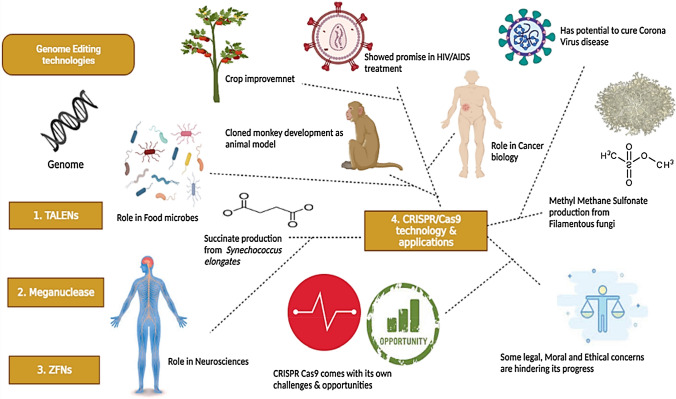
Abstract
Genome-editing technology has enabled scientists to make changes in model organisms’ DNA at the genomic level to get biotechnologically important products from them. Most commonly employed technologies for this purpose are transcription activator like effector nucleases (TALENs), homing-endonucleases or meganucleases, zinc finger nucleases (ZFNs), and clustered regularly interspaced short palindromic repeats (CRISPR) associated protein 9 (Cas9). Among these tools, CRISPR/Cas9 is most preferred because it's easy to use, has a small mutation rate, has great effectiveness, low cost of development, and decreased rate of advancement. CRISPR/Cas9 has a lot of applications in plants, animals, humans, and microbes. It also has applications in many fields such as horticulture, cancer, food biotechnology, and targeted human genome treatments. CRISPR technology has shown great potential for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic to provide early and easy detection methods, possible treatment, and vaccine development. In the present review, genome-editing tools with their basic assembly and features have been discussed. Exceptional notice has been paid to CRISPR technology on basis of its structure and significant applications in humans, plants, animals, and microbes such as bacteria, viruses, and fungi. The review has also shed a little light on current CRISPR challenges and future perspectives.
Graphical abstract
Keywords: CRISPR/Cas9, Gene editing, Meganuclease, SARS-CoV2, TALENs
Introduction
Paul Berg’s work on recombinant DNA technology in the 1970s laid the foundation for a new era of science. These advancements enabled scientists to make changes in model organisms’ DNA to get biotechnologically important products from them [1, 2]. In Recombinant DNA technology, insertions, deletions, and other such specific changes are made into the DNA to get desired end product [3]. For a long time, genome-editing procedures were mostly limited to some specific organisms such as the study of homologous recombination in mice and yeast. The use of targeted genome editing in living organisms was a turning point in the biological world [1, 4]. In the last 10 years, much progress has been made in genome-editing technology [5]. In the present review, diverse genome-editing tools with their basic assembly and features have been discussed. Exceptional notice has been paid to clustered regularly interspaced short palindromic repeats (CRISPR) technology on the basis of its structure and significant applications in humans, plants, animals, and microbes such as bacteria, viruses, and fungi. The review has also shed a little light on current CRISPR challenges and future perspectives.
Modern Targeted Genome-Editing Technologies
Genome-editing-tools has enabled scientists to make changes in their choice of organisms at the genomic level. Most commonly employed technologies for this purpose are transcription activator like effector nucleases (TALENs), homing-endonucleases or meganucleases, zinc finger nucleases (ZFNs), and clustered regularly interspaced short palindromic repeats (CRISPR) CRISPR associated protein 9 (Cas9) [6].
ZFNs
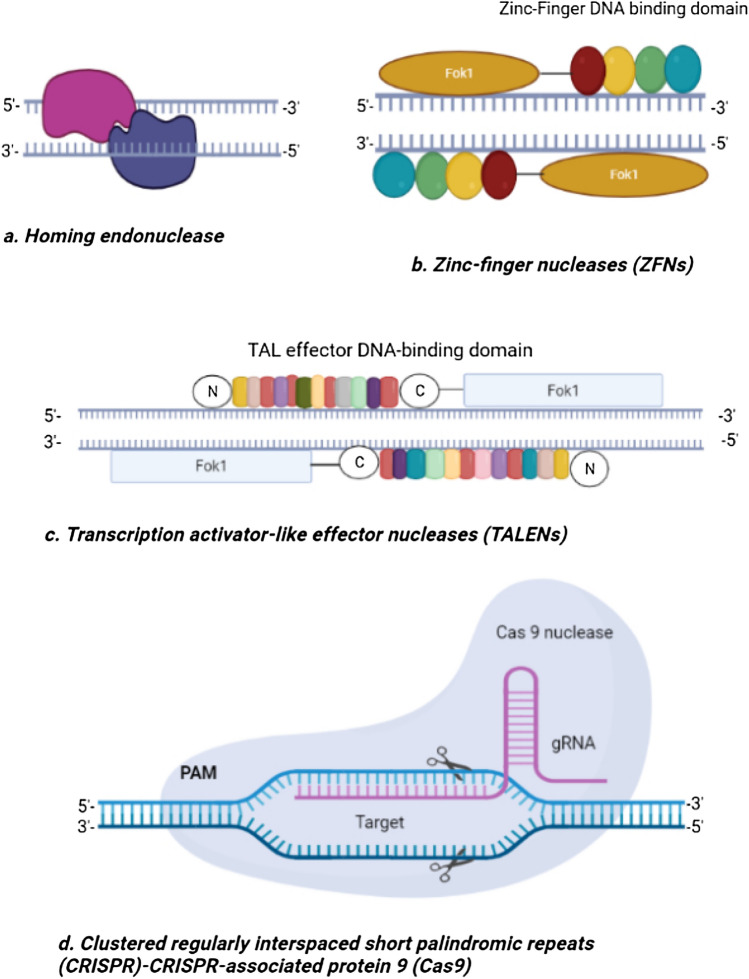
These artificially made restriction enzymes are the most generally utilized endonucleases. They have zinc finger proteins known as ZFPs, which also act as transcription factors in eukaryotes, and work as DNA binding domains. ZFNs additionally have nucleotide cleavage area (Folk1) derived from F. okeanokoites. Based on the target side, the cleavage domain is usually surrounded by 4–6 zinc finger proteins. For efficient gene editing at the target site, ZFPs have a target specificity of 18 base pair (bp). A typical, ZFP is 30 amino acids in length with an alpha-helix structure that is opposite to two antiparallel β-sheets. This technology uses Homology-directed-repair or Homologous-recombination and non-homologous end joining repair mechanism, for successful gene editing in prokaryotes and eukaryotes [7]. In 1996, the first report on ZFNs was published, and since then they are continuously being used in many organisms. The examples included are gene modifications in maize for herbicide resistance and direct endogenous gene inactivation in Arabidopsis. ZFNs have many benefits over other gene-editing technologies in terms of their effectiveness, target specificity, and a low number of non-target effects, etc. (Fig. 1) [2].
Fig. 1.
General assembly of major genome-editing technologies. (a) Homing endonuclease, target DNA as homo-dimers without having any clear-cut DNA binding and cleavage domains. (b) Zinc finger nucleases (ZFMs) have a Zinc finger DNA binding domain and Fok1-cleavage-domains. (c) Transcription activator like effector Nucleases (TALENs) have also similar domains in it like ZFNs (d) Clustered regularly interspaced short palindromic repeats (CRISPR) and associated protein9 (Cas9), is consists of Cas9 nuclease, a single guide RNA (sgRNA) which is located upstream to PAM (Proto spacer-adjacent-motifs). Single guide RNA (sgRNA) guides Cas9 to select DNA sequences that are relatively complementary to the target (Proteins and DNA are not drawn exactly to scale) [6].
TALENs
TALE (transcription activator like effector) proteins which have been originated from the Xanthomonas bacteria’s gene. These TALE proteins are capable of binding with very short sequences (up to 1–2 nucleotides long) due to carrying tandem repeats of 34 amino acids. TALE proteins can bind to plant promoters with the help of 34 amino acids tandem repeats, this role of TALE proteins gives birth to TALENs gene-editing technology [7, 8]. TALENs also have two domains just like ZFNs, one for the DNA cleavage target site and the other works as a DNA binding domain. This domain system is what makes this technique a powerful tool for gene editing in many eukaryotes such as rats, zebra fish, chicken, frogs, some mammalian cells, arabidopsis, tomato, wheat, rice, and potato [7, 9].
