Abstract
Background: Fast and computationally efficient strategies are required to explore genomic relationships within an increasingly large and diverse phage sequence space. Here, we present PhageClouds, a novel approach using a graph database of phage genomic sequences and their intergenomic distances to explore the phage genomic sequence space.
Methods: A total of 640,000 phage genomic sequences were retrieved from a variety of databases and public virome assemblies. Intergenomic distances were calculated with dashing, an alignment-free method suitable for handling massive data sets. These data were used to build a Neo4j® graph database.
Results: PhageClouds supported the search of related phages among all complete phage genomes from GenBank for a single query phage in just 10 s. Moreover, PhageClouds expanded the number of closely related phage sequences detected for both finished and draft phage genomes, in comparison with searches exclusively targeting phage entries from GenBank.
Conclusions: PhageClouds is a novel resource that will facilitate the analysis of phage genomic sequences and the characterization of assembled phage genomes.
Keywords: phage genomics, comparative genomics, genomic graph database
Introduction
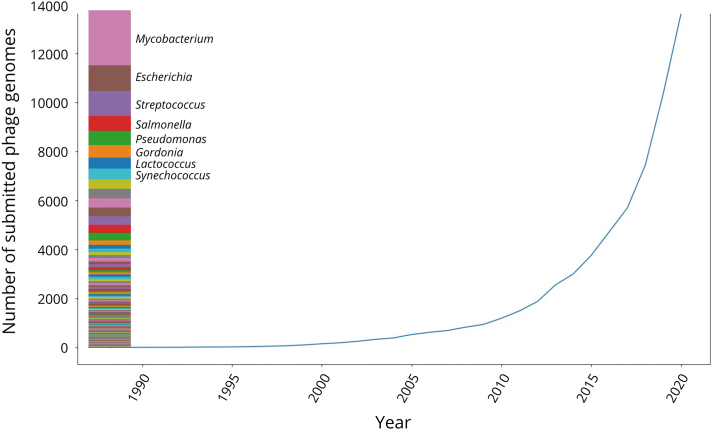
Phages are viruses that target bacteria and were first described independently by Frederick Twort and Felix D'Herelle in the early 20th century.1 Since then, phages have been regarded as a potential therapeutic approach to treat bacterial infections, particularly in recent years due to a higher incidence of antibiotic-resistant pathogens.2,3 This has encouraged the isolation and characterization of novel phages that target clinically relevant bacteria, leading to the accumulation of isolated phage genomes in reference databases (Fig. 1).
FIG. 1.
The number of complete phage genomes deposited in GenBank across time. The introduction of NGS technologies in the early 2000s was followed by an exponential increase in the number of phage genomes deposited in GenBank. The stacked bar plot on the left indicates the proportion of phage genomes that target different bacterial genera. The top eight targeted bacterial genera account for half of the phage genomes currently available in GenBank.
In addition to their potential as therapeutic agents, various studies have demonstrated the impact that phages have on biogeochemical cycling, bacterial ecology and evolution, and even potential roles in human health.4–6 Such studies, along with increasingly cheaper and accessible high-throughput sequencing technologies, have prompted the discovery of a myriad of phage genomic sequences.7 These sequences have primarily been derived from the identification of putative prophages in bacterial genomes or the detection of phage genomic sequences in metagenomic data sets or viral-enriched samples.8–10
This explosion of phage genomic data has uncovered an astonishing spectrum of genetic diversity.11 However, as the volume of phage genomic data grows, the estimation of genomic relatedness by alignment-based methods becomes increasingly impractical as these are slow and require large amounts of computing memory. Several alternative approaches have recently been developed to circumvent these limitations, among which the use of the MinHash algorithm became a popular choice for a range of tools.12–14 For instance, Mash uses MinHash to calculate the distance between a pair of genomes by estimating the Jaccard index for their combined kmer content, and produces a result that strongly correlates with the average nucleotide identity (ANI).12
Mash's optimal use of computational resources was leveraged for estimating the distances between all genome pairs among 2333 phages from NCBI's RefSeq.15 The study reported that using kmer and sketch sizes (Mash's main parameters) of 15 and 25,000 resulted in the correlation of Mash distances and ANI values for the analyzed genomes, up to a Mash distance of ∼0.33.15 This demonstrated that tools such as Mash can be used to quickly and reliably estimate close evolutionary relationships between phages in massive data sets, where alignment-based methods become computationally impractical.16
In addition to estimating intergenomic distances for large phage data sets, it is imperative to select appropriate visualization strategies that aid the identification of clusters of related phages. Phylogenetic trees are commonly used to illustrate evolutionary relationships between biological entities, and they have been extensively used for phages when the inferred relationships are based on single or small sets of genes.17–19
More recently, a phylogenetic tree of phages in the order Caudovirales was generated with a collection of prevalent single-copy genes, which proved to be highly consistent with the taxonomy defined by the International Committee on Taxonomy of Viruses (ICTV).20 However, their use for showing relationships between phages has been argued against, as they fail to depict intercluster connections that result from the horizontal exchange of gene modules.21,22 Instead, other studies have showcased the use of networks as an alternative for representing phage evolutionary relationships.23,24 In fact, the algorithm used by the phage taxonomic classification tool vConTACT2 is based on the construction and analysis of gene-sharing networks.25
Despite the progress made so far, there is a need for resources that efficiently place novel phage genomic sequences within the context of the currently known phage diversity. Here we present PhageClouds, a novel approach based on the creation of a graph database that stores phage genomic sequences from a range of databases and their intergenomic distances. PhageClouds has a user-friendly interface allowing the user to search a database of 635,850 phage genomic sequences quickly and efficiently, and retrieve clusters of related phages (from hereon in referred to as phage clouds) based on a user-defined query.
