Abstract
The number of sequenced bacteriophage genomes is growing at an exponential rate. The majority of sequenced bacteriophage genomes are annotated by one or more of several freely available gene identification programs (Glimmer, GeneMark, RAST, Prodigal, etc.). No program has been shown to consistently outperform the others; thus, the choice of which program to use is not obvious. We present the Phage Commander application for rapid identification of bacteriophage genes using multiple gene identification programs. Phage Commander runs a bacteriophage genome sequence through nine gene identification programs (and an additional program for identification of tRNAs) and integrates the results within a single output table. Phage Commander also generates formatted output files for direct export to National Center for Biotechnology Information GenBank or genome visualization programs such as DNA Master. Users can select the threshold for which genes to export (genes identified by at least one program, genes identified by at least two programs, etc.). Phage Commander was benchmarked using eight high-quality bacteriophage genomes whose genes are backed by experimental data. Our results show that the most accurate annotations are obtained by exporting genes identified by at least two or three programs. Many groups opt to manually curate the annotations obtained from gene identification programs, and Phage Commander was designed to facilitate manual curation of genome annotations. Our benchmarking results show that manual curation does indeed produce more accurate annotations than any individual gene identification program. The authors thus recommend manually curating the output of Phage Commander to generate maximally accurate annotations. Phage Commander is currently being used in the corresponding author's bacteriophage genome annotation class and has reduced the labor cost and improved the quality of genome annotations.
Keywords: bacteriophages, genome annotation, genomics, gene identification
Introduction
Each year, antibiotic-resistant bacteria cause an estimated 2.8 million infections and 35,000 deaths in the United States according to the U.S. Center for Disease Control.1 This figure is set to increase, as preliminary evidence suggests that part of the high mortality rate of coronavirus disease 2019 could be due to opportunistic bacterial infections.2,3 Bacteriophages, or phages for short, are attractive as an alternative to antibiotics because they are effective at lysing their host rapidly, are highly specific to their host and therefore harmless to humans and gut flora, cause few if any side effects, and coevolve with their host, thereby reducing the chance of their host evolving resistance.4 Inconsistent early results and the development of highly successful antibiotics such as penicillin led to phages being eclipsed as treatment agents in the West. However, several recent high-profile cases of successful use of phages to resolve antibiotic-resistant infections in the United States and United Kingdom have generated significant attention and renewed interest in phages as treatment agents.5–7 Phages are also becoming increasingly important in agricultural and commercial applications as treatments for infections in livestock and honeybees.8,9 They are also the source of a large number of commercial enzymes used in molecular biology, and have featured in many seminal discoveries in molecular biology.10,11
Owing to the growing interest in phages and the constantly decreasing cost of sequencing, the number of sequenced phage genomes is growing at an exponential rate.12 The sequencing of a novel phage genome is followed by annotation, which consists of (1) identifying genes, (2) identifying gene starts, and (3) assigning putative function to genes.13 Genes and gene starts are usually identified using one or more gene identification programs, such as Glimmer,14 GeneMark,15 GeneMark.hmm,16 GeneMark with Heuristics,17 GeneMarkS,18 GeneMarkS2,19 RAST,20 BASys,21 Prodigal,22 Prokka,23 MetaGene,24 and PhANOTATE.25 Although designed for bacterial genomes (with the exception of PhANOTATE), these programs rapidly produce phage genome annotations with roughly 80–90% accuracy.12 Each of these programs uses a different algorithm and produces unique results. Preliminary study has shown that no program consistently outperforms the others,12 and, thus the choice of which program to use is not obvious. Many groups, therefore, combine results from multiple programs and manually interpret their findings to achieve higher accuracy.13,26
The process of manual curation of an annotated genome can be time- and labor-intensive, particularly in the case of a large or novel phage genome, or a large batch of phage genomes, which is increasingly common. To this end, we have designed Phage Commander to accelerate running a phage genome through multiple gene identification programs simultaneously. Rather than trying to decide which program(s) to use, our philosophy is to combine multiple programs within a single user interface and integrate the results. We have thus included as many gene identification programs as possible within a single interface. The various programs have different strengths and weaknesses; thus, combining their results aims to increase sensitivity and specificity, provided the output is integrated appropriately.
