Abstract
Background: Bacteriophages that infect Escherichia coli are relatively easily isolated, with >600 coliphage genomes sequenced to date. Despite this there is still much to be discovered about the diversity of coliphage genomes.
Materials and Methods: Within this study, we isolated a coliphage from cattle slurry collected from a farm in rural England.
Results: Transmission electron microscopy identified the phage as member of the Siphoviridae family. Phylogenetic analysis and comparative genomics further placed it within the subfamily Tunavirinae and forms part of a new genus.
Conclusions: Characterization of the lytic properties of vB_Eco_SLUR29 reveals that it is rapidly able to lyse its host when infected at high multiplicity of infection, but not at low multiplicity of infection.
Keywords: coliphage, phages, genomes
Introduction
Bacteriophages infecting Escherichia coli (coliphages) are readily isolated from a variety of sources, with >600 complete or near complete coliphage genomes publicly available (May 2019). Coliphage genomes range in size from 3.39 kb1 to 386.44 kb2 and are represented in a number of phage families including the Leviviridae, Siphoviridae, Podoviridae, Myoviridae, and the Microviridae. Currently, within these families, there are 37 genera and 157 species that contain coliphages, with many taxa being poorly sampled.3 Therefore, there is still much to be discovered by the continued isolation and sequencing of coliphages, with many new phage types and taxa still to be discovered.3 Building on our previous research that has isolated coliphages from animal slurries,4,5 we aimed to isolate further phages from this system and characterize the phenotypic properties.2.5
Materials and Methods
Bacteriophage vB_Eco_SLUR29 was isolated from cattle slurry that was collected from a farm in the East Midlands, in the United Kingdom. The double-agar overlay method was used for phage isolation and the subsequent three rounds of purification using E. coli K-12 MG1655 as host, as has been described previously.6 High titer lysate was produced by infection of ∼50 mL of exponentially growing E. coli MG1655 (∼0.3–0.4) and incubated at 37°C with shaking at 300 rpm, until lysis had occurred. Phage growth parameters (burst size, eclipse, and latent period) were determined by performing one-step growth experiments as described by Hyman and Abedon, with free phages being removed from the culture by pelleting the host cells by centrifugation at 10,000 g for 1 min, removing the supernatant and resuspending cells in fresh medium.7
Three independent replicates were carried out for each experiment. The virulence index was calculated using the method described by Storms and Sauvageau using a SPECTROstar Omega (BMG) plate reader. For genome sequencing, DNA was extracted from 1 mL of bacteriophage lysate as previously described.8 One nanogram of DNA was used as input for Nextera XT library preparation, following the manufacturer's instructions. Sequencing was performed on an Illumina MiSeq (250 bp paired end) with both the genome sequence (accession: LR596614) and raw reads (accession: ERR3385641) submitted to the European Nucleotide Archive (ENA) under Project accession: PRJEB32519.
Bioinformatics and comparative genomics
Reads from sequencing were trimmed with Sickle v1.33 using default parameters.9 Assembly was carried out with SPAdes v.3.6.0 using “—only-assembler” option.10 Genome assembly errors were corrected with two rounds of checking and polishing with Pilon v1.23 using default parameters.11 The genome was annotated with Prokka 1.12 using a protein database constructed from accession LR027385.12 The start of the genome was arbitrarily set at the gene encoding for the large terminase subunit, for ease of comparison. For rapid comparison against all other phage genomes, a Mash database was constructed of all complete bacteriophage genomes available at the time of analysis (∼11,000, April 2019) using the following Mash setting: “–s 10000” and a previously described method.13
Closely related genomes were identified using this database and the “dist” function. From this initial set of genomes, the terL gene was used to construct a phylogenetic tree using IQ-TREE.14 After this, a more detailed analysis of the most closely related genomes was carried out. Phage genomes that were found to be similar were reannotated with Prokka to ensure consistent gene calling between genomes for comparative analysis12 and the GET_HOMOLOGUES pipeline used to identify core genes.15 For calculation of phage average nucleotide identity (ANI), pyani was used with default settings.16 Core gene analysis for phages within the putative genus “Swanvirus” was carried out with ROARY using “—e—mafft -p 32 –i 90.”17
Transmission electron microscopy
Transmission electron microscopy (TEM) was carried out at the University of Leicester Core Biotechnology Services Electron Microscopy using a previously described method.13 Digital images were collected with a Megaview III digital camera using iTEM software. Phage images were processed in ImageJ using the measure tool, with the scale bar present used as a calibration to measure phage particle size.18 The data presented are the mean of 15 phage particles.