Methods
Data set selection
The PhageClouds' graph database was built using phage genomic sequences obtained from the following sources: NCBI's GenBank database, the Gut Phage Database (GPD),10 phages from the Global Ocean Viromes (GOV) reported by the TARA Oceans consortium,9 phages in the Integrated Microbial Genome/Virus (IMG/VR) database7 from samples categorized as terrestrial, prophages from bacterial genomes in the Genome Taxonomy Database (GTDB),26 non-GOV entries in the Phages and Integrated Genomes Encapsidated Or Not (PIGEON) database,27 the Cenote Human Virome Database (CHVD),28 The Gut Virome Database (GVD),29 phages from the cattle slurry virome,30 phages from horse feces viromes,31 and phages from marine samples taken at the U.K.'s south coast (MarineUKSouth)32 (Table 1).
Table 1.
List of Reference Databases Used for Building Our Graph Database
| Source | Sequence count | Date accessed | % targeted host | % country |
|---|---|---|---|---|
| GenBank | 17,062 | July 29, 2021 | 87.5 | 62.1 |
| GPD | 142,809 | January 1, 2021 | 23 | 92.7 |
| TARA Oceans | 195,728 | November 2, 2020 | 0 | 0 |
| GTDB prophages | 64,180 | July 17, 2020 | 98.6 | 0 |
| IMG/VR terrestrial | 45,364 | January 29, 2021 | 9.82 | 0 |
| PIGEON | 95,047 | June 5, 2021 | 0 | 0 |
| GVD | 15,330 | June 23, 2020 | 0 | 0 |
| CHVD | 42,142 | June 5, 2021 | 0 | 0 |
| Horse virome | 1640 | June 5, 2021 | 0 | 0 |
| MarineUKSouth | 9915 | June 5, 2021 | 0 | 0 |
| Slurry | 6633 | June 5, 2021 | 0 | 0 |
Data on the number of sequences provided by each database, the last date they were accessed, and the percentage of entries that have information on targeted hosts and countries of isolation/detection are provided.
CHVD, Cenote Human Virome Database; GPD, Gut Phage Database; GTDB, Genome Taxonomy Database; GVD, Gut Virome Database; IMG/VR, Integrated Microbial Genome/Virus; PIGEON, Phages and Integrated Genomes Encapsidated Or Not.
Genomes of dsDNA phages were retrieved from NCBI's GenBank database using the following Entrez query: db = “nucleotide”, term = “gbdiv PHG[prop]”. To filter out entries corresponding to incomplete genomes or genomic segments, the following terms were searched in each record definition field: cds, gene, fragment, region, protein, partial, left end. These terms are present in the definition fields of GenBank entries related to incomplete phage genomes (e.g., GenBank accessions MW929183 and MZ573924) and phage genes (e.g., GenBank accessions MZ148154, MT769248, and MZ209176).
Prophages were identified from bacterial genomes in the GTDB database using PhageBoost.33 FASTA files from release 95 of the GTDB database were used as input and the prophage prediction was conducted using default parameters, accepting prophage predictions that contained at least 20 genes. Overall, 635,850 phage genomic sequences were collected from the aforementioned sources.
Calculation of intergenomic distances
Intergenomic distances were calculated for all pairs of genomic sequences with dashing v0.4.7.34 Sketches were built for all genomic sequences using dashing's sketch command with a kmer size of 15 and a log2 sketch size of 14.61, which corresponds to a sketch size of 25,000. Using the calculated sketches, intergenomic distances between all pairs of genomic sequences were calculated using dashing's cmp command and forcing the output to report Mash distances instead of dashing distances. Henceforth, intergenomic distance values and thresholds are referring to Mash distances calculated with dashing. In total, ∼400 billion intergenomic distances were calculated for the complete set of 635,850 phage genomic sequences. This calculation took ∼5 h using 10,000 CPUs and 128 GB RAM, which corresponds to 50,000 CPU hours.
Building of the graph database
A Neo4j® graph database v.4.3.2 was built using the collected phage genomic data and the calculated intergenomic distances. Neo4j is an open-source native graph database that, unlike traditional relational databases, has a flexible structure that directly stores the relationships between the data. This type of database was selected as it allowed the modeling of our collection of phages and intergenomic distances as a network of nodes and relationships, hence enabling large-scale searches of phage clusters based on a predefined query.
Three types of nodes were used for building the graph database: PhageGenome, Host, and Country. Table 1 indicates the percentage of entries in each reference database for which there is information available on the hosts targeted by the corresponding phages and the countries where they were isolated or detected. PhageGenome nodes represent each of the collected phage genome sequences with additional optional attributes such as lifestyle, genome size, and environment. Host nodes represent the bacterial hosts targeted by the phages, and Country nodes refer to the place where they were isolated or detected.
After filtering with a maximum intergenomic distance threshold of 0.3 that corresponds to an ANI of ∼60%,15 800 million of the 4 × 1011 calculated intergenomic distances were kept to provide the basis for the graph. In total, the PhageClouds graph database occupied more than half a terabyte of disk space.
