Abstract
Background: Phage therapy (PT), as a method to treat bacterial infections, needs identification of bacteriophages targeting specific pathogenic host. Enterococcus faecalis, a Gram-positive coccus resident in the human gastrointestinal tract, may become pathogenic in hospitalized patients showing acquired resistance to vancomycin and thus representing a possible target for PT.
Materials and Methods: We isolated four phages that infect E. faecalis and characterized them by host range screening, transmission electron microscopy, and genome sequencing. We also identified and three-dimensional modeled a new hyaluronidase enzyme.
Results: The four phages belong to Siphoviridae family: three Efquatrovirus (namely vB_EfaS_TV51, vB_EfaS_TV54, and vB_EfaS_TV217) and one Saphexavirus (vB_EfaS_TV16). All of them are compatible with lytic cycle. vB_EfaS_TV16 moreover presents a gene encoding for a hyaluronidase enzyme.
Conclusions: The identified phages show features suggesting their useful application in PT, particularly the Saphexavirus that may be of enhanced relevance in PT because of its potential biofilm-digestion capability.
Keywords: phage isolation, host range, phage therapy cocktails, 3D model of hyaluronidase
Introduction
Antibiotic treatments can lose efficiency because bacteria can develop resistance and create a spread of infection not easily controlled. Multidrug resistance (MDR) and hospital break-down diffusion have dangerous increase. It was estimated that 700,000 deaths per year may be actually caused by MDR and that this amount may rise to 106 by 2050.1,2
Phage therapy (PT) has yet a low application in occidental countries although a renewed interest is caused by the concern of increasing diffusion of bacteria with antibiotic resistance.3 The quality of PT has been described widely by efficient applications and experimental data. Recent studies suggest the best characteristics of phage necessary for the application to the PT.4,5 The risk of bacterial escape to phage infection and resistance was successfully resolved with the adoption of phage cocktails, since the risk of multiple resistance can be excluded because of bacterial vitality loss.6 The safety of PT was largely demonstrated in mouse model and with many human experiments in adult subjects to target antibiotic resistant superbacteria.7,8 Natural and engineered bacteriophages seem to be naturally optimized to circulate in mammalian bodies escaping rapid neutralization.9,10 The advantage of a low immune response or tolerance to the phage is relevant and confirmed also by the presence of phages in human microbiota.11,12
The problem of resistance selection was challenged by Schooley and his team that asseverated the utility of phage cocktails.13 Available phage must be implemented and tested for this aim.14,15 The assumption that more phage species are necessary to prepare phage cocktails prompted us to isolation of new Enterococcus faecalis phages.8 A general effort should be pursued with the integration of the available collections with the new phage isolated worldwide and described with genome characteristics in peer reviewed journals.4,16 It was demonstrated that the therapeutic application of phage on microbiota as anti-inflammatory agent conducts to positive results in equilibrating gut dysbiosis.17 Bacteriophages with digestive enzymes can also depolymerize biofilm, which is another form of defense of bacteria against chemical disinfectants and antibiotics.18 Hyaluronidase is an enzyme with possible phage utility, and is present in several species of Streptococcus pyogenes phage probably necessary to digest the bacterial capsule. Here we report the isolation and analysis of four new E. faecalis bacteriophages: vB_EfaS_TV16, vB_EfaS_TV217, vB_EfaS_TV51, and vB_EfaS_TV54 (hereinafter referred as TV16, TV217, TV51, and TV54).
Three phages are Efquatrovirus with lysins potentially active also versus bacterial biofilm. TV16 Saphexavirus instead harbors a hyaluronidase, although its host has no capsule that can be targeted by this catalytic protein. The presence of hyaluronidase gene suggests that in phages there could also be depolymerizing enzymes, other than those to infect bacterial species, potentially useful to lysate bacterial biofilms.18,19
Materials and Methods
Isolation and propagation of bacteriophages
To select phages, an aliquot of 50 mL of municipal raw sewages (Rome, Italy) was centrifuged at 4700 × g for 10 min at 25°C and then filtered through syringe filter with 0.2 μm pore. A 500 μL volume of this suspension was then mixed with 0.2 mL of indicator bacteria and plated by standard double-layer agar method, as detailed in the bacterial strains and growth conditions method section. To obtain pure bacteriophages preparations, three rounds of infection and picking of isolated plaques were performed. The E. faecalis strain ATCC2912 was used as host for isolation and propagation of TV16 and TV217 phages, while the clinical strain 5/5477 was used for phages TV51 and TV54. Also the two indicator bacteria were prepared by standard double-layer agar method. The isolated phages were named according to the systematic naming scheme proposed by Kropinski et al.20 Phage propagation was performed as previously described.21
Bacterial strains and growth conditions
A collection of 15 E. faecalis and 5 Enterococcus faecium clinical strains were obtained from the Careggi University Hospital (Florence, Italy) (Table 1). Additional reference strains were obtained from American Type Culture Collection (ATCC) or Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ). Tryptic soy broth (TSB) or tryptic soy agar (Liofilchem srl, Italy) were used to grow bacterial strains. Soft agar for double-layer plating was composed of TSB plates solidified with 0.7% w/v agar. Collected bacteria were employed to determine the host range of the four isolated phages. Suspension medium (SM) buffer (10 mM Tris-HCl, pH 7.5; 100 mM NaCl; 10 mM MgSO4) was used for suspending and titrating bacteria and phages.