Homing Endonucleases or Meganucleases
Homing endonucleases, a monomer, are enzymes that belong to the family of the LAGLIDADG group of endonucleases. They are named due to conserved amino acid sequences present in them. This enzyme cut DNA into 14–40 bp after recognizing it. Homing endonucleases are extremely target-specific and only interact with their target DNA. Nonetheless, homing endonucleases' binding and cleavage domains are not similar to TALENs and ZFNs [10]. This ultimately limited their use for quality genome-editing. Recently, Mega TALs (made by joining of TALE-binding domains and a rare homing endonuclease) are reported for their high target specificity in gene editing. They have applications in tumor and HIV treatment therapies where TALENs have made it possible to integrate anti-tumor and anti-HIV factors into the host’s CCR5 gene (in this case human) present in primary T cells and hematopoietic stem cells [10]. In a study, T cells have shown promise for improved immune-therapies after gene modification study in endogenous T-cell receptor-components [6].
CRISPR Associated Protein 9 (Cas9)
After 10 years of research on acquired immunity mechanisms in bacteria, scientists discovered the CRISPR system, which is present in all bacteria and archaea [1]. In 1987, CRISPR sequences were first seen while contemplating the lap enzyme in Escherichia coli [1]. Scientists also found that CRISPR repetitive sequences work actually work as a mechanism of immunity in bacteria and have originally been derived from plasmids and bacteriophages (Table 1) [11]. In 1987, with the accidental discovery in E. coli, scientists started looking for these sequences in other similar organisms such as Salmonella, Shigella and Mycobacterium tuberculosis. Initially, it was thought that CRISPR work in genotyping, whereas their actual role was revealed after the discovery of Cas proteins and with the advancement in recombinant DNA technology. In 2012, it was also identified as a gene-editing tool after the documentation of its DNA cleaving abilities [16].
Table 1.
A brief comparison of genome-editing technologies
| Functions | ZFNs | TALENs | Meganucleases | CRISPR-Cas9 | References |
|---|---|---|---|---|---|
| Origin | Eukaryotes | Bacteria | Bacteria/plant/animal | Bacteria/archaea | [12] |
| Structure | Dimer | Dimer | Dimer | Monomer | [12] |
| Modification pattern | Foki nuclease | Foki nuclease | Endonuclease | Cas 9 nuclease | [12] |
| Target recognition efficiency | Low | Higher | Relatively low | Highest | [13, 14] |
| Recognition location | Typically, 9–18 bp per monomer, 18–36 bp per pair | Typically, 14–20 bp per monomer, 28–40 bp per pair | Between 14 and 40 bp | Typically, 20 bp guide sequence + PAM sequence | [15] |
| Rate of mutation | High | Middle | Middle | Low | [13] |
| Cloning | Needed | Needed | Not needed | Not needed | [13] |
| Difficulties of engineering | Protein engineering is required | Molecular cloning methods are needed | Protein engineering is needed | Using simple cloning methods and oligo synthesis | [5, 15] |
| Difficulties of in vivo delivery | Easy, small size expression systems needed for viral vectors | Difficult for requiring large size components | Easy, small size expression systems needed for viral vectors | Use spcas (with large size, viral vectors, i.e., AAV may suffer packaging harms | [5, 15] |
| Cost of development | High | Higher | High | Low | [13] |
CRISPR Structure
Structurally, it has Cas9 helicase which can bind to RNA transcribed using palindromic repeats of host DNA and slice RNA-spacers matched foreign DNA. In this arrangement, trans-activating CRISPR-RNA (tracrRNA) is named for the transcript of palindromic repeats of DNA while the spacers’ transcript is known as crRNA (CRISPR-RNA). The trans-activating CRISPR-RNA and crRNA form an individual solitary-guide-RNA (sgRNA) that can direct Cas9 to locate target DNA and cleave it, producing a double stranded DNA break (DSB) at PAMs (Proto spacer-adjacent-motifs) area. PAMs are small protein motifs of 3-6 bp residing in the foreign DNA and are recognized by Cas proteins [16].
There are two functioning domains in Cas9-nuclease for DNA cleavage, i.e., HNH (histidine asparagine histidine) as well as RuvC. The HNH domain cuts target DNA at the site where sgRNA base pairs with targeted DNA, however, the non-targeted strand is cleaved by domain RuvC [11]. DSB is repaired either by non-homologous end joining (NHEJ) method (with no template) or by homology-directed repair (HDR) method, where a template DNA is provided. If this DNA will come to the cell, homology arms flanking the locus focused by Cas9, and utilized as a template. NHEJ repair mechanism adds insertion, deletion, and substitution in target DNA. But in the case of homology-directed repair (HDR), donor DNA is used as a template, and thus desired DNA sequence is added here [11, 17]. However, NHEJ method is used more commonly as it is more efficient. Many factors play their role in making NHEJ more efficient such as cell cycle stage, cell type, type of CRISPR/Cas system used, donor DNA template, DNA concentration, type of delivery approach used, and length of the homologous arms [16]. CRISPR/Cas9 is not only a gene-editing tool but also has other important functions. For instance, any mutation in two nuclease domains of Cas9 will inactivate Cas9 (dCas9), a protein important for locus-specific DNA binding. This dCas9 protein helps regulate gene expression by binding to a transcription activator or repressor domain [11].
CRISPR/Cas Framework Classification
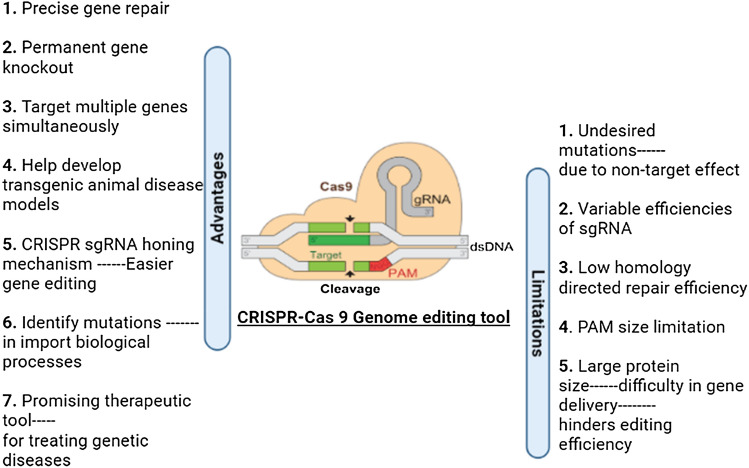
CRISPR framework has two major classes and six significant types [18]. Class 1 and 2 both use protein effector complexes, the only difference is that while Class1 uses a multi-protein complex, Class 2 uses a single protein complex. Class 1 is additionally separated into various kinds I, III, and IV, while class 2 incorporates types II, V, and VI. It can likewise be separated into 19 diverse sub-types, and it is probably going to keep on extending as new CRISPR/Cas frameworks are continuously being discovered [19]. Numerous Cas-proteins included in Type I and III CRISPR loci form a complex with crRNA. These complexes are essential for target nucleic acid recognition and destruction [1]. There are very few Cas proteins in the type II systems, amid these kinds, the class 2 type II CRISPR/Cas 9 framework is the most well-developed and well-studied gene-editing tool [18]. Some of the major advantages and challenges of the CRISPR genome-editing tool (Fig. 2) [4, 20, 21].
Fig. 2.
A brief account of some major advantages and limitations of CRISPR/Cas9 technology
Latest CRISPR Systems
With continuous research in the CRISPR gene-editing systems, new CRISPR tools are being discovered such as type II Cas9 ortholog and Nm2Cas9 which are currently being studied as an alternative for genetic engineering and gene therapy. Similarly, Cas12a orthologs have shown promise in human gene editing. BhCas12b is another powerful CRISPR tool showing potential in gene editing. Other than the currently available CRISPR sub-types, sub-types such as Cas (12 h), Cas (12i), and Cas (12 g) have been identified from metagenome and are included in the type-V CRISPR system [22]. Some sub-types can be used in in vitro as programmable endonuclease to cut ssDNA, dsDNA, ssRNA, or dsRNA. Cas X, designated Cas12e, is found to be effective for gene editing in human cells. In addition to this, Cas14, part of the Cas12f family, is also a human gene-editing tool but it has low efficiency [18]. Class 1 CRISPR system has used many strategies such as native nuclease effector or fused FokI domain for DNA cleavage, compared to the Class 2 CRISPR system [23–25]. Previously, a few individuals from (type I F) frameworks were proposed as an editing tool for targeted DNA modification [26]. All this work on CRISPR and its frameworks urges biologists to explore more into this field and develop more human-friendly technologies (Table 2).
