Abstract
Background: Pseudomonas syringae are ubiquitous epiphytic plant pathogens infecting a wide range of important agricultural plant species. Bacteriophages has been proposed as biocontrol agents against plant pathogens, however, in order to utilize this approach, a deeper understanding of phage diversity and phage-host interactions is required.
Materials and Methods: Phages targeting P. syringae GAW0113 were isolated from organic waste samples. Three distinct phage isolates were purified and subjected to whole-genome sequencing, comparative genomics, transmission electron microscopy and host-range assay using a wide selection of diverse P. syringae isolates.
Results: The three phage isolates, Pseudomonas phage Bertil, Misse, and Strit, were shown to have podovirus morphology with a short tail stub and isometric head. They had double-stranded DNA ranging from 41,342 to 41,374 bp in size comprising 50–51 open reading frames. The three phage genomes were highly similar and genomic comparison analyses showed that they all belong to the Autographiviridae family of the order Caudovirales. All three phages were shown to have a narrow host-range.
Conclusions: The three phages were shown to share morphological and genomic features with other phages in the Autographiviridae family, however, based on the limited nucleotide similarity we propose that the phages constitute a novel genus. All three phages were found to infect multiple strains of P. syringae covering several phylogroups.
Keywords: bacteriophage, biocontrol, Pseudomonas phage, Pseudomonas syringae, phage therapy
Introduction
Pseudomonas syringae is a gram-negative rod-shaped bacterium. Strains of P. syringae are ubiquitous epiphytic plant pathogens and can infect a wide range of important agricultural plant species. P. syringae has been known to cause diseases such as bacterial canker of wild cherries1 and kiwifruit,2 bacterial speck of tomato3 and leaf blight in wheat.4 Many P. syringae strains isolated from nonagricultural environments have functional type 3 secretion systems (T3SS).5 The T3SS has been linked to the virulence of P. syringae. The presence of strains with functional T3SS suggests that there is a reservoir of plant-pathogenic bacteria in other environmental niches than plants. The widespread presence of pathogenic P. syringae strains in agricultural plants calls for an inexpensive and sustainable method for preventing or treating bacterial infections. Bacteriophage therapy is a promising approach to disease management, but requires an extensive library of relevant phages and understanding of phage–host interaction between phages and the P. syringae species.6 In this study, we present three phages isolated against P. syringae strain GAW0113: Pseudomonas phage Bertil, Pseudomonas phage Misse, and Pseudomonas phage Strit. No reports of phages isolated against GAW0113 were available at the time of this study. Even though GAW0113 has not been associated with disease in crops, characterization of novel phages targeting strains of P. syringae can help in understanding the diversity of P. syringae phages and phage–host interactions, including bacterial resistance mechanisms.7 Furthermore, the host-range assay of the three phages shows that they are able to target other strains within the P. syringae species complex. Increased diversity within the known phages targeting P. syringae strains also allow for development of more complex phage cocktails. One of the potential flaws of using phages as antimicrobial agents is that bacteria can develop resistance relatively quickly against specific phages.8 The use of therapeutic cocktails of diverse phages makes it harder for the target bacteria to adapt and for resistant cells to survive.6
Materials and Methods
Isolation
Samples of primarily household organic waste (OW) from a waste management facility were collected, aliquoted, and filtered with 0.45 μm PVDF syringe filters (Merck Millipore, Darmstadt, Germany). The filtrates were tested for phage activity against P. syringae GAW0113 using a double agar overlay assay with 0.6% soft agarose top agar.9 To obtain pure isolates, plaques with distinct appearance or from different OW samples were collected with a sterile pipette tip and transferred to 900 μL SM buffer. The phage lysate was then diluted and replated using the same double agar method. This purification process was performed three times to obtain phage isolates from single plaques. The final phage isolates were stored at 4°C.