Table 1.
Bacterial Strains and Host Range
| Bacterial strainsa |
Origin |
|
EOPb |
|
|
|---|---|---|---|---|---|
| TV16 | TV217 | TV51 | TV54 | ||
| Enterococcus faecalis | ATTC 19433 | ||||
| ATCC 29212 | 1.00 | 0.70 | 2.5 × 10−2 | 1.0 × 10−2 | |
| E. faecalis | 1/1112 | ||||
| E. faecalis | 2/2353 | ||||
| E. faecalis | 3/3866 | ||||
| E. faecalis | 4/4692 | ||||
| E. faecalis | 5/5477 | 1.00 | 1.00 | 1.00 | |
| E. faecalis | 6/8037 | ||||
| E. faecalis | 7/8085 | ||||
| E. faecalis | 8/8096 | ||||
| E. faecalis | 9/8097 | ||||
| E. faecalis | 10/8168 | ||||
| E. faecalis | 11/8181 | ||||
| E. faecalis | 12/8186 | 0.75 | |||
| E. faecalis | 13/8231 | 0.30 | 2.5 × 10−3 | ||
| E. faecalis | 14/8237 | 2.5 × 10−3 | |||
| E. faecalis | 15/8300 | 5.0 × 10−5 | 2.0 × 10−3 | 5.0 × 10−4 | |
| Enterococcus faecium | DSM 20477 | ||||
| E. faecium | 16/2933 | ||||
| E. faecium | 17/4303 | ||||
| E. faecium | 18/4988 | ||||
| E. faecium | 19/5288 | ||||
| E. faecium | 20/5323 | ||||
| Staphylococcus aureus | ATCC 29213 | ||||
The Enterococcus faecalis and the Enterococcus faecium strains, except ATCC and DSM strains, are clinically isolated at Careggi University Hospital of Florence, Italy.
EOP was calculated as the titer (PFU/mL) on the test strain/titer (PFU/mL) on E. faecalis ATCC29212 for TV16 or 5/5477 for the other three phages. Blank cells indicate that no plaques were detected (EOP <1 × 10−7).
ATCC, American Type Culture Collection; DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH; EOP, efficiency of plating.
Determination of bacteriophages' host range
The plaquing host range was determined by efficiency of plating (EOP) analyses. To determine the EOP, the phages were tested at different dilutions ranging from 108 to 103 PFU/mL against all the strains listed in Table 1. For this purpose, bacterial strains were grown O/N in TSB at 37°C and, after incubation, 200 μL of bacterial suspension was infected with 100 μL of diluted phage and plated. Plates were incubated O/N at 37°C and PFUs were enumerated. Spot test and EOP were repeated three times for each bacterial strain. The EOP was finally computed as the ratio between average PFU on target bacteria and average PFU on E. faecalis ATCC29212 for TV16 or on 5/5477 for the other three phages.
Electron microscopy
Phage morphology was examined by transmission electron microscopy of negatively stained preparations. In detail, a phage suspension was centrifuged for 100 min at 40,000 g in Beckman JA-20 rotor and pellet was resuspended at ∼1012 PFU/mL in SM buffer. 10 μL of the concentrated phage suspension was adsorbed on a carbon-coated matrix and then stained with 1% uranyl acetate for 15 s. The grid was subsequently washed two times with ddH2O, air dried, and observed by an FEI Tecnai G2 (FEI, Eindhoven, The Netherlands) transmission electron microscopy at 100 kV. Images were captured using a Veleta (Olympus Soft Imaging System) digital camera.
Bacteriophages genome sequencing and analysis
The DNA extraction was performed with the Phage DNA isolation kit (Norgen Biotek Corp., Canada). DNA samples were processed using Nextera sample prep kit from Illumina (Illumina, Inc., CA, USA) and sequenced at paired-end 2 × 300 bp setup on Illumina MiSeq (Illumina, Inc.) at IGA Technology Services (Udine, Italy). Raw reads were deposited to NCBI databases (Bioproject: PRJNA729411; BioSamples: SAMN19115498, SAMN19115499, SAMN19115500, SAMN19115501). Depth of coverage was evaluated at 253X, 7X, 135X, and 205X for TV16, TV217, TV51, and TV54, respectively. Raw FASTQ sequences were trimmed with erne-filter22 using default parameters and assembled using SPAdes 3.11.1.23 Phage genomes were annotated by using the rapid annotation using Subsystem Technologies (RAST) web service v.1.8.1.24 Automatic annotation was manually reviewed based on evidence from BLASTP analysis against the NCBI nonredundant protein database (Blast +2.11.0).25 The tRNAscan-SE 2.0 and ARAGORN v1.2.41 web tools were used to predict phage and host tRNA genes.26,27 The assembled and annotated genomes of TV16, TV217, TV51, and TV54 were deposited in the GenBank database under accession numbers MN939408.1, MT627482.1, MT661597.1, and MT661598.1, respectively.
