Abstract
Despite the abundance and significance of bacteriophages to microbial ecosystems, no broad ecological frameworks exist within which to determine “bacteriophage types” that reflect their ecological strategies and ways in which they interact with bacterial cells. To address this, we repurposed the well-established Grime's triangular CSR framework, which classifies plants according to three axes: competitiveness (C), ability to tolerate stress (S), and capacity to cope with disturbance (R). This framework is distinguished from other accepted schemes, as it seeks to identify individual characteristics of plants to understand their biological strategies and roles within an ecosystem. Our repurposing of the CSR triangle is based on phage transcription and the observation that typically phages have three major distinguishable transcription phases: early, middle, and late. We hypothesize that the proportion of genes expressed in these phases reflects key information about the phage “ecological strategy,” namely the C, S, and R strategies, allowing us to examine phages in a similar way to how plants are projected onto the triangle. In the “phage version” of this scheme, we suggest: (1) that some phages prioritize the early phase of transcription that shuts off host defense mechanisms, which reflects competitiveness; (2) other phages prioritize tuning resource management mechanisms in the cell such as nucleotide metabolism during their “mid” expression profile to tolerate stress; and (3) a further subset of phages (termed Ruderals) survive disturbance by investing significant resources into regeneration so they express a higher proportion of their genes during late infection. We examined 42 published phage transcriptomes and show that they fall into discrete CSR categories according to their expression profiles. We discuss these positions in the context of their biology, which is largely consistent with our predictions of specific phage characteristics. In this opinion article, we suggest a starting point to ascribe phages into different functional types and thus understand them in an ecological framework. We suggest that this may have far-reaching implications for the application of phages in therapy and their exploitation to manipulate bacterial communities. We invite further use of this framework via our online tool; www.PhageCSR.ml.
Keywords: ecology, bacteriophage types, bacteriophage ecology transcriptional profiling, Grimes CSR triangle
Introduction
Environments select for specific types of organisms with optimal survival strategies. This is well understood across most domains of life, but remains largely unexplored within bacteriophages (or phages), despite them being the most abundant organisms on earth. Although phages significantly impact microbial ecology, physiology, evolution, and community composition,1,2 no frameworks exist to contextualize the different ways in which they do this. Instead, phages are classified as being temperate if they integrate into bacteria, obligately lytic if they exclusively promote lytic infections, chronic if they promote the continuous release of phage virions, or pseudolysogenic if they infect cells and reside in their genomes but do not integrate.3 Although this four-way classification is a good starting point, it is over-simplistic and limits our understanding of the different ways that phages drive bacterial dynamics in specific environments.
Often, phages are classified in ad hoc ways such as “T4-like” or “Lambda-like”. Sometimes, this means that the phages have one or two genes in common, or that they share a roughly similar morphology to their “-like” phage. Although the use of such terms is misleading, it reflects a desire to have recognizable “phage types.” The strong and incredibly valuable community-led effort to genomically classify phage taxa by using the binomial Linnaean system4 is based on genomic similarity; although it helps us understand where phages have evolved from, and their genetic relations, it does not reflect the ecological behavior of phages.
Understanding ecology relies on a robust taxonomy but phages are hard to classify into recognizable taxa5 and understanding functional ecology requires linking traits (properties that facilitate survival) directly to environmental gradients, which is also difficult in phages. These difficulties stem from not knowing the function of most phage genes (thus not knowing what their traits are), and from phages not fitting standard classifications schemes by virtue of having small and highly diverse genomes. Further, unlike the situation in bacteria, there are no universal markers such as the 16S ribosomal RNA subunits.
Excitingly, many novel viruses have recently been isolated and sequenced and phage metagenomes have provided enormous amounts of novel genomic data.6–16 However, the uncharacterized functions encoded by so much novel phage diversity limit our ability to go beyond a genome/image-based classification. Therefore, phage sequence data cannot be translated directly into tangible knowledge of the impacts of phages on microbial ecology.
One study that recognized ecological phage types within a genus focused on two phages that infect the marine bacterium Pseudoaltermonas.17 This work examined the concept of a “virocell” and showed that two phages infected their bacterial host using contrasting strategies such that the infected bacteria had very different properties. The authors suggest that the complementarity between the two genomes (phage and host) drives the amount of cellular reprogramming that is needed, and the phage that was most distant from the bacteria in terms of nucleotide composition needed to carry our more bacterial transcriptional modification. They suggest that this work lays the foundation to identifying “functional guilds” as seen in other organisms. We agree with their conclusion that understanding the reprogramming of cellular mechanisms is crucial to understanding the impact of phages on ecological systems, and we outline a suggested theoretical framework within which to examine this, not based on guilds per se, but on a similar, slightly broader concept of ecological strategies.
In contrast to phages, plants have been extensively studied in terms of their ecological strategies and different “vegetation types” are known to perform specific functions within an environment that do not map to taxonomy. Many adaptations are critical to allow plants to thrive in specific environments. Therefore, there is vast knowledge and validated frameworks to determine, for example, which types of plant features are consistent stabilize and colonize environments, what adaptations benefit plants when ambient conditions are optimal for growth, or allow them to survive stress and disturbance. Indeed, when ecologists examine key physiological processes they often ignore the species and instead look for functional plant categories to quantify processes of interest.
The CSR Scheme
Several different schemes have been developed to understand and interpret plant ecology, for example, Raunkiaer looked at life forms,18 Praeger19 studied succession, Tansley looked at plant communities,20 and Greig-Smith attempted to make mathematical assessments of plant communities by removing a subjective assessment.21
Very influential in ecology was work on r-K strategies that looked at the species' ability to rapidly colonize and reproduce in disturbed environments (r strategy) and species' ability to exploit more efficiently the carrying capacity of the environment (K strategy).22 Although this scheme has been used or discussed for bacteriophages,1,23 it may be overly simplistic.
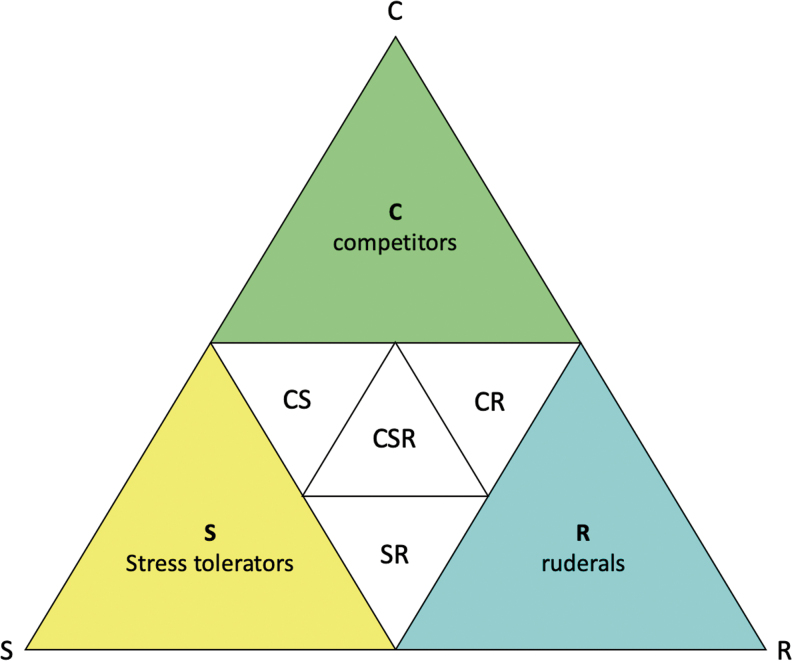
One framework that has been extensively developed and is still used to provide key insights into the functioning of plant-dominated natural environments was developed by John Philip Grime in 1977.24,25 Grime showed that plants can be classified according to the combination of their individual “traits” or biological characteristics into three primary strategies: their competitiveness (C), their ability to tolerate stress (S), and how good they are at coping with disturbance—which is termed ruderal (R), derived from the Latin word rudus meaning “rubble” as such plants colonize these newly formed environments. This CSR framework has also been interpreted quantitatively with demographic parameters.26
The CSR strategies are shown in Figure 1on Grime's triangle. All plants are mapped within these three dimensions and are positioned at the apices of the triangle with all their features consistent with them being primary competitors, stress tolerators, ruderals, or within other intermediate strategies. This framework provides a clear way to understand plant specializations and can be used to map species' positions according to their traits to see which strategy they conform to. Plant strategies are translated into functional types by identifying their ecological functions.27 We expand on the philosophy of this scheme, with reference to examples of well-known plants to provide the rationale as to how we envisage this scheme can be applied to phages.
FIG. 1.
Grime's CSR triangle showing that plants can be classified according to three axes: their competitiveness (C), their ability to tolerate stress (S), and their ability to cope with disturbance (R).
We acknowledge that this is a first attempt at providing such a broad scheme in phages, and similar to the evolution of plant ecological frameworks in general and the CSR framework specifically, this scheme is likely to evolve or develop as researchers embrace it and as more phage systems are studied in detail. We hope, however, that the ideas presented here provide a starting point for a broadly applicable way to determine how phages are impacting microbial communities.
Of interest, the CSR scheme has also been used by microbial ecologists to classify bacteria and archaea into functional types.28–31 However, there are no particular refinements of this scheme within the bacterial work that specifically render it more adaptable to phages than the original scheme, and as such we apply the plant-constructed framework directly to phages. Thus, we link plant ecology to phage ecology without further reference to the CSR insights from bacterial work.