DNA extraction and sequencing
The phage DNA was extracted from lysate, as previously described.10 Five units of DNAse (A&A Biotechnology, Gdynia, Poland) were added to 200 μL lysate and incubated at 37°C for 1 h followed by heat inactivation at 70°C for 10 min. Next, 6 U of Proteinase k (A&A Biotechnology, Gdynia, Poland) and sodium dodecyl sulfate–polyacrylamide [final concentration of 0.1% (w/v)] were added followed by another step of heat inactivation at 70°C for 10 min. The DNA solution was then purified with DNA Clean and Concentrator-5 kit (Zymo Research, California, USA). Phage DNA libraries were prepared using NEBNext Ultra II FS library Prep and Kit for Illumina (New England Biolabs, Ipswich, USA). The sequencing was performed on the Illumina iSeq platform as part of a flowcell (2 × 151, 300 cycles; Illumina). Sequence trimming and de novo assembly was performed in CLC Genomics Workbench version 12.0.3. Blastn was used to search for sequence similarity against the nonredundant National Center for Biotechnology Information (NCBI) databases.11
Gene annotation and sequence similarity
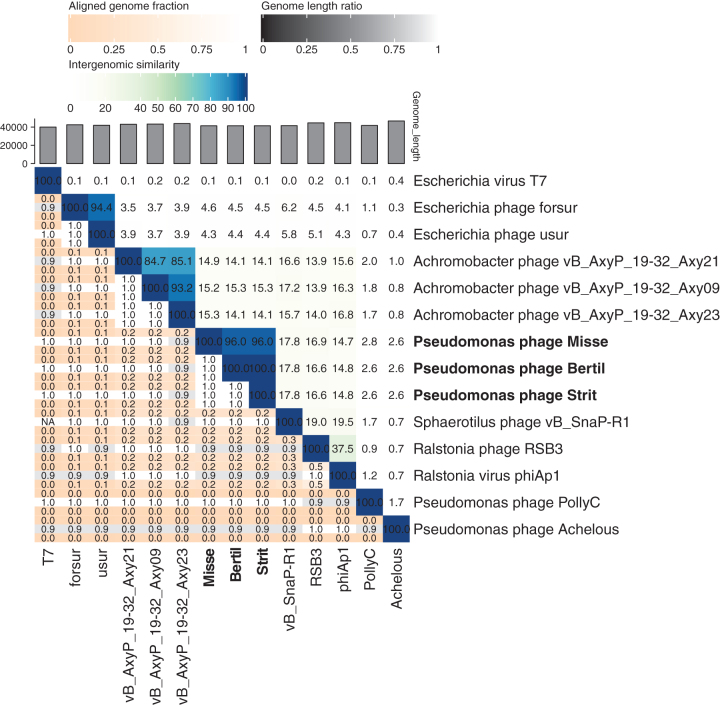
Open reading frames (ORFs) were identified automatically with GeneMark and Glimmer and revised manually in DNA Master.12–14 The genomes were searched for tRNA sequences using Aragorn and tRNAscan-SE.15,16 The ORFs were manually annotated using both HHPred against three downloaded databases (pFam 32.0, SCOP70 1.75 and PDB70) and blastp against the NCBI nonredundant databases.17 The nucleotide similarity heatmap between Bertil, Misse, and Strit and Autographiviridae phages was made with viridic (default settings).18 Gene synteny and nucleotide similarity were compared with Easyfig version 2.2.2. (default blastn settings with 2.9.0+ NCBI blast).19 Phylogenetic analyses of the predicted RNA polymerase, major capsid protein and large terminase were performed with MEGAX version 10.1.8.20 The 20 most similar proteins from other phages, found by blastp, were included in the trees. The RNA polymerase, major capsid protein and large terminase proteins of the Autographiviridae phages included in Figure 2 were also included in the phylogenetic analyses. The proteins were aligned with MUSCLE, and the phylogenetic distances were inferred using a maximum likelihood method. The reliability of the trees was tested using bootstrap with 100 replicates. The assembled and annotated genomes of Bertil, Misse, and Strit were uploaded to the NCBI with accession numbers MW286266, MW286267, and MW286268, respectively.
FIG. 2.