Materials and Methods
Phage Commander is freely available for download, along with the source code and instructions, from GitHub (https://github.com/sarah-harris/PhageCommander). Phage Commander was coded in Python 3.6+ and runs on Windows, Mac OS, and Linux. Linux users need to have Python installed on their systems and can install and run Phage Commander following the instructions on GitHub. For Windows and Mac, a stand-alone executable is available on GitHub.
Phage Commander incorporates the following nine programs for gene identification: Glimmer, GeneMark, GeneMark.hmm, GeneMark S, GeneMark with Heuristics, GeneMark S2, Prodigal, RAST, and MetaGene. The list of gene identification programs included in Phage Commander is given in Table 1. In addition to these, Phage Commander also includes the program Aragorn (www.ansikte.se/ARAGORN) for identification of phage tRNAs.27
Table 1.
Gene Identification Programs Included in Phage Commander
| Name (version) | Algorithm | URL |
|---|---|---|
| Glimmer (3.02) | Interpolated Markov model | ccb.jhu.edu/software/glimmer/index.shtml |
| GeneMark (2.5) | Markov chain/Bayesian | exon.gatech.edu/GeneMark/gm.cgi |
| GeneMark.hmm (3.25) | Host-trained hidden Markov model | exon.gatech.edu/GeneMark/gmhmmp.cgi |
| GeneMarkS (4.28) | Self-trained hidden Markov model | exon.gatech.edu/GeneMark/genemarks.cgi |
| GeneMark with Heuristics (3.25) | Hidden Markov with heuristics for short genomes | exon.gatech.edu/GeneMark/heuristic_gmhmmp.cgi |
| GeneMarkS2 | Self-trained hidden Markov model with heuristics | exon.gatech.edu/GeneMark/genemarks2.cgi |
| Prodigal (2.6.3) | Dynamic programming | github.com/hyattpd/Prodigal |
| RAST (2.0) | Subsystems | rast.nmpdr.org |
| MetaGene | RBS statistical model | metagene.nig.ac.jp |
The input to Phage Commander is a phage genome sequence in fasta format. By clicking “New” in the File menu, users select which programs and genome file to use. If using GeneMark.hmm, users must select the bacterial host, as this is required by GeneMark.hmm. If using RAST, users should create an account on the RAST server (https://rast.nmpdr.org/rast.cgi) and enter their RAST credentials when prompted by Phage Commander.
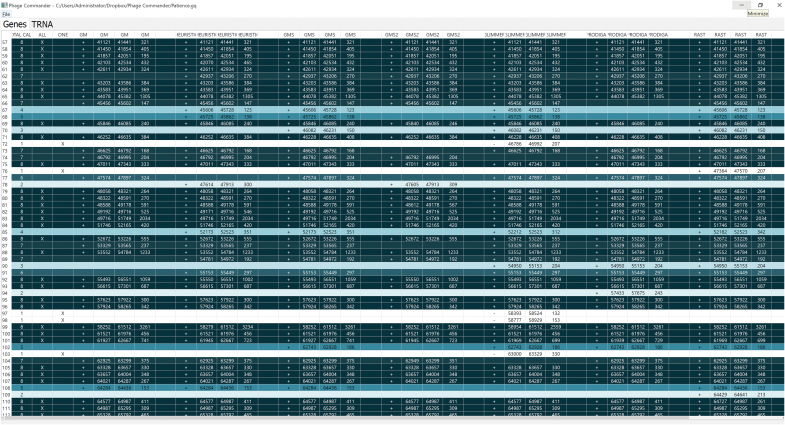
The output of Phage Commander is a list of genes predicted by each program in spreadsheet format. A sample screenshot is shown in Figure 1. Each row represents a gene and each set of four columns corresponds to one of the programs used. For each gene identified by a program, the strand (indicated by “+” or “−”), gene start coordinate, gene stop coordinate, and gene length are listed. Gene rows are shaded based on how many programs identify that particular gene, with darker shading corresponding to more programs. The leftmost column is the number of programs that identify a gene; however, which programs identify a particular gene may vary. For example, in Figure 1, gene 70 and gene 90 are both identified by three programs, but not by the same three programs. The “ALL” and “ONE” columns indicate that a gene is called by all programs or by only one program, respectively. The tRNA genes identified by Aragorn are given in the separate “TRNA” tab.