Bacteriophages and the CSR scheme
Being able to group phages into their “strategies” or “functional types” would allow the exploration of key questions such as how environmental conditions dictate which phage types dominate. Specific bacteria inhabit ecological niches where a multitude of diverse phages with different ecological strategies have the potential to infect them. A framework for understanding which phages survive under particular conditions would help to understand fundamental microbial processes and in an applied context help us to formulate phage cocktails for specific purposes.
Examples of the diversity of ecological strategies can be seen by considering the wide array of phages that target Escherichia coli, five of which were analyzed here. Of these, the model T4 myovirus has a large cargo of auxiliary metabolic genes in addition to the genes needed to complete the phage life cycle.32 T7, in contrast, has a much smaller genome,33 and Mu, which also has a small genome, can also function as a transposon and has a further interesting strategy.34 These phages were chosen for in-depth study by virtue of being among the first of their “types” to be isolated and the fact that they were easy to propagate. Little is known about the environments that these well-known phages are associated with, or the conditions in which they would flourish. However, the decades of study that such phages have had, means that they form a key mechanistic reference point for starting to decipher such information.
The CSR Concept and the Early, Middle, and Late Genes
The C, S, and R categories in plants refer to those that devote the majority of their resources to growth, maintenance, and regeneration, respectively. We hypothesize that the major three temporal categories of phage gene transcripts that can be discerned in most studies (early, middle, and late) each contribute to an aspect of phage behavior that is analogous to these three categories. We also suggest that this is a good proxy to understanding phage “functional types” without having specific knowledge of what the genes encode. In general, early genes encode for proteins that interact with and co-opt host systems, middle genes encode phage genome replication, and late genes are for viral morphogenesis.35,36
To test the applicability of the CSR to phages, we compiled a list of phage transcriptomic studies where early, middle, and late gene phases had or could be determined (Supplementary Table S1). We then recreated a Grime CSR triangle plot by using the proportion of genes expressed in each category as indicators for putative Competitor, Stress, and Ruderal traits, respectively. Thus, we use this one quantitative trait to characterize phage types, in a manner similar to the demographic approaches used in plants.26 Regardless of genome size, the proportion of the phage genome allocated to growth, maintenance, and dispersal is likely to be a critical trait since genome size is limited by storage capacity. Unlike for plants, its predictive functional power remains unexplored.
Although the transcriptional profiles for 42 phages had previously been established17,33,37–67—no previous work has attempted to broadly compare these profiles across different phage groups. However, when analyzed as we have done here, these data present an excellent opportunity for comparative studies.
CSR traits of known plant species and phage analogies
To show how contextualizing phages within the CSR framework could be paradigm-shifting in terms of our understanding of their biology, we summarized the biology of key well-known plants that are representative of each life-strategy, and we show how we can infer biologically useful information from this and infer how phages in these categories would function. We summarize our interpretation of the different CSR phage categories in Table 1.
Table 1.
Phage-Centric Summary of CSR Strategies
| Category | Strengths | Phage centric summary of strategies |
|---|---|---|
| C | Competition | Effective when bacterial conditions are good. Their large proportion of early genes can “tune” the cell to establish the conditions that they need to replicate efficiently. |
| S | Stress | After successful infection—when the phage is in the cell, it can sense the environment and can respond to a wide array of conditions. |
| R | Fluctuating conditions | Can cope with fluctuating conditions and do not need to invest in reprogramming the cell to the same extent—potentially they are quick in-and-out phages. |
| CS | Competition and stress | Can cope with competition from other phages even when their environment is stressed. |
| CR | Competition and fluctuating conditions | Can cope with competition from other phages even when their environment is disturbed. |
| SR | Stress and fluctuating conditions | After successful infection—when a phage is in a cell, it can sense the environment and can respond to cope with a wide array of conditions even if they are disturbed. |
| CSR | Good all-rounder | This is your “jack of all trades but master of none” phage—it can tolerate many conditions but would not fare well against the single or dual phages in their preferred environment. |
Competitors
An example of a “Competitor plant” is the stinging nettle, Urtica dioica. The traits that make it a competitor are large fleshy leaves that deter animals through its sting, sizable underground storage roots and an ability to thrive on nutrient-rich ground exploiting undisturbed environments. These features mean that they can rapidly withdraw resources from the environment. They also have the plasticity to change their leaves if needed, for example, when growing in shade. They are not competitive when the ground is disturbed, because they delay their reproduction to make more biomass, which means that other plant types (ruderals) will flower first. It takes a lot of resources to actively forage what limits their success in disturbed environments. In summary, the important traits of competitors are that they can monopolize resource capture, especially in undisturbed environments.
We speculate that this Competitor trait, with respect to phages, would manifest as phages with multiple gene products that hijack the bacterial cell machinery—directing the cellular metabolism to suit its needs—in a way that is akin to monopolizing plant resource capture. Products encoded by these genes would also prevent competition from other phages during coinfections. To do this, such phages would have a substantial proportion of early genes. By analogy to plants, a competitor phage would thrive when bacterial hosts are in high numbers within a stable environment and would do less well when their bacteria are stressed, or disturbed.
Stress tolerators
A classic example of a Stress tolerator plant is the wild thyme, Thymus praecox that grows on droughted grasslands, with minimal water and nutrients. The plants and flowers are tiny, as they do not waste resources. Other stresses include mineral shortage, especially for phosphorus and nitrogen. Stress tolerator plants are efficient with nutrient capture and sparse with their usage. They are more specialist than competitor plants, so they are only found in specific stressful environments where they can compete against plants with other strategies.
Conceptually, a stress-tolerant phage would infect bacterial hosts that are starved of nutrients or in suboptimal nutritional states. To do this, such phages would need a high proportion of their genes activated during the middle phase. They would infect hosts, then express an array of genes needed to allow them to survive, and finally propagate in the prevailing hostile conditions.
Ruderals
Finally, we illustrate the classic “R” or ruderal plant by using the poppy Papaver somniferum, which can survive high levels of disturbance. In contrast to stresses, such as nutrient deprivation, ruderal plants thrive in environments that are regularly disturbed through processes such as ploughing, or fire. Poppies thrive in disturbed landscapes and are a symbol of war, because they are among the first plants to colonize ravaged trench-ridden soils. Ruderal plants survive disturbance by having high growth rates as seedlings and an early onset of reproduction and, thus, seed production. They focus on seed production instead of leaves and root production, so they make seeds despite having small leaves, stems, and roots.
By analogy to plants, a classic R phage would make a lot of offspring despite a relatively short latent period. It would infect bacteria in fluctuating disturbed physical conditions. The main operational mode for a ruderal phage would be to quickly reproduce, create structural proteins, assemble, and leave the cell. Doing this requires fewer early and middle genes compared with the late genes that are needed to build and assemble structural proteins.
Intermediate strategies
We anticipate that similar to plants, some phages will have strategies based on multiple traits and thus we briefly discuss the characteristics of plants within mixed categories later.
Competitor-Stress plants combine traits of competitive ability with stress tolerance to enable them to thrive in a stressed environment. A typical CS plant is the blueberry, Vaccinium myrtillus. A CS phage would have a higher proportion of early and middle genes. Competitive-Ruderal plants combine the competitive traits with those that enable them to survive in disturbed environments, such as the creeping buttercup (Ranunculus repens). CR phages would have a higher proportion of early and late genes. The final two-strategy plant is the Stress-Ruderal, which copes with stressed disturbed environments; this is typified by the carline thistle, Carlina acaulis. Finally, the classic CSR plant is typified by the wild strawberry, Fragaria vesca, that combines all three traits. By analogy, a CSR phage would have the same proportion of genes expressed at the early, middle, and late stages of infection.
Grime initially developed his theory to test the existence of primary strategies in plants. After about 20 years, he reinterpreted and expanded his thoughts with reference to the intervening body of literature, and to better understand the ecosystem structure and dynamics.25 A benefit of this system is that it is possible to map “functional traits”—key aspects of plant physiology that allow plants to thrive in specific environments—to different CSR categories. This is extremely helpful to understand the biology of individual plants and how and why they play specific roles in a community. We outline our first attempts to repurpose this scheme to further our understanding of phages by observing the relative proportion of genes expressed at different parts of the phage life cycle. We hypothesize that this framework will be a helpful hook from which to expand, adapt, and inform our knowledge of phage biology.
Results and Discussion
We intend to bring the CSR theory to phage biology on the basis that phages need to compete with bacterial cells for resources, and with additional phages, survive stressful environments, and tolerate disturbance. Phages have three major gene expression profiles: early, middle, and late, which we hypothesize correlate with phages being good competitors (early genes), having the ability to survive stress (middle genes) and disturbance (late genes).