Results
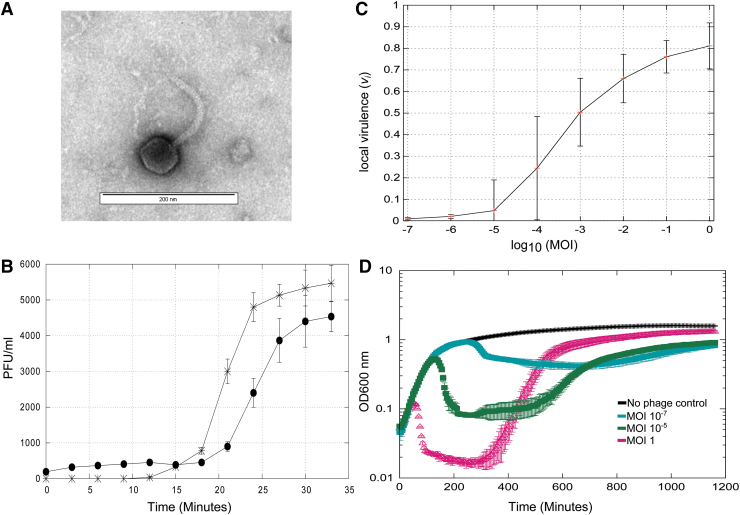
Bacteriophage vB_Eco_SLUR29 was isolated from animal slurry collected from a farm in the East Midlands, United Kingdom, using E. coli K-12 MG1655 as a host. The plaque morphology was small (<3 mm) clear plaques, suggestive of an obligately lytic phage. Imaging of phage vB_Eco_SLUR29, using TEM, revealed a polyhedral head with a long flexible noncontractile tail. The head was 57 nm (±2.8) and 56.7 nm (±6.7) in width and length, respectively, with a tail that was 12 nm (±1.4) wide and 142 nm (±23.2) long. The long noncontractile tail allowed its classification within the Siphoviridae and the head length–width ratio further classified vB_Eco_SLUR29 within subgroup B119 (Fig. 1A).
FIG. 1.
Phenotypic properties of phage vB_Eco_SLUR29. (A) TEM image of phage vB_Eco_SLUR29, identifying the morphological features representative of siphoviruses. (B). One-step growth experiment (n = 3), samples treated with CHCl3 are closed black circles, untreated samples are stars. (C) Local virulence index of vB_Ecol_SLUR29 at MOIs ranging from 1 to 1 × 10−7. (D) Killing curves of vB_Eco_SLUR29 against Escherichia coli K-12 MG1655 (n = 3). MOI, multiplicity of infection; TEM, transmission electron microscopy.
