Abstract
Introduction: Bacteriophage plaque enumeration is a critical step in a wide array of protocols. The current gold standard for plaque enumeration on Petri dishes is through manual counting. However, this approach is not only time-consuming and prone to human error but also limited to Petri dishes with countable number of plaques resulting in low throughput.
Materials and Methods: We present OnePetri, a collection of trained machine learning models and open-source mobile application for the rapid enumeration of bacteriophage plaques on circular Petri dishes.
Results: When compared against the current gold standard of manual counting, OnePetri was ∼30 × faster. Compared against other similar tools, OnePetri had lower relative error (∼13%) than Plaque Size Tool (PST) (∼86%) and CFU.AI (∼19%), while also having significantly reduced detection times over PST (1.7 × faster).
Conclusions: The OnePetri application is a user-friendly platform that can rapidly enumerate phage plaques on circular Petri dishes with high precision and recall.
Keywords: plaque enumeration, computer vision, machine learning, Petri dish assays
Introduction
Bacteriophage (phage) enumeration is central to many assays and experiments, including the production of phage-based products and therapies, detection of bacterial infections, and biocontrol of foodborne pathogens.1 Many methods to quantify phage particles exist and include transmission electron microscopy (time-intensive, costly), flow cytometry (specialized equipment and high titers required), quantitative PCR (rapid, requires prior knowledge of phage genomic sequence), and epifluorescence microscopy (low throughput, high experimental variability, commonly used with environmental samples), among others.1–3
Despite the diverse repertoire of quantification techniques, the classical double agar overlay plaque assay protocol has long been the gold standard for phage enumeration, yielding visible phage plaques on a solid lawn of susceptible host bacteria,4 and where plaque formation is usually a direct result of phage infection and bacterial death.5 However, in addition to being time-intensive, this method is often limited to plates with a countable number of plaques, typically <300, and results may be inconsistent upon recount by different individuals.4
To this end, several image processing techniques for automating plaque counts have been created in recent years, many of which rely on contour or edge detection to identify plaques.6–9 Some of these tools require specific types of images, such as those obtained through fluorescence microscopy, to obtain plaque counts, increasing experimental complexity for the benefit of automation. Despite being designed to automate plaque counts, these tools often require user intervention and fine-tuning of image processing parameters to improve detection results and avoid false positives. Furthermore, most tools created for this purpose are designed to run on a desktop computer, breaking the workflow where counts would be followed by calculations and immediate experimental continuation.
We thus developed OnePetri, a mobile application using a collection of trained machine learning object detection models for the rapid enumeration of phage plaques on circular Petri dishes. Using images provided by the Howard Hughes Medical Institute's (HHMI) Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program,10 we successfully trained Petri dish and phage plaque object detection models, which have high recall and precision. Using machine learning and computer vision, we were able to build a flexible solution that can detect diverse plaque morphologies on different types of agar media, regardless of lighting conditions, without requiring any special image capture devices or fluorescent labeling.
When benchmarked against two other similar tools, CFU.AI (Apple App Store) and Plaque Size Tool (PST),8 OnePetri was significantly faster and more accurate, reproducibly detecting hundreds of overlapping and nonoverlapping phage plaques within a few seconds.
Materials and Methods
Image data set description
Over 12,000 image files were generously provided by the HHMI SEA-PHAGES program from the PhagesDB database10 for use in training machine learning models, most of which were of plaque assays in circular Petri dishes. Files that were not images (such as Microsoft Word documents, text files, and PDF files) were excluded from our data set. Some images were not Petri dish images, but rather transmission electron micrographs of phage isolates—these were excluded from our data set. Information on the size of the Petri dishes, growth media, bacterial host, and phage in each image was not provided. No other inclusion or exclusion criteria were applied.
Image data set curation, annotation, and preprocessing
After manual curation to remove images smaller than 1024 × 1024 pixels in size, as the plaques in these images would be too low resolution for model training, 10,261 images remained. A random subset of 98 images (75 training +23 validation) and 38 images (29 training +9 validation) were manually selected and used in preparing the Petri dish detection and plaque detection models, respectively. Most Petri dish images in the training and validation data sets had more than one Petri dish, while most plaque assay images had at least 100 plaques.
Images were manually annotated (bounding boxes drawn around each Petri dish or plaque to train the model with) using the Roboflow online platform (Roboflow, Inc., Des Moines, IA, USA). Before export for training, annotated images for the Petri dish detection model were preprocessed to fit within 1024 × 1024 pixels (maintaining aspect ratio). Annotated images for the plaque detection model were automatically preprocessed on the Roboflow platform as follows: tiled into 5 rows and 5 columns, tiles resized to fit within 416 × 416 pixels (maintaining aspect ratio). Tiling can help with detection of small objects (such as phage plaques) while decreasing training times by fragmenting a large image into multiple smaller tiles, resulting in each small object taking up a larger proportion of the tiled image than it did pretiling.