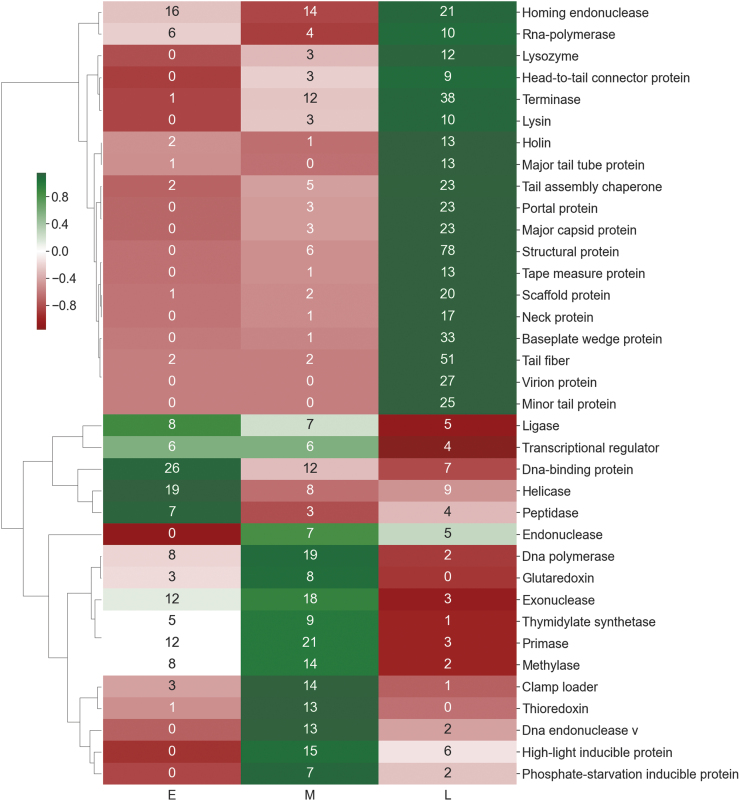
To test this, we collected and analyzed 42 publically available phage transcriptomes. We extracted temporal gene expression data from the literature and analyzed them in terms of their profile commonalities. Figure 2shows that when the functional annotations of the protein-encoding genes within the 42 phage genomes are clustered according to expression profiles, their clusters make biological sense. Most early genes of known function are involved in nucleotide metabolism, and thereby likely to have a role in repurposing the transcriptional machinery of the host. The “middle” genes encode for an array of proteins involved in DNA modification such as methylation, and for proteins involved in “protecting” the cell such as high light-inducible proteins and those expressed under phosphate starved conditions. The majority of late genes have functions involved in regeneration, building the virion shells and lysing bacteria.
FIG. 2.
Z-score clustering of protein annotations that occur at least in 10 transcriptome phases: early (E), middle (M), and late (L) phases (the number in parenthesis is the total observations for a particular function).
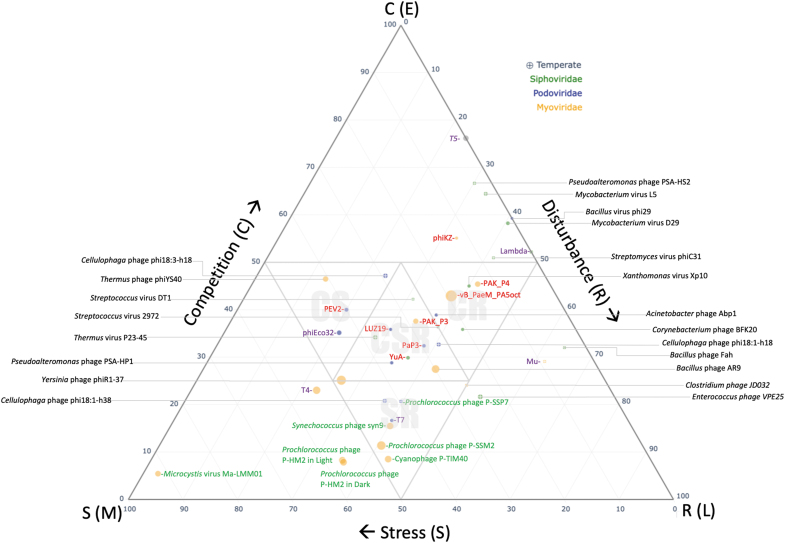
The analysis of gene functions, therefore, gave us confidence that a comparative transcriptomic analysis fits into our proposed framework where the proportion of genes expressed at the different phases of a phage cycle are indicative of the three categories of growth. By reducing each phage transcriptional profile to one value (based on the relative proportion of early, middle, and late genes), we mapped their transcriptional profiles to the seven categories of the CSR framework as shown in Figure 3.
FIG. 3.
Shows the position of 42 phages based on the proportions of early, middle, and late genes within the CSR triangle, which reflects their ability to tolerate competition, stress, or disturbance. The color and size of the dots represent the phage classification into myoviruses, siphoviruses, and podoviruses and their genome size. Although the phages were all examined during their lytic cycle, those with a cross have access to the temperate cycle. Associated labels of phages that target Pseudomonas are shown in red, Escherichia in purple, and cyanobacteria in green.
There is significant biological knowledge for some phages, but less for others, which limits our interpretation of them in our analysis. We, therefore, focus our interpretations on well-understood phages. As more phages are analyzed according to their transcriptional profiles, they can be added to the triangle, to further test ideas. To facilitate this, we have created a free to use and access, interactive online tool, termed PhageCSR, to allow researchers to explore existing phages and add any additional phages to the CSR framework. This can be found at www.PhageCSR.ml The framework uses transcriptomic data as input, as this is the only way to achieve the temporal separation. Although genomic data based on promoter analysis could be used as a proxy, there are inherent difficulties in identifying and validating promoters, and some genes have multiple promoters so it will not be clear where the primary expression is. However, promoters can sometimes be identified, and their temporal class assigned with some certainty based on genomic data or genomic similarity, such as with Enquatroviruses. If this is the case, then either the number of promoters in each class or better yet the number of genes in putative operons could be used in our tool to provide a tentative CSR assignment.
Phages Occupying All Seven Categories
The 42 studied phages fall into all seven CSR categories and are discussed according to their ecology and biology with respect to this position. A few phages have decades of mechanistic study, in particular T4, which we discuss in detail as it shows how our theory fits with an understanding of the stages of a phage life cycle where such information is known.
Competitor phages
Seven C phages are identified with a large proportion of early genes: the marine Pseudoalteromonas phage PSA-HS2; two mycobacterial phages, L5 and D29; the Bacillus phage phi29; the Streptomyces phage phiC31; the Pseudomonas aeruginosa phage phiKZ; and the E. coli phage T5. Our prediction by analogy to C plants, such as the nettle, is that these phages would thrive when bacteria are in stable high numbers in nutrient replete conditions. We would also predict that they would spend initial resources tuning their environment to their needs and that they would be effective at co-opting agency over the infected cell and be resistant to host defenses and competition from other phages. Although there is little commonality between these seven phages, it is conceivable that they all infect bacteria that could flourish when conditions are good, and they are able to resist bacterial defenses and interference from other phages.
Mycobacterium phages L5 and D29
Although more than 13,000 mycobacterial phages have been sequenced, only L5 and D29 have had their transcriptomes analyzed. These closely related phages were isolated in 1954 from soil and infect the saprophytic Mycobacterium smegmatis, the pathogenic Mycobacterium tuberculosis, and other mycobacterial species with D29 having a broader host range than L5. It will be interesting to see where the multitude of other mycobacterial phages fall in the CSR triangle when they are analyzed and if broad host range as a trait correlates with a “C” strategy.
E. coli phage T5
The coliphage T5 encodes for a distinctive progression of its obligately lytic lifecycle. After irreversible binding to the host receptor FhuA, around 8% of the T5 genome enters the cell, which is believed to inhibit host RNA and protein synthesis68 and promote restriction insensitivity.69,70 It also encodes for proteins with nuclease activity that degrades the genome to mononucleotides that are further broken down and excreted by the cell.71,72 This seemingly curious waste of valuable nucleotide substrates appears to be necessary, because high concentrations of nucleotides may be toxic and there is a delay between degrading cellular DNA and making phage DNA. This situation contrasts with T4 and T7 infections where host DNA degradation and phage DNA synthesis occur simultaneously and thus there is no build-up of such substrates.
This dramatic destruction and ejection of the host genome fits with our understanding of a “C” trait, redundant to the inhibition of host RNA and protein synthesis, for excluding adaptive phage defense mechanisms as well as co-infecting phages. The waste inherent to maintaining it may also serve as an instructive lesson as to why so many other phages might encode for elaborate middle-mode nucleotide metabolism. Indeed, phages that efficiently conserve energy and nucleotide substrates while avoiding osmotic stresses with a phage-encoded nucleotide metabolism might be more resilient to nutrient limiting and other stressful conditions.
Pseudoalteromonas phage PSA-HS2
The siphovirus HS2 has multiple genes involved in lysogeny. Interestingly, during infection, it does not significantly reprogram the transcriptome of the host cell, and instead maintains host transcription during its infection cycle while directing the machinery to phage propagation. The latent period of this phage is 60 min: 67% of H2S genes are expressed early, 3% in the middle, and 30% are expressed late. Many of these early genes are used for DNA metabolism, such as DNA helicase and DNA recombination and repair. It seems that this phage by tuning itself so well to the metabolism of the host replicates well (burst size of ∼120) with a minimal energetic cost to the cell.
Pseudomonas phage phiKZ
The jumbo phage PhiKZ spends the early phase of its infection constructing an elaborate virocell with a metabolism and structure that is radically distinct from that of its host. Indeed, with the use of two phage-encoded RNA polymerases, one of which is packaged into the virion, PhiKZ completes its infection cycle in the absence of any transcription from the host RNA polymerase.61 PhiKZ constructs a nucleus-like structure73 that broadly renders related phages immune to DNA-based phage exclusion mechanisms.74 PhiKZ also profoundly reorganizes the metabolism of the infected cell to suit its needs, enhancing stocks of metabolic resources.74,75
Stress-tolerant phages
The two stress-tolerant cyanophages (one phage grown in two conditions) discussed later make biological sense by analogy to the classic stress-tolerant plant thyme; they infect cyanobacteria that inhabit nutrient-poor and high UV conditions. Further, T4 is known to be able to infect E. coli in a range of conditions and thus it makes sense to have a suite of genes to facilitate this.76
Microcystis Ma-LMM01 and Prochlorococcus phage P-HM2
The large Microcystis myovirus Ma-LMM01 is the most extreme S-phage with 169 middle genes, 10 early, and 5 late. The host Microcystis species, M. aeruginosa has many antiviral mechanisms, including a novel way of escaping host defenses.41 Interestingly, there is no change in host promoter activity during any of the expression phases, so it appears that the phage infects in an incognito manner, maintaining host photosynthesis throughout its life cycle. The relatively small change in host expression suggests that this phage does not significantly impact host transcription, despite expressing what it needs to cope with the stressful environment.