Results and Discussion
Fast and structured illustration of host-specific phage sequence space
One of the main functionalities offered by PhageClouds is the ability to explore the phage genomic sequence space associated with a user-defined host. This approach will allow users to identify clusters of closely related phages that target a specific host, which provides an opportunity to rapidly explore and get a visual overview of the known phage space and diversity associated with that host.
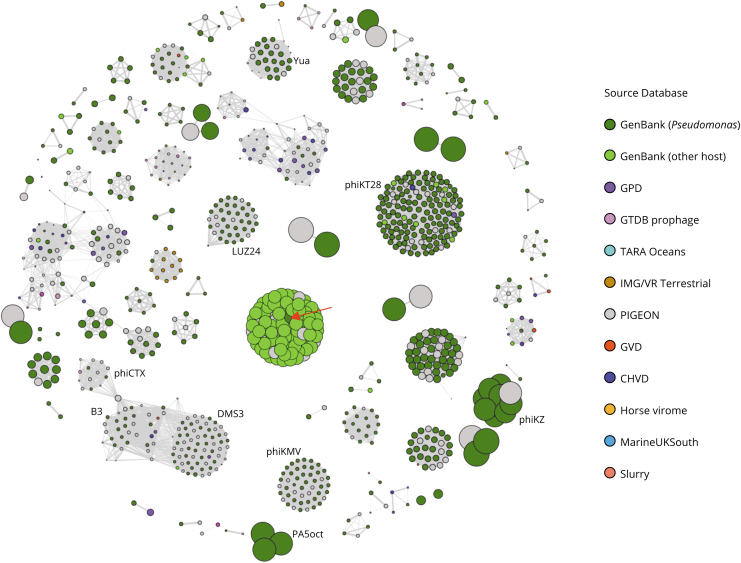
To illustrate this, we searched for all phage clouds containing phages that target bacteria from the genus Pseudomonas and whose members are connected by intergenomic distances of no >0.15. The query used for searching these clouds identified all phage nodes whose metadata indicated they target Pseudomonas and all nodes connected to them with an intergenomic distance below the set threshold. Thus, the resulting clouds also included genomic sequences of phages either targeting other bacterial genera or lacking data on the host they target. After 7 min and 26 s, the search generated the graph illustrated in Figure 2, where nodes are proportionally sized to the genome size and colored according to the database where the corresponding phage was retrieved from.
FIG. 2.
Clouds of phages targeting Pseudomonas. The graph database was queried to retrieve all phages that target the genus Pseudomonas and all phages to which they are connected at a maximum intergenomic distance of 0.15. Node colors indicate the database where the corresponding phages were collected from and their size is proportional to the phages' genome sizes. The red arrow points to a cloud that largely comprised GenBank phages targeting other genera, which is further discussed in the text. Names of some representative Pseudomonas aeruginosa phages are displayed next to the clouds that contain them.
The individual clouds displayed in Figure 2 may potentially be contrasting phage types that use different ecological strategies. For example, several jumbo phage clouds such as those containing the myoviruses phiKZ and PA5oct are easily identifiable, as well as other clouds that contain smaller phages such as the siphovirus, YuA, and the podovirus LUZ24. These clouds are not connected to each other at the selected intergenomic distance threshold, but many do contain well-characterized phages with very different infection strategies.
For example, YuA is an all-rounder phage that pursues a “leeching” strategy by actively depleting host metabolites rather than manipulating the host, whereas phiKZ tunes the host cell to establish the conditions it needs to replicate efficiently. By contrast, PA5oct hijacks the host's transcriptional environment by directing the cellular metabolism to suit its needs.35,36 While we do not know for sure, it may be that other related phages in the clouds interact in similar ways with their bacterial hosts to the better understood phages.
Most of the obtained clouds consisted largely of entries from GenBank corresponding to phages that target members of the genus Pseudomonas. However, there was an instance of a phage cloud in which the number of GenBank phages targeting other hosts was higher than the number of those targeting Pseudomonas. This cloud, highlighted with a red arrow in Figure 2, contained phages from the PIGEON database, many GenBank phages targeting bacteria from the genus Salmonella, and a single GenBank phage that in our graph database is annotated as a Pseudomonas phage (accession number MN871475).
At the time this article was prepared, this GenBank entry was described as a Pseudomonas phage, although it was labeled as an unverified entry. Thus, this might be a Salmonella phage instead, and we expect that the phages from the PIGEON database in this cloud also target this genus. This observation demonstrates how PhageClouds can be leveraged to identify incorrect annotations among entries in reference phage databases, which would help prevent the propagation of such errors when these reference databases are used in phage sequence characterization pipelines.
Rapid search of phage clouds related to user-defined query phages
Our graph database allows users to analyze a custom set of query phages to identify all phage clouds that they are associated with. To showcase the simplest scenario, we searched our graph database using the complete genome sequence of Enterobacter phage IME278 (GenBank accession number MW748991). The search was conducted using 20 cores and setting the distance threshold to 0.15 and 0.21. These values were selected because both favor primarily the selection of connections between phages from the same genus, while largely excluding connections between phages from different genera (Supplementary Fig. S1).
One search was performed by limiting the search space to reference phages from GenBank, and another search was conducted using all entries in our graph database. This focus on GenBank stems from our decision to regard this database as our gold standard for annotations, as its entries generally provide more metadata and are more carefully curated than entries in the other selected databases. We acknowledge that there can be errors in the annotations linked to entries from GenBank, but this should be minimal in comparison with the remaining databases. The running times for all these searches are indicated in Table 2, including the search of the complete graph database using an intergenomic distance threshold of 0.21 that took only 15 s.