Table 2.
A list of CRISPR-Cas systems with their functions in different microbial hosts
| CRISPR system | Organism | Size (bp) | PAM | sgRNA size (bp) | Target/gene target | Functions/Findings | References |
|---|---|---|---|---|---|---|---|
| Cas3 |
E. coli, S. enterica |
– | – | – | ftsA, asd, msbA, nusB, fucP, ogr, groL, arpA, PentC, phoH, PppsR | Selectively removes individual strains in both pure and mixed cultures | [27] |
| Cas9 | S. aureus | – | – | – | aph-3, mecA | Phagemid delivery for selective killing of virulent and avirulent Staphylococcus aureus, elimination of mecA gene without harming or killing the host cells | [28] |
| Cas9 | HSV-1 | – | – | – | ICP0, ICP4, ICP27 | Type II -Cas9, ICP4, ICP27 Limited HSV infection cycle in human oligodendroglioma cells | [29] |
| Cas9 | EBV | – | – | – | EBNA-1 | Targets Burkitt cancerous cells which are infected with EBV through 2 gRNAs against EBNA-1 has resulted in 95% loss of EBV genome | [30] |
| Cas9 | JCV | – | – | – | T-antigen | Suppressed JCV replication in HJC-2 cells using lentivirus vector deliver | [31] |
| SpCas9 | Streptococcus pyogenes | 1368 | 5ʹ-NGG-3ʹ | 20 | Target dsDNA | Here PAM corresponds to an NGG consensus sequence containing two G: C base pairs | [32] |
| FnCas9 | Francisella novicida | 1629 | 5ʹ-NGG-3ʹ | 20 | Target DNA | Identifies 5ʹ-NGG-3ʹ PAM, uses structural information to create a variant that can identify more relaxed 5ʹ-YG-3ʹ PAM | [33] |
| St1Cas9 | Strep. Thermophilus | 1121 | NNAGAAW | 20 | Target DNA | Both St1Cas9 and SaCas9 play a critical role in the bacterial positive selection system. St1Cas9 works as a nuclease in human cells | [34] |
| CjCas9 | C. jejuni | 984 | NNNNACAC and NNNRYAC | 22 | Target DNA | Target DNA CjCas9 cleaves only a limited number of sites in the mouse or human genome | [35] |
| AacC2c1 | Alicyclobacillus acidoterrestris | 1277 | T-rich PAM | 20 | Target DNA | AacC2c1 is a C2c1-a type V-B CRISPR/Cas endonuclease, has a bilobed architecture that consists of a REC and NUC lobe | [36] |
| Cas14 | Uncultivated archaea | 400–700 | – | – | Target ssDNA | Cas14 proteins have 400 to 700 aminoacids, can cut ssDNA with no need of restrictive sequence | [37] |
| Cpf1 (AsCpf1) | Acidaminococcus sp. | 1307 | 5ʹ-TTTN-3ʹ | 24 | Target DNA | Identifies crRNA scaffold and the 5ʹ-TTTN-3ʹ PAM in structure and sequence-dependent manners has two domains located at positions suitable to generate staggered double stranded breaks (DSBs) in DNA | [38] |
Recent Eminent Applications of CRISPR Technology
CRISPR applications have not only grown very quickly but have also revolutionized life sciences [22]. At present, this technology is being used in different fields of science such as horticulture, food biotechnology, medicine, cancer, and targeted human genome treatments [39].
In Humans
Role in Biotherapy
Correction or replacement of faulty or undesirable genes in /from a cell can be done through gene therapy. Usually, gene therapy refers to humans, although it can be done in plants and animals equally. Lots of diseases in humans are due to genetic defects either through gene mutation which may result in over, under, or no expression of the genes. Such conditions result in genetic disorders which cannot be cured through medication and their best treatment option is gene therapy. Hence gene therapy has gotten a lot of attention from the scientific community and pharmaceutical companies since its first discovery in the later 1980s and early 1990s [40, 41]. Gene therapy can be categorized into two types; ex vivo and in vivo gene therapy.
Ex vivo gene therapy: Genetic diseases of hematopoietic system cells can be treated if genetic mutations which occur in human hematopoietic stem and progenitor cells (HSPCs) are corrected. A potential strategy to treat β-thalassemia and sickle-cell-disease is by disturbing the production of erythroid enhancers of human BCL11A induced fetal hemoglobin production [42, 43]. In vivo gene therapy: In vivo gene therapy treatment is difficult, because of the necessity of competent and tissue explicit in Vivo rescue strategies. AAV vector is mainly a famous choice in many ongoing investigations due to its high recurrence of vector addition into the cell genome, which has raised some wellbeing concerns. Furthermore, the possible immunogenicity of SpCas9 and SaCas9 is another area and should be measured for an in vivo treatment. The most exceptional medical trials of CRISPR treatment are found in a pre-clinical report completed by Edits, which demonstrates complete recovery from a mutation inside the intron 26 of the CEP290 gene utilizing AAV-saCas9 [44].
CRISPR/Cas9-Based Strategies of Gene Therapy
After being established as a gene-editing tool in 2012, CRISPR/Cas technology was employed in gene therapy such as gene edition, gene correction, and gene replacement. In the past decade, CRISPR technology has developed remarkably and many strategies for its use have been developed, among which prime editing, base editing, gene knockin, and gene knockout were found to be most effective and efficient in gene therapy [41, 45].
Gene Knockout Strategy
CRISPR/Cas9-based gene knockout strategy is the first well developed method to remove undesirable and disease causing genes. After CRISPR/Cas system is introduced in a cell, the Cas nuclease will locate and cut the target DNA to form double stranded DNA break (DSB). gRNAs guide Cas nuclease in detecting the target DNA sequence along with its PAM sequence (complementary to gRNA). A common repair mechanism is NHEJ which links 2 DSB molecules together. In the NHEJ mechanism, frameshift change is usually produced because it works by deleting or adding a few nucleotides while combing two DSBs together. NHEJ repair mechanism always has two major outcomes; one is the silence of edited gene due to frameshift change and the other is the induction of nonsense mutation [41].
Gene Knockin Strategy
Here CRISPR/Cas9 system plus a functional DNA template is introduced in a cell. Once CRISPR/Cas cuts the target DNA sequence, the cell uses transfer DNA as a template to repair DSB, and thus template DNA is inserted into the genome [41]. This method is especially efficient in adding a desirable gene in place of an unwanted gene. This method has a low chance of success and is employed only in divided cells [46].
Edition Strategy
There are two functional domains of Cas9; HNH and Ruvc, which cut one strand of target DNA and forms A DSB [47, 48]. Cas proteins that bind to DNA depend on gRNA and are not related to their activities. Hence either one or both of its domains can be inactivated through the change in their amino acids or structural modification without affecting their binding activities. Using this principle, scientists have produced modified Cas endonuclease which can deactivate HNH and Ruvc domains to form a deactivated Cas (dCas) or Cas nikase (nCas). All Cas proteins like dCas or nCas can be fused with other molecules like enzymes without having any effect on their own binding and cleave functions [41]. Using this principle, Komor et al. [49] constructed a cytosine base editor (CBE) by fusing cytidine deaminase enzyme with nCas9. This CBE was able to successfully convert cytidine (C) into uridine (U), which can then become thymine (T). Eventually, the group was able to obtain the base edition from G to A or C to T successfully. DNA base editing has potential applications in treating genetic diseases associated with single‐nucleotide polymorphism or point mutation [41].
Prime Edition Strategy
This application of CRISPR genome editing is newly developed in 2019 and it has added two changes to the traditional CRISPR/Cas system. Firstly, Cas9 nickase was fused with reverse transcriptase and secondly, traditional gRNA was replaced with prime editing guide RNA (pegRNA). pegRNA not only contains single gRNA (contain both spacer and tracrRNA) but also gRNA which is linked to a gene specific RNA sequence (containing a primer binging site, PBS). This PBS site has a complementary sequence to the target region of the edited DNA sequence [41]. CRISPR/Cas9 system binds with target DNA and nCas9 cuts opposite DNA strand to produce a single stranded DNA break. This broken DNA sequence serves as a primer and binds the PBS site in the sgRNA to produce a new DNA fragment using Cas9 fused reverse transcriptase. This newly synthesized DNA will then replace the targeted DNA. This is how prime editing can replace target DNA sequences without needing a DNA template. This system showed great efficiency in mouse cortical neurons and many human cell lines [41].