The lytic properties of the phage vB_Eco_SLUR29 were determined using a one-step experiment (Fig. 1B). The latent period was determined to be ∼21 min, the eclipse period 17 min, with a burst size of 25 (±6). To further characterize the infection properties, killing curves were used to investigate the ability of vB_Eco_SLUR29 to kill its host over a range of multiplicity of infections (MOIs) and also determine the recently described virulence index.20 At an MOI of 1, there was rapid lysis of the culture with near complete lysis after 100 min, followed by a steady increase in growth after 300 min. When a very low MOI of 1 × 10−5or lower was used, these cultures displayed growth comparable with an uninfected control until the onset of lysis was observed at ∼300 min (MOI 1 × 10−7). After 300 min, a decrease in growth was observed, before a steady increase from 700 min onward. For calculation of the virulence index, the local virulence values at MOIs from 1 to 10−7 were determined (Fig. 1C). This further highlighted that phage vB_Eco_SLUR29 was inefficient at lysing its host at very low MOIs (Fig. 1C). The virulence index of phage vB_Eco_SLUR29 was calculated to be 0.37 at 37°C in Lysogeny Broth (LB) medium.
Genome sequencing
To further classify vB_Eco_SLUR29 beyond morphological similarity to other Siphoviruses, the genome of vB_Eco_SLUR29 was sequenced. Genome sequencing resulted in a single chromosome with an average coverage of 426 × . The genome was 48,466 bp in length with a G + C content of 44.7%, with 78 predicted genes and no tRNAs. Of the 78 predicted genes, functions could only be predicted for 25 of the proteins they encode and the majority of these were phage structural proteins. Comparing the genome sequence of vB_Eco_SLUR29 against current phage genomes identified that it had the greatest similarity (Mash distance <0.05) to the coliphages SECphi27, vB_Eco_swan01, vB_EcoS-95, vB_Eco_mar001J1, and vB_Eco_mar002J2. These phages have previously been found to form a monophyletic cluster, which represents a putative genus within the subfamily Tunavirinae.13 In addition, vB_Eco_SLUR29 showed some similarity to a group of phages infecting Campylobacter (Mash distance <0.25), which are not currently classified by the International Committee on Taxonomy of Viruses (ICTV).
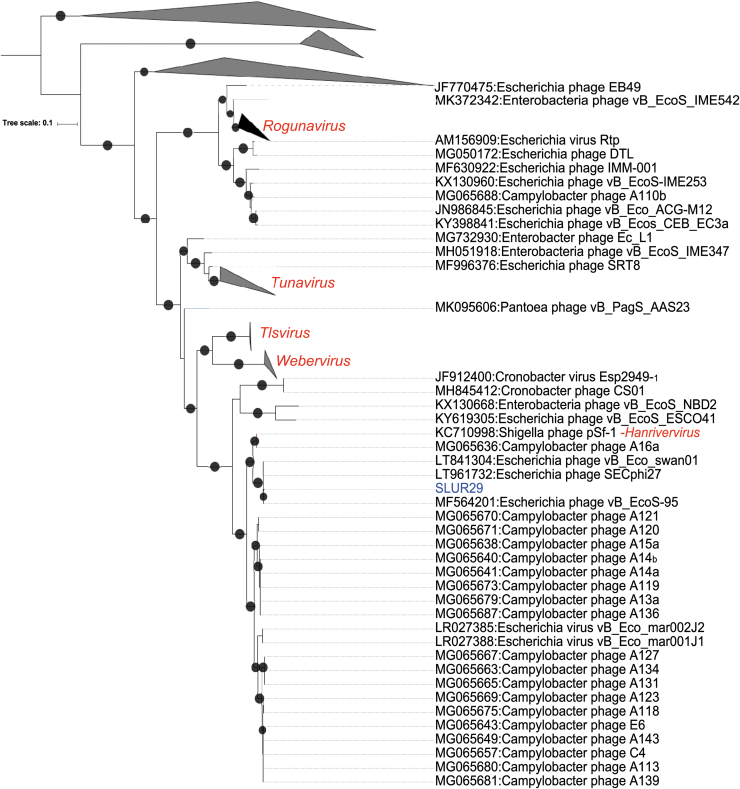