The Prochlorococcus phage P-HM2 has 137 middle, 19 early, and 96 late genes. This cyanophage shows transcriptional rhythms; it cannot infect host cells at night and has a large burst size perhaps by initiating infection during the daytime when cyanobacteria are metabolically active. Presumably, it needs its suite of genes to do this. Our analysis showed that it did not change places in the triangle when it infected Prochloroccus under light or under dark conditions and was then moved to the light.
E. coli phage T4
As so much is known about T4, we are able to expand more on its mechanism of infection with reference to its ecology. The analysis of T4 transcription effectively brought transcriptomics to the phage world.40 This work largely confirmed detailed historical work on T4 transcription and comprehensively showed that its gene expression could be divided into early, middle, and late “modes,” with some genes expressed in more than one “mode.” T4 is close to CSR, and thus it also has a significant proportion of the C and R genes. Interestingly, the function of many “middle” or “S” genes is not known.
We base our arguments for the suitability of repurposing “CSR” on early, middle, and late genes. However, in most phages, we do not know the exact function of the proteins encoded by the genes during both of the phases, or how the “switch” from the three phases is triggered, but in T4 we do have this knowledge so we expand on this as it supports our hypothesis of the genes having functions that are of direct use to the distinct phases of the phage life cycle.77
An example of the type of stress where “middle” genes are expressed is nutrient limitation that T4 detects and responds to by initiating “hibernation” mode. Indeed, under the conditions examined, T4-infected cells produced over 40 times more progeny when nutrients were added to the media after infection compared with if it “scavenges” nutrient-depleted cells. This persistent but reversible T4 hibernation mode is initiated by T4 during nutrient stress at the beginning of middle mode transcription and continues until nutrient stress resolves. Interestingly, the host stationary phase sigma factor is not needed for T4 hibernation, suggesting that a potentially elaborate array of phage-encoded machinery is responsible, and that T4 is appropriately placed in the S apex of our triangle.
In addition to the multiple S-related genes, this phage has many early (C) and late (R) genes, which makes sense as it is a borderline CSR. A major C trait of interest is that to infect many of its hosts, T4 modifies its bases to avoid degradation by the host and its own nucleases. It fully substitutes glucosylated 5-hydroxymethylcytosine (5hmCyt) for cytosine in its DNA, and it partially substitutes 6mAde by using primarily middle mode genes.78–80 Phage enzymes slowly restrict host DNA (but not the protected phage DNA) that are further degraded to provide the phage-pregnant cell with host-derived nucleotides via a scavenging pathway.79
The 5hmCyt in T4 DNA also allows the phage to block elongation during transcription of host DNA and all other cytosine-containing DNAs. To accomplish this, T4 uses the phage-encoded protein gp Alc, which modifies the specificity of host RNA polymerase complex, to only recognize 5hmCyt containing DNA. The effect of alc expression is extremely host lethal, but it further powerfully excludes co-infection by any competing phages that do not also incorporate 5hmCyt.78,80,81 Thus, crucial for our understanding within a CSR context, this nucleotide modification strategy both protects phage DNA from degradation and prevents transcription from either the host or other competing phages.
The relatively large number of genes involved in excluding host nucleotide metabolism, replacing it with a phage-encoded metabolism, scavenging nucleotide substrates by slowly degrading the host genome, and incorporating the resulting non-canonical nucleotides into progeny T4 genomes are largely expressed in the middle mode that we hypothesize promotes “S” traits. As stated earlier, many of the T4 middle genes are hypothetical; thus, despite this phage being so well characterized, further study is needed to fully reveal the mechanisms by which this phage can cope with the multitude of stresses that we know it can replicate in.
Ruderal phages
We identified four “Ruderal” phages that by analogy to plants are like poppies in terms of coping with disturbance. These are the Bacillus “Fah,” the E. coli phage “Mu,” Enterococcus phage VPE25, and Clostridium phage JD032.
Bacillus phage “Fah”
Bacillus phage Fah is a 37,974 bp well-studied siphovirus that is used extensively in the former USSR to identify anthrax.44 The evolutionary history of Fah is lost in the mists of time; it has some similarities to prophage found in both Bacillus anthracis and Bacillus thurengenis prophage, so it may have originally been induced from one of these bacterial species. Although predicted to access the lysogenic cycle, it forms tight clear plaques and it is not known whether the repressor proteins are functional.
To determine the transcriptional profile, the authors used the non-harmful Bacillus cereus as a bacterial host. Of note, Fah encodes nine recognizable transcriptional regulators. These may modify Fah's biology to bias it to having a large proportion of genes expressed late during transcription. The phage does not cause a transcriptional shutdown, and it exclusively uses the host RNA polymerase.
A further interesting component of this phage is that it encodes a sigma factor. Although the role of this sigma factor is unknown, the best-known phage-encoded sigma factor gp55 in T4 directs transcription to the late promoters. The Fah sigma factor is closely related to the Bacillus transcription factors, which, similar to gp55, directs transcription to late promoters. The structural proteins in this phage can be transcribed by either the host or the phage-encoded sigma factor. Strangely, this Fah sigma factor is negatively regulated by a host sigma factor that controls sporulation! Thus, this ruderal phage appears to be regulated by the host sigma factor, the phage-encoded host-modulated sigma factors, and other phage-encoded putative transcriptional regulators. Combined, this suggests that this phage may have multiple strategies to regulate its transcription and cope with its “disturbed” environment.
E. coli phage Mu
Isolated in the 1950s, Mu can be a phage or a transposon—it can express transposase and multiply by lysogenizing its bacterial cell. When existing as a phage, it is a myovirus of 36,717 base pairs encoding 55 genes. This unusual biology makes biological sense that it is of a “ruderal” disposition and needs to cope with different environments.
Enterococcus phage VPE25
The transcriptome of the siphovirus VPE25 was recently determined along with work to establish the key genes involved in phage interactions by generating and screening a transposon library. The phage was shown to regulate novel aspects of bacterial physiology involved in quorum sensing. The phage is dependent on many host-encoded metabolism genes such as fructose kinase, suggesting that it has a reliance on the maintenance of cellular energy maintenance. This fits with its strategy as a ruderal phage, as it does not overly upset bacterial metabolism to produce lots of progeny.
CS phages
Plants in this “blueberry” category tend to both lay down resources in terms of their physical structure and can cope with flexibility when times are less optimal. Four phages are in this category: PhiYS40, which infects the thermo-tolerant Thermus hermophilus; the E. coli phage phiEco32; the Cellulophaga phage phi18:3; and the Pseudomonas phage PEV2.
PEV2
PEV2 is closely related to the well-studied type phage E. coli N4. Both phages are notable for their elegant transcriptional scheme where they progressively express phage genes by using three distinct RNA polymerases, including a virion-associated RNA polymerase (vRNAP) packaged into the capsid along with its DNA.82,83 N4 infection begins when the phage injects its vRNAP and a short DNA sequence into the cell, which is then transcribed in the forward direction by using host factors and the vRNAP, both creating early transcripts and pulling the rest of the genome into the cell (Demidenko and Rothman-Denes, unpublished data).82,84 The second virally encoded RNA polymerase (N4 RNAPII, gp15–16), as well as two virally encoded transcription factors (gp01–02), are then expressed from these early transcripts and together promote the middle mode of transcription involving gene features associated with DNA replication.82,83,85 This includes a single-stranded DNA-binding protein that interacts with the host sigma70 sigma factor to promote late phage transcription of structural and lysis genes.86
PEV2 fits into our view of a classic CS phage, as it invests heavily into optimizing the phage-infected host on both a transcriptional and metabolic level.75,86 It is also able to infect its host under anaerobic respiration and aerobic conditions,75,86,87 so it really needs an array of genes to modify host metabolism to facilitate this.
Cellulophaga baltica phage phi18:3
The detailed ecological characterization of two phages that target the oceanic bacterium Cellulophaga baltica88 has allowed us to compare a known generalist and specialist to see how they sit within our CSR framework. The authors characterized two phages, the specialist phage phi18:3 and the generalist phage phi 38:1; part of this work was the transcriptome profiling of two hosts. The podovirus phi18:3 is defined as a specialist, as it has a narrow range of host species that it can target.88 As more phage sets with similar host-range metadata are examined according to their transcriptomes and other phenotypic characteristics, it will be interesting to see whether this SR position in the triangle is a common feature.
CR phages
Five phages fall into this “buttercup” category that sits between the C and R. These are Brevibacterium phage BFK20, Xanthomonas phage Xp10, Acinetobacter phage Abp1, and Pseudomonas phages PAK_P4 and vB_PaeM_PA5oct.
Acinetobacter phage Abp1
The well-characterized Abp1 has a large burst size of 350, and a short latent period of 10 min. This is consistent with it being able to infect quickly, reproduce efficiently, and finally lyse. Abp1 also exhibits high thermal and pH stability, implying that it could survive an inhospitable environment. It has a low frequency of lysogeny, suggesting that this is not its “usual” method of survival—instead, it makes lots of propagules and a subset will survive.