FIG. 1.
Sample Phage Commander output. Programs shown include GeneMark, GeneMark with heuristics, GeneMark S, GeneMark S2, Glimmer, Prodigal, and RAST (GeneMark.hmm and MetaGene not shown). Each gene is a row, and shaded based on how many programs identify it. tRNA, transfer RNAs.
Two export options exist: Excel format (.xlsx) and GenBank format (.gb). GenBank formatted files can be directly uploaded to the NCBI GenBank genome repository. In our workflow, we export the genes predicted from Phage Commander in .gb format and import them into the DNA Master software,13 which we use to assign putative functions. The fully annotated genomes can then be again exported from DNA Master in .gb format for upload to NCBI.
When exporting in GenBank format, users have the ability to set the threshold for exporting genes, in terms of the number of programs that identify genes. For example, users can select to export genes identified by at least one program (i.e., equivalent to logical ANY/OR), all the way up to genes identified by all programs used (equivalent to logical ALL/AND). Users have the option of exporting gene starts chosen by (1) the majority of programs (in case of a tie, the program will choose the one with the longer open reading frame), (2) a specific program (e.g., export the starts chosen by Glimmer), or (3) the start that generates the longest open reading frame for each gene. The gene start chosen by the majority of programs is shown in white or black font, with gene starts chosen by a minority of programs shown in alternate font colors.
Results
To benchmark Phage Commander, we searched for phage genomes whose genes are known with a high degree of certainty through experiments. We identified eight phages genomes that meet this criterion, shown in Table 2.
Table 2.
Phages Used in Testing Phage Commander
| Name | Host | Genome length (bp) | No. of genes | GenBank accession no. |
|---|---|---|---|---|
| Lambda | Escherichia coli | 48,502 | 71a | NC_001416 |
| Mu | E. coli | 36,717 | 55 | NC_000929 |
| Patience | M. smegmatis | 70,506 | 110 | NC_023691 |
| Kampy | M. smegmatis | 51,378 | 89b | NC_024141 |
| Giles | Mycobacterium smegmatis | 53,746 | 85c | NC_0099933 |
| PAK_P3 | Pseudomonas aeruginosa | 88,097 | 165 | NC_022970 |
| YuA | P. aeruginosa | 58,663 | 79b | NC_010116 |
| API480 | Paenibacillus larvae | 45,026 | 77 | MK533143 |
Two duplicate genes not counted.
Includes genes added.
Based on the annotation of LilHazelnut.
The Escherichia coli phage Lambda was isolated in 1950 and is perhaps the most studied phage in existence.28 Phage Mu is another E. coli phage that has been extensively studied since it was isolated in the early 1960s, and its genome has been carefully annotated.29 Phage Patience is a somewhat atypical Mycobacterium smegmatis phage whose genome has been extensively studied through RNA-Seq transcriptomics and mass spectrometry.30 Phage Kampy is a cluster A4 M. smegmatis phage that has also been studied through transcriptomics and mass spectrometry.31 Phage Giles is a cluster Q M. smegmatis phage that has been studied through mass spectrometry and whose protein interactome has been mapped out.32 Phage PAK_P3 is a Pseudomonas aeruginosa phage that has been studied through transcriptomics experiments.33 Phage YuA is a P. aeruginosa phage that has been studied through mass spectrometry and other experiments.34 Phage API480 is a phage that infects the bacterium Paenibacillus larvae, a pathogen of the honeybee Apis mellifera, and has been the subject of transcriptomics experiments.35 We consider the published annotations of these phage genomes in NCBI GenBank highly trustworthy and we used them as references to benchmark the performance of Phage Commander. A gene that is detected by transcriptomics or mass spectrometry is likely to be real, so we did not remove any genes from the reference annotations. However, even transcriptomics might miss some genes that are rarely expressed. We thus inspected each genome carefully and added one gene to the reference genome of Kampy and two genes to YuA that were identified by multiple (at least seven) programs, had many statistically significant homologs, and filled a coding gap (evidence provided in Supplementary Table S1). For phage Giles, we used the annotation of the closely related phage LilHazelnut (99.99% nucleotide identity), whose annotation is more recent and appears more complete.36 Lambda has two cases of two genes within the same reading frame, differing only by their start (genes nu3 and D, and genes S and R); we thus counted the duplicate genes as a single gene (i.e., we counted nu3 and D as one gene, and S and R as one gene), because no program or method has the ability to identify two overlapping genes within the same reading frame.