Role in Neurosciences
The present comprehension of brain function and its problems has been incredibly developed with the help of currently available devices and machinery. Among these CRISPR/Cas9 technology is a powerful innovation and hence is being utilized to produce new models for testing and understanding neurological diseases. CRISPR can progress both fundamental and translational neuroscience research [50]. Schizophrenia is a psychological issue with a worldwide predominance of around 1.5 million individuals [51]. Recently, the gene which encodes for protein DISC1 (disturbed in schizophrenia) has been recognized as an applicant gene for the illness by CRISPR.
Role in Cancer Biology
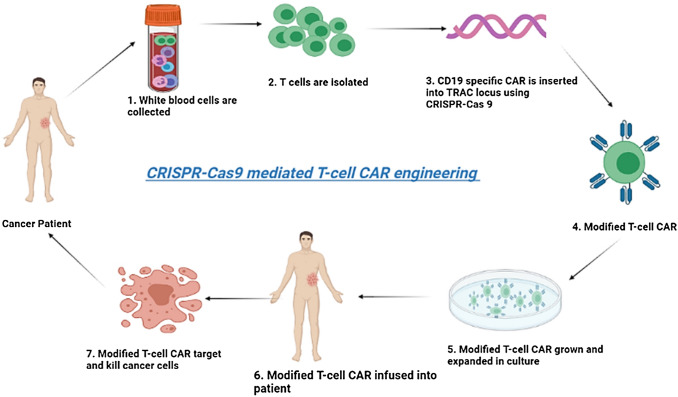
Cancer, in comparison to other diseases, is one of the major causes of human death each year around the world, but no powerful treatment has been found/discovered yet [50]. CRISPR technology was developed with the aim of cancer treatment and locating its source genes inside the host so that effective animal models can be generated for disease study [52]. Chen and his associates [53] proposed that CRISPR technology might be valuable for making malignant growth organism models for better comprehension of cancer biology. In 2016, a Chinese research group has turned out to be the first worldwide group to provide an individual, suffering from destructive cellular breakdowns in the lungs, with cells containing CRISPR/Cas9 edited genes. They simply took the patient’s immune cells, and knocked out the PD-1 gene responsible for immune response, on the basis that now these new cells can fight against cancer more effectively after their multiplication. Similarly, T cells design based on CRISPR illustrates great potential as a treatment for cancer and has shown to be more compelling than the original T-cell design (the whole process is shown in Fig. 3) [5, 54].
Fig. 3.
CRISPR/Cas mediated CAR (chimeric antigen receptors) T-cell engineering (Step 1–7). Cancer patients’ blood is collected and T cells are isolated. Then CD19 specific CAR is inserted into TRAC (T-cell receptor α constant) with the use of CRISPR/Cas9 technology. Thus modified or CAR engineered T cells are developed and grown in culture. These cells are then infused in the same patient where they attack their target cancer cells and cause apoptosis
Other types of cancers such as Epstein-Barr virus (EBV)-positive gastric cancer, glioblastoma, colorectal cancer, ovarian cancers, urinary bladder cancer, and melanoma can also be potentially treated through CRISPR/Cas9 disruption of PD-L1/PD-1 gene (a regulator molecule, expressed in T-cell and has a role in T-Cell mediated immunity) [16]. For example, genes involved in colorectal cancer are MLH1, MSH2, SMAD4, APC, Trp53, KRAS, and PIK3CA. Scientist conducted in vivo studies using genetically engineered mouse model to correct genes tumor suppressing genes APC, Trp53 along with SMAD4, KRAS, PIK3CA with the help of CRISPR/Cas (9 technology which showed promising results. In another study, genes PANDAR, LncRNA-UCA1 were found to increase expression of urinary bladder cancer. CRISPR/Cas9 technology was also employed here on Cell lines (T24, 5637 bladder cell lines) to knockout LncRNA-UCA1 genes to treat urinary bladder cancer [40]. Gao et al. [55] studied the role of AKT1 E17K mutation against TP53-null background. Results revealed that β-catenin signaling was abrogated which resulted in AKT1 E17K inhibiting cancer cell migration. Similar to this, Bungsy et al. [56] found that chromosome instability (CIN) is associated with the expression of the RBX1 gene. Knocking out of the RBX1 gene can be a cause of high-grade serous ovarian cancer (HGSOC). It is because once RBX1 is knocked out, CIN phenotypes increase, and Cyclin E1 levels and growth in fallopian tube secretory epithelial cells also increase (a precursor for HGSOC).
In Plants
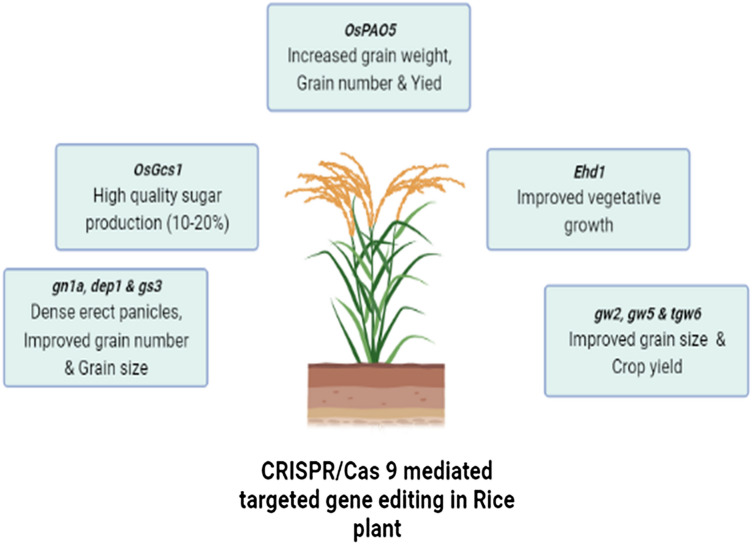
In plants, one of the great applications of CRISPR/Cas9 technology is found in crop improvement. CRISPR/Cas system was first time established in two major food crops, wheat, and rice. Crops have genes called negative regulators which have negative effects on crop yield. In rice, negative regulators like Gn1a, dep1, and gs3 negatively affect grain number, seed size, and crop yield. Through CRISPR/Cas 9 system, all 3 genes were knocked out which had helped in increased grain number, improved grain size, and dense, erect panicles in rice (Fig. 4). Such similar applications of CRISPR are also found in other plants such as in rapeseed, where gene BnaMAX1 was targeted which improved overall plant architecture and yield [16]. CRISPR technology has also been utilized in different plant researches, such as in biosynthesis and activity of the plant cell outer wall. Nowadays, people are very much reliant on plant cells wall based by-products like asylum, food, garments, and fuel. Genes involved in plant cell wall congregation are frequently very repetitive. With the help of hereditary crossing, these repetitive genes can be reorganized and their knock-out copies can be produced. In comparison to hereditary crossing, CRISPR base gene editing will not only reduce the production time of high mutants but also fasten its screening process which will help in the characterization of those cell wall genes which need more time for growth [39].
Fig. 4.
CRISPR/Cas9 mediated gene editing in rice plant
Furthermore, CRISPR is an excellent tool that can also help improve crop storage and post-harvest quality. Fruits like tomatoes become soft once fully ripe which makes their storage and transportation difficult to deal with. Scientists targeted both ALC gene mutations and replacement in tomato fruit which helped improved its storage and shelf life without compromising plant size and fruits firmness [57]. In another study, genes lncRNA, and lnRNA1459 related to fruit ripening in tomatoes were knocked out using the CRISPR/Cas 9 system which successfully resulted in a new variety of tomato with altered fruit ripening [58]. Zachary B. Lippmann and his associates used CRISPR technology to develop a new variety of minimized, acquiescent tomato plants appropriate to cultivate in metropolitan cities for future horticulture [59]. In wheat crop, genes α‐gliadin, TaGW2, and TaSBEIIa were targeted to produce varieties of wheat with low gluten, increased plant weight and protein content, and amylose and resistant starch content [16].
In Animals
CRISPR framework is broadly utilized in animal reproduction and disease displaying models due to being a precise, quicker, and productive genome-editing tool. In addition to gene-editing tools, there are a few essential approaches to creating animal models. Gene-altered organisms can be made by exploiting embryonic stem (ES) cells from gene-altered donor cells to produce genetically modified animals. For efficient and effective delivery of genome components, viral particles and nanoparticles are currently being exploited as vectors [60–62]. In the past, animal experimentation was carried out by using different models keeping in mind the species of the animal used. For instance, small animals and rodents were used to get ES cells infusion to create chimera gene-altered models because of their ease of use in research. Later on, through regular mating, homozygous mutated models were established. Yet, for large animals such as monkeys, the most productive approach through a straightforward infusion of the CRISPR Cas9 framework into zygote and embryo transfer is performed later on.