Pair-wise nucleotide comparisons.
Transmission electron microscopy
To obtain pure phage lysates for electron microscope pictures, 10 μL of phage lysates containing 108 plaque forming units (PFU)/mL were further propagated in 1 L lysogeny broth (LB) medium (10 mM of MgCl2 and CaCl2)) with 100 μL host overnight culture and left on a shaking table at 200 rpm overnight. The concentrated lysates were centrifuged (5500 × g, 10 min, 4°C). Fifty-eight grams NaCl followed by 100 g PEG 8000 were added directly to the supernatant. The resulting phage-PEG suspension was shaken overnight (200 rpm, 5°C). The PEG suspension was centrifuged again, and the pellet was resuspended in 5 mL SM-buffer and incubated overnight. The resuspended PEG pellet was centrifuged (10,000 × g, 10 min, 4°C) and the pellet discarded. The phage particles were then purified in a cesium chloride gradient, as described elsewhere.21 For obtaining electron micrographs, 100 μL of the CsCl phage suspension were dialyzed against SM-buffer on a Millipore VSWP 0.025 μm filter membrane (Merck, Darmstadt, DE) for 30 min. The dialyzed phages were allowed to absorb to ultrathin carbon films for 30 min and then fixed with 1% (v/v) glutaraldehyde (EM-grade) for an additional 30 min. Negative staining was performed with 2% (w/v) uranyl acetate. The ultrathin carbon films were picked up with standard 400-mesh copper grids (3 mm diameter). The transmission electron microscopy was performed using an accelerating voltage of 80 kV (Tecnai 10; FEI Thermo Fisher Scientific, Eindhoven, The Netherlands) and micrographs were taken with a MegaView G2 CCD camera (Emsis, Muenster, Germany).
Host-range screening and efficiency of plating
The phages were originally isolated with P. syringae GAW0113. The plaque forming ability of the three phages were additionally tested against all 13 strains of P. syringae: GAW0113, CC0663, CC1417,CC1513, CC1532, CC1557 CC1630, CC94, CFBP2341, CFBP4407, CLA0302, R4_A29-2, and UB246.22 The original isolation host, GAW0113, was included as a positive control and for comparison. The phages were propagated as stated earlier to a final titer of 108–109 PFU/mL. The phage lysates were used in a double agar overlay assay with 13 strains of P. syringae. The lysates were spotted (10 μL/spot) on LB plates with 100 μL overnight culture of the 13 different hosts and the isolation host and incubated overnight at room temperature. The relative efficiency of plating was measured for the sensitive strains and the original isolation host. The phage lysates were serially diluted 10-fold to 10–10 and a 5 μL aliquot of each dilution step was spotted on three separate LB plates for each host strain. Efficiency of plating (EOP) was calculated as the titer relative to that of the original isolation host.
Results
Genomics
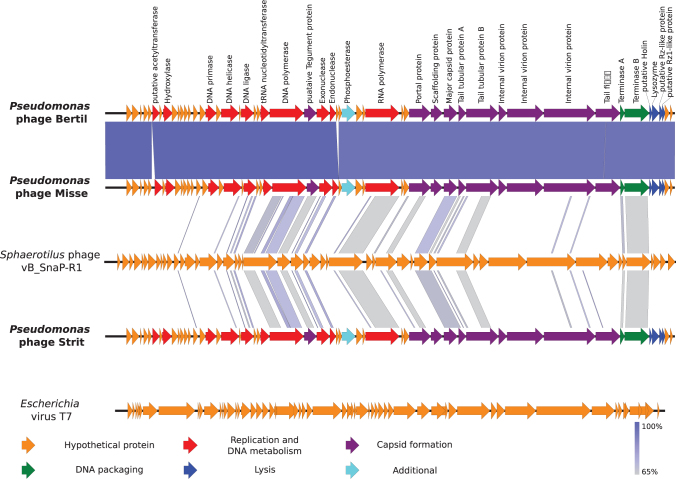