Xp10
Xp10 targets the plant pathogen, rice blight, and is essentially a siphovirus but with chimerism and some podovirus characteristics. This phage encodes a single-subunit RNAP. Interestingly, part of its genome is jointly transcribed by both this phage and the host RNA polymerase.89
PAK_P4
PAK_P4 is a distant relative of PAK_P3 (discussed below in CSR phages) sharing recognizable synteny but displaying only limited DNA similarity outside of the structural genes. Its close position next to PAK_P3 in the CSR triangle (Fig. 3) might be remarkable if it were not for the conserved ancestral infectious strategy described by Blasdel et al.63
SR phages
The plant that typifies the SR strategy is the carline thistle that copes with both stressful and disturbed conditions. The cyanophages in this category have a relatively low number of early genes and a significant number of middle genes: 100, 160, and 113, respectively, for Syn9 (Synechococcus), P-SSM2 (Prochlorococcus), and P-TIM40, respectively. The relatively large number of late gene expression likely reflects their slightly larger size, although interestingly the ∼40 kb E. coli phage T7 is also an SR phage. The C. baltica phage phi38:1 is also in this category, with 43 middle genes out of 101 in total when grown on its optimal host. Later, we see this phage again when it is grown on its less optimal host.
T7
Similar to T4, it is worth considering the well-characterized model E. coli podovirus T7 in some detail. The phage has a short latent period. And the first genes enter the host with the E. coli RNA polymerase pulling the genome into the cell through attraction to the early promoters. Among the genes transcribed is the T7 RNA polymerase and a protein that inhibits host polymerase. The phage polymerase also requires a bacterial protein TrxA for effective transcription. As expected, the genes expressed during the late stages of transcription are responsible for progeny formation and host lysis; T7 produces holins, lysins, and spanins as part of its escape strategy. Essentially this small, well-characterized phage has a complex and sophisticated way of re-purposing the bacterial transcriptional machinery and having a complex escape strategy, earning itself a place in the SR part of our triangle.
Cyanophages Syn9, P-TIM40, and P-SSM2
The cyanobacterial myoviruses Syn9 and P-TIM40 are broad host-range cyanophages that infect Synechococcus and Prochloroccus, respectively. For Syn9, early transcription is controlled by a novel regulatory element, whereas middle transcription is controlled by the host. This infection method where there is a crossover between phage and host involvement to ensure transcription is reminiscent of T7 and could perhaps be a feature of SR phages.
P-SSM2 and P-SSP7 are T4-like and T7-like phages, respectively, that infect marine cyanobacterium Prochloroccus. The striking thing about these phages is that they have such an interlinked physiology with their bacterial hosts.
Cellulophaga baltica phi38:1
The generalis podovirus C. baltica phi38:1 is one of the most abundant phage genera in the global oceans.88,90 In our analysis, we showed that this phage presents as an SR phage when grown on its preferred host-38 and as a CSR phage when it is grown on its less optimal host CB18. On this 38 host it evades restriction modification systems, represses all of the host machinery, and synchronizes phage transcription with translation. It will be of interest to see whether phages that repress host machinery such as T7 and this phi38:1 grown on 38 are common in SR.
CSR phages
There are 11 phages that are categorized as CSR phages: 2 Streptococcus phages, 2972 and DTI; 4 Pseudomonas phages PaP3, LUZ19, PAK_P3, and Yua; the Pseudoalteromonas phage PSA-HP1; the Thermus phage P23-45; Bacillus phage AR9; Yersinia phage phiR1-37; and the Cellulophaga phage 38:1 when it is grown on CB18.
Streptococcus thermophilus phages DT1 and 2972
These were originally chosen for transcriptomic analysis, as they packaged their genomes by using cos-type and pac-type strategies, respectively. Phages with cos-type packaging have concentric cohesive ends within the phage genomes that mark the junction of the different phages: These proteins also form the basis of the termination sites for the phages, and thus there is no slippage in the genomes; all packaged phages will have the same genome. In contrast, phages with the PAC mechanism package until their heads are full may result in slippage and slightly different transcripts packaged. Interestingly, for the purposes of our analysis, despite these differences between the phages both of them cluster as CSR phages.
YuA
The type species for the Yuavirus, YuA is a siphovirus resembling phage M6 with a circularly permuted 58,663 bp genome.91 Even on PAO1, the host it was isolated on, YuA forms small plaques and displays inefficient binding kinetics. It remains unclear whether YuA has an obligately lytic or temperate, pseudo-temperate nature, or indeed does not fit these categories.91 YuA is predicted to encode for a hydroxymethyl-dUMP transferase that appears to replace the thymidylate synthase function in its host to create a pool of 5-hydroxymethyluridine that is incorporated into DNA replication instead of thymidine.91,92 De Smet et al.75,91,92 found that YuA pursues a “leeching” strategy of actively depleting host metabolites, rather than manipulating the host into maintaining steady-state levels in the face of phage exploitation. This is the only known Pseudomonas phage to have this strategy. Seventeen percent of all measured metabolites, including 14 out of 20 amino acids, begin to be exhausted immediately after infection. The large increase in available nucleotide monophosphates in YuA-infected cells seen rapidly after infection proved to be the exception to this rule, possibly explained by the presence of an exonuclease actively degrading the host genome. To summarize, YuA incorporates aspects of both the C and S strategies. Although not as clear cut, the tendency to immediately take resources from the host cell is reminiscent of the R strategists in plants that quickly exploit resources while they can; it, therefore, makes biological sense that this is a CSR phage.
Pseudomonas PAK_P3
This is a relatively newly described genus of Pseudomonas phage known as Kpp10virus. It has a particularly interesting method of the host taking over whereby it actively degrades host transcription products with its own nucleases. Unlike many of the viruses discussed in this analysis, its transcriptome was studied very recently and used RNAseq to determine the impact that the phage had on bacterial metabolism.66 Interestingly, the phage increased several facets of bacterial metabolism—such as increasing pyrimidine metabolism to bolster the amount of phages that can be produced. Small non-coding RNAs and antisense RNAs also appear to be used to modify transcription. This suggests that multiple mechanisms exist to manipulate expression within the virocell and overcome adaptive forms of phage defense. In addition, PAK_P3 completes a productive infection in a remarkably short amount of time for a large myovirus (eclipse period: 12.3 ± 0.4 min).66 With its combination of seemingly effective C, S, and R traits, PAK_P3 appears to make a good canonical CSR phage.
Cellulophaga baltica phi38:1
When infecting this less preferred host (strain 18) did not repress the bacterial transcriptome or proteome, unlike the situation when it infected 38. When phi38:1 infects its preferred host, it needs only 21 (12 genes less) early genes to establish itself, but 43 middles genes (16 genes more) than when it infects a less compatible host. This begs the question as to how many other CSR phages are propagated on less optimal hosts.
Pseudoalteromonas phage PSA-HP1
This podovirus phage completely takes over the metabolism of the host: It strongly represses host transcription and the virocell and reprograms it for its own purposes. It contrasts with the other Pseudoaletermonas phage where the virocell barely differs from that of the host. This phage has 17, 22, and 20 genes expressed early, middle, and late, respectively, compared with the siphovirus HS2 that has 42, 2, and 19, rendering it a C phage. In contrast to HS2, HP1 does not have any putative phage integrases.
Next Steps: Going Forward with CSR
Having discussed the CSR categories, we turn to the limitations and merits of our analysis before covering how we think our model is useful going forward, and how it can be applied to key areas of phage therapy.
Limitations and strengths of the analysis
We showed that phages, for which transcriptomic data are available, can be grouped according to their gene expression profiles into “ecological types” that are consistent with known aspects of their biology. Our intention was not to present a “set” or full classification scheme, but to provide a starting point to test theories of phage biology that our analysis supports.
We used the datasets available in the literature and, thus, our analysis is vulnerable to bias from different perspectives on what constitutes each phase, designation of the most strongly expressed (if genes are expressed during two phases), and the possibility that phages may have more or less than three phases. There is also potential for bias and in the use of different experimental methods (arrays, RNA-seq, classical methods) as well as different analytical methods for assigning temporal differential expression. However, as our major focus is on the phages, we have included data based on either method.
We have used the proportion of genes that contribute to the strategy rather than the actual numbers because, regardless of size, the proportion represents the amount of investment a phage puts into different parts of the life cycle. Further, this allowed us to compare the “ecological strategy” in a unified way across all phage types. Arguably, phages with large genomes could cluster together and thereby bias the data purely due to large genome size attraction. Reassuringly, however, the positioning of large phage genomes in the CSR triangle shows that they do not cluster but are found within several different categories.
Although 42 transcriptomes is a small fraction of the ∼10,000 sequenced phage genomes, together they provide a useful start to promote the idea of a unifying ecological theory. The advantage of using the different gene expression phases is that they are a measurable quantitative trait. Further work, particularly when additional phages are studied under multiple physiological conditions and with a larger body of better-understood metadata, will facilitate the identification of additional specific “functional traits” that can be mapped onto the phage lifestyles. This is what is currently done to better understand specific plants.
Functional predictions based on our model
Although there are challenges to comparing phage host ranges across independent datasets, we suggest that C phages are likely to have broad host ranges given their predicted ability to effectively overcome a wide array of host restriction mechanisms.93 Phage receptors exist in an evolutionary tension between selection for narrow adsorption ranges that prevent suicide by adsorption into non-susceptible hosts and wide adsorption ranges that prevent phage particles from missing out on susceptible hosts that they encounter. We predict that C phages will experience less of the first pressure and more of the second, which will select for receptors that bind less selectively and support broader host ranges.