However, the disadvantage of the CRISPR framework is that a comparable gRNA results in different gene types in different cells and people, so it's difficult to get indistinguishable models for disease mutation [22]. A breakthrough in clone animal model research was seen while studying animal models development in monkeys, where gene-editing technology was used with somatic cell nuclear transfer (SCNT) [63]. An animal model produced this way will share similar genome typing as its donor cells. Scientists have started research on Huntington’s disease pigs and a BMAL1-ablation macaque based on this above-mentioned approach to creating animal models [11, 64, 65].
In Microbes
The CRISPR framework is a prokaryotic defense mechanism in opposition to DNA and RNA viruses in different microorganisms [66, 67]. Up until this point, the CRISPR framework has been embraced as a technology for important studies, metabolic designing, and genome functioning. In this discipline of molecular biology, the Cas9 framework has been used in various model bacteria, for example, E. coli, Mycobacterium spp., Saccharomyces cerevisiae, Clostridium spp., Lactobacillus spp., Bacillus spp., Pseudomonas spp., and Streptomyces spp. There remain many microbes for them Cas framework has not been used yet.
Role in Bacteria
Even though the major focus of CRISPR technology is on eukaryotes genome engineering, there are very high chances to use Cas9 in bacteria in common and particularly in food microbes. Even though CRISPR-based gene editing is particularly difficult in bacterial systems, overall, some breakthroughs have also been achieved as exemplified in Bacillus smithii, and various Lactobacillus spp. [68]. Type I CRISPR framework in L. crispatus was effectively used to complete genome editing in terms of addition, deletion, and single nucleotide replacement [69], outlining how Cas frameworks are promptly and programmable bridled in Lactobacillus spp. for gene altering. In their lab research, Qi and his associates developed a plasmid pNICK clos using CRISPR technology which can carry out numerous rounds of genome altering in C. acetobutylicum and C. beijerinckii with effectiveness shifting from 6.8 to 100 percent (%) and 18.7 to 100 percent (%), individually [70]. Cas9-based genome-editing tools have also been used for the creation of mass synthetic compounds, for example, succinate in Synechococcus elongates [71].
Role in Viruses
CRISPR methodology was primarily tried in the early 2000s for HIV1/AIDS treatment. Ebina and his associates effectively utilized the Cas9 gene to smother the appearance of AIDS genes in the Jurkat cell. Objective locales were the NF(κB) restricting tapes situated in the U-3 district of LTR, also TAR series in the region R, individually. This brought about proficient hindrance of HIV (1) provirus record. Significantly, it also illustrated that CRISPR can dispose of interior incorporated viral qualities just like the tainted host chromosome, which proposed that Cas9 might be, a possible option for AIDS treatment [11].
Role in Filamentous Fungi
Genome editing of fungi has not progressed very much in recent decades due to problems such as lack of suitable promoters, poor editing efficacy, plasmid hindrance, and gene delivery issues. CRISPR has been a successful technology in this regard as it has helped in genome editing in filamentous fungi after overcoming these issues. Conventional hereditary gene-editing techniques tried to advance the productivity of HR (Homologous recombination) by erasing KU80 and KU70 dimers ultimately making these fungi more sensitive to certain chemicals [72] for example, phleomycin and methyl methane sulfonate [73]. Conquering this impediment, the CRISPR framework in the filamentous mold of Trichoderma reesei uses a specialized codon in vitro RNA transcription [74]. The Cas 9 gene-editing tool not only effectively locate targeted gene but can also add multiple genes which have made this technology a powerful weapon for genome editing in filamentous organisms.
Role in SARS-CoV-2 Detection and Possible Treatment
In late 2019, a novel coronavirus disease (known as severe-acute-respiratory-syndrome-coronavirus-2 (SARS-CoV-2) in Wuhan, China had emerged which has resulted in deaths across the globe and is declared a pandemic by WHO in early 2020 [75]. There is a lack of effective diagnostic methods and treatments available against this viral disease. It is highly transmissible among humans and takes between 5 and 14 days upon infection to show symptoms. The most prevalent symptoms of the disease reported were muscle pain, fever, diarrhea, difficulty in breathing, dry cough, chest pain, elevated levels of cytokines in the bloodstream, and death in severe disease cases [76]. CRISPR/Cas technology has shown promise in providing methods for coronavirus detection and possible treatments (Table 3) [11].
Table 3.
CRISPR-based methods for severe-acute-respiratory-syndrome-coronavirus-2 (SARS-CoV-2) detection in human patients
| Method | Introduced by | Year | Time (min) | Working principle | References |
|---|---|---|---|---|---|
| DETECTR (DNA Endonuclease-Targeted CRISPR Trans Reported) | Mammoth Biosciences group | 2018 | 30 | Uses CRISPR/Cas12a nucleases system to detect targeted DNA sequences | [77] |
| FELUDA (FNCAS9 Editor Linked Uniform Detection Assay) | Debjyoti Chakraborty group from the Institute of Genomics and Interactive Biology (IGIB) | – | 45 | Uses CRISPR/Cas9 nucleases system to detect targeted genetic material of coronavirus | [78] |
| SHERLOCK (Specific High Sensitivity Enzymatic Reported unlocking) | Feng Zhang's group | 2017 | 60 | Uses CRISPR/Cas13 nucleases system to detect targeted RNA sequences | [79] |
CRISPR Cas protein orthologs like Cas9, 12, 13 and many more are currently being explored for their potential to act as an antiviral agent for coronavirus. The working strategy is to target SARS-CoV-2 housekeeping genes through different orthologs of Cas protein. Among these Cas protein orthologs, Cas 13 nuclease, which can cleave ssRNA of coronavirus, is the most promising antitherapeutic tool for antiviral agent development [80, 81]. Cas 13 nucleases are preferred because they can be programmed to cleave target ssRNA in humans with high precision without producing any off-target effects. Furthermore, it has no requirement for PAM or other sequences to cleave target sequences unlike other Cas nucleases such as Cas9 and Cas12 [76].
To control this viral pandemic, so far WHO has allowed the administration of 17 COVID-19 vaccines globally. Normally the development of a vaccine is a long and laborious process and takes somewhere between 5 and 10 years for commercialization. However, all of these vaccines were made commercially available within 1 year. Although these vaccines were made after valid pre-clinical and clinical experimentations but still there is a need for extensive research on these vaccines in terms of their long-term efficacy, safety and regulations, etc. With the emergence of SARS-CoV-2 mutants, there is a threat that these vaccines may lose their effectiveness against these mutants. Hence, it is needed to develop vaccines that can work against such variants effectively. CRISPR technology has shown promising results in this regard and researchers are utilizing it fully to produce engineered B cells as well as safe, recombinant, and multivalent viral vaccines [2]. Johnson et al. [82] had used CRISPR/Cas9 nucleases to engineer B Cells to produce monoclonal antibodies (mAbs) against various antigens. This approach can be used to express SARS-CoV-2 specific antibodies in the host by inserting engineered nucleic acid sequence in host B cells [82, 83]. A lot of such studies are currently going on, however, there is much work to be done in terms of immunogenicity, Cas protein expression in the host, CRISPR components delivery, off-target effects, etc. to design and produce an effective, efficient, safe, and multivariate vaccine against SARS-CoV-2.
CRISPR Ethics
Moral choices, especially in biomedicine, imply assessing possible profit-loss analysis. Toward exploring moral dynamics, it is better to think about the scope of potentially bad situations, the likelihood of every start-up, and the potential reasoning for every possible outcome [84]. CRISPR/Cas9 is not only a cheap, proficient, and precise technique to make changes at the single nucleotide level, but also assists in investigating numerous scientific and research inquiries [85]. Human germ line modifications through CRISPR base gene editing and lack of complete knowledge regarding human germ line mutagenesis have raised many bioethical concerns and have given birth to a variety of viewpoints [14]. Janssens and Cecile talked about the technical feasibility of this technology for the enhancement of certain desirable human traits. According to them, this technology is more beneficial in the case of single mutation genetic disorders such as sickle cell anemia rather than a polygenic disease controlled by many genes [86]. Li and QIAN argued that the technology is not developed to the extent where it can be employed for fine tuning humans. The reasons they stated include probable off-target mutations and unknown consequences of gene editing (may transfer to the next generation) associated with gene editing [87].