The genomes of the four phages were aligned by blastn versus GenBank databases to identify closest relative and evaluate intergenomic distances/similarities among viral genomes, getting a species/strain classification.25 Selected proteins (hyaluronidase, lysins, and holins) of the four phages were aligned by blastp versus GenBank databases and Blast Tree View 3.6.0 was used to detect hypothetical phylogenetic relationships.25 ViPTree 1.9 was used to construct a viral tree, finding the closest relative of the four phages in the ViPTree phages database.28 MUSCLE 3.8, Clustal Omega 1.2.2, and MAFFT 7 were run to multialign amino acidic sequences of the selected proteins, whereas Jalview 2.11.1.4 was run to calculate and draw average distance trees.29–32 VIRIDIC v1 was used to compute pairwise intergenomic distances/similarities among viral genomes and verify species/strain classification.33
Modeling of the TV16 putative hyaluronidase
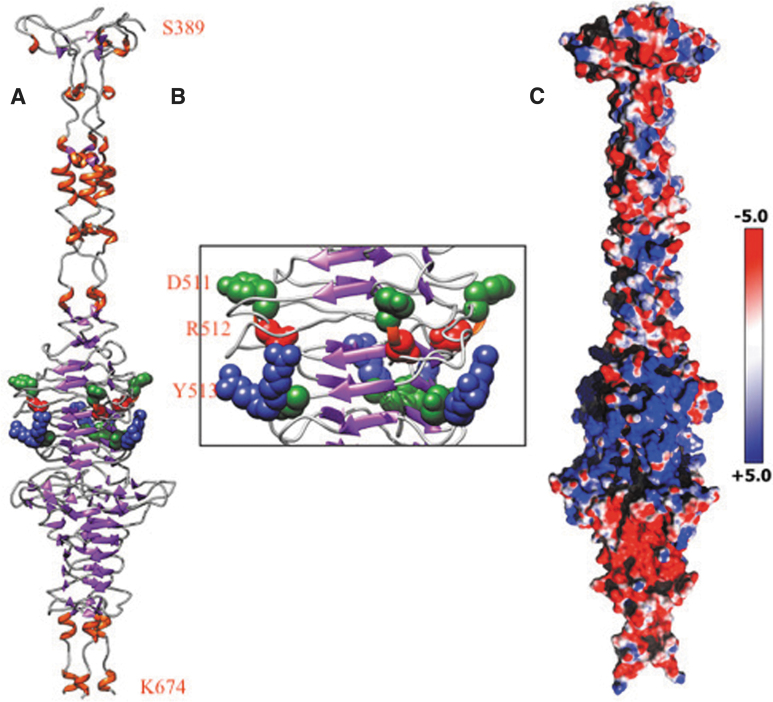
The crystal structure of HylP1 from S. pyogenes, deposited in the Protein Data Bank (PDB; www.rcsb.org/pdb) with the PDB ID:2C3F, has been used as a template to generate a TV16 putative hyaluronidase model structure (the TV16_33; Fig. 1A).34 The model structure has been produced using the SWISS-MODEL protein modeling tool (swissmodel.expasy.org) and, after a structural check, the monomeric unit has been superimposed on the 2C3F biological assembly to generate the TV16_33 trimer.35
FIG. 1.
TV16 hyaluronidase core catalytic domain (S389-K674): (A) 3D model showing β-helix; (B) enhanced view of DRY motif; (C) 3D model showing electrostatic surface potential. 3D, three-dimensional.
Results
Isolation of four phages of E. faecalis
We isolated four new phages that belong to the Siphoviridae family from hospital sewage. Three phages were Efquatrovirus, namely vB_EFaS_TV217 (TV217; MT627482.1, 41,486 bp), vB_EFaS_TV51 (TV51; MT661597.1, 41,821 bp), and vB_EFaS_TV54 (TV5; MT661598.1, 42,644 bp). The fourth was a Saphexavirus, namely vB_EFaS_TV16 (TV16; MN939408.1, 58,127 bp).