Each genome was run through Phage Commander using all nine gene identification programs. The threshold for exporting genes was varied from one to nine programs, and the results were compared with the reference GenBank annotations. Figures 2 and 3 show the performance of Phage Commander compared to the reference GenBank annotations. A false positive is a gene that is identified by one or more programs but that is not present in the reference annotation. A false negative is a gene present in the reference annotation but not detected by any of the programs used.
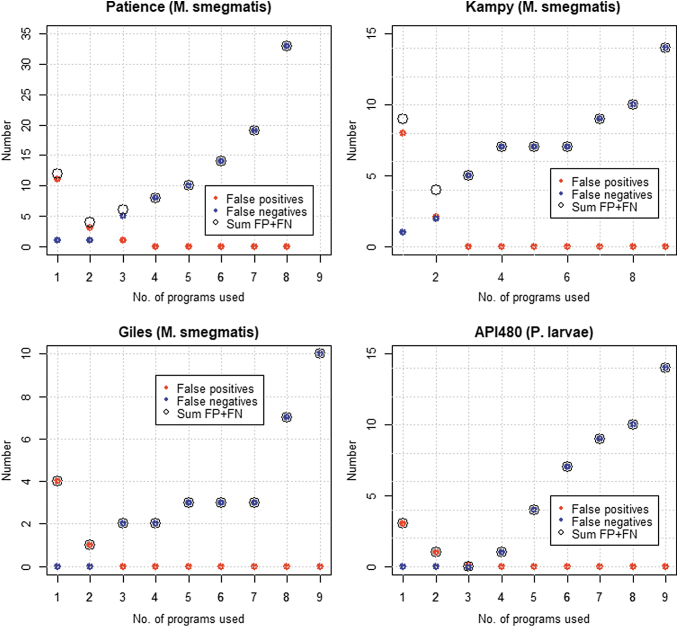
FIG. 2.
False positives, false negatives, and their sum, based on the number of programs used to export genes in Phage Commander for phages Patience, Kampy, Giles, and API480. FN, false negative; FP, false positive.
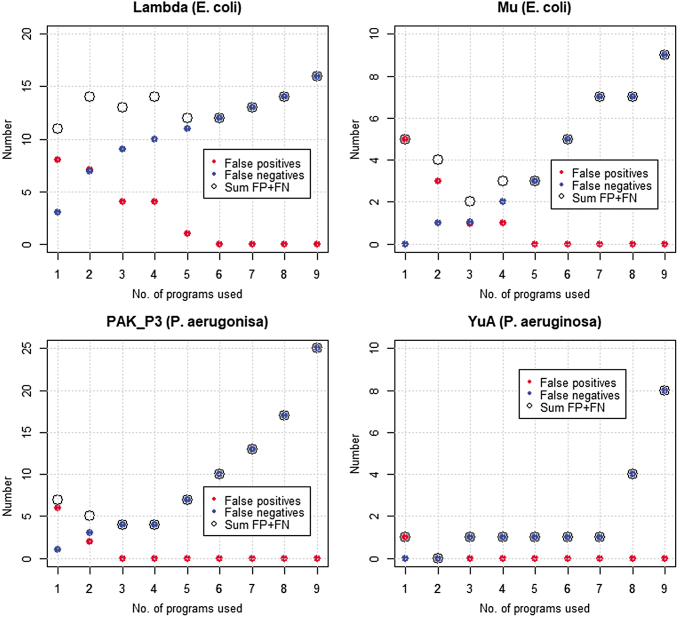
FIG. 3.
