A1, two main lineages of Tn1546 derivatives may have evolved. Lineage I includes the types A2 (not found in this study), A5, and A6 and is characterized by the point mutation at 8234 in vanX. Types A5, encountered twice in Germany, and A6, present in one isolate from France, are closely related. They both contain, in addition to the G→T point mutation in the vanX gene at position 8234, a small deletion at the left end of the transposon. Although the exact size of the deletion in the types A5 and A6 could not be determined, the RFLP and PCR results revealed that the deletion in type A5 has to be close to the HaeIII restriction site at position 467, while in type A6, the deletion is probably somewhat smaller than the deletion in A5. Lineage II types include the B and E types and are characterized by IS1216V insertions in the vanX-vanY intergenic region, often accompanied by small deletions adjacent to the insertion site, and insertions of IS1216V at the left end of the transposon associated with large deletions encompassing the orf1 and orf2 region (Fig. 1 and 2). Transposon types of lineage II were found in isolates from Belgium (B4), the United Kingdom (types E4 and E12), and Israel (E2, E4, and E13). Strains 7 (Enterococcus faecalis) and 8 (Enterococcus faecium) from Israel both possess type E13, suggesting horizontal transfer of this transposon type between two different enterococcal species. Interestingly, in these two strains, the insertion element IS1542 was inserted in the VanA transposon at exactly the same position as in the group H VanA transposon described by Woodford and colleagues (29). Nevertheless, type E13 is probably not identical to the group H transposon, because the left-end deletion in type E13 appears to be much larger than in the group H transposon. So far, IS1542 has been found frequently in clinical and poultry VRE isolates from the United Kingdom and Ireland and sporadically in glycopeptide-sensitive enterococci from the United Kingdom and Brazil. The finding of lineage II transposon types in the United Kingdom is in agreement with results published previously by Woodford et al., who found that the majority of their human United Kingdom strains possessed a transposon that would most likely fit into our groups B and E (29). Finally, in one isolate from the United Kingdom, transposon type C was found. This transposon type was described previously (27).
FIG. 1.
Genetic maps of 10 Tn1546 types. The thick horizontal lines represent the different Tn1546 types. The positions of genes and open reading frames (orf1 and orf2) and the direction of transcription are depicted with open arrows. Dotted boxes represent IS elements. The positions of the first nucleotide upstream and the first nucleotide downstream from the IS insertion sites are depicted. Solid arrows indicate the transcriptional orientation of the inserted IS elements. Deletions (del) are indicated by dotted lines. The position of the base pair mutation at 8234, G→T (K→N), is indicated above the A5 and A6 Tn1546 types.
FIG. 2.
Hypothetical evolutionary scheme for the various Tn1546 derivatives characterized in this study from the archetypal transposon Tn1546 (type A1) as described by Arthur et al. in 1993 (1). The types A2, B1, E1, and E3 were described previously by Willems et al. (27) and not in this study. They were included for better understanding of the different Tn1546 type evolutionary relationships. Boxes represent the different Tn1546 types. Solid arrows indicate the transition of Tn1546 type A1 to the other Tn1546 types. The two different lineages, I and II, are indicated. The names of the countries where the different transposon types were found are indicated in parentheses. This scheme was partly based on the evolutionary scheme described by Willems et al. (27).
It is interesting that the lineage II transposon types B and E, which are predominantly found in the United Kingdom and Israel, are also the types frequently found in poultry isolates (22, 24, 27, 29), while, e.g., in The Netherlands, type A2, a lineage I type, is the most prevalent type encountered in human and pig isolates (27). In Israel and the United Kingdom, the per capita consumption of poultry meat (34.8 and 19.2 kg, respectively) is about twofold higher than in The Netherlands (15.6 kg). On the other hand, the Dutch consume twice as much pork per capita as people in the United Kingdom (45.6 versus 25.6 kg, respectively) (http://usda.mannlib.cornell.edu/data-sets/food/91004/). Although no exact figures for the consumption of pork in Israel were available, the level is expected to be very low as a result of the kosher lifestyle. These data suggest that the geographical distribution of different transposon types may be a result of the differences in meat consumption in the different European countries, thus indicating that vanA transposons found in humans in Europe originate from farm animals.