A phylogeny was reconstructed using terL, with the top 100 hits to the vB_Eco_SLUR29 terL based on Basic Local Alignment Search Tool Protein (BLASTP) analysis. The resultant phylogeny placed vB_Eco_SLUR29 within the subfamily Tunavirinae, forming a clade with the coliphages vB_Eco_swan01 (accession: LT841304),21 SECphi27 (accession: LT961732), vB_Eco_mar001J1 (accession: LR027388), vB_Eco_mar002J2 (accession: LR027385), and vB_EcoS-95 (accession: MF564201),22 which is a sister group to the clade containing phage pSF-1, the sole representative of the genus Hanrivervirus.23
To further clarify the position of vB_Eco_SLUR29 within the subfamily Tunavirinae, a core gene phylogeny was constructed for phages that were closest to vB_Eco_SLUR29. This included phages of the genera Tlsvirus, Webervirus, and Hanrivervirus and the large group of unclassified phages that infect Campylobacter (Fig. 2). Four core genes were identified in this set of phages (representative homologues are SLUR29_0019, SLUR29_0039, SLUR29_0053, and SLUR29_0059) using the GET_HOMOLOGUES pipeline.15 The resultant four genes were concatenated and used in further phylogenetic analysis. This phylogeny confirmed that phage vB_Eco_SLUR29 formed a clade with the coliphages vB_Eco_swan01 SECphi27, vB_EcoS-95, vB_Eco_mar001J1, and vB_Eco_mar002J2, all of which were isolated on E. coli and likely represent a new genus. The Shigella infecting phage pSf-1 again grouped with phage A16a, with this cluster being more closely related to the group of unclassified phages infecting Campylobacter than it is to vB_Eco_SLUR29.
FIG. 2.
Phylogenetic analysis of vB_Eco_SLUR29. The phylogeny was created using the terL gene as marker. Sequences were aligned with MAFTT25 and trees were constructed with IQ-TREE with 1000 bootstrap replicates.14 Bootstrap values >70 are marked with a black circle, with increasing size proportional to the bootstrap values. The clades containing phages of the genera Tlsvirus and Webervirus were collapsed for clarity.
Comparative genomics
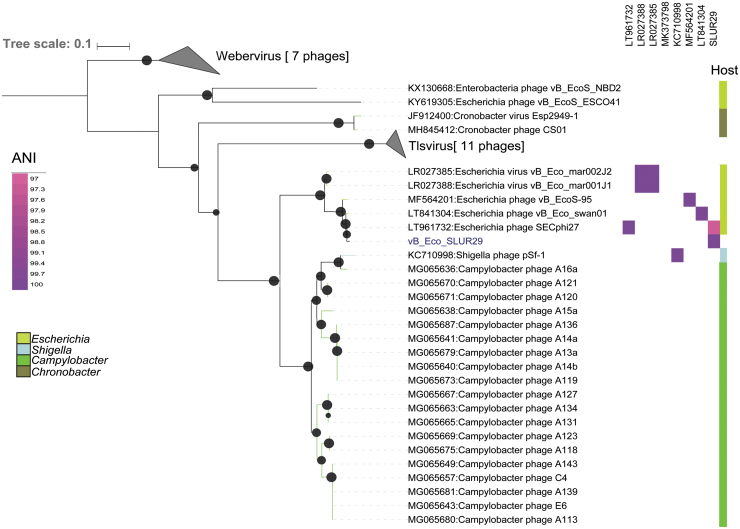
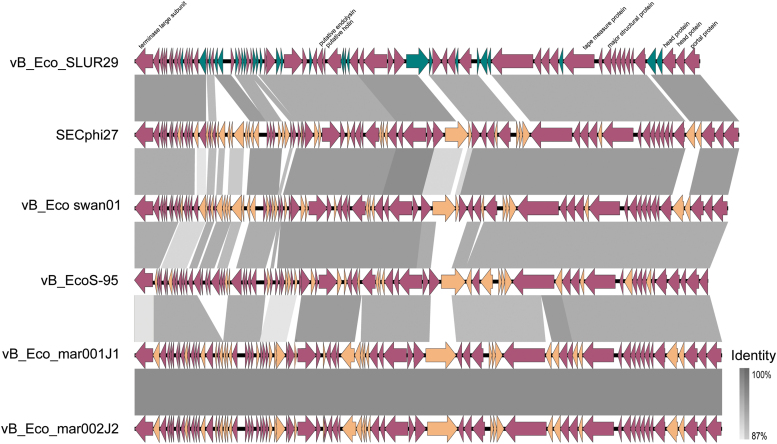
Phages vB_Eco_SLUR29, vB_Eco_swan01, SECphi27, vB_EcoS-95, vB_Eco_mar001J1, and vB_Eco_mar002J2 all have ANI >90% with each other and an ANI of <80% with pSF1 (Fig. 3). Whole genome comparisons of vB_Eco_SLUR29, vB_Eco_swan01, SECphi27, vB_EcoS-95, vB_Eco_mar001J1, and vB_Eco_mar002J2 reveal they have 51 core genes (Fig. 4) and have high degree of synteny across the genomes (Fig. 4). Inclusion of the phage pSF-1 in this analysis results in only one core gene.