To date, very few adequately large and comparable host range analyses exist that could be used to validate this prediction, and none that we are aware of that include phages with transcriptional data. We are hoping to inspire researchers to collect comparable host range data for phages with transcriptional data, or collect transcriptional data for phages with host range data, and look forward to seeing our prediction either confirmed or refuted. We would also predict that phages closer to the C apex would be more likely to exclude replication by other phages during the latent phase of co-infected virocells and produce unimpeded bursts of their own viral particles.
Although nearly all phage experiments are performed on bacteria growing exponentially in rich media, it is becoming clear that some phages encode for elaborate mechanisms for adapting to hosts growing or persisting under diverse metabolic conditions.75,76,86,87 We predict that S phages will be more likely to perform well in cells experiencing nutrient-depleted conditions or other metabolic and toxin-related stresses as well as in bacterial hosts undergoing diverse forms of energy metabolism as they cause infection. The overrepresentation of cyanophages in the S and SR apexes would seem to validate our prediction, as they infect cyanobacteria that for much of the year inhabit UV-stressed and nutrient-depleted waters.
We predict that R phages, having been stripped of genetic machinery for C and S strategies, will be more likely to have relatively high burst sizes and short latent periods and high manufacturing titers under optimal metabolic conditions in lab-adapted hosts that have lost phage defense systems. Currently, only one R phage in our data has a known burst size; we, therefore, look forward to other R phages being further characterized.
Generalist versus specialist phages
The positions of the phages in the CSR triangle are not fixed but are a function of their interaction with their bacterial host. Specialist phages, by definition, infect very few hosts and may be relatively fixed in their strategy. Generalist phages, in contrast, could be more flexible. The example we discussed is the C. baltica phage 38:1 that moves from an SR phage to a CSR phage when it grows on a less optimal bacterial strain. The ramifications of this are interesting, as a generalist phage grown on one bacterial host may present as a particular ecological phage type; however, when it is used to treat an infection, it may then use a different strategy. Therefore, transcriptomic analysis of phages on multiple hosts could provide useful information for future phage development as phages that are less (or possibly more) flexible in strategy could be more useful as therapeutic phages.
Applications of CSR to phage-bacterial interactions
The categorization of phages into functional or ecological niches/groups leads to a question of the bacterial phage load, and the extent and diversity of a collective set of phages for a given bacterial species. If one considers bacteria to be the environment for a set of phages, clearly many different “phage types” target each bacterial species and perhaps the different phages dominate when the bacteria are growing in different niches. This information could inform future “phage hunts.”
In our analysis, the diversity of phage types infecting one species is illustrated by the fact that the eight Pseudomonas phages we studied are found in four ecological categories. PhiKZ is a C phage; Pak-P4 and PA5oct are CR phages; PEV2 is a CS phage; and Yua, Pap3, Pak-P3, and LUz19 are CSR phages. Similarly, of the five E. coli phages analyzed, one is found in each of the primary strategies; lambda is C, Mu, is R and T4 is S.
In contrast to the situation for Pseudomonas and E. coli, bacteria that are found in more specialized environments could perhaps have a lower diversity of phages associated with them, for example the cyanophages are all clustered in the S and SR area.
Applications of CSR to phage therapy
Phages offer a potentially vast array of novel approaches and gene products by which to tackle antibiotic-resistant bacteria, for human health, agriculture, and animal husbandry. A major priority is to identify which phages are most suited to overcoming the significant manufacturing, commercial, and biological challenges. There is already a wide recognition of the value of the traits that we predict for “C” and “R” phages, given the centrality of both broad host ranges and high manufacturing titers under ideal conditions for the logistical and commercial feasibility of phage therapy. However, the CSR model would suggest that heavy selection for “C” and “R” phages might cause us to miss out on the critical advantages of “S” traits.
Both “hunts” for prospective therapeutic phages and “optimization” to establish propagation conditions for phages are typically limited to bacteria growing exponentially and aerobically in rich media. However, this bears almost no resemblance to the in situ conditions where we hope these phages will be effective. Indeed, in the two recently published phage therapy trials94–96 the phages selected had not been demonstrated to kill or infect bacteria under the metabolic or stress conditions present in the systems being treated. This framework could improve the efficacy of future phage selection by providing an understanding of the importance of “S” traits that may function optimally under conditions where bacteria causing infection are stressed.
Our model also suggests a framework through which, given more data, interactions between phages in a cocktail might be predicted. The overall efficacy of a phage cocktail is the sum of all the phages in the solution and their ability to target and kill all the bacterial strains of interest. This efficacy will be reduced by bacteria becoming phage resistant or by the phages competing with each other. The question of what is the most effective combination of phages in a given cocktail may be related to the ecological niches of the individual phages. Is a combination of closely related phages preferred, or should one strive to have a combination of phages with different ecological lifestyles? In a CSR-based scheme, we can perhaps achieve a broader diversity of phages by maximizing the area between the different phages or by focusing on a specific place within the CSR triangle.
Future perspectives and developments
Phages are currently categorized by either genome-based taxonomy or morphological classification and they are also given a designation based on their ability to follow lytic or temperate life cycles. The CSR scheme may allow us to classify them as ecological types and allow us to utilize this information to advance our understanding of phage biology and optimize their exploitation.
Genome-wide comparisons of phage genomes are difficult, as phage genes are extremely diverse, where typically less than 25% of genes within individual phage genomes have sequence similarities, and generally only within closely related groups. As each phage type will differentially impact the metabolic state of bacterial cells,17 organizing phage genomes into functional niches will help us to identify certain genome and gene-wise features that are associated with lifestyles and “phage types.” This is what is widely done in plants where functional traits are associated with plants in specific environments. Although species traits respond to environmental pressures, some of these traits also affect their environment, and thus we can better understand the functional role of plant communities.
Ultimately, we hope that some of the following questions will be answered by applying an ecological framework to phages: (1) Are the same ecological strategies used within groups of related phages or have they diverged within phage groups? (2) Are particular environments (so both natural and anthropomorphic influenced) dominated by certain phage types? (3) Is there a finite collection of phages or phage types for a given bacteria? (4) Are specific functional types particularly suited for specific exploitation such as therapy or are particular combinations more effective? (5) Which phages or combinations of phages are particularly good at combating specific bacterial pathogens?
“To do science is to search for repeated patterns and not accumulate facts”97 (ecologist Robert H. MacArthur's opening in Geographical Ecology, 1972). As thousands of phages have now been isolated and studied, phage biologists are in a good position to look for repeated patterns as to how phages impact the microbial communities they inhabit. Previous studies have not attempted to unify phage ecology in a predictive way, largely because most researchers focus on specific environments or particular phage groups and it is not obvious how to make quantitative measurements to compare phage taxa. Further, there is limited cross-talk between the fields of botany and phage biology. Although we are aware of the challenges associated with attempting to unify the highly diverse phage sphere, we hope that this article promotes further thoughts and insights from interpreting the new wealth of “omic” data into well-understood frameworks that have provided a robust understanding of ecology in the botanical world.
Materials and Methods
From literature searches, we compiled a list of 42 phages where transcriptomic studies had been performed and the early, middle, and late gene phases were determined and extracted or resolved the temporal expressions from the Supplementary Data from the publications listed in Supplementary Table S1. For each phage, we collected or identified the number of genes labeled as early, middle, or late. Genes that were expressed neither early nor late were assigned as “middle” on the basis that they represent an additional pool of resources that the phage could use, for example when infecting a less compatible host.
Protein annotations for all phage genomes were extracted from the GenBank files listed in Supplementary Table S2 by using Biopython.98 All annotations were grouped with their transcriptome phases (E, M, and L) and plotted in a clustered heatmap by using z-score clustering in Seaborn.99
Phages genomes were labeled as temperate phages if their gene annotations matched at least one protein associated with a temperate lifestyle of integrase, ParA, ParB, site-specific recombinase, excise, cro, or c-repressor (listed in Supplementary Table S2).
For the CSR analysis, each phage was plotted into a ternary plot by using Plotly,100 depicting the ratios of early, middle, and late genes as coordinates in an equilateral triangle.
The list of early, middle, and late genes for each phage genome is available in Supplementary Table S2. An interactive online version was built with Python, Plotly100 and that allows for adding phages based on their transcriptomic expression profiles and it can be found at www.phageCSR.ml
Supplementary Material
Acknowledgments
The authors would like to thank Jill Turner for drawing the plant and phage images in the graphical abstract that encapsulates the concept of the article. They would like to thank Julian Clokie for his thoughts and discussions on the philosophy of the CSR in a plant context and for his insights into other ecological classification schemes.
Authors' Contributions
M.R.J.C. and T.S.P. conceived the idea, carried out the analysis, and drafted the article. B.O.L.D. provided expertise in the applications of CSR to plant ecology, B.G.B. provided expertise in phage transcriptional profiling and mechanisms of interaction, and both contributed to the article. All authors read and approved the final article.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
No funding was received for this article.