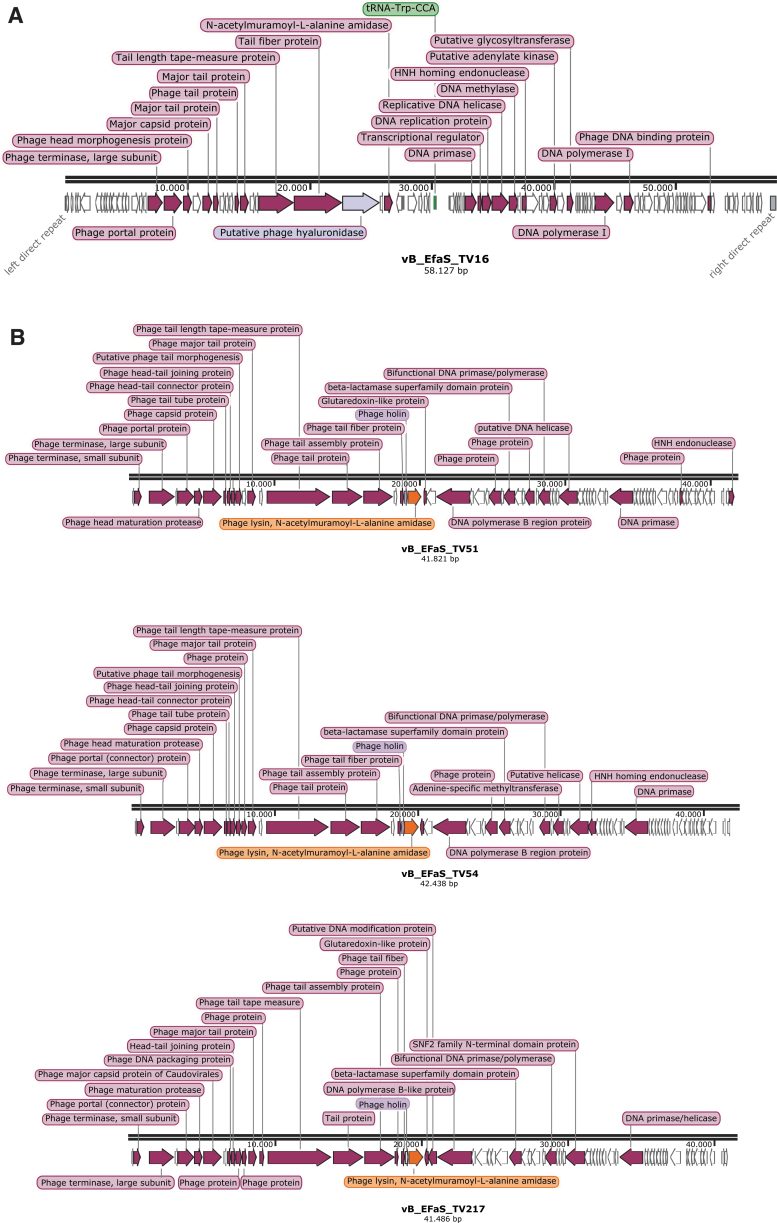
The genomic description of the four phages is summarized in Figure 2: 97, 66, 68, and 66 putative coding sequences were predicted for TV16, TV217, TV51, and TV54, respectively. A tRNA-Trp (CCA) was detected in TV16.
FIG. 2.
Genetic maps reporting the ORF directions and the tags of the genes identified by RAST. White arrows illustrate position of hypothetical proteins ORFs. (A) Saphexavirus. (B) Efquatroviruses. ORF, open reading frame; RAST, rapid annotation using Subsystem Technologies.
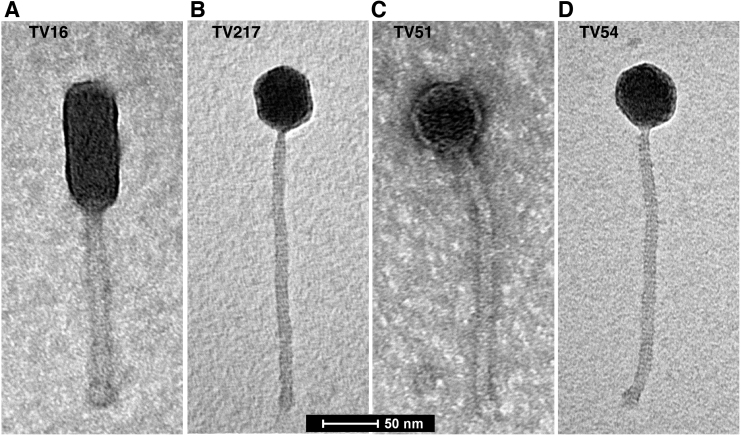
The TEM analysis of the four phages is reported in Figure 3. TV16 is 287 ± 5 nm in length and is characterized by a long noncontractile tail (170 ± 5 nm in length, 17 ± 2 nm in width) and an elongated cylindrical head (118 ± 4 nm in length, 49 ± 2 nm in width) with a length-to-width ratio of 2.4 that is the hallmark of B3 phage morphology.36 TV217, TV51, and TV54 phages are similar in size with a total length of 310 ± 10 nm. They have a long noncontractile tail (250 ± 10 nm in length, 12 ± 2 nm in width) and isometric head (57 ± 3 nm). For these characteristics, they can be attributed to the morphological group B1.36 The bacteria–host analysis results of the four isolated phages are reported on Table 1.
FIG. 3.
Electron microscopy analysis of the four phages: (A) TV16, (B) TV51, (C) TV54, and (D) TV217.