Some scientists also supported human gene editing such as Sharma and Scot under the title of ‘appropriate and justified use’. According to them, germ line editing should only be allowed in a human embryo under 14 days of culture as per standard ethical guidelines [88]. However, Lander [89] completely opposed the idea of human genome editing and strongly showed support for the ban of technology. Lander states that the use of technology should only be allowed in case of a genetic disease that has no other possible medical cure. He further argued that as we have no complete knowledge of possible genetic changes and their consequence, thus this technology should not be applied to the human germ line [46]. In 2017, the US National Academies on Human Gene Editing issued reviews concerning recent development in gene-editing technology. The report stated that germ line modification to create new human beings is not allowed however in certain medical situations it can be negotiated [84].
Environmental Impact of CRISPR/Cas9 Technology
No doubt CRISPR/Cas technology is of the best discovery of the twentieth century. With its everyday advancement, it is not only facing issues like legal issues, ethical issues, and social issues but also environmental issues in fields such as livestock, agriculture, and medicine. CRISPER does not affect the environment directly but indirectly through CRISPR modified microbes, plants, animals, and humans [90]. One of the major obstacles in CRISPR/Cas9 technology is off target mutations which may have a great impact on environmental integrity. As there is always a possibility that CRISPR modified organisms may and can transfer the acquired genes between each other through the process of gene drift. There are also chances of CRISPR related bioterrorism by using experimental organisms, modified through CRISPR technology, which can affect the environment very badly. All of the previously available gene-editing technology such as ZFNs or TALENs need specially designed proteins to bind with the targeted DNA, a process that may take a few years. However, CRISPR is most efficient and fast than these gene-editing technologies as it only needs a commentary RNA to bind with the targeted DNA. And this complementary RNA can be made in only a few days. This ability of CRISPR technology not only makes it a fast gene-editing technology but also a freighting gene-editing tool at the same time in terms of its irresponsible use [91].
Genetic drift which occurs due to CRISPR-based off target mutations may persist in a population and will likely continue in the next generation. No one can predict how this mutation will affect the next generation. Genetically modified organisms such as humans, animals, plants, and microbes not only will transfer these mutations within their specie but also with species where they may cause serious negative effects and can be the potential cause of an ecosystem disturbance [90]. Another concerning point is the use of genetic enhancement in the military, as some countries may use CRISPR technology for their soldiers’ genetic enhancement to increase their combat power, in that scenario it can be a serious threat to world peace and may cause long-lasting damages to the environment [90]. Although various small-scale research labs are researching possible negative impacts of CRISPR on the environment. No lab setting can accurately predict and forecast natural ecosystem future problems. Another concern is the maintenance of a peaceful environment among the common public regarding genetically modified foods. As the public has always had a native attitude toward any genetically modified product in the past. Therefore, the only way to move forward is to use CRISPR technology by keeping in mind all the safety and security measures [90].
Conclusion and Future Perspective
CRISPR has been observed as rather conceivably the valuable treasure for targeted genome editing in the light of its remarkable effectiveness, moderately minimal effort, low mutation rate & cost of development, and high target efficiency in contrast to other genome altering methods, like ZFNs and TALENs. Gene-editing technology has improved rapidly in this decade owing to the achievements of scientists. As a result, the CRISPR gene-editing technique has been used to eradicate genetic diseases from the planet, increase agricultural yields, make plants resistant to environmental pressures, develop sustainable goods, and create individuals with the potential to pass on desirable traits to future generations. This might indicate people with a higher IQ and a greater ability to fight a variety of diseases. As a result, addressing ethical and societal concerns remains a challenge for this genome-editing approach. While science has traveled great distances in developing new perspectives to enhance life on our planet, there is always the option of modifying life in the least harmful way possible.
For efficient gene editing, several factors are important and need to be taken into consideration such as CRISPR/Cas9 delivery method, target site selection, Cas 9 system activity, single guide RNA (sgRNA) design, and DNA repair mechanism efficiency. Major challenges associated with CRISPR development are off-target effects, CRISPR/Cas 9 delivery, and the efficiency of the repair mechanism. CRISPR/Cas system has a high chance of producing off-target effects (> 50%) which may affect the normal function of genes making them genetically unstable. These off-target effects can be dealt with through strategies like the selection of a suitable CRISPR/Cas delivery system, delivery of CRISPR/Cas system only to the target cells, designing of the most appropriate sgRNA, and selection of the right Cas enzyme.
Efficient delivery of the CRISPR/Cas9 system into the target is another hurdle that needs to be passed. CRISPR-based delivery systems are of three types, physical, viral, and non-viral vectors. Designing and selecting an appropriate vector along with full knowledge of its cons and pros is the only way to go forward. Another major obstacle is the DNA repair mechanism. Although NHEJ is the most efficient and commonly used DNA repair mechanism, its chances of producing a mutation are as high as 20–60%. Whereas in the case of HDR, chances of mutation are very low, as template DNA is provided here, however, its repair efficiency is low (0.5–20%). A way to deal with this obstacle is to improve HDR efficiency or suppress NHEJ's possible mutations. Although major critical advancement has been made in CRISPR/Cas9 system to enhance its target specificity, efficient gene delivery, and DNA repair mechanism, much work is still needed to be done to make it a ‘super’ gene-editing technology.
There is also a need to further explore and study the relationship between Cas9 protein activity and the ratio of Cas9 to sgRNA over time. For rapid progress in CRISPR technology, ethical, moral, and environmental issues regarding human genome editing are still under debate and need to be concluded. This debate has already come to an end in the USA but is still a very intense discussion in other European and Asian countries and this has caused an obstacle in CRISPR due to progress worldwide in medicine, biotechnological, and pharmaceutical industries. Similar to the unnaturally conceived first test-tube baby that showed up in 1978, CRISPR/Cas9 technology has also raised some concerns. And it is expected that if laws are completely made for it, very rapid progress will be seen in many fields of biological sciences such as genomics, livestock, medicine, and agriculture.
Acknowledgements
This work is carried out with the help of prestigious material of the institute’s libraries.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
The authors declare that they have no competing interests. We assure the integrity and quality of our research work. It is also stated that there is no plagiarism in this work and all points taken from other authors are well cited in the text. This study is completely independent and impartial.
Informed Consent
This research did not involve human participants.