FIG. 3.
Phylogenetic analysis of vB_Eco_SLUR29. The phylogeny was created using four concatenated core genes. Genes were aligned with MAFFT25 and tress were constructed with IQ-TREE,14 using a SYM+R4 model of evolution. Bootstrap values >70 are marked with a black circle, with increasing size proportional to the boostrap values. The clades containing phages of the genera Tlsvirus and Webervirus were collapsed for clarity in the presenting tree.
FIG. 4.
Comparative genomics of vB_Eco_SLUR29 with phages SECphi27, vB_Eco_swan01, vB_EcoS-95, vB_Eco_mar001J1, and vB_Eco_mar002J2. Alignments were constructed with EasyFig using BLASTn.26 Selected genes are annotated for vB_Eco_SLUR29. Core genes are in purple, noncore genes are in green in vB_Eco_SLUR29, and orange in all other phages.
Discussion
We have previously isolated coliphages from the same slurry tank and have found phages that are representatives of the genera T4virus and Seuratvirus.4,5 This is the first report of a phage from within the subfamily Tunavirinae from this particular slurry tank environment. Whether this is a reflection of their abundance or due to small sample sizes remains to be seen. Comparison of vB_Eco_SLUR29 with its closest relatives reveals they have all been isolated on E. coli K-12 MG1655, although many can infect other bacteria within the Enterobacteriaceae (Fig. 3).13,22
Given the high sequence identity between vB_Eco_SLUR29 and its closest relatives, it was unsurprising that it has similar morphological properties when examined by TEM. Phylogenetic analysis clearly places vB_Eco_SLUR29 within the subfamily Tunavirinae, in a clade that is sister to a clade that contains phages of the genus TLsvirus and contains the phages vB_Eco_mar001J1, vB_Eco_mar002J2, vB_EcoS-95, vB_Eco_swan01, and SECphi27. Previously, we have suggested that SECphi27, vB_Eco_mar001J1, vB_Eco_mar002J2, vB_Eco_swan01, and pSf1 are members of the same genus.13
Since then, Shigella phage pSf1 has been formally classified as the sole member of the genus Hanrivervirus. The addition of phages vB_Eco_SLUR29, vB_EcoS-95, and Campylobacter infecting phages to the database further clarifies the position of Shigella phage pSf1 into a separate clade from vB_Eco_mar001J1, vB_Eco_mar002J2, and SECphi27. The current starting point for classification of phage species is >95% ANI.24 Using this criterion, vB_Eco_SLUR29, vB_Eco_mar001J1, vB_Eco_mar002J2, vB_EcoS-95, vB_Eco_swan01, SECphi27, and vB_EcoS-95 would form a single species. This is inconsistent with the phylogeny observed and a higher ANI cutoff of >97% might be more suitable for this group of phages, as has previously been suggested for this and other phage groups.5,13 Thus, we propose this group of phages represents a new genus with four species, represented by the type phages vB_Eco_mar001J1, vB_EcoS-95, vB_Eco_swan01, and SECphi27. With vB_Eco_SLUR29 being the same species as phage SECphi27, which was isolated first. We propose the genus is named “Swanvirus” after the phage vB_Eco_swan01, which was the first isolated phage in the genus.