The genomes of the three phages are all double-stranded DNA, with sizes of 41,373, 41,342, and 41,374 bp for Bertil, Misse, and Strit, respectively. The GC-content is 58% for all three phages. All the genomes contain inverted terminal repeats of slightly varying length (307, 302, and 305 bp, respectively). The positions of the terminal repeats, and thus the genome start sites, were identified by the presence of two distinct spikes in read-start coverage at two nearby sites in the genome, encompassing areas with approximately double coverage of reads for that position compared with the rest of the genome. Bertil and Strit are almost identical except one deletion and nine single nucleotide mutations including a missense mutation (p.His97Gln) in the DNA polymerase gene. The other eight mutations and the deletion are either silent or in noncoding regions. Misse lacks a 195 bp ORF located in the middle of the genomes of Bertil and Strit, but contains an additional 100 bp noncoding region (Fig. 1). No function could be assigned for the ORF only found in Bertil and Strit. In total the phage genomes contain either 50 or 51 ORFs, all on the same strand and in the same reading direction. As many as 27 ORFs could be annotated with a putative function for each of the phages (Fig. 1). There was limited sequence similarity between the newly isolated phages and other phages of Autographiviridae on the nucleotide level (Fig. 2), but gene synteny and gene size seems to be conserved for most of the predicted core genes involved with replication, DNA packaging, and capsid-assembly and lysis (Fig. 2). The similarity in gene synteny is, however, limited for many other putative ORFs with no predicted product (Fig. 1). No integrases or other genes related to a lysogenic phase were found in any of the three genomes. No tRNA sequences were found in any of the three genomes.
FIG. 1.
Comparison between Pseudomonas phages Bertil, Misse, and Strit and two Autographiviridae phages with Easyfig software (default blast settings). Sphaerotilus phage vB_SnaP-R1 has the highest nucleotide similarity with Bertil, Misse, and Strit and Escherichia virus T7 is shown for comparison of gene synteny as one of the most studied Autographiviridae phages. Open reading frames are represented by arrows indicating the reading direction. Pairwise nucleotide similarity between the different genomes is shown by color (from gray to blue). The predicted gene products for Bertil, Misse, and Strit are shown above the genome of Bertil. Colors denote protein classes as described in the figure.
Virion morphology
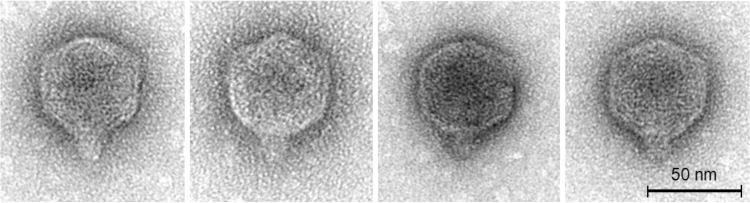
Because of high genetic similarity between Bertil, Misse, and Strit, only one phage, Strit, was selected for transmission electron microscopy. Although the function of the missing gene in Misse is unknown, it is presumed that Misse and Bertil share the same or very similar capsid structure as Strit. Transmission electron microscopy (with seven phage particles) shows that Strit possesses an isometric head (diameter: 58 nm) and a short cone-shaped tail stub (length: 13 nm) (Fig. 3).
FIG. 3.
Transmission electron micrographs.
Nucleotide similarity to other phages
A nucleotide comparison using blastn, to available sequences in the NCBI NR database, shows that the three genomes have the largest similarity with Sphaerotilus phage yB_SnaP_R1 (21% query coverage and 67.02% identity) belonging to the Autographiviridae family. The blastn resulted in 126 out of 128 hits against phages in the Caudovirales order with 123 of the hits against Autographiviridae phages. Phylogenetic trees of the RNA polymerase, major capsid protein, and large terminase show that Bertil, Misse, and Strit also form their own cluster apart from other Autographiviridae phages.