Genomic analysis of the Efquatrovirus phages TV51, TV54, and TV217
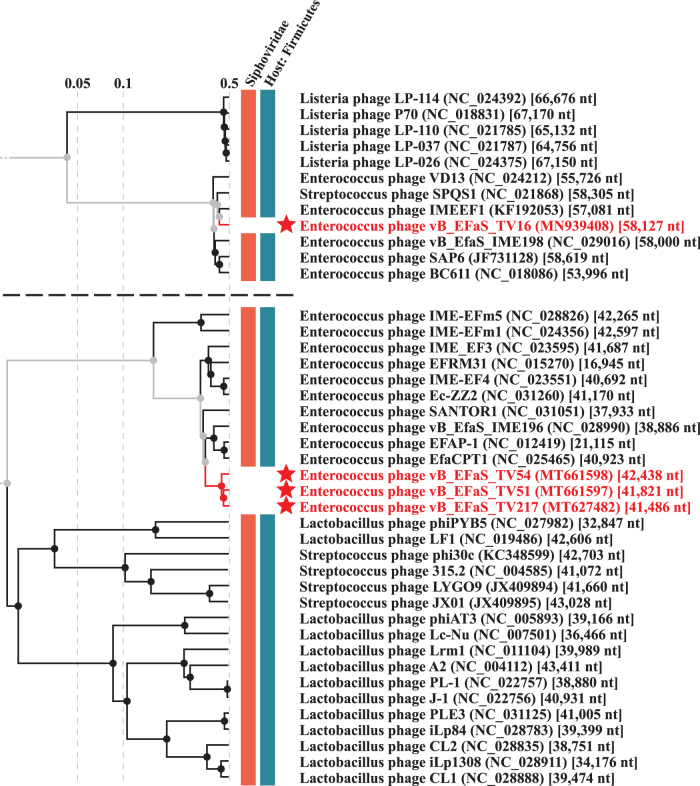
The three phages showed sequence and characteristics of the Efquatrovirus phages and we confirmed the classification by ViPtree analysis (Fig. 4). We performed a similarity search in GenBank databases by bacteriophage whole genomes (Supplementary Table S1A). Considering the 95% similarity threshold rule, TV217 is a new strain of the same species to which phiSHEF2 belongs, whereas TV51 and TV54 are new species (Supplementary Table S1A).37 These findings were confirmed by performing VIRIDIC analyses. Supplementary Table S1A also summarizes similarity search by selected proteins (i.e., lysins, holins, and phage tail tape measure proteins). Holins form pores in the host's cell membrane, allowing lysins to reach and degrade peptidoglycan, a component of bacterial cell walls, so both are essential to trigger the final step of the lytic cycle. In contrast, the tail tape measure proteins are implicated in phage genome injection in the host cytoplasm. Although different as genomic sequences, the three phages seem to belong to the same subgroups of Efquatrovirus when considering the lysin genes, sharing high similarity versus the same set of proteins from genomes of the same genus (level of gray in Supplementary Table S1A). Also tail tape measure proteins clustered the three phages together in the same subgroup, but with a different set of relatives with respect to the lysins' one. Finally, their holins clustered ascribing TV54 and TV217 to a distinct cluster subgroup than TV51.
FIG. 4.
ViPTree viral tree branches including the phages TV16, TV51, TV54, and TV217 (stars).
Genomic analysis of the Saphexavirus phage TV16
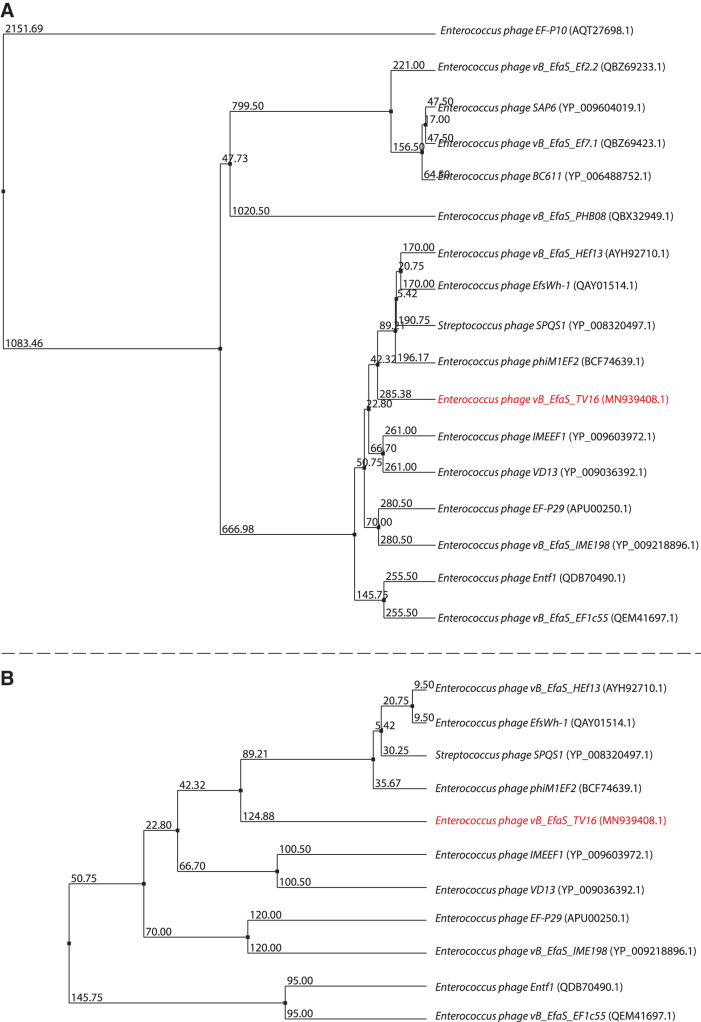
TV16 showed sequence and characteristics of the Saphexavirus phages, including left and right direct tandem repeats. We performed comparison of both the TV16 phage genome and annotated putative hyaluronidase (Hyal_TV16) versus genomes and proteins of Saphexavirus that share the same host E. faecalis. Genome similarity among TV16 and known Saphexavirus phages is lower than 95%, that is the threshold value to label TV16 as a new phage species instead of a strain of a known species (Supplementary Table S1B). The ViPtree analysis produced a viral tree confirming the classification (Fig. 4).