Research Involving Human Participants and/or Animals
This research did not involve human participants and/or animals.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;5:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamburova VS, Nikitina EV, Shermatov SE, Buriev ZT, Kumpatla SP, Emani C, Abdurakhmonov IY. Genome editing in plants: An overview of tools and applications. International Journal of Agronomy. 2017 doi: 10.1155/2017/731535113. [DOI] [Google Scholar]
- 3.Moon SB, Kim DY, Ko JH. Recent advances the CRISPR genome editing tool set. Experimental and Molecular Medicine. 2019;51:1–11. doi: 10.1038/s12276-019-0339-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad HI, Ahmad MJ, Asif AR, Adnan M, Iqbal MK, Mehmood K, Muhammad SA, Bhuiyan AA, Elokil A, Du X, Zhao C, Liu X, Xie S. A review of CRISPR-based genome editing: Survival, evolution and challenges. Current issues in Molecular Biology. 2018;28:47–68. doi: 10.21775/cimb.028.047. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduction and Targeted Therapy. 2020;3:5–1. doi: 10.1038/s41392-019-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaj T, Sirk SJ, Shui SL, Liu J. Genome-editing technologies: Principles and applications. Cold Spring Harbor Perspective Biology. 2016;1:8–12. doi: 10.1101/cshperspect.a023754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaiswal S, Singh DK, Shukla P. Gene editing and systems biology tools for pesticide bioremediation: A review. Frontiers in Microbiology. 2019;13:10–87. doi: 10.3389/fmicb.2019.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palpant NJ, Dudzinski D. Zinc finger nucleases: Looking toward translation. Gene Therapy. 2013 doi: 10.1038/gt.2012.2. [DOI] [PubMed] [Google Scholar]
- 9.Xiong J, Ding J, Li Y. Genome-editing technologies and their potential application in horticultural crop breeding. Horticulture Research. 2015 doi: 10.1038/hortres.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sather BD, Romano Ibarra GS, Sommer K, Curinga G, Hale M, Khan IF, Singh S, Song Y, Gwiazda K, Sahni J. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Science Translational Medicine. 2015;7:307. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao Q, Guo D, Chen S. Application of CRISPR/Cas9-based gene editing in HIV-1/AIDS therapy. Frontiers in Cellular and Infection Microbiology. 2019;22:9–69. doi: 10.3389/fcimb.2019.00069.PMID:30968001;PMCID:PMC6439341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khalil AM. The genome editing revolution. Journal of Genetic Engineering and Biotechnology. 2020;18:1–16. doi: 10.1186/s43141-020-00078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmar S, Saeed S, Khan MHU, Ullah Khan S, Mora-Poblete F, Kamran M, Jung KH. A revolution toward gene-editing technology and its application to crop improvement. International Journal of Molecular Sciences. 2020;21(16):5665. doi: 10.3390/ijms21165665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulvihill JJ, Capps B, Joly Y, Lysaght T, Zwart HA, Chadwick R. Ethical issues of CRISPR technology and gene editing through the lens of solidarity. British medical bulletin. 2017;122(1):17–29. doi: 10.1093/bmb/ldx002. [DOI] [PubMed] [Google Scholar]
- 15.Li Q, Qin Z, Wang Q, Xu T, Yang Y, He Z. Applications of genome editing technology in animal disease modeling and gene therapy. Computational and Structural Biotech Journal. 2019 doi: 10.1016/j.csbj.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C, Brant E, Budak H, Zhang B. CRISPR/Cas: A nobel prize award-winning precise genome editing technology for gene therapy and crop improvement. Journal of Zhejiang University-SCIENCE B. 2021;22(4):253–284. doi: 10.1631/jzus.B2100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature Protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding W, Zhang Y, Shi S. Development and application of CRISPR/Cas in microbial biotechnology. Frontiers in Bioengineering and Biotechnology. 2020 doi: 10.3389/fbioe.2020.00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford K, McDonald D, Mali P. Functional genomics via CRISPR–Cas. Journal of Molecular Biology. 2019;431:48–65. doi: 10.1016/j.jmb.2018.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guitart JR, Johnson JL, Chien WW. Research techniques made simple: The application of CRISPR-Cas9 and genome editing in investigative dermatology. The Journal of Investigative Dermatology. 2016;136:e87–e93. doi: 10.1016/j.jid.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manghwar H, Lindsey K, Zhang X, Jin S. CRISPR/Cas system: Recent advances and future prospects for genome editing. Trends in plant science. 2019;24(12):1102–1125. doi: 10.1016/j.tplants.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Chen B, Niu Y, Wang H, Wang K, Yang H, Li W. Recent advances in CRISPR research. Protein & Cell. 2020;11(11):786–791. doi: 10.1007/s13238-020-00704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolan AE, Hou Z, Xiao Y, Gramelspacher MJ, Heo J, Howden SE, Freddolino PL, Ke A, Zhang Y. Introducing a spectrum of long-range genomic deletions in human embryonic stem cells using type I CRISPR-Cas. Molecular Cell. 2019;74(5):936–95032. doi: 10.1016/j.molcel.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morisaka H, Yoshimi K, Okuzaki Y, Gee P, Kunihiro Y, Sonpho E, Xu H, Sasakawa N, Naito Y, Nakada S. CRISPR-Cas3 induces broad and unidirectional genome editing in human cells. Nature Communications. 2019;10:5302. doi: 10.1038/s41467-019-13226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cameron P, Coons MM, Klompe SE, Lied AM, Smith SC, Vidal B, Donohoue PD, Rotstein T, Kohrs BW, Nyer DB. Harnessing type I CRISPR-Cas systems for genome engineering in human cells. Nature Biotechnology. 2019;37(12):1471–1477. doi: 10.1038/s41587-019-0310-0. [DOI] [PubMed] [Google Scholar]
- 26.Klompe SE, Vo PLH, Halpin-Healy TS, Sternberg SH. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature. 2019;571:219–225. doi: 10.1038/s41586-019-1323-z. [DOI] [PubMed] [Google Scholar]
- 27.Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio. 2014 doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nature Biotechnology. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roehm PC, Shekarabi M, Wollebo HS, Bellizzi A, He L, Salkind J, Khalili K. Inhibition of HSV-1 replication by gene editing strategy. Science and Reports. 2016 doi: 10.1038/srep23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Diemen FR, Kruse EM, Hooykaas M, Bruggeling CE, Schurch AC, van Ham PM, Imhof SM, Nijhuis M, Wiertz EJ, Lebbink RJ. CRISPR/Cas9- mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathogens. 2016;12:e1005701. doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wollebo HS, Bellizzi A, Kaminski R, Hu W, White MK, Khalili K. CRISPR/Cas9 system as an agent for eliminating polyomavirus JC infection. PLoS ONE. 2015;10:e0136046. doi: 10.1371/journal.pone.0136046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity y. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirano H. Structure and engineering of Francisella novicida Cas9. Cell. 2016;164:950–961. doi: 10.1016/j.cell.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleinstiver BP. Engineered CRISPRCas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim E. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nature Communications. 2017;8(14500):102. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L. C2c1-sgRNA complex structure reveals RNA-guided DNA cleavage mechanism. Molecular Cell. 2017;65(310–322):103. doi: 10.1016/j.molcel.2016.11.040. [DOI] [PubMed] [Google Scholar]
- 37.Harrington LB. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamano T. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016;165:949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Showalter AM. CRISPR/Cas9 genome editing technology: A valuable tool for understanding plant cell wall biosynthesis and function. Frontiers in Plant Science. 2020 doi: 10.3389/fpls.2020.589517.PMID:33329650;PMCID:PMC7714752.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akram F, Ul Haq I, Ahmed Z, Khan H, Ali MS. CRISPR-Cas9, a promising therapeutic tool for cancer therapy: A review. Protein and peptide letters. 2020;27(10):931–944. doi: 10.2174/0929866527666200407112432. [DOI] [PubMed] [Google Scholar]
- 41.Zhang B. CRISPR/Cas gene therapy. Journal of Cellular Physiology. 2021;236(4):2459–2481. doi: 10.1002/jcp.30064. [DOI] [PubMed] [Google Scholar]
- 42.Vertex. (2018a) A safety and efficacy study evaluating CTX001 in subjects with severe sickle cell disease. ClinicalTrial.gov Identifier: NCT03745287.
- 43.Vertex. (2018b). A safety and efficacy study evaluating CTX001 in subjects with transfusion-dependent β-thalassemia. ClinicalTrial.-gov Identifier: NCT03655678.
- 44.Allergan. 2019. Single ascending dose study in participants with LCA10. ClinicalTrial.gov Identifier: NCT03872479.