Phage host range
The three phages were able to lyse and form plaques with similar efficiency against 3 out of 13 P. syringae strains including GAW0113 against which it was originally isolated (Table 1). The variation in EOP between the different host strains were <10-fold for all three phages, despite a starting titer between 108 and 109 PFU/mL (Table 1). None of the phages had a significantly increased EOP for either of the tested strains compared with the isolation host (Wilcoxon Rank-Sum test, p-values >0.05).
Table 1.
Host Range Assay
| Host strain | Phylogroup | Phage |
||
|---|---|---|---|---|
| Bertil | Misse | Strit | ||
| CC1630 | 01 | — | — | — |
| CC94 | 02 | — | — | — |
| R4_A29-2 | 03 | — | — | — |
| CC1513 | 04 | — | — | — |
| CFBP2341 | 05 | — | — | — |
| CC0663 | 07 | — | — | — |
| CC1532 | 09 | 5 ± 1.3 | 3.9 ± 0.6 | 1.6 ± 0.6 |
| CC1417 | 09 | 2.3 ± 1.4 | 2.1 ± 0.2 | 1.6 ± 1.1 |
| CC1557 | 10 | — | — | — |
| CFBP4407 | 11 | — | — | — |
| GAW0113 | 12 | 1 ± 0.2 | 1 ± 0.6 | 1 ± 1.0 |
| UB246 | 13 | — | — | — |
| CLA0302 | 13 | — | — | — |
Discussion
Phage genomes are difficult to compare on a nucleotide level because of their high diversity. Gene synteny and core proteins are, however, often conserved even in phages with low nucleotide similarity.23 Most genes required for assembly of the procapsid, capsid, tail fibers, and DNA packaging have been predicted in Misse, Bertil, and Strit, but no proteins with homology to coat proteins have been found. Neighboring holin and endolysin genes were predicted in all three phages (gp49 and gp50, respectively). gp51 shows a weak similarity to a Rz-like protein from Escherichia virus T7 (highest E-value = 0.003) based on blastn. Interestingly, gp51 also shows a high similarity to several genes placed directly upstream of a gene encoding a lysozyme in other Autographiviridae phages. A small potential ORF was found overlapping both gp51 and gp52 in all three phage genomes. The product of these genes shows a similarity to putative Rz1-like proteins. Based on this, the phage genomes contain lysis cassettes similar to that of Escherichia virus T7 and found in many phages targeting gram-negative bacteria.23
Phages have originally been classified based on the morphology of phage particles or host range, but the basis of phage taxonomy has gradually shifted to genetic analyses.24 Based on this, the International Committee on Taxonomy of Viruses (ICTV) recently changed the taxonomy of the Caudovirales order, so that the Autographiviridae is now a family within the Caudovirales order. Bertil, Misse, and Strit have basic characteristics, such as a genome size of 41 kb with inverted terminal repeats, as well as morphological traits similar to the known characteristics of the Autographiviridae phages.25 The three phages also present the highest nucleotide level similarity to phages within the Autographiviridae family. However, the low similarity to other phages makes it difficult to place the phages within an existing subfamily or genus. The three phages could, therefore, constitute a new genus within the Autographiviridae family. This is corroborated by phylogenetic analysis of the predicted RNA polymerase, major capsid protein, and large terminase (Supplementary Fig. S1). For all three proteins, Bertil, Misse, and Strit form their own cluster among other Autographiviridae phages. The separate branching of Bertil, Misse, and Strit is most reliable for the major capsid protein and the large terminase.
The P. syringae species complex has been classified in 13 phylogroups as well as additional subgroups based on the multilocus sequence typing of four housekeeping genes.22 Although not all strains have been tested for pathogenicity against crops, many strains of P. syringae from nonagricultural origin are nevertheless potential pathogens.5 The original isolation strain, GAW0113, belongs to phylogroup 12, which comprises only a small number of strains. The two other sensitive strains tested in the host-range assay, CC1417 and CC1532, belong to phylogroup 9 (Table 1). Although none of the sensitive strains have been tested for pathogenicity on plants, CC1417 can induce a hypersensitive reaction on tobacco leaves unlike CC1532 and GAW0113.26 Successful infection replication of phage is influenced by many factors such as host cell receptors and compatibility with the host replications machinery. Bacteria have also developed an array of different antiphage defense mechanisms, such as CRISPR-Cas, that influence the susceptibility of a bacteria toward phage infection.27 Despite the genetic diversity of the hosts, the difference in EOP was <10-fold between the three sensitive P. syringae. Berge et al.22 have previously shown that the different phylogroups share many phenotypic traits despite being genetically distinct and this seems to include sensitivity toward specific phages.