False positives, false negatives, and their sum, based on the number of programs used to export genes in Phage Commander for phages Lambda, Mu, PAK_P3, and YuA.
The results show that the sum of false positives and false negatives is usually a minimum when exporting genes called by at least two programs (Patience, Kampy, Giles, and YuA), or genes called by at least three programs (Mu, PAK_P3, and API480). The results for Lambda are somewhat anomalous due to the high number of false negatives. Thus, either of the “at least 2 programs” or “at least 3 programs” settings offer the best trade-off between sensitivity and specificity, with the “at least 2 programs” setting favoring sensitivity (fewer false negatives), and the “at least 3 programs” setting favoring specificity (fewer false positives). Exporting genes called by at least one program (i.e., all genes identified) will likely produce a large number of false positives, although it will generate the fewest false negatives. This setting should thus (and only thus) be used if the results are subsequently manually curated to remove false positives. Exporting genes only called by four or more programs is in the opinion of the authors too stringent and will result in an unacceptably high number of false negatives (typically between 3 and 10 per genome). These results also show variability between phages, with Lambda having the largest number of false positives and false negatives, and YuA the fewest, even when adjusted for genome length.
Given that no gene identification program is 100% accurate, many research groups opt to manually curate the annotations obtained from gene identification programs.12,13 Phage Commander was developed to simplify and accelerate the manual curation process. A major component of manual curation is tabulating the number of programs that identify a candidate gene when making a gene identification decision (in addition to using additional information such as homologs, operons, overlap, ribosome binding score, and synteny).12 Before the development of Phage Commander, phage genomes had to be run through multiple gene identification programs separately, and the output of each program was manually collated into a single spreadsheet.12 Phage Commander was designed to accelerate manual curation by automating the process of running phage genomes through the gene identification programs, and outputting the results into a single spreadsheet automatically. By our estimate, using Phage Commander accelerates our workflow by 25–30% (given that compiling the results of multiple gene identification programs is one of the four main components of the manual curation process).
Of interest is the relative performance of each of the nine programs used and manual curation, which integrates all the programs. In a previous publication presenting a manual curation method developed in the Tsourkas Lab, we benchmarked the accuracy of the method on phages Lambda and Patience.12 In Figures 4 and 5, we compare the performance of the nine programs and the manual curation method for the eight phages in this study. Given that the manual curation method is somewhat subjective (although we have designed it so as to minimize subjectivity to the extent possible), these results may vary slightly depending on the annotator.12 We are in the process of including PhANOTATE in Phage Commander and have included it in Figures 4 and 5, as this program is designed specifically for phage genomes, and is the most directly comparable with manual curation.25