- 45.Adli M. The CRISPR tool kit for genome editing and beyond. Nature Communications. 2018;9:9. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devkota S. The road less traveled: Strategies to enhance the frequency of homology-directed repair (HDR) for increased efficiency of CRISPR/Cas-mediated transgenesis. BMB Reports. 2018;51:437–443. doi: 10.5483/BMBRep.2018.51.9.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang FG, Taylor DW, Chen JS, Kornfeld JE, Zhou KH, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh V, Gohil N, RamírezGarcía R, Braddick D, Fofié CK. Recent advances in CRISPR-Cas9 genome editing technology for biological and biomedical investigations. Jouranl of Cellular Biochemistry. 2018 doi: 10.1002/jcb.26165. [DOI] [PubMed] [Google Scholar]
- 51.Harrison PJ. The neuropathology of schizophrenia. A critical review of the data and their interpretation. Brain. 1999;122:593–624. doi: 10.1093/brain/122.4.593. [DOI] [PubMed] [Google Scholar]
- 52.Sanchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nature Reviews Cancer. 2015;15:387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen S, Sun H, Miao K, Deng CX. CRISPR-Cas9: From genome editing to cancer research. International Journal of Biological Sciences. 2016;12:1427–1436. doi: 10.7150/ijbs.17421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eyquem J, Mansilla-Soto J, Giavridis T, Van Der Stegen SJ, Hamieh M, Cunanan KM, Odak A, Gonen M, Sadelain M. Targeting a CAR to the € TRAC locus with CRISPR/Cas9 enhances tumor rejection. Nature. 2017;543(7643):113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao SP, Kiliti AJ, Zhang K, Vasani N, Mao N, Jordan E, Wise HC, Bhattarai TS, Hu W, Dorso M, Rodrigues JA. AKT1 E17K inhibits cancer cell migration by abrogating β-catenin signaling. Molecular Cancer Research. 2021;19(4):573–584. doi: 10.1158/1541-7786.MCR-20-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bungsy M, Palmer MCL, Jeusset LM, et al. Reduced RBX1 expression induces chromosome instability and promotes cellular transformation in high-grade serous ovarian cancer precursor cells. Cancer Letters. 2021;500:194–207. doi: 10.1016/j.canlet.2020.11.051. [DOI] [PubMed] [Google Scholar]
- 57.Yu QH, Wang B, Li N, Tang Y, Yang S, Yang T, Xu J, Guo C, Yan P, Wang Q, Asmutola P. CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Scientific Reports. 2017;7(1):1–9. doi: 10.1038/s41598-017-12262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li R, Fu D, Zhu B, Luo Y, Zhu H. CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. The Plant Journal. 2018;94(3):513–524. doi: 10.1111/tpj.13872. [DOI] [PubMed] [Google Scholar]
- 59.Kwon CT, Heo J, Lemmon ZH, Capua Y, Hutton SF, Van Eck J, Park SJ, Lippman ZP. Rapid customization of Solanaceae fruit crops for urban agriculture. Nature Biotechnology. 2019;38:182–188. doi: 10.1038/s41587-019-0361-2. [DOI] [PubMed] [Google Scholar]
- 60.Amoasii L, Hildyard JCW, Li H, Sanchez-Ortiz E, Mireault A, Caballero D, Harron R, Stathopoulou TR, Massey C, Shelton JM. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362:86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao X, Tao Y, Lamas V, Huang M, Yeh WH, Pan B, Hu YJ, Hu JH, Thompson DB, Shu Y. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217–221. doi: 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nelson CE, Wu Y, Gemberling MP, Oliver ML, Waller MA, Bohning JD, Robinson-Hamm JN, Bulaklak K, Castellanos Rivera RM, Collier JH. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nature Medicine. 2019;25:427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Z, Cai Y, Wang Y, Nie Y, Zhang C, Xu Y, Zhang X, Lu Y, Wang Z, Poo M. Cloning of macaque monkeys by somatic cell nuclear transfer. Cell. 2018;172(881–887):e887 41. doi: 10.1016/j.cell.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 64.Qiu PY, Jiang J, Liu Z, Cai YL, Huang T, Wang Y, Liu QM, Nie YH, Liu F, Cheng JM. BMAL1 knockout macaque monkeys display reduced sleep and psychiatric disorders. National Science Review. 2019;6:87–100. doi: 10.1093/nsr/nwz002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan SZ, Tu Z, Liu Z, Fan N, Yang H, Yang S, Yang W, Zhao Y, Ouyang Z, Lai C. A Huntingtin Knockin PIG model capitulates features of selective neurodegeneration in Huntington’s disease. Cell. 2018;173(989–1002):e1013. doi: 10.1016/j.cell.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Research. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annual Review of Biochemistry. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 68.Pan M, Barrangou R. Combining omics technologies with CRISPR-based genome editing to study food microbes. Current Opinion in Biotechnology. 2020;61:198–208. doi: 10.1016/j.copbio.2019.12.027. [DOI] [PubMed] [Google Scholar]
- 69.Hidalgo-Cantabrana C, Goh YJ, Pan M, Sanozky-Dawes R, Barrangou R. Genome editing using the endogenous type I CRISPR-Cas system in Lactobacillus crispatus. Proceedings of the National Academy of Sciences. 2019;116:15774–15783. doi: 10.1073/pnas.1905421116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Q, Chen J, Minton NP, Zhang Y, Wen Z, Liu J, Yang H, Zeng Z, Ren X, Yang J, Gu Y, Jiang W, Jiang Y, Yang S. CRISPR-based genome editing and expression control systems in Clostridium acetobutylicum and Clostridium beijerinckii. Biotechnology Journal. 2016 doi: 10.1002/biot.201600053. [DOI] [PubMed] [Google Scholar]
- 71.Li H, Shen CR, Huang CH, Sung LY, Wu MY, Hu YC. CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production. Metabolic Engineering. 2016;38:293–302. doi: 10.1016/j.ymben.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 72.Weld RJ, Plummer KM, Carpenter MA, Ridgway HW. Approaches to functional genomics in filamentous fungi. Cell Research. 2006;16:31–44. doi: 10.1038/sj.cr.7310006. [DOI] [PubMed] [Google Scholar]
- 73.Liu R, Chen L, Jiang Y, Zhou Z, Zou G. Efficient genome editing in filamentous fungus Trichodermareesei using the CRISPR/Cas9 system. Cell Discovery. 2015;1:15007. doi: 10.1038/celldisc.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hao Z, Su X. Fast gene disruption in Trichoderma reesei using in vitro assembled Cas9/gRNA complex. BMC Biotechnology. 2019;19:2. doi: 10.1186/s12896-018-0498-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khan KA, Duceppe MO. Cross-reactivity and inclusivity analysis of CRISPR-based diagnostic assays of coronavirus SARS-CoV-2. PeerJ. 2021;9:e12050. doi: 10.7717/peerj.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gupta R, Kazi TA, Dey D, Ghosh A, Ravichandiran V, Swarnakar S, Syamal R, Swades RB, Ghosh D. CRISPR detectives against SARS-CoV-2: A major setback against COVID-19 blowout. Applied Microbiology and Biotechnology. 2021;105(20):7593–7605. doi: 10.1007/s00253-021-11583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Azhar, M., Phutela, R., Ansari, A.H., Sinha, D., Sharma, N., Kumar, M., Aich, M., Sharma, S., Rauthan, R., Singhal, K., Lad, H., Patra, P.K., Makharia, G., Chandak, G.R., Chakraborty, D., Maiti, S. (2020) Rapid, felddeployable nucleobase detection and identification using FnCas9. bioRxiv
- 79.Abbott TR, Dhamdhere G, Liu Y, Lin X, Goudy L, Zeng L, Chemparathy A, Chmura S, Heaton NS, Debs R, Pande T, Endy D, Rudda MFL, Lewis DB, Qi LS. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181(4):865–876. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuan C, Tian T, Sun J, Hu M, Wang X, Xiong E, Cheng M, Bao Y, Lin W, Jiang J, Yang C, Chen Q, Zhang H, Wang H, Wang X, Dengm X, Liaom X, Liu Y, Wang Z, Zhou X. Universal and naked-eye gene detection platform based on the clustered regularly interspaced short palindromic repeats/Cas12a/13a system. Analytical Chemistry. 2020 doi: 10.1021/acs.analchem.9b05597. [DOI] [PubMed] [Google Scholar]
- 81.Straiton J. CRISPR vs COVID-19: How can gene editing help beat a virus? BioTechniques. 2020;69:327–329. doi: 10.2144/btn-2020-0145. [DOI] [PubMed] [Google Scholar]
- 82.Johnson MJ, Laoharawee K, Lahr WS, Webber BR, Moriarity BS. Engineering of primary human B cells with CRISPR/ Cas9 targeted nuclease. Scientific Reports. 2018 doi: 10.1038/s41598-018-30358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Faiq MA. B-cell engineering: A promising approach towards vaccine development for COVID-19. Medical Hypotheses. 2020;144:109948. doi: 10.1016/j.mehy.2020.109948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brokowski C, Adli M. CRISPR ethics: Moral considerations for applications of a powerful tool. Journal of Molecular Biology. 2019;431:88–101. doi: 10.1016/j.jmb.2018.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sugarman J. Ethics and germline gene editing. EMBO Reports. 2015;16:879–880. doi: 10.15252/embr.201540879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Janssens AC. Designing babies through gene editing: Science or science fiction? Genetics in Medicine. 2016;18:1186–1187. doi: 10.1038/gim.2016.28. [DOI] [PubMed] [Google Scholar]
- 87.Li CX, Qian HL. A double-edged sword: CRISPR-Cas9 is emerging as a revolutionary technique for genome editing. Military Medical Research. 2015;2:25. doi: 10.1186/s40779-015-0054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sharma A, Scot CT. The ethics of publishing human germline research. Nature Biotechnology. 2015;33:590–592. doi: 10.1038/nbt.3252. [DOI] [PubMed] [Google Scholar]
- 89.Lander ES. Brave New Genome. New England Journal of Medicine. 2015;373:5–8. doi: 10.1056/NEJMp1506446. [DOI] [PubMed] [Google Scholar]
- 90.Ayanoğlu FB, Elçin AE, Elçin YM. Bioethical issues in genome editing by CRISPR-Cas9 technology. Turkish Journal of Biology. 2020;44(2):110–120. doi: 10.3906/biy-1912-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shinwari ZK, Tanveer F, Khalil AT. Ethical issues regarding CRISPR mediated genome editing. Current Issues in Molecular Biology. 2018;26(1):103–110. doi: 10.21775/cimb.026.103. [DOI] [PubMed] [Google Scholar]