Supplementary Material
References
- 1. Suttle CA. Marine viruses—Major players in the global ecosystem. Nature Rev Microbiol. 2007;5:801–812. [DOI] [PubMed] [Google Scholar]
- 2. Clokie MR, Millard AD, Letarov AV, et al. . Phages in nature. Bacteriophage. 2011;1:31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Howard-Varona C, Hargreaves KR, Abedon ST, et al. . Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017;11:1511–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adriaenssens EM, Sullivan MB, Knezevic P, et al. . Taxonomy of prokaryotic viruses: 2018–2019 update from the ICTV bacterial and archaeal viruses subcommittee. Arch Virol. 2020;165:1253–1260. [DOI] [PubMed] [Google Scholar]
- 5. Roux S, Adriaenssens EM, Dutilh BE, et al. . Minimum information about an uncultivated virus genome (MIUViG). Nat Biotechnol. 2019;37:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zrelovs N, Cernooka E, Dislers A, et al. . Isolation and characterization of the novel virgibacillus-infecting bacteriophage Mimir87. Arch Virol. 2020;165:737–741. [DOI] [PubMed] [Google Scholar]
- 7. Matamp N, Bhat SG. Genome characterization of novel lytic Myoviridae bacteriophage φVP-1 enhances its applicability against MDR-biofilm-forming Vibrio parahaemolyticus. Arch Virol. 2020;165:387–396. [DOI] [PubMed] [Google Scholar]
- 8. Imam M, Alrashid B, Patel F, et al. . vB_PaeM_MIJ3, a novel jumbo phage infecting Pseudomonas aeruginosa, possesses unusual genomic features. Front Microbiol. 2019;10:2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Z, Chen F, Chu X, et al. . Diverse, abundant, and novel viruses infecting the marine roseobacter RCA lineage. mSystems. 2019;4:e00494-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caruso SM, deCarvalho TN, Huynh A, et al. . A novel genus of Actinobacterial Tectiviridae. Viruses. 2019;11:1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morozova V, Babkin I, Kozlova Y, et al. . Isolation and characterization of a novel Klebsiella pneumoniae N4-like bacteriophage KP8. Viruses. 2019;11:1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawato Y, Istiqomah I, Gaafar AY, et al. . A novel jumbo Tenacibaculum maritimum lytic phage with head-fiber-like appendages. Arch Virol. 2020;165:303–311. [DOI] [PubMed] [Google Scholar]
- 13. Chatterjee A, Sicheritz-Pontén T, Yadav R, et al. . Genomic and metagenomic signatures of giant viruses are ubiquitous in water samples from sewage, inland lake, waste water treatment plant, and municipal water supply in Mumbai, India. Sci Rep. 2019;9:3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thanki AM, Brown N, Millard AD, et al. . Genomic characterization of jumbo salmonella phages that effectively target United Kingdom pig-associated salmonella serotypes. Front Microbiol. 2019:10:1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rashid SJ, Barylski J, Hargreaves KR, et al. . Two novel myoviruses from the north of iraq reveal insights into Clostridium difficile phage diversity and biology. Viruses. 2016;8:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rashid SRJ, Clokie MRJ, Millard AD. Draft genome sequences of three novel clostridium isolates from Northern Iraq. Genome Announc. 2016;4:e00033-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Howard-Varona C, Lindback MM, Bastien GE, et al. . Phage-specific metabolic reprogramming of virocells. ISME J. 2020;14:881–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Raunkiaer C. The life forms of plants and statistical geography: Being the collected papers of C. Raunkiaer. Geog J. 1934;84:455. [Google Scholar]
- 19. Praeger RL, et al. Natural history of Ireland. A sketch of its flora and fauna. London: Collins; 1950:350. [Google Scholar]
- 20. Tansley AG, Adamson RS. Studies of the vegetation of the English chalk: III. The chalk grasslands of Hampshire-Sussex border. J Ecol. 1925;13:177–223. [Google Scholar]
- 21. Greig-Smith P. Data on pattern within plant communities: I. The analysis of pattern. J Ecol. 1961;49:695–702. [Google Scholar]
- 22. MacArthur RH, Wilson E. The theory of island biogeography. Princeton, NJ: Princeton University Press; 1967. [Google Scholar]
- 23. Keen EC. Tradeoffs in bacteriophage life histories. Bacteriophage. 2014;4:e28365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grime JP. Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Am Nat. 1977;111:1169–1194. [Google Scholar]
- 25. Grime JP. Plant strategies, vegetation processes, and ecosystem properties, second edition. Environ Conserv. 2002;29:263–270. [Google Scholar]
- 26. Silvertown J, Franco M, McConway K. A demographic interpretation of grime's triangle. Funct Ecol. 1992;6:130–136. [Google Scholar]
- 27. Smith TM, Smith TM, Shugart HH, et al. Plant Functional Types: Their Relevance to Ecosystem Properties and Global Change. Cambridge: Cambridge University Press; 1997:20–43. [Google Scholar]
- 28. Wood JL, Tang C, Franks AE. Competitive traits are more important than stress-tolerance traits in a cadmium-contaminated rhizosphere: A role for trait theory in microbial ecology. Front Microbiol. 2018;9:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ho A, Kerckhof F-M, Luke C, et al. . Conceptualizing functional traits and ecological characteristics of methane-oxidizing bacteria as life strategies. Environ Microbiol Rep. 2013;5:335–345. [DOI] [PubMed] [Google Scholar]
- 30. Krause S, Le Roux X, Niklaus PA, et al. . Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front Microbiol. 2014;5:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ho A, Di Lonardo DP, Bodelier PLE. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol Ecol. 2017;93. [DOI] [PubMed] [Google Scholar]
- 32. Wood WB, Revel HR. The genome of bacteriophage T4. Bacteriol Rev. 1976;40:847–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. [DOI] [PubMed] [Google Scholar]
- 34. Harshey RM. The Mu story: How a maverick phage moved the field forward. Mob DNA. 2012;3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guttman B, Raya R, Kutter E. Basic phage biology. In: Kutter E, Sulakvelidze A; eds. Bacteriophages: Biology and Application. Boca Raton, FL: CRC Press; 2005:29–66. [Google Scholar]
- 36. Roucourt B, Lavigne R. The role of interactions between phage and bacterial proteins within the infected cell: A diverse and puzzling interactome. Environ Microbiol. 2009;11:2789–2805. [DOI] [PubMed] [Google Scholar]
- 37. Smith MC, Burns RN, Wilson SE, et al. . The complete genome sequence of the Streptomyces temperate phage straight phiC31: Evolutionary relationships to other viruses. Nucleic Acids Res. 1999;27:2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hirofumi Y, Ito J. Nucleotide sequence of the major early region of bacteriophage ϕ29. Gene. 1982;17:323–335. [DOI] [PubMed] [Google Scholar]
- 39. Dedrick RM, Mavrich TN, Ng WL, et al. . Expression and evolutionary patterns of mycobacteriophage D29 and its temperate close relatives. BMC Microbiol. 2017;17:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luke K, Radek A, Liu X, et al. . Microarray analysis of gene expression during bacteriophage T4 infection. Virology. 2002;299:182–191. [DOI] [PubMed] [Google Scholar]
- 41. Morimoto D, Kimura S, Sako Y, et al. . Transcriptome analysis of a bloom-forming Cyanobacterium Microcystis aeruginosa during Ma-LMM01 phage infection. Front Microbiol. 2018;9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thompson LR, Zeng Q, Chisholm SW. Gene expression patterns during light and dark infection of Prochlorococcus by Cyanophage. PLoS One. 2016;11:e0165375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mioduser O, Goz E, Tuller T. Significant differences in terms of codon usage bias between bacteriophage early and late genes: A comparative genomics analysis. BMC Genomics. 2017;18:866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Minakhin L, Semenova E, Liu J, et al. . Genome sequence and gene expression of Bacillus anthracis bacteriophage Fah. J Mol Biol. 2005;354:1–15. [DOI] [PubMed] [Google Scholar]
- 45. Sevostyanova A, Djordjevic M, Kuznedelov K, et al. . Temporal regulation of viral transcription during development of Thermus thermophilus bacteriophage phiYS40. J Mol Biol. 2007;366:420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pavlova O, Lavysh D, Klimuk E, et al. . Temporal regulation of gene expression of the Escherichia coli bacteriophage phiEco32. J Mol Biol. 2012;416:389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Howard-Varona C, Hargreaves KR, Solonenko NE, et al. . Multiple mechanisms drive phage infection efficiency in nearly identical hosts. ISME J. 2018;12:1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Semenova E, Nagornykh M, Pyatnitskiy M, et al. . Analysis of CRISPR system function in plant pathogen Xanthomonas oryzae. FEMS Microbiol Lett. 2009;296:110–116. [DOI] [PubMed] [Google Scholar]
- 49. Majtan T, Halgasova N, Bukovska G, et al. . Transcriptional profiling of bacteriophage BFK20: Coexpression interrogated by “guilt-by-association” algorithm. Virology. 2007;359:55–65. [DOI] [PubMed] [Google Scholar]
- 50. Yang Z, Yin S, Li G, et al. . Global transcriptomic analysis of the interactions between phage ϕAbp1 and extensively drug-resistant Acinetobacter baumannii. mSystems. 2019;4:e00068-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Doron S, Fedida A, Hernández-Prieto MA, et al. . Transcriptome dynamics of a broad host-range cyanophage and its hosts. ISME J. 2016;10:1437–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lin X, Ding H, Zeng Q. Transcriptomic response during phage infection of a marine cyanobacterium under phosphorus-limited conditions. Environ Microbiol. 2016;18:450–460. [DOI] [PubMed] [Google Scholar]
- 53. Lindell D, Jaffe JD, Coleman ML, et al. . Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature. 2007;449:83–86. [DOI] [PubMed] [Google Scholar]
- 54. Duplessis M, Russell WM, Romero DA, et al. . Global gene expression analysis of two Streptococcus thermophilus bacteriophages using DNA microarray. Virology. 2005;340:192–208. [DOI] [PubMed] [Google Scholar]
- 55. Leskinen K, Blasdel BG, Lavigne R, et al. . RNA-sequencing reveals the progression of phage-host interactions between ϕR1–37 and Yersinia enterocolitica. Viruses. 2016;8:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao X, Chen C, Shen W, et al. . Global transcriptomic analysis of interactions between Pseudomonas aeruginosa and Bacteriophage PaP3. Sci Rep. 2016;6:19237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lavysh D, Sokolova M, Slashcheva M, et al. . Transcription profiling of Bacillus subtilis cells infected with AR9, a giant phage encoding two multisubunit RNA polymerases. mBio. 2017;8:e02041-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Berdygulova Z, Westblade LF, Florens L, et al. . Temporal regulation of gene expression of the Thermus thermophilus bacteriophage P23-45. J Mol Biol. 2011;405:125–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li T, Zhang Y, Dong K, et al. . Isolation and characterization of the novel phage JD032 and global transcriptomic response during JD032 infection of Clostridioides difficile ribotype 078. mSystems. 2020;5:e00017-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chatterjee A, Willett JLE, Nguyen UT, et al. . Parallel genomics uncover novel Enterococcal-Bacteriophage interactions. MBio. 2020;11:e03120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ceyssens P-J, Minakhin L, Van den Bossche A, et al. . Development of giant bacteriophage φKZ is independent of the host transcription apparatus. J Virol. 2014;88:10501–10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wagemans J, Blasdel BG, Van den Bossche A, et al. . Functional elucidation of antibacterial phage ORFans targeting Pseudomonas aeruginosa. Cell Microbiol. 2014;16:1822–1835. [DOI] [PubMed] [Google Scholar]
- 63. Blasdel BG, Chevallereau A, Monot M, et al. . Comparative transcriptomics analyses reveal the conservation of an ancestral infectious strategy in two bacteriophage genera. ISME J. 2017;11:1988–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lood C, Danis-Wlodarczyk K, Blasdel BG, et al. . Integrative omics analysis of Pseudomonas aeruginosa virus PA5oct highlights the molecular complexity of jumbo phages. Environ Microbiol. 2020;22:2165–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Blasdel B. Whole transcriptome analyses of Pseudomonas aeruginosa after infection by representatives of different phage clades. 2016. Available at https://lirias.kuleuven.be/1717205?limo=0. (last accessed August 25, 2020).