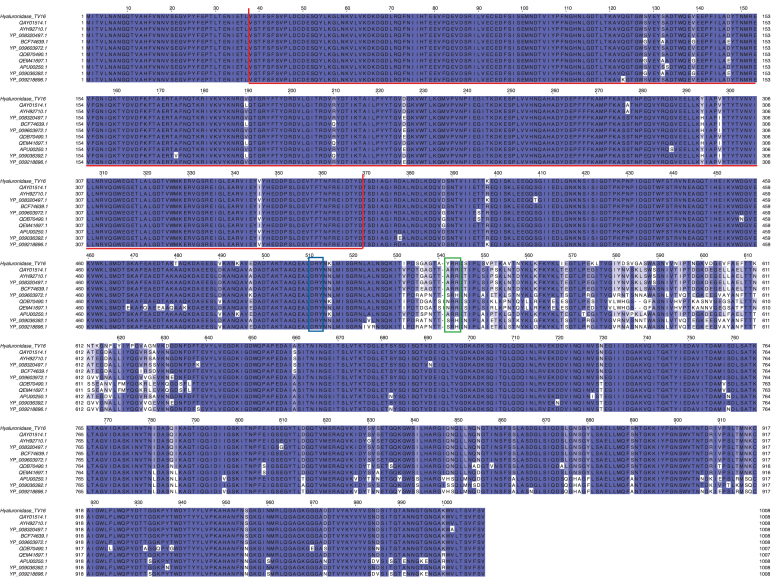
Bacteriophage hyaluronidases may allow phages possessing such enzymes to gain access to appropriate bacterial cell receptors.38 Comparison of Hyal_TV16 detected two groups of sequences: hyaluronidases belonging to 10 phages showed a high level of coverage and identity versus Hyal_TV16, whereas 6 phages showed lower level of both the parameters (Supplementary Table S1B). The most conserved region is the N-terminal section of the protein (Fig. 5), where a protein domain search recognized a prophage-tail domain that is shared among the 17 phages. The C-terminal part is instead more variable (Supplementary Table S1B). A comparison among the 17 hyaluronidases focused on the N-terminal of the enzyme produces the phylogenetic tree shown in Figure 6A, whereas considering the full length of the 11 more similar hyaluronidases produces the phylogenetic tree shown in Figure 6B.
FIG. 5.
Multialignment of hyaluronidase genes of Saphexavirus. Highlighted regions: prophage tail domain (red box), Asp-Arg-Tyr motif (blue box), and Tyr-Arg-Arg motif (green box).
FIG. 6.
TV16 hyaluronidase phylogenetic trees (A) on the entire enzyme, (B) focused on the N-terminal part of the enzyme.
The structure of the TV16 putative hyaluronidase
The structure of the Hyal_TV16 has been modeled to validate its supposed ability in disrupting the biofilms produced by E. faecalis clinical strains (Fig. 1A–C). The obtained model reveals a catalytically active triple-stranded β-helix (Fig. 1B) that maintains the HylP1 hyaluronidase active site, compatible with the function observed for the template structure, indicating that Hyal_TV16 can be a hyaluronate lyase.34 In fact, HylP1 is a hyaluronidase phage tail fiber protein responsible for the digestion of the S. pyogenes hyaluronan capsule during phage infection and shares ∼29% of sequence with the protein coded by TV16_33 CDS, making possible the use of the homology modeling technique.34 The three-dimensional model structure provides insight into the role of the Hyal_TV16 protein in virulence. The triple-stranded β-helix assembly of Hyal_TV16, as observed for the structure of HylP1 from S. pyogenes, showed three copies of the active center on the triple-helical fiber itself without the need for an accessory catalytic domain. This structural scaffold provides an extended groove suitable to bind long stretches of hyaluronic acid (HA) for the optimal reduction in hyaluronan viscosity to aid phage penetration of the capsule.34 In fact, every three to four β-strands, positively charged Arg or Lys residues protrude into this grove, spaced between 9 and 14 Å apart, consistent with the spacing of negatively charged glucuronic acid of the alternating GlcA-GlcNAc polymer in HA.34 Closer inspection of the Hyal_TV16 structure reveals Asp-Arg-Tyr triplets (Asp511-Arg512-Tyr513; Fig. 1A, B), adjacent to a loop, reminiscent of similar pairings at the center of structurally unrelated hyaluronate and chondroitin lyases, in which Tyr residues act as the catalytic base for proton abstraction from C5 of the uronic acid with the Asp involved in pKa maintenance.39
Blast alignment of the protein Hyal_TV16 with hyaluronidase from both Enterococcus phage vB_EfaS_EF1c55 (entry QEM41697.1:1-1007) and Enterococcus phage vB_EfaS_IME198 (entry YP_009218896.1:1-1008) showed the conservation of these residues (Fig. 5, DRY motif in the blue rectangle). Other three residue groups (Tyr541-Arg542-Arg543, Fig. 5 green rectangle) are also reported in proximity of the mentioned Asp-Arg-Tyr triplets, which, in partnership with the latter, could have a catalytic role or a substrate recognition role only (Fig. 1B). Moreover, the electrostatic surface potential of Hyal_TV16 confirms the presence of an extended substrate binding cleft that is predominantly positively charged, complementing the negative charge of the HA substrate (Fig. 1C).