Table 2.
Running Times for Searching Phage Clouds Related to Different Sets of Query Phages, Using Different Combinations of Search Parameters and 20 Computing Cores
| 0.15a, GBb | 0.21, GB | 0.15, WGD | 0.21, WGD | |
|---|---|---|---|---|
| Single-query phage | 00:00:10c | 00:00:10 | 00:00:11 | 00:00:15 |
| 79 new GenBank phages | 00:13:42 | 00:25:09 | 00:39:37 | 02:22:11 |
Selected intergenomic distance threshold.
Graph database entries included in the search.
Time format h:min:s.
GB, GenBank; WGD, Whole graph database.
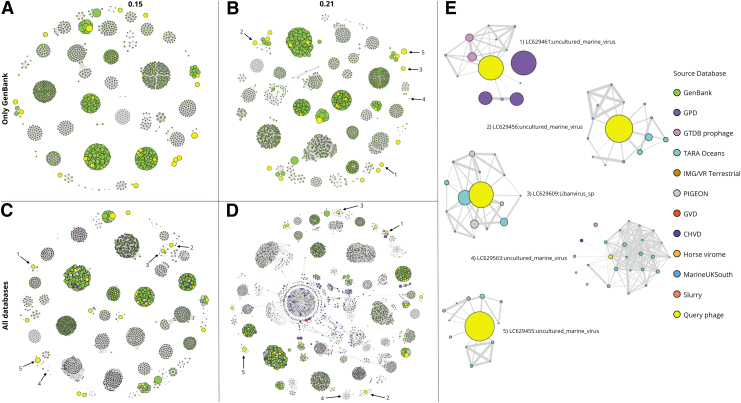
A more complex scenario is the search of related sequences for a larger set of query phages. As an example, we searched the graph database using 79 phages from GenBank that had not been included among the records used to build our database. The search of phage clouds was first restricted to entries from GenBank and was carried out using intergenomic distance thresholds of 0.15 and 0.21. Table 2 indicates the time that these searches took, and the corresponding results are depicted in Figure 3A and B. At least one related phage from GenBank was identified for 56 out of the 79 query phages for an intergenomic distance threshold of 0.15, but this number increased to 63 when the search was conducted with a threshold of 0.21.
FIG. 3.
Searching phage clouds for a set of input query phages and user-defined intergenomic distance thresholds. A set of 79 phage genomes from the GenBank that were not included in the graph database were used for searching phage clouds using two different intergenomic distance thresholds, 0.15 (A, C) and 0.21 (B, D). (A, B) Show the result of searching clouds composed exclusively of GenBank phages, while the clouds in (C, D) include phages from all reference databases. (E) Illustrates some examples of query phages shown as singletons in (B), but captured within some of the clouds present in (D). Arrows in (B, C, D) point to the query nodes present in the clouds found in (E). Node colors indicate the database where the corresponding phages were collected from and their size is proportional to the phages' genome sizes.
Therefore, these results indicate that limiting the search to entries from GenBank and a maximum intergenomic distance of 0.21 does not lead to the identification of at least one related phage for all 79 query phages. Perhaps extending the search space to all entries in our graph database would help achieve the goal of finding a closely related genomic sequence for all 79 query phages.
We repeated the search of phage clouds for the same set of 79 query phages but including all entries in the graph database and testing the same combinations of search parameters. Table 2 indicates the running times required for each search, and the corresponding results are depicted in Figure 3C and D. Using an intergenomic distance threshold of 0.15 resulted in the identification of closely related sequences for 10 of the 16 singletons from the GenBank-constrained search. Furthermore, increasing the threshold to 0.21 led to the identification of closely related sequences for all of the singletons.
Table 3 lists the best matches from the graph database that were identified for each of the 16 singletons. Our results demonstrated that, despite a significant increase in running time, searching the whole graph database led to the identification of closely related sequences for all the query phages.
Table 3.