All phages of the proposed genus Swanvirus have a conserved and syntenous genome structure. In contrast to the conservation of genes between phages, there is greater variation in phenotypic properties. The burst size of vB_Eco_SLUR29 is 22, which is smaller than the reported values of 115, 78, and 51 for vB_Eco-S59, vB_Eco_swan01, and vB_Eco_mar002J2, respectively.13,22 There is also considerable variation in the latent period for these phages, with vB_Eco_SLUR29 having a substantially longer latent period (21 min) than the reported 4, 12, and 15 min for vB_Eco-S59, vB_Eco_swan01, and vB_Eco_mar002J2.13,22 However, the reported 4 min latent period of vB_Eco-S59 seems extraordinarily rapid and maybe an artifact of the method used.
Phage vB_Eco_SLUR29 was found to rapidly lyse its host when used at an MOI of 1, but was less effective at lower MOIs. This was apparent in the local virulence index that is very close to zero at MOIs of 1 × 10−6 and 1 × 10−7 when the integrated area of the curve of infected and control samples is compared. To overcome this, we could have chosen a later point where lysis had occurred for cells infected at low MOIs. However, this would be well into the stationary phase of growth in the uninfected control, so we chose to integrate from time zero to the onset of stationary phase as described in the original method.20 When compared with the virulence index of phages T7, T5, and T4, it has a lower virulence index of 0.3720 than any of these phages under any condition, suggesting it is not a particularly virulent phage.
A further observation of the properties of vB_Eco_SLUR29 infection was the rapid recovery of infected cells. When infected at high MOIs, there was rapid lysis of host cells, with recovery of number of cells close to an uninfected control after 13 h. Although we did not explicitly test cells at the end of the killing assay, it is most probable that this recovery in cell growth is due to the rapid selection of resistant cells. The rapid development of resistance in E. coli to the closely related phage vB_Eco_S59 has been reported, suggesting that there might be a minimal cost to developing resistance for this type of phage.22 However, the mechanism behind the development of resistance still remains to be elucidated.
The sequencing of vB_Eco_SLUR29 has expanded and helped to further clarify the phylogeny of phages within the genus Tunavirinae, with vB_Eco_SLUR29 a member of putative new genus that is clearly separate from the phage pSf-1 (Fig. 4). Furthermore, the characterization of phenotypic properties reveals that phages that are similar at the genomic level have very different phenotypic properties. This highlights the need to assess the lytic properties of phages that are genetically similar, as it cannot always be assumed they will have similar infection properties. With increasing interest in the use of phages as therapeutics, the lytic properties are important factors that will need to be considered. In part, differences in lytic properties may result from the method used for carrying out one-step experiments. Therefore, we utilized the recently developed virulence index20 that should allow more consistent comparison of the lytic properties of phages from different laboratories in the future.
Acknowledgment
Bioinformatics analysis was carried out using MRC CLIMB Infrastructure MR/L015080/1.
Authors' Contributions
J.L.H. and A.M. conceived the idea. I.B., P.S., C.H., L.G., T.R., S.M., and S.P.H. were involved in practical aspects of sample collection, phage isolation, genome sequencing, one-step growth experiments, virulence assays, and analysis of raw data. L.G., C.H., T.R., and A.M. wrote the article. All authors have reviewed and approved of the article before submission.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
A.M., J.L.H., and S.P.H. were funded by NERC AMR-EVAL FARMS (NE/N019881/1). T.R. and S.M. were in receipt of PhD studentships funded by the Natural Environment Research Council (NERC) CENTA DTP.