Acknowledgments
We thank all of our colleagues from the European VRE Study Group who collected enterococcal strains for us. Furthermore, we thank Willem Melchers and Hannie Roelofs-Willemse, Department of Medical Microbiology, University Hospital St. Radboud, Nijmegen, The Netherlands, for technical support.
REFERENCES
- 1.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Depardieu F, Gerbaud G, Galimand M, Leclercq R, Courvalin P. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J Bacteriol. 1997;179:97–106. doi: 10.1128/jb.179.1.97-106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darini A L D, Palepou M F I, Woodford N. Nucleotide sequence of IS1542, an insertion sequence identified within vanA glycopeptide resistance elements of enterococci. FEMS Microbiol Lett. 1999;173:341–346. doi: 10.1111/j.1574-6968.1999.tb13523.x. [DOI] [PubMed] [Google Scholar]
- 4.Descheemaeker P R M, Chapelle S, Devriese L A, Butaye P, Vandamme P, Goossens H. Comparison of glycopeptide-resistant Enterococcus faecium isolates and glycopeptide resistance genes of human and animal origins. Antimicrob Agents Chemother. 1999;43:2032–2037. doi: 10.1128/aac.43.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fines M, Perichon B, Reynolds P, Sahm D F, Courvalin P. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob Agents Chemother. 1999;43:2161–2164. doi: 10.1128/aac.43.9.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goossens H. Spread of vancomycin-resistant enterococci: differences between the United States and Europe. Infect Control Hosp Epidemiol. 1998;19:546–551. doi: 10.1086/647871. [DOI] [PubMed] [Google Scholar]
- 8.Haaheim H, Dahl K H, Simonsen G S, Olsvik O, Sundsfjord A. Long PCRs of transposons in the structural analysis of genes encoding acquired glycopeptide resistance in enterococci. BioTechniques. 1998;24:432–437. doi: 10.2144/98243st02. [DOI] [PubMed] [Google Scholar]
- 9.Handwerger S, Skoble J, Discotto L F, Pucci M J. Heterogeneity of the vanA gene cluster in clinical isolates of enterococci from the northeastern United States. Antimicrob Agents Chemother. 1995;39:362–368. doi: 10.1128/aac.39.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirakata Y, Yamaguchi T, Izumikawa K. In vitro susceptibility studies and detection of vancomycin resistance genes in clinical isolates of enterococci in Nagasaki, Japan. Epidemiol Infect. 1997;119:175–181. doi: 10.1017/s0950268897007954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen L B. Internal size variations in Tn1546-like elements due to the presence of IS1216V. FEMS Microbiol Lett. 1998;169:349–354. doi: 10.1111/j.1574-6968.1998.tb13339.x. [DOI] [PubMed] [Google Scholar]
- 12.Jensen L B, Ahrens P, Dons L, Jones R N, Hammerum A M, Møller Aarestrup F. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J Clin Microbiol. 1998;36:437–442. doi: 10.1128/jcm.36.2.437-442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 14.MacKinnon M G, Drebot M A, Tyrrell G J. Identification and characterization of IS1476, an insertion sequence-like element that disrupts VanY function in a vancomycin-resistant Enterococcus faecium strain. Antimicrob Agents Chemother. 1997;41:1805–1807. doi: 10.1128/aac.41.8.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martone W J. Spread of vancomycin-resistant enterococci: why did it happen in the United States? Infect Control Hosp Epidemiol. 1998;19:539–545. doi: 10.1086/647870. [DOI] [PubMed] [Google Scholar]