Best Reference Matches for 16 Singleton Phage Genomes That Resulted from a GenBank-Constrained Search of the Graph Database
| Query phage | Best match | Source database | Intergenomic distance |
|---|---|---|---|
| LC629455:uncultured_marine_virus | Station58_DCM_ALL_assembly_NODE_127_length_41467_cov_96.610693 | TARA Oceans | 0.108736 |
| LC629456:uncultured_marine_virus | Station58_DCM_ALL_assembly_NODE_174_length_37974_cov_33.677971 | TARA Oceans | 0.101924 |
| LC629458:Caudovirales_sp | Station56_SUR_ALL_assembly_NODE_61_length_88758_cov_21.840028 | TARA Oceans | 0.064562 |
| LC629459:uncultured_marine_virus | Station18_DCM_ALL_assembly_NODE_544_length_22784_cov_15.229311 | TARA Oceans | 0.166993 |
| LC629461:uncultured_marine_virus | Flavobacteriales_bacterium__NHFO01000063_phage12__64kb | GTDB prophages | 0.128538 |
| LC629467:uncultured_marine_virus | Bacterium_isolate__PAZJ01000031_phage54__57kb | GTDB prophages | 0.077934 |
| LC629474:uncultured_marine_virus | Station31_SUR_ALL_assembly_NODE_1914_length_19272_cov_27.587397 | TARA Oceans | 0.168987 |
| LC629494:Caudovirales_sp | Station30_DCM_ALL_assembly_NODE_1322_length_15351_cov_25.741828 | TARA Oceans | 0.181003 |
| LC629500:Siphoviridae_sp | Station58_DCM_ALL_assembly_NODE_568_length_22502_cov_143.650866 | TARA Oceans | 0.037512 |
| LC629563:uncultured_marine_virus | Station100_SUR_ALL_assembly_NODE_1036_length_14465_cov_49.298959 | TARA Oceans | 0.123668 |
| LC629575:uncultured_marine_virus | Station123_SUR_ALL_assembly_NODE_2449_length_10403_cov_69.936799 | TARA Oceans | 0.129898 |
| LC629600:uncultured_marine_virus | Station76_DCM_ALL_assembly_NODE_3213_length_10752_cov_25.308030 | TARA Oceans | 0.108739 |
| LC629609:Libanvirus_sp | Station18_DCM_ALL_assembly_NODE_18_length_107714_cov_7.163711 | TARA Oceans | 0.058169 |
| LC629612:uncultured_marine_virus | PIGEON_EarthsVirome_17958 | PIGEON | 0.160845 |
| MT025940:Enquatrovirus_sp | Station168_IZZ_ALL_assembly_NODE_477_length_89554_cov_52.179924 | TARA Oceans | 0.208478 |
| MW822601:Synechoccus_phage_S-SRP02 | IMGVR_UViG_3300035703_000158 | IMG/VR (Terrestrial) | 0.204246 |
Figure 3E illustrates the clouds retrieved for five of the singletons by the search conducted over the complete database with an intergenomic distance threshold of 0.21. As indicated in the figure, most of these singletons are annotated in GenBank as uncultured marine viruses (Fig. 3E). The figure revealed that all phage sequences connected to each of the singletons correspond to entries from databases other than GenBank. In fact, most of these sequences corresponded to entries from the TARA Oceans reference data set. This is expected as this data set consisted entirely of marine viral sequences.
Moreover, four of the singletons ended up connected to significantly smaller sequences from the TARA Oceans data set, and many of these were not connected to each other. Supplementary Figure S2 illustrates the alignment of all reference phage genomic sequences connected to the query phage (GenBank accession LC629609) in phage cloud 3 from Figure 3E, some of which were derived from the same sample (e.g., Station18_DCM and Station31_SUR). Thus, some of these reference sequences might correspond to different fragments of a phage genome that was not fully assembled when this data set was generated. This suggests that PhageClouds might be a useful resource for detecting genomic fragments that are derived from the same phage genome.
Phage clouds reflect taxonomic groups of phages
To demonstrate that the phage clouds reliably represent groups of closely related phages, we analyzed clouds that included phages of the family Herelleviridae. Until recently, members of this family were classified in the Spounavirinae subfamily within the family Myoviridae, but a series of complementary genomic/proteomic analyses demonstrated that the spounaviruses were markedly distinct from other members of the family Myoviridae.37 Therefore, those analyses supported the creation of the family Herelleviridae and the definition of its internal structure, meaning that subfamilies and genera within it have been defined based on genomic/proteomic relationships between member phages.37 Thus, we considered that this family would be an ideal example to illustrate that phage clouds depict genuine relationships between closely related phages.
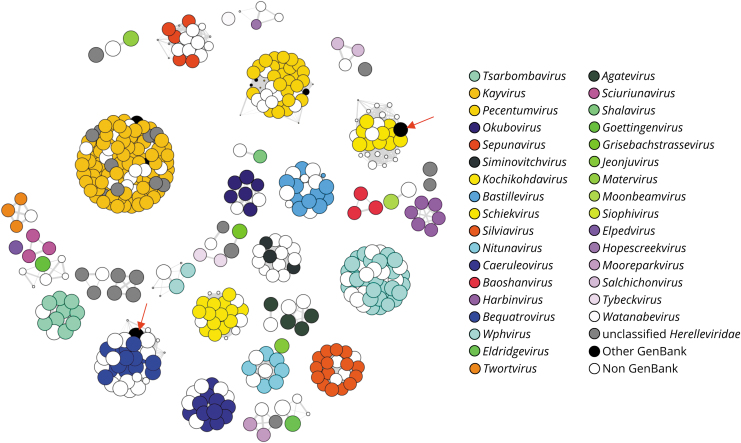
Figure 4 shows the clouds extracted with an intergenomic distance threshold of 0.15 and that contain at least one known member of the family Herelleviridae. The figure shows that all phages with known genus membership clustered together with phages from the same genus. The figure also highlights several instances of phages not classified in the family Herelleviridae that were tightly connected to clouds that represent different genera within this family.
FIG. 4.
Herelleviridae clouds colored by genus. Clouds containing at least one member of the family Herelleviridae were retrieved from the graph database, using an intergenomic distance threshold of 0.15. Nodes are colored based on genus memberships, according to the annotations in the corresponding GenBank files. Gray nodes correspond to Herelleviridae phages without genus affiliation. Black nodes correspond to GenBank phages classified in other families or not currently classified at the family level. White nodes refer to phages from any of the other databases used to create the graph database. The red arrows highlight examples discussed in the text of GenBank phages classified in other phage families.
The majority of these correspond to entries in NCBI currently labeled as “unclassified bacterial viruses,” such as the entries with accession numbers MW528836 and MN935200. However, the nodes highlighted with red arrows in Figure 4 are two examples of phages in our graph database that are classified in the family Myoviridae. While the record of the phage connected to the Schiekviruses was recently removed from GenBank (accession number LR760131), the phage connected to the Bequatroviruses is Bacillus phage BM5 that is currently classified in the family Myoviridae (accession number KT995479).