- 66. Chevallereau A, Blasdel BG, De Smet J, et al. . Next-generation “-omics” approaches reveal a massive alteration of host RNA metabolism during Bacteriophage infection of Pseudomonas aeruginosa. PLoS Genet. 2016;12:e1006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu X, Jiang H, Gu Z, et al. . High-resolution view of bacteriophage lambda gene expression by ribosome profiling. Proc Natl Acad Sci U S A. 2013;110:11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Davison J. Pre-early functions of bacteriophage T5 and its relatives. Bacteriophage. 2015;5:e1086500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Davison J, Brunel F. Restriction insensitivity in bacteriophage T5 I. Genetic characterization of mutants sensitive to EcoRI restriction. J Virol. 1979;29:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Davison J, Brunel F. Restriction insensitivity in bacteriophage T5. II. Lack of EcoRI modification in T5 and T5ris mutants. J Virol. 1979;29:17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Warner HR, Drong RF, Berget SM. Early events after infection of Escherichia coli by bacteriophage T5. Induction of a 5’-nucleotidase activity and excretion of free bases. J Virol. 1975;15:273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mozer TJ, Thompson RB, Berget SM, et al. . Isolation and characterization of a bacteriophage T5 mutant deficient in deoxynucleoside 5’-monophosphatase activity. J Virol. 1977;24:642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chaikeeratisak V, Nguyen K, Egan ME, et al. . The phage nucleus and tubulin spindle are conserved among large Pseudomonas phages. Cell Rep. 2017;20:1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Malone LM, Warring SL, Jackson SA, et al. . A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat Microbiol. 2020;5:48–55. [DOI] [PubMed] [Google Scholar]
- 75. De Smet J, Zimmermann M, Kogadeeva M, et al. . High coverage metabolomics analysis reveals phage-specific alterations to Pseudomonas aeruginosa physiology during infection. ISME J. 2016;10:1823–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bryan D, El-Shibiny A, Hobbs Z, et al. . Bacteriophage T4 infection of stationary phase E. coli: Life after log from a phage perspective. Front Microbiol. 2016;7:1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Miller ES, Kutter E, Mosig G, et al. . Bacteriophage T4 genome. Microbiol Mol Biol Rev. 2003;67:86–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kutter EM, Bradley D, Schenck R, et al. . Bacteriophage T4 alc gene product: General inhibitor of transcription from cytosine-containing DNA. J Virol. 1981;40:822–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Williams KP, Kassavetis George A, Herendeen Daniel R, et al. . Regulation of Late-Gene Expression. In: Karam JD, Drake JW; eds. Molecular Biology of Bacteriophage T4. Washington, DC: American Society of Microbiology; 1994:161–175. [Google Scholar]
- 80. Kutter E, Beug A, Sluss R, et al. . The production of undegraded cytosine-containing DNA by bacteriophage T4 in the absence of dCTPase and endonucleases II and IV, and its effects on T4-directed protein synthesis. J Mol Biol. 1975;99:591–607. [DOI] [PubMed] [Google Scholar]
- 81. Kashlev M, Nudler E, Goldfarb A, et al. . Bacteriophage T4 Alc protein: A transcription termination factor sensing local modification of DNA. Cell. 1993;75:147–154. [PubMed] [Google Scholar]
- 82. Lenneman BR, Rothman-Denes LB. Structural and biochemical investigation of bacteriophage N4-encoded RNA polymerases. Biomolecules. 2015;5:647–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Willis SH, Kazmierczak KM, Carter RH, et al. . N4 RNA polymerase II, a heterodimeric RNA polymerase with homology to the single-subunit family of RNA polymerases. J Bacteriol. 2002;184:4952–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Choi KH, McPartland J, Kaganman I, et al. . Insight into DNA and protein transport in double-stranded DNA viruses: The structure of bacteriophage N4. J Mol Biol. 2008;378:726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Carter RH, Demidenko AA, Hattingh-Willis S, et al. . Phage N4 RNA polymerase II recruitment to DNA by a single-stranded DNA-binding protein. Genes Dev. 2003;17:2334–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lindberg G, Kowalczykowski SC, Rist JK, et al. . Purification and characterization of the coliphage N4-coded single-stranded DNA binding protein. J Biol Chem. 1989;264:12700–12708. [PubMed] [Google Scholar]
- 87. Ceyssens P-J, Brabban A, Rogge L, et al. . Molecular and physiological analysis of three Pseudomonas aeruginosa phages belonging to the “N4-like viruses.” Virology. 2010;405:26–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dang VT, Howard-Varona C, Schwenck S, et al. . Variably lytic infection dynamics of large Bacteroidetes podovirus phi38:1 against two Cellulophaga baltica host strains. Environ Microbiol. 2015;17:4659–4671. [DOI] [PubMed] [Google Scholar]
- 89. Djordjevic M, Semenova E, Shraiman B, et al. . Quantitative analysis of a virulent bacteriophage transcription strategy. Virology. 2006;354:240–251. [DOI] [PubMed] [Google Scholar]
- 90. Holmfeldt K, Howard-Varona C, Solonenko N, et al. . Contrasting genomic patterns and infection strategies of two co-existing Bacteroidetes podovirus genera. Environ Microbiol. 2014;16:2501–2513. [DOI] [PubMed] [Google Scholar]
- 91. Ceyssens P-J, Mesyanzhinov V, Sykilinda N, et al. . The genome and structural proteome of YuA, a new Pseudomonas aeruginosa phage resembling M6. J Bacteriol. 2008;190:1429–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kutter EM, Skutt-Kakaria K, Blasdel B, et al. . Characterization of a ViI-like phage specific to Escherichia coli O157:H7. Virol J. 2011;8:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hyman P, Abedon ST. Bacteriophage host range and bacterial resistance. Adv Appl Microbiol. 2010;70:217–248. [DOI] [PubMed] [Google Scholar]
- 94. Kieser S, Sarker SA, Sakwinska O, et al. . Bangladeshi children with acute diarrhoea show faecal microbiomes with increased Streptococcus abundance, irrespective of diarrhoea aetiology. Environ Microbiol. 2018;20:2256–2269. [DOI] [PubMed] [Google Scholar]
- 95. Sarker SA, Sultana S, Reuteler G, et al. . Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: A randomized trial in children from Bangladesh. EBioMedicine. 2016;4:124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jault P, Leclerc T, Jennes S, et al. . Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): A randomised, controlled, double-blind phase 1/2 trial. Lancet Infect Dis. 2019;19:35–45. [DOI] [PubMed] [Google Scholar]
- 97. MacArthur RH. Geographical Ecology: Patterns in the Distribution of Species. Princeton: Princeton University Press; 1972. [Google Scholar]
- 98. Cock PJA, Antao T, Chang JT, et al. . Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Seaborn. GitHub. Available at: https://github.com/mwaskom/seaborn (last accessed August 25, 2020).
- 100. Plotly: The front-end for ML and data science models. Available at: https://plot.ly (last accessed August 25, 2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.