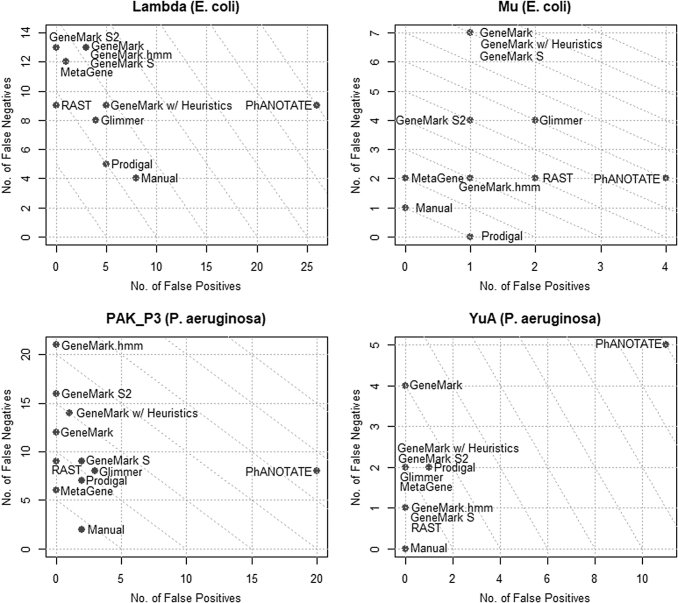
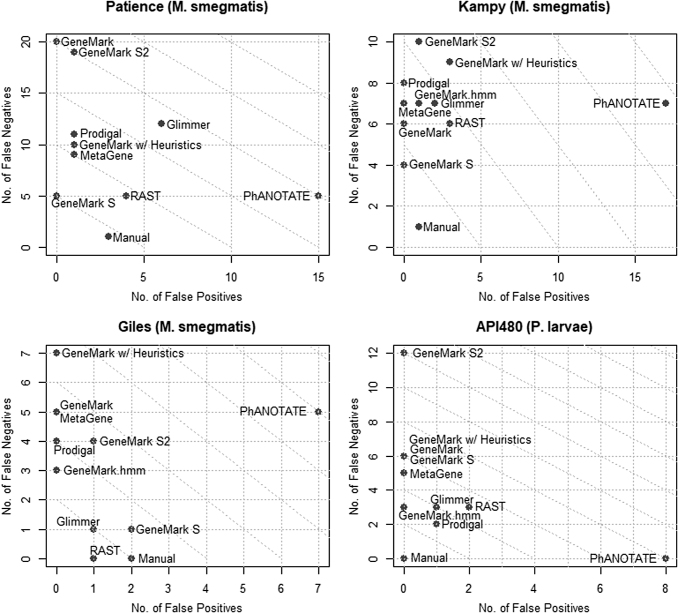
FIG. 4.
Plots of false positives versus false negatives for Patience, Kampy, Giles, and API 480. Diagonal lines represent equal numbers of false positives and false negatives.
FIG. 5.
Plots of false positives versus false negatives for Lambda, Mu, PAK_P3, and YuA. Diagonal lines represent equal numbers of false positives and false negatives.
Manual curation produces the best results (fewest false positives and false negatives) in all phages except Lambda and Giles, and is tied with Prodigal in phage Mu. Also of note is that manual curation produced the fewest false negatives in all phages except Mu. Manual curation has zero false negatives for phages Giles, API480, and YuA, and achieved perfect results (zero false positives and false negatives) for phages API480 and YuA.
Consistent with our earlier results, no program unambiguously outperformed the others. RAST marginally appears to be the most sensitive (fewest false negatives), whereas the GeneMark suite of programs (GeneMark, GeneMark with heuristics, and GeneMark S2) are the least sensitive on average (most false negatives). MetaGene consistently generates the fewest false positives (never more than one, and usually zero). Of the GeneMark suite, GeneMark S and GeneMark.hmm appear to have the best performance. GeneMark.hmm is host trained and cannot be used when the host is not known (e.g., metagenomics) or when the host is unusual. For example, GeneMark.hmm was not used for phage Patience, as this phage is an atypical M smegmatis phage that appears to be transitioning from another host to M. smegmatis,31 and thus GeneMark.hmm produced highly anomalous results for this phage.
PhANOTATE consistently identified a large number of genes not found in the reference genomes in GenBank, or identified by other programs (i.e., false positives). The number of false positives identified by PhANOTATE ranged from 4 (Mu) to 26 (Lambda) and was in the double digits for most phages. These results are consistent with those of our earlier study, in which PhANOTATE identified >200 genes in an artificial randomly generated 40 kbp DNA sequence.12 In terms of false negatives, PhANOTATE performed at a level similar to RAST.
Discussion
Phage Commander is an application designed to accelerate the labor-intensive and time-consuming manual curation of phage genomes. It incorporates nine gene identification programs (plus an additional program for identification of tRNA genes) that are widely used in phage genomics within a single interface and then summarizes the results within a single spreadsheet to display the results in a visually appealing manner. Users have the ability to select which programs to use and which genes to export based on the number of programs identifying a gene. Phage Commander has the ability to export files in GenBank format (.gb) for direct deposition to NCBI GenBank, or for further processing through DNA Master.