The taxonomic classification of Bacillus phage BM5 was not established by the ICTV. Thus, our results suggest that this phage represents an instance of a misannotated record within the GenBank, which might have resulted from the fact that members of the Herelleviridae were formerly classified as Myoviridae based on virion morphology.37 This observation is supported by results obtained using VIRIDIC,38 which demonstrated that the Bacillus phage BM5 is more closely related to members of the genus Bequatrovirus than to phages from all the genera currently classified within the family Myoviridae in NCBI (Supplementary Fig. S3).
Nevertheless, the ICTV established a minimum of 70% nucleotide identity over the full genome as the criterion for classifying phages in the same genus.16 As the largest percent identity observed between the Bacillus phage BM5 and a member of genus Bequatrovirus was 50.4% (Supplementary Fig. S3), our results do not support the inclusion of this phage within this genus. Nonetheless, the data in Supplementary Figure S3 demonstrated that Bacillus phage BM5 is more likely to be a member of the Herelleviridae rather than the Myoviridae family.
Figure 4 also illustrates many examples of phage genomic sequences from the other data sets in our graph database that connect to clouds representing a variety of genera within the family Herelleviridae. Among these are 31 entries from IMG/VR that are indeed currently classified as members of this family. The presence of the remaining phage genomic sequences in the retrieved clouds suggests that they could be members of the corresponding genera within the family Herelleviridae.
However, PhageClouds was not designed to be a tool for taxonomic classification of phage sequences. Based on the search of Herelleviridae clouds described here, it seems feasible that PhageClouds could help with the identification of phage clusters that correspond to taxa at the species or genus rank. Nevertheless, we would highly recommend the use of additional tools designed to provide taxonomic annotations of phage genomic sequences at the mentioned taxonomic ranks, especially if their use is advised by the ICTV (e.g., VIRIDIC and vConTACT2).16
Rapid searching of assembled viromes
To demonstrate the capacity to rapidly compare a large number of phages against the database, 971 previously assembled viral operational taxonomic units (vOTUs) from the U.K. coastal waters (MarineUKSouth data set) were used as a query.32 The wall time for comparison of all 971 vOTUs against the complete graph database with an intergenomic distance threshold of 0.20 was 1 h, 41 min, and 11 s (40 cores used). Previously only 75 (7.7%) of the MarineUKSouth vOTUs had been identified to cluster with a database of ∼14,000 known phages using vConTACT2 (wall time ∼48 h with 32 cores).32 Here, we were able to associate 517 (53.2%) vOTUs from the MarineUKSouth data set with similar phages in other environmental databases (Supplementary Fig. S4).
It could be theoretically possible to run this analysis with vConTACT2, but the practical aspects of comparing ∼640,000 phages using all-versus-all protein alignments prevent this from being run by an average user in a reasonable time frame. Instead, vConTACT2 could be used as a complementary analysis that attempts to identify more distant relationships between phage clouds, based on the genes shared between them. Furthermore, if a user is interested in analyzing a very large set of query phages (e.g., thousands of vOTUs), a sensible approach could entail the use of PhageClouds to quickly dereplicate and identify very close relatives from our graph database, thereby reducing the number of query phages to be analyzed with more sensitive alignment-based tools.
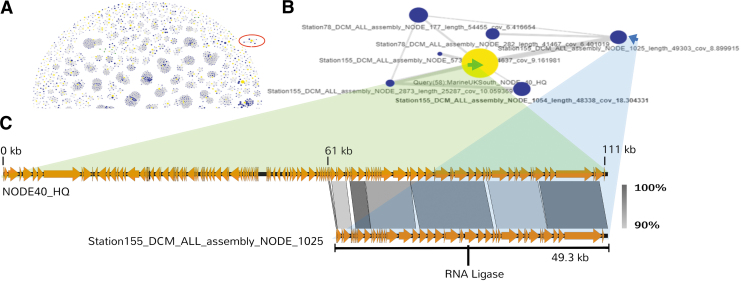
Of the 517 vOTUs that were associated with other phages, the closest relatives based on the calculated intergenomic distances were exclusively found in other marine viromes (Supplementary Fig. S4). Given that both viromes were constructed from seawater samples, this was not entirely unexpected. However, it was possible to quickly establish that very similar phages were found in geographically distant places. The vOTU NODE40_HQ was previously found to be present at station L4 and Plymouth Sound surface water.
PhageClouds identified a cluster of similar contigs in two different stations (78 and 155) from TARA Oceans (Fig. 5A, B). Three of the four contigs from station 155 could be aligned to NODE40_HQ with an ANI of between 96% and 98%. The alignment against NODE40_HQ demonstrated that both phages had a conserved synteny in gene order and content (Fig. 5C). The use of PhageClouds helped to rapidly establish that a vOTU found in the water samples off Plymouth coast was very similar to viral contigs from TARA station 155 (lat 54.5742, long −16.8345) that is ∼1000 km apart. Furthermore, the station 155 contigs are likely fragments from a larger more complete phage genome.
FIG. 5.