As HylP1, the Hyal_TV16 structure seems to be part of the bacteriophage tail assembly, with the “enzymatic needle” function that digests the HA capsule during phage invasion of the host.34 As HylP1, Hyal_TV16 is not an efficient hyaluronan degrader but an efficient hyaluronan viscosity reducer in proximity to a single phage particle, to allow phage approach to the cell wall. Hyal_TV16 acts as a long structural protein whose extended binding surface provides the unusual properties demanded for phage dissemination in biofilm and penetration of the bacterial capsule.
Discussion
Electron microscopy evidenced an elongated capsid belonging to TV16 Saphexavirus phage and an icosahedral isometric capsid of the TV51, TV54, and TV217 Efquatrovirus phages. The four viruses show various strains of E. faecalis as target bacteria in our host range assays, ensuring the high specificity of the phages infection competence. These phages were isolated and studied to add new phages to the available choices for the PT of E. faecalis. These bacteria are considered part of the normal healthy microbiota but can cause opportunistic infections with severe pathologies and is often present in hospitals, also in antibiotic-resistant forms.40 In the microbiota of mammals, the presence of E. faecalis species is constant, nevertheless it can induce a pathologic status in conditions such as immune depression or chronic illness and nosocomial chronic stay.41 The increasing frequency of detection of antibiotic-resistant E. faecalis, including Vancomycin that is often used in secondary prophylaxis in patients with persistent bacterial infection, is enhancing the interest for these phages. The interest in PT is also due to the absence of side effects but a selective depletion of microbiota members that a simple administration of probiotics could efficiently treat.
The variability of phage genomes explains the high number of phages that can target each specific bacteria host, giving a wide range of alternatives to be part of PT cocktails. Horizontal exchange of genome material contributes to the phages variability.42 The TV16 Saphexavirus here investigated has a hyaluronidase protein conserved in phages of the same genus, but at least two different levels of coverage and similarity value (Supplementary Table S1B and Fig. 6). The other three phages have similar trends when investigated for either lysins, holins, or phage tail tape measure proteins conservation versus other Efquatrovirus. They showed to target the same host.
The spreading mechanism of phages has been studied leading to the description of the particular mode adopted by lytic phages that was defined “mosaic pattern.”43 Digestion of biofilm turns out to be a useful feature for the spreading of phages, and thus for PT, in case where bacteria produce extracellular secretions. This activity is a double advantage in PT, indeed digesting biofilm can help in the penetration of both phages and antibiotics/antimicrobial in the inner microbiota environment. In fact, anaerobic bacteria are often protected by biofilm and hardly affected by chemical agents. TV16 hyaluronidase seems to play this essential role for the Saphexavirus success and could strongly contribute to the E. faecalis depletion in PT.
Conclusion
This study of the phages TV16 (Saphexavirus), TV217, TV51, and TV54 (Efquatrovirus) increases the collection of E. faecalis phages that could be subject of trial in a mouse model for all kinds of dysbiosis and be considered for application in PT.8 This study also gives a first insight into the structure of Saphexavirus phages hyaluronidases.
Supplementary Material
Authors' Contributions
G.D.L. isolated the bacteriophages and determined their host range. M.F. and F.I. have performed the EF1 molecular modeling and the structural analysis of the hyaluronidase enzyme coded in the genome of EF1. D.F. and P.D. performed bioinformatics analyses of genome sequences. All authors contributed to the discussion of the experiments and writing of the article.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
No funding was received for this article.
Supplementary Material
References
- 1. Balasegaram M, Piddock LJV. The Global Antibiotic Research and Development Partnership (GARDP) not-for-profit model of antibiotic development. ACS Infect Dis. 2020;6(6):1295–1298. [DOI] [PubMed] [Google Scholar]
- 2. Tagliabue A, Rappuoli R. Changing priorities in vaccinology: Antibiotic resistance moving to the top. Front Immunol. 2018;9:1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abedon ST. Use of phage therapy to treat long-standing, persistent, or chronic bacterial infections. Adv Drug Deliv Rev. 2019;145:18–39. [DOI] [PubMed] [Google Scholar]
- 4. Paule A, Frezza D, Edeas M. Microbiota and phage therapy: Future challenges in medicine. Med Sci (Basel). 2018;6(4):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abedon ST. Phage therapy: Various perspectives on how to improve the art. Methods Mol Biol (Clifton). 2018;1734:113–127. [DOI] [PubMed] [Google Scholar]