An important feature of Phage Commander is the ability for users to set the threshold for exporting genes. Users can export genes on a sliding scale, starting from exporting all genes called by at least one program (logical ANY/OR) to only exporting genes called by all nine programs (logical ALL/AND). Phage Commander was tested on eight high-quality annotated phage genomes, the majority of whose genes are experimentally verified. Results show that the optimal settings are to export genes called by at least two or three programs. Exporting genes called by one program resulted in few false negatives (typically 0 or 1), but a high number of false positives (typically 5–10). Exporting genes called by more than three programs produced an unacceptably high number of false negatives (typically 3–10). No matter which setting is used, the authors recommend manually inspecting and curating the output from Phage Commander, using a method such as in Salisbury and Tsourkas.12
Of the nine gene identification programs used in Phage Commander, results with the eight phage genomes used in this study showed RAST to be the most sensitive (fewest false negatives on average), and MetaGene the most specific (fewest false positives on average). Manual curation,12 which relies on integrating the output of all nine programs through Phage Commander, produced the best results (fewest combined false positives and false negatives on average) for six of the eight phages tested, showing that combining the output from multiple programs, in combination with additional information, reduces the number of false positives and false negatives.
Phage Commander is currently being used in an undergraduate class on phage genome annotation taught by the corresponding author as part of the Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science program.37 The setting used in the class is to export all genes identified by at least one program, and the results are manually curated by the students using the method described in Salisbury and Tsourkas.12 Phage Commander has demonstrably reduced the laboriousness of the manual curation process, and improved results.
Future directions include adding more gene identification programs (e.g., PhANOTATE and BASys), and homology search results (BLAST, CD-Search, HMMer, etc.) so as to include putative gene product function information. In addition, we plan to integrate the results not just using simple logical rules (ANY, ALL), but by developing a machine learning algorithm to do so.
Conclusion
We present Phage Commander, an application for rapid identification of genes in phage genomes using multiple gene identification programs. By combining different existing programs within a single interface, Phage Commander achieves better gene detection than any single gene-identification program, while also reducing the labor cost of manual curation of genome annotations. Phage Commander is freely available for download from GitHub and runs on Windows, Mac, and Linux.
Supplementary Material
Authors' Contributions
M.L. and G.R. developed the software. S.L.H. secured funding for the project, revised the code base, and edited the article. P.K.T designed the project, secured funding, and wrote the article. All coauthors have reviewed and approved of the article for submission. The article has been submitted solely to PHAGE and is not published, in press, or submitted elsewhere.
Author Disclosure Statement
The authors do not have financial interests to disclose.
Funding Information
P.K.T. and S.L.H. wish to acknowledge support from the UNLV Faculty Opportunity Award program for funding.
Supplementary Material
References
- 1. Antibiotic/Antimicrobial Resistance (AR/AMR). https://www.cdc.gov/drugresistance/index.html (last accessed April 27, 2021).
- 2. Cevik M, Bamford C, Ho E. COVID-19 pandemic—A focused review for clinicians. Clin Microbiol Infect. 2020;7:842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020;395:1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Altamirano G, Barr JJ. Phage therapy in the postantibiotic era. Clin Microbiol Rev. 2019;32:143–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schooley RT, Biswas B, Gill JJ, et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother. 2017;61:e00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan BK, Turner PE, Kim S, et al. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Pub Health. 2018;1:60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dedrick RM, Guerrero-Bustamante CA, Garlena RA, et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med. 2019;25:730–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cooper IR. A review of current methods using bacteriophages in live animals, food and animal products intended for human consumption. J Microbiol Meth. 2016;130:38–47. [DOI] [PubMed] [Google Scholar]
- 9. Brady TS, Merrill BD, Hilton JA, et al. Bacteriophages as an alternative to conventional antibiotic use for the prevention or treatment of Paenibacillus larvae in honeybee hives. J Invertebr Pathol. 2017;150:94–100. [DOI] [PubMed] [Google Scholar]