Searching phage clouds for a set of 971 marine vOTUs. The search was conducted over the complete graph database using an intergenomic distance threshold of 0.20. (A) Illustrates a section of the obtained phage clouds (the complete set is illustrated in Supplementary Figure S3). Yellow nodes represent vOTUs from the U.K. coastal water viromes, and blue nodes represent entries from the TARA Oceans data set included in the PhageClouds' graph database. (B) Shows a detailed view of the cloud highlighted in red from (A). The green and blue arrows point to the phage sequences present in the sequence alignment depicted in (C). vOTUs, viral operational taxonomic units.
Conclusion
PhageClouds is a computational resource that exploits the power and versatility of graph databases to offer a way of exploring the phage genomic diversity encompassed by published viromes. This tool will enable scientists to analyze their complete and draft phage genomes easily and efficiently by comparing them to a massive data set of ∼640,000 reference phage sequences. In addition, PhageClouds is suitable for exploring phage diversity under a host-centric perspective, facilitating the identification of different groups of phages with different infection strategies.
Furthermore, inspection of phage clouds searched for a specific host simplifies the identification of potentially incorrect host annotations, which are easily propagated by the association with closely related phage genomic sequences. PhageClouds is also suitable for identifying genomic fragments from viromes that might be part of the same phage genome and ultimately supports the recovery of more complete genomes.
Currently, PhageClouds' underlying algorithm and assumptions encompass equally all phage entries in our graph database, regardless of the type of nucleic acid in their genomes [i.e., dsDNA, ssDNA, ssRNA(+), ssRNA(−), dsRNA]. We acknowledge that phages with different types of nucleic acids might be associated with very different mutation rates, which suggests that the intergenomic distance thresholds used for finding clusters of related phages must differ between varying nucleic acid types.
Up to the time this article was prepared, and probably for the foreseeable future, most of the genomic sequences in public databases correspond to dsDNA phages, as evidenced by GenBank and IMG/VR, in which this type of phages correspond to ∼97% of their entries.7 With this in mind and considering that one of PhageClouds' main goals is to provide a resource for the quick and efficient comparison of a massive set of reference phage genomic sequences, we thought that the current functionality of our tool would constitute a good starting point. Nonetheless, future versions of PhageClouds could look deeper into defining bespoke intergenomic distance thresholds for groups of phages with different types of nucleic acids.
PhageClouds is hosted on our online server and is accessible from any web browser, and thus, users of this resource do not require any experience running software from the command line. PhageClouds is part of our online infrastructure at https://phagecompass.dk and https://phageclouds.dk. The tool accepts phage genomic sequences in FASTA format as input to query the graph database and allows users to examine precalculated phage clouds filtered by a specific host.
Supplementary Material
Acknowledgment
All calculations were performed on the Danish National Life Science Supercomputer Computerome.
Authors' Contributions
T.S.P. and M.R.J.C. conceived the idea of PhageClouds, and G.R.-P. generated and analyzed the data. A.M., A.R., and K.S. contributed to the design and content of the graph database. B.P. established the PhageClouds' computing and testing infrastructure. S.M., D.S., and A.M. generated the marine virome assemblies used for testing PhageClouds. All authors reviewed, proofread, and approved the final article. They hereby confirm that all the coauthors have reviewed and approved the submission of this article. They also confirm that this article has been submitted solely to this journal and is not published, in press, or submitted elsewhere.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
The work done by G.R.-P. was funded by the Foundation Idella project “Advancing the Medical Future Through Unlocking the Complex Genetic Material of the Amazon Rainforest.” The work done by K.S. was funded by the Mælkeafgiftsfonden project “Metacheese.”
Supplementary Material
References
- 1. Sulakvelidze A, Alavidze Z, Morris JG Jr. Bacteriophage therapy. Antimicrob Agents Chemother. 2001;45(3):649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carlet J, Collignon P, Goldmann D, et al. . Society's failure to protect a precious resource: Antibiotics. Lancet. 2011;378(9788):369–371. [DOI] [PubMed] [Google Scholar]
- 3. Rios AC, Moutinho CG, Pinto FC, et al. . Alternatives to overcoming bacterial resistances: State-of-the-art. Microbiol Res. 2016;191:51–80. [DOI] [PubMed] [Google Scholar]
- 4. Roux S, Brum JR, Dutilh BE, et al. . Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537(7622):689–693. [DOI] [PubMed] [Google Scholar]
- 5. Braga LPP, Soucy SM, Amgarten DE, et al. . Bacterial diversification in the light of the interactions with phages: the genetic symbionts and their role in ecological speciation. Front Ecol Evol. 2018;6:431. [Google Scholar]
- 6. Norman JM, Handley SA, Baldridge MT, et al. . Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160(3):447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roux S, Páez-Espino D, Chen I-MA, et al. . IMG/VR v3: An integrated ecological and evolutionary framework for interrogating genomes of uncultivated viruses. Nucleic Acids Res. 2021;49(D1):D764–D775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roux S, Hallam SJ, Woyke T, et al. . Viral dark matter and virus–host interactions resolved from publicly available microbial genomes. Elife. 2015;4:e08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gregory AC, Zayed AA, Conceição-Neto N, et al. . Marine DNA Viral Macro- and Microdiversity from Pole to Pole. Cell. 2019. [Epub ahead of print]; DOI: 10.1016/j.cell.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Camarillo-Guerrero LF, Almeida A, Rangel-Pineros G, et al. . Massive expansion of human gut bacteriophage diversity. Cell. 2021;184(4):1098–1109.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dion MB, Oechslin F, Moineau S. Phage diversity, genomics and phylogeny. Nat Rev Microbiol. 2020;18(3):125–138. [DOI] [PubMed] [Google Scholar]