- 6. Oechslin F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses. 2018;10(7):351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zuo T, Ng SC. The Gut Microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front Microbiol. 2018;9:2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bolocan AS, Upadrasta A, Bettio PHA, et al. Evaluation of phage therapy in the context of Enterococcus faecalis and its associated diseases. Viruses. 2019;11(4):366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mutti M, Corsini L. Robust approaches for the production of active ingredient and drug product for human phage therapy. Front Microbiol.. 2019;10:2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dedrick RM, Guerrero-Bustamante CA, Garlena RA, et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med. 2019;25(5):730–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hodyra-Stefaniak K, Lahutta K, Majewska J, et al. Bacteriophages engineered to display foreign peptides may become short-circulating phages. Microb Biotechnol. 2019;12(4):730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zalewska-Piątek B, Piątek R. Phage therapy as a novel strategy in the treatment of urinary tract infections caused by E. coli. Antibiotics (Basel). 2020;9(6):304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schooley RT, Biswas B, Gill JJ, et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother. 2017;61(10):e00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Galtier M, De Sordi L, Sivignon A, et al. Bacteriophages targeting adherent invasive Escherichia coli strains as a promising new treatment for Crohn's Disease. J Crohns Colitis. 2017;11(7):840–847. [DOI] [PubMed] [Google Scholar]
- 15. Sabino J, Hirten RP, Colombel JF. Review article: Bacteriophages in gastroenterology-from biology to clinical applications. Aliment Pharmacol Ther. 2020;51(1):53–63. [DOI] [PubMed] [Google Scholar]
- 16. Cui CH, Zhu Y, Jia Z, et al. Identification of two novel anti-HCV E2 412–423 epitope antibodies by screening a Chinese-specific phage library. Acta Virol. 2019;63(2):149–154. [DOI] [PubMed] [Google Scholar]
- 17. Febvre HP, Rao S, Gindin M, et al. PHAGE Study: Effects of supplemental bacteriophage intake on inflammation and gut microbiota in healthy adults. Nutrients. 2019;11(3):666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. D'Andrea MM, Antonelli A, Brenciani A, et al. Characterization of Tn6349, a novel mosaic transposon carrying poxtA, cfr and other resistance determinants, inserted in the chromosome of an ST5-MRSA-II strain of clinical origin. J Antimicrob Chemother. 2019;74(10):2870–2875. [DOI] [PubMed] [Google Scholar]
- 19. Boas DV, Almeida C, Azevedo N, et al. Techniques to assess phage-biofilm interaction. Methods Mol Biol (Clifton). 2019;1898:137–146. [DOI] [PubMed] [Google Scholar]
- 20. Kropinski AM, Prangishvili D, Lavigne R. Position paper: The creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ Microbiol. 2009;11(11):2775–2777. [DOI] [PubMed] [Google Scholar]
- 21. Di Lallo G, Evangelisti M, Mancuso F, et al. Isolation and partial characterization of bacteriophages infecting Pseudomonas syringae pv. actinidiae, causal agent of kiwifruit bacterial canker. J Basic Microbiol. 2014;54(11):1210–1221. [DOI] [PubMed] [Google Scholar]
- 22. Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS One. 2013;8(12):e85024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bankevich A, Nurk S, Antipov D, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aziz RK, Bartels D, Best AA, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. [DOI] [PubMed] [Google Scholar]
- 26. Lowe TM, Chan PP. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44(W1):W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32(1):11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishimura Y, Yoshida T, Kuronishi M, et al. ViPTree: The viral proteomic tree server. Bioinformatics. 2017;33(15):2379–2380. [DOI] [PubMed] [Google Scholar]
- 29. Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sievers F, Higgins DG. The Clustal Omega Multiple Alignment Package. Methods Mol Biol (Clifton). 2021;2231:3–16. [DOI] [PubMed] [Google Scholar]
- 31. Katoh K, Rozewicki J, Yamada KD. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinformatics. 2019;20(4):1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Waterhouse AM, Procter JB, Martin DM, et al. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moraru C, Varsani A, Kropinski AM. VIRIDIC—A Novel Tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses. 2020;12(11):1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith NL, Taylor EJ, Lindsay AM, et al. Structure of a group A streptococcal phage-encoded virulence factor reveals a catalytically active triple-stranded beta-helix. Proc Natl Acad Sci U S A. 2005;102(49):17652–17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Biasini M, Bienert S, Waterhouse A, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ackermann HW. Frequency of morphological phage descriptions in the year2000. Brief review. Arch Virol. 2001;146(5):843–857. [DOI] [PubMed] [Google Scholar]
- 37. Turner D, Kropinski AM, Adriaenssens EM. A Roadmap for genome-based phage taxonomy. Viruses. 2021;13(3):506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hynes WL, Walton SL. Hyaluronidases of Gram-positive bacteria. FEMS Microbiol Lett. 2000;183(2):201–207. [DOI] [PubMed] [Google Scholar]
- 39. Lunin VV, Li Y, Linhardt RJ, et al. High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: An enzyme-substrate complex defines the catalytic mechanism. J Mol Biol. 2004;337(2):367–386. [DOI] [PubMed] [Google Scholar]
- 40. Lewis CM, Zervos MJ. Clinical manifestations of enterococcal infection. Eur J Clin Microb Infect Dis. 1990;9(2):111–117. [DOI] [PubMed] [Google Scholar]
- 41. Ceci M, Delpech G, Sparo M, et al. Clinical and microbiological features of bacteremia caused by Enterococcus faecalis. J Infect Dev Ctries. 2015;9(11):1195–1203. [DOI] [PubMed] [Google Scholar]
- 42. Gabashvili E, Kobakhidze S, Koulouris S, et al. Bi- and multi-directional gene transfer in the natural populations of polyvalent bacteriophages, and their host species spectrum representing foodborne versus other human and/or animal pathogens. Food Environ Virol. 2021;13(2):179–202. [DOI] [PubMed] [Google Scholar]
- 43. Mavrich TN, Hatfull GF. Bacteriophage evolution differs by host, lifestyle and genome. Nat Microbiol. 2017;2:17112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.