- 16.McDonald L C, Kuehnert M J, Tenover F C, Jarvis W R. Vancomycin-resistant enterococci outside the health-care setting: prevalence, sources, and public health implications. Emerg Infect Dis. 1997;3:311–317. doi: 10.3201/eid0303.970307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKessar S J, Berry A M, Bell J M, Turnidge J D, Paton J C. Genetic characterization of vanG, a novel vancomycin resistance locus of Enterococcus faecalis. Antimicrob Agents Chemother. 2000;44:3224–3228. doi: 10.1128/aac.44.11.3224-3228.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palepou F I M, Adebiyi A M A, Tremlett C H, Jensen L B, Woodford N. Molecular analysis of diverse elements mediating VanA glycopeptide resistance in enterococci. J Antimicrob Chemother. 1998;42:605–612. doi: 10.1093/jac/42.5.605. [DOI] [PubMed] [Google Scholar]
- 19.Patel R, Piper K, Cockerill III F R, Steckelberg J M, Yousten A A. The biopesticide Paenibacillus popilliae has a vancomycin resistance gene cluster homologous to the enterococcal VanA vancomycin resistance gene cluster. Antimicrob Agents Chemother. 2000;44:705–709. doi: 10.1128/aac.44.3.705-709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schouten M A, Voss A, Hoogkamp-Korstanje J A A the European VRE Study Group. Antimicrobial susceptibility patterns of enterococci causing infections in Europe. Antimicrob Agents Chemother. 1999;43:2542–2546. doi: 10.1128/aac.43.10.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonsen G S, Haaheim H, Dahl K H, Kruse H, Lovseth A, Olsvik O, Sundsfjord A. Transmission of VanA-type vancomycin-resistant enterococci and VanA resistance elements between chicken and humans at avoparcin-exposed farms. Microb Drug Resist Mech Epidemiol Dis. 1998;4:313–318. doi: 10.1089/mdr.1998.4.313. [DOI] [PubMed] [Google Scholar]
- 22.Stobberingh E E, van den Bogaard A, London N, Driessen C, Top J, Willems R. Enterococci with glycopeptide resistance in turkeys, turkey farmers, turkey slaughterers, and (sub)urban residents in the south of The Netherlands: evidence for transmission of vancomycin resistance from animals to humans? Antimicrob Agents Chemother. 1999;43:2215–2221. doi: 10.1128/aac.43.9.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van den Bogaard A E, Jensen L B, Stobberingh E E. Vancomycin-resistant enterococci in turkeys and farmers. N Engl J Med. 1997;337:1558–1559. doi: 10.1056/NEJM199711203372117. [DOI] [PubMed] [Google Scholar]
- 24.van den Braak N, van Belkum A, van Keulen M, Vliegenthart J, Verbrugh H A, Endtz H P. Molecular characterization of vancomycin-resistant enterococci from hospitalized patients and poultry products in The Netherlands. J Clin Microbiol. 1998;36:1927–1932. doi: 10.1128/jcm.36.7.1927-1932.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werner G, Klare I, Witte W. Arrangement of the vanA gene cluster in enterococci of different ecological origin. FEMS Microbiol Lett. 1997;155:55–61. doi: 10.1111/j.1574-6968.1997.tb12685.x. [DOI] [PubMed] [Google Scholar]
- 26.Willems R J L. Vancomycineresistente enterokokken; epidemiologie en transmissie van resistentie. Ned Tijdschr Med Microb. 1999;1:5–8. [Google Scholar]
- 27.Willems R J L, Top J, van den Braak N, van Belkum A, Mevius D J, Hendriks G, van Santen-Verheuvel M, van Embden J D A. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob Agents Chemother. 1999;43:483–491. doi: 10.1128/aac.43.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodford N, Watson A P, Chadwick P R. Investigation by long PCR of the genetic elements mediating VanA glycopeptide resistance in enterococci from uncooked meat in south Manchester. Adv Exp Med Biol. 1997;418:409–412. doi: 10.1007/978-1-4899-1825-3_98. [DOI] [PubMed] [Google Scholar]
- 29.Woodford N, Adebiyi A M A, Palepou M I, Cookson B D. Diversity of VanA glycopeptide resistance elements in enterococci from humans and nonhuman sources. Antimicrob Agents Chemother. 1998;42:502–508. doi: 10.1128/aac.42.3.502. [DOI] [PMC free article] [PubMed] [Google Scholar]