- 12. Ondov BD, Treangen TJ, Melsted P, et al. . Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17(1):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pierce NT, Irber L, Reiter T, et al. . Large-scale sequence comparisons with sourmash. F1000Res. 2019;8:1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jain C, Rodriguez-RLM, Phillippy AM, et al. . High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9(1):5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mavrich TN, Hatfull GF. Bacteriophage evolution differs by host, lifestyle and genome. Nat Microbiol. 2017;2:17112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Turner D, Kropinski AM, Adriaenssens EM. A roadmap for genome-based phage taxonomy. Viruses. 2021;13(3). [Epub ahead of print]; DOI: 10.3390/v13030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brüssow H, Desiere F. Comparative phage genomics and the evolution of Siphoviridae: Insights from dairy phages. Mol Microbiol. 2001;39(2):213–222. [DOI] [PubMed] [Google Scholar]
- 18. Breitbart M, Miyake JH, Rohwer F. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol Lett. 2004;236(2):249–256. [DOI] [PubMed] [Google Scholar]
- 19. Dorigo U, Jacquet S, Humbert J-F. Cyanophage diversity, inferred from g20 gene analyses, in the largest natural lake in France, Lake Bourget. Appl Environ Microbiol. 2004;70(2):1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Low SJ, Džunková M, Chaumeil P-A, et al. . Evaluation of a concatenated protein phylogeny for classification of tailed double-stranded DNA viruses belonging to the order Caudovirales. Nat Microbiol. 2019;4(8):1306–1315. [DOI] [PubMed] [Google Scholar]
- 21. Corel E, Lopez P, Méheust R, et al. . Network-thinking: graphs to analyze microbial complexity and evolution. Trends Microbiol. 2016;24(3):224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iranzo J, Krupovic M, Koonin EV. A network perspective on the virus world. Commun Integr Biol. 2017;10(2):e1296614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lima-Mendez G, Van Helden J, Toussaint A, et al. . Reticulate representation of evolutionary and functional relationships between phage genomes. Mol Biol Evol. 2008;25(4):762–777. [DOI] [PubMed] [Google Scholar]
- 24. Shapiro JW, Putonti C. Gene co-occurrence networks reflect bacteriophage ecology and evolution. MBio. 2018;9(2). [Epub ahead of print]; DOI: 10.1128/mBio.01870-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jang H, Bolduc B, Zablocki O, et al. . Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat Biotechnol. 2019;37:1–8. [DOI] [PubMed] [Google Scholar]
- 26. Parks DH, Chuvochina M, Chaumeil P-A, et al. . A complete domain-to-species taxonomy for Bacteria and Archaea. Nat Biotechnol. 2020;38(9):1079–1086. [DOI] [PubMed] [Google Scholar]
- 27. ter Horst AM, Santos-Medellín C, Sorensen JW, et al. Minnesota peat viromes reveal terrestrial and aquatic niche partitioning for local and global viral populations. bioRxiv. 2020 [cited 2021 Jul 8]. p. 2020.12.15.422944. https://www.biorxiv.org/content/10.1101/2020.12.15.422944v1 (accessed July 8, 2021). [DOI] [PMC free article] [PubMed]
- 28. Tisza MJ, Buck CB. A catalog of tens of thousands of viruses from human metagenomes reveals hidden associations with chronic diseases. Proc Natl Acad Sci U S A. 2021;118(23). [Epub ahead of print]; DOI: 10.1073/pnas.2023202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gregory AC, Zablocki O, Zayed AA, et al. . The Gut Virome Database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe. 2020;28(5):724–740.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cook R, Hooton S, Trivedi U, et al. . Hybrid assembly of an agricultural slurry virome reveals a diverse and stable community with the potential to alter the metabolism and virulence of veterinary pathogens. Microbiome. 2021;9(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Babenko VV, Millard A, Kulikov EE, et al. . The ecogenomics of dsDNA bacteriophages in feces of stabled and feral horses. Comput Struct Biotechnol J. 2020;18:3457–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Michniewski S, Rihtman B, Cook R, et al. Identification of a new family of “megaphages” that are abundant in the marine environment. bioRxiv. 2021. http://biorxiv.org/lookup/doi/10.1101/2021.07.26.453748 (accessed August 3, 2021). [DOI] [PMC free article] [PubMed]
- 33. Sirén K, Millard A, Petersen B, et al. . Rapid discovery of novel prophages using biological feature engineering and machine learning. NAR Genom Bioinform. 2021;3(1):lqaa109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baker DN, Langmead B. Dashing: Fast and accurate genomic distances with HyperLogLog. Genome Biol. 2019;20(1):265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lood C, Danis-Wlodarczyk K, Blasdel BG, et al. . Integrative omics analysis of Pseudomonas aeruginosa virus PA5oct highlights the molecular complexity of jumbo phages. Environ Microbiol. 2020;22(6):2165–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Clokie MRJ, Blasdel BG, Demars BOL, et al. . Rethinking phage ecology by rooting it within an established plant framework. PHAGE. 2020;1(3):121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barylski J, Enault F, Dutilh BE, et al. . Analysis of spounaviruses as a case study for the overdue reclassification of tailed phages. Syst Biol. 2020;69(1):110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moraru C, Varsani A, Kropinski AM. VIRIDIC—a novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses. 2020;12(11). [Epub ahead of print]; DOI: 10.3390/v12111268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.