- 10. Grose, JH, Casjens SR. Understanding the enormous diversity of bacteriophages: The tailed phages that infect the bacterial family Enterobacteriacae. Virol. 2014;468–470:421–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kropinski AM, Clokie MRJ. Introduction. In: Clokie MRJ, Kropinski AM; eds. Bacteriophages: Methods and Protocols, Volume 2: Molecular and Applied Aspects. Valley Stream, NY: Humana Press; 2019: xiii–xxii. [Google Scholar]
- 12. Salisbury A, Tsourkas PK. A method for improving the accuracy and efficiency of bacteriophage genome annotation. Int J Mol Sci. 2019;20(14):3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pope WH, Jacobs-Sera D. Annotation of bacteriophage genome sequences using DNA Master: An overview. In: Clokie MRJ, Kropinski AM; eds. Bacteriophages: Methods and Protocols, Volume 3: Molecular and Applied Aspects. Valley Stream, NY: Humana Press; 2018: 217–229. [DOI] [PubMed] [Google Scholar]
- 14. Delcher AL, Harmon D, Kasif S, et al. Improved microbial gene identification with GLIMMER. Nucl Acid Res. 1999;27(23):4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Borodovsky M, McIninch J. GeneMark: Parallel gene recognition for both DNA strands. Comput Chem. 1993;17:123–133. [Google Scholar]
- 16. Lukashin AV, Borodovsky M. GeneMark.hmm: New solutions for gene finding. Nucl Acids Res. 1998;26(4):1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Besemer J, Borodovsky M. Heuristic approach to deriving models for gene finding. Nucl Acids Res. 1999;27:3911–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Besemer J, Lomsadze A, Borodovsky M. et al. A self-training method for gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucl Acids Res. 2001;29:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lomsadze A, Gemayel K, Tang S, et al. Modeling leaderless transcription and atypical gene results in more accurate gene prediction in prokaryotes. Genome Res. 2018;20:1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aziz RK, Bartels D, Best AA, et al. The RAST server: Rapid annotations using subsystems technology. BMC Genomics. 2008:8(9):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Domselaar GH, Stothard P, Shrivastava S, et al. BASys: A web server for automated bacterial genome annotation. Nucl Acid Res. 2005;33:W455–W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hyatt D, Chen GW, LoCascio PF, et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. [DOI] [PubMed] [Google Scholar]
- 24. Noguchi H, Park J, Takagi T. MetaGene: Prokaryotic gene finding from environmental genome shotgun sequences. Nucl Acids Res. 2006;34(19):5623–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McNair K, Zhou C, Dinsdale EA, et al. PHANOTATE: A novel approach to gene identification in phage genomes. Bioinformatics. 2019;35:4537–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Philipson CW, Voegtly LJ, Lueder MR, et al. Characterizing phage genomes for therapeutic applications. Viruses. 2018;10(4):188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32(1):11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Casjens SR, Hendrix RW. Bacteriophage lambda: Early pioneer and still relevant. Virology. 2015;479–480:310–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morgan GJ, Hatfull GF, Casjens S, et al. Bacteriophage Mu genome sequence: Analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J Mol Biol. 2002;317(3):337–359. [DOI] [PubMed] [Google Scholar]
- 30. Pope WH, Jacobs-Sera D, Russell DA, et al. Genomics and proteomics of mycobacteriophage Patience, an accidental tourist in the Mycobacterium neighborhood. mBio. 2014;5(6):e02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Halleran A, Clamons S, Saha M. Transcriptomic characterization of an infection of Mycobacterium smegmatis by the cluster A4 Mycobacteriophage Kampy. PLoS One. 2015;10(10):e0141100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mehla J, Dedrick RM, Caufield JH, et al. The protein interactome of mycobacteriophage giles predicts functions for unknown proteins. J Bacteriol. 2015;197(15):2508–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blasdel BG, Chevallereau A, Monot M, et al. Comparative transcriptomics analyses reveal the conservation of an ancestral infectious strategy in two bacteriophage genera. ISME J. 2017;11(9):1988–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ceyssens PJ, Mesyanzhinov V, Sykilinda N, et al. The genome and structural proteome of YuA, a new Pseudomonas aeruginosa phage resembling M6. J Bacteriol. 2008;190(4):1429–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ribeiro HG, Melo LDR, Oliveira H, et al. Characterization of a new podovirus infecting Paenibacillus larvae. Sci Rep. 2019;9(1):20355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shanks RA, Hazel AN, Jones WH, et al. Genome Sequence of Mycobacterium Phage LilHazelnut. Microbiol Resource Announc. 2019;8(19):e00431-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jordan TC, Burnett SH, Carson S, et al. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. mBio. 2014;5(1):e1051-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.