All three sensitive strains have been isolated from aquatic habitats in either France or the United States.22 This indicates that the characteristics influencing phage susceptibility are shared by some P. syringae strains independent of phylogroups or geographic distribution. It has been suggested that bacterial metabolic membrane-proteins are often used by phages for adsorption and the environment of the host bacteria could, therefore, influence phage host range.28 However, populations of P. syringae mix between different habitats, so this parameter alone does not explain the shared phage susceptibility and more detailed analyses are needed to elucidate the phage–host mechanisms.29 The lack of genes associated with the lysogenic cycle and the ability to target multiple strains within the P. syringae species make Bertil, Misse, and Strit relevant as antimicrobial therapeutic agents. The low relatedness to other phages could be an advantage for using one of the three phages as part of a larger phage cocktail. However, as they are mostly identical it is probably redundant to apply more than one of the three phages together.
Conclusion
The three isolated phages are highly similar, with Misse being the most distinct on a nucleotide and protein level. They all share many genomic and morphological features with other phages of the newly established Autographiviridae family, but limited nucleotide similarity. Based on morphology, nucleotide similarity and homology of predicted core proteins, Bertil, Misse, and Strit seem to be three phage isolates within their own genus of distinct Autographiviridae phages. All three phages were able to infect and lyse multiple strains of P. syringae across different phylogroups.
Supplementary Material
Acknowledgment
Angela Back (Max Rubner-Institut) is thanked for her technical assistance with the electron microscopy.
Authors' Contributions
Conceptualization by J.B.J., A.M.D., A.B.C., and L.H.H.; methodology by J.B.J., A.M.D., A.B.C., W.K., H.N., and L.H.H.; validation by J.B.J., A.M.D., A.B.C., W.K., and L.H.H.; formal analysis by J.B.J., H.N., and W.K.; investigation by J.B.J., A.M.D., and A.B.C.; resources by W.K. and L.H.H.; data curation by J.B.J., A.M.D., and A.B.C.; writing—original draft preparation by J.B.J.; writing—review and editing by J.B.J., A.M.D., A.B.C., W.K., H.N., C.M., and L.H.H.; visualization by J.S.P. and H.N.; supervision by A.M.D., A.B.C, W.K., and L.H.H.; project administration by W.K. and L.H.H.; funding acquisition by W.K. and L.H.H. All coauthors have reviewed and approved of the article before submission.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This study was part of the “Phytoprotect” project funded by the Danish Research Council for Technology and Production Grants (Grant No. DFF-4184-00070), the Human Frontier Science Program (Grant No. RGP0024/2018), and the Villum Experiment (Grant No. 17595).