The following augmentations were applied to the plaque detection training data set, with a total of three outputs being produced per training example (tile): grayscale applied to 35% of images, hue shift between −45° and +45°, blur up to 2 pixels, mosaic. Image augmentations can increase the performance of object detection models by artificially increasing the diversity of images in a given training data set. After augmentations (training set only) and tiling (training + validation sets), the total number of tiles used for training and validating the plaque detection model was 2175 and 225, respectively.
Machine learning model training and validation
The trained PyTorch models were generated using the annotated, preprocessed, and augmented data set and the Ultralytics YOLOv5 training script (“You Only Look Once” version 5; Ultralytics, Los Angeles, CA, USA).11,12 The YOLO family of models was designed to rapidly detect and identify objects in images by drawing boxes (bounding boxes) around those that resemble objects the model was trained on.13 The YOLOv5 models use a modified Cross-Stage Partial Networks (CSPNet) backbone, which extracts useful features from images for downstream machine learning or inference.14
When working with object detection machine learning models, the training and inference processes are generally faster and uses less video random access memory (VRAM) on the graphics processing unit (GPU) when the training images are of lower resolution. While it would be ideal to train a model using images with their native multimegapixel resolution, this is not usually feasible given the VRAM limits on many GPUs. The following parameters were used to train the Petri dish detection model: 320 × 320 pixel image resolution (scale to fit each Petri dish image), 500 epochs, batch size of 16, YOLOv5s model (yolov5s.pt weights file), cache images enabled, default hyperparameters (hyp.scratch.yaml file).
The following parameters were used to train the plaque detection model: 416 × 416 pixel image resolution (scale to fit each tile), 500 epochs, batch size of 128, YOLOv5s model (yolov5s.pt weights file), cache images enabled, default hyperparameters (hyp.scratch.yaml file).
The generated “best.pt” weights files for the trained YOLOv5 models were converted to the Apple Core ML “mlmodel” file format using the coremltools Python package (version 4.1; https://github.com/apple/coremltools) in conjunction with a custom script provided by Hendrik Kueck (Pocket Pixels, Inc., Vancouver, Canada), which is available at the following GitHub repository link: https://github.com/pocketpixels/yolov5/blob/better_coreml_export/models/coreml_export.py.
iOS mobile application development and benchmarking
The mobile application for iOS was developed with the Swift programming language (version 5) using the Xcode 12.5.1 (build 12E507) integrated development environment (Apple, Inc., Cupertino, CA, USA) with a target SDK of iOS 13. Benchmarking of the application was carried out on the iPhone 12 minisimulator available within Xcode 12 (iOS 14.5, build 18E182), as well as on a physical iPhone 12 Pro running the same operating system build as the simulator. The iOS development simulator was running on a 2020 MacBook Air with M1 chip (8 CPU core and 8 GPU core variants) and 16 GB RAM, and which was connected to the power adapter.
Benchmarking OnePetri
Fifty images were randomly selected from the original unprocessed data set provided by the SEA-PHAGES team to be used in the benchmarking analysis. These images were not included in any of the model training or validation data sets for either the Petri dish or plaque YOLOv5 models, meaning this is the first time the models encounter these images for inference. Images were processed sequentially in OnePetri (version 1.0.1-8) and compared with the manual counting gold standard and with two other software programs. These include CFU.AI (version 1.4), a free mobile application on iOS and Android originally developed in 2019 to count bacterial CFU, and PST, a recently published Python tool for desktop computers that detects plaques and measures their size.8
Various methods were used to determine the speed at which the final output is obtained, depending on the tool. OnePetri uses code embedded within the compiled application to report runtime statistics to the debug console when running in debug mode on-device and in simulator. PST runtime was measured using the “time” command from the command line. CFU.AI runtime was approximated with a stopwatch, as the source code is not publicly available and there is no way to natively measure application runtime on iOS. All statistical analyses and data visualizations were performed using R (version 4.1.0, 2021-05-18, aarch64),15 ggplot2 (version 3.3.5), ggpubr (version 0.4.0),16 ggsignif (version 0.6.2),17 reshape2 (version 1.4.4),18 and tidyverse (version 1.3.1).19
Code and data availability
The Swift source code and Xcode project for OnePetri for iOS is available under the GNU General Public License v3.0 (GPL-3.0) at the following link: https://github.com/mshamash/OnePetri. The trained machine learning models (PyTorch and Apple MLModel formats) are available at the following link: https://github.com/mshamash/onepetri-