References
- 1. Friedman SD, Genthner FJ, Gentry J, et al. . Gene mapping and phylogenetic analysis of the complete genome from 30 single-stranded RNA male-specific coliphages (family Leviviridae). J Virol. 2009;83:11233–11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chinnadurai L, Eswaramoorthy T, Paramachandran A, et al. . Draft genome sequence of Escherichia coli phage CMSTMSU, isolated from shrimp farm effluent water. Microbiol Resour Announc. 2018;7:4–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Korf IHE, Meier-kolthoff JP, Adriaenssens EM, et al. . Still something to discover: Novel insights into Escherichia coli phage diversity and taxonomy. Viruses. 2019;1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith R, O'Hara M, Hobman JL, et al. . Draft genome sequences of 14 Escherichia coli phages isolated from cattle slurry. Genome Announc. 2015;3:e01364-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sazinas P, Redgwell T, Rihtman B, et al. . Comparative genomics of bacteriophage of the genus Seuratvirus. Genome Biol Evol. 2017;10:72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kropinski AM, Waddell TE, Franklin K, et al. . Enumeration of bacteriophage by double agar overlay plaque assay. Methods Mol Biol. 2009;501:287–292. [DOI] [PubMed] [Google Scholar]
- 7. Hyman P, Abedon ST. Practical methods for determining phage growth parameters. Methods Mol Biol. 2009;501:175–202. [DOI] [PubMed] [Google Scholar]
- 8. Rihtman B, Meaden S, Clokie MRJ, et al. . Assessing Illumina technology for the high-throughput sequencing of bacteriophage genomes. PeerJ. 2016;4:e2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Joshi NA, Fass JN. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) [Software]. 2011. Available at https://github.com/najoshi/sickle.
- 10. Bankevich A, Nurk S, Antipov D, et al. . SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walker BJ, Abeel T, Shea T, et al. . Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 2014;9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [DOI] [PubMed] [Google Scholar]
- 13. Michniewski S, Redgwell T, Grigonyte A, et al. . Riding the wave of genomics to investigate aquatic coliphage diversity and activity. Environ Microbiol. 2019;21:2112–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nguyen L, Schmidt HA, von Haeseler A, et al. . IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Contreras-Moreira B, Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 2013;79:7696–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pritchard L, Glover RH, Humphris S, et al. . Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8:12–24. [Google Scholar]
- 17. Page AJ, Cummins CA, Hunt M, et al. . Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015;31:3691–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 2012;9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ackermann HW, Krisch HM. A catalogue of T4-type bacteriophages. Arch Virol. 1997;142:2329–2345. [DOI] [PubMed] [Google Scholar]
- 20. Storms ZJ, Teel MR, Mercurio K, et al. PHAGE 2019;1:1X. Available at https://www.liebertpub.com/doi/10.1089/phage.2019.0001 [DOI] [PMC free article] [PubMed]
- 21. Michniewski S, Redgwell T, Scanlan DJ, et al. . Draft genome sequence of bacteriophage vB_Eco_swan01. Genome Announc. 2017;5; pii: e00501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Topka G, Bloch S, Nejman-Falenczyk B, et al. . Characterization of bacteriophage vB-EcoS-95, isolated from urban sewage and revealing extremely rapid lytic development. Front Microbiol. 2019;10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jun JW, Kim JH, Shin SP, et al. . Characterization and complete genome sequence of the Shigella bacteriophage pSf-1. Res Microbiol. 2013;164:979–986. [DOI] [PubMed] [Google Scholar]
- 24. Adriaenssens E, Brister JR. How to name and classify your phage: An informal guide. Viruses 2017;9: pii: E70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sullivan MJ, Petty NK, Beatson SA. Easyfig: A genome comparison visualizer. Bioinformatics 2011;27:1009–10. [DOI] [PMC free article] [PubMed] [Google Scholar]