Supplementary Material
References
- 1. Scortichini M, Biocca M, Rossi MP. Pseudomonas syringae pv Morsprunorum on wild cherry for timber production: Outbreak and field susceptibility. Eur J For Pathol. 1995;25(6–7):343–350. [Google Scholar]
- 2. Vanneste JL. The scientific, economic, and social impacts of the New Zealand outbreak of bacterial canker of kiwifruit (Pseudomonas syringae pv. actinidiae). Annu Rev Phytopathol. 2017;55(1):377–399. [DOI] [PubMed] [Google Scholar]
- 3. Sahin F. Severe outbreak of bacterial speck, caused by Pseudomonas syringae pv. tomato, on field-grown tomatoes in the eastern Anatolia region of Turkey. Plant Pathol. 2001;50(6):799. [Google Scholar]
- 4. Otta J. Wheat leaf necrosis incited by Pseudomonas syringae. Phytopathology. 1972;62(10):1110. [Google Scholar]
- 5. Morris CE, Monteil CL, Berge O. The life history of Pseudomonas syringae: Linking agriculture to earth system processes. Annu Rev Phytopathol. 2013;51(1):85–104. [DOI] [PubMed] [Google Scholar]
- 6. Svircev A, Roach D, Castle A. Framing the future with bacteriophages in agriculture. Viruses. 2018;10(5):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roux S, Hallam SJ, Woyke T, et al. Viral dark matter and virus–host interactions resolved from publicly available microbial genomes. eLife. 2015;4:e08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loc-Carrillo C, Abedon ST. Pros and cons of phage therapy. Bacteriophage. 2011;1(2):111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kropinski AM, Mazzocco A, Waddell TE, et al. Enumeration of bacteriophages by double agar overlay plaque assay. Chap. 3, 69–76 Methods Mol Biol. 2009;501:69–76. [DOI] [PubMed] [Google Scholar]
- 10. Carstens AB, Djurhuus AM, Kot W, et al. A novel six-phage cocktail reduces Pectobacterium atrosepticum soft rot infection in potato tubers under simulated storage conditions. FEMS Microbiol Lett. 2019;366(9):fnz101. [DOI] [PubMed] [Google Scholar]
- 11. Altschul S, Madden T, Schaffer A, et al. Gapped blast and psi-blast: A new generation of protein database search programs. FASEB J. 1998;12(8):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29(12):2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delcher AL, Bratke KA, Powers EC, et al. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23(6):673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pope WH, Jacobs-Sera D. Annotation of bacteriophage genome sequences using DNA Master: An overview. Bacteriophages: Methods and Protocols, Volume 3. Methods Mol Biol. 2018:217–229. [DOI] [PubMed] [Google Scholar]
- 15. Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32(1):11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chan PP, Lowe TM. tRNAscan-SE: Searching for tRNA genes in genomic sequences. Methods Mol Biol. 2019;1962:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(suppl_2):W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moraru C, Varsani A, Kropinski AM. VIRIDIC—A novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses. 2020;12(11):1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sullivan MJ, Petty NK, Beatson SA. Easyfig: A genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 2013;30(5):1229–1235. [DOI] [PubMed] [Google Scholar]
- 21. Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual. 4th ed. Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- 22. Berge O, Monteil CL, Bartoli C, et al. A user's guide to a data base of the diversity of Pseudomonas syringae and its application to classifying strains in this phylogenetic complex. PLoS ONE. 2014;9(9):e105547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dion MB, Oechslin F, Moineau S. Phage diversity, genomics and phylogeny. Nat Rev Microbiol. 2020;18(3):125–138. [DOI] [PubMed] [Google Scholar]
- 24. Barylski J, Enault F, Dutilh BE, et al. Analysis of spounaviruses as a case study for the overdue reclassification of tailed phages. Syst Biol. 2020;69(1):110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adriaenssens EM, Sullivan MB, Knezevic P, et al. Taxonomy of prokaryotic viruses: 2018-2019 update from the ICTV Bacterial and Archaeal Viruses Subcommittee. Arch Virol. 2020;165(5):1253–1260. [DOI] [PubMed] [Google Scholar]
- 26. Stopelli E, Conen F, Guilbaud C, et al. Ice nucleators, bacterial cells and Pseudomonas syringae in precipitation at Jungfraujoch. Biogeosciences. 2017;14(5):1189–1196. [Google Scholar]
- 27. Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317–327. [DOI] [PubMed] [Google Scholar]
- 28. Hyman P, Abedon ST. Bacteriophage host range and bacterial resistance. In: Adv Appl Microbiol. 2010;70:217–248. [DOI] [PubMed] [Google Scholar]
- 29. Morris CE, Sands DC, Vanneste JL, et al. Inferring the evolutionary history of the plant pathogen Pseudomonas syringae from its biogeography in headwaters of rivers in North America, Europe, and New Zealand. mBio. 2010;1(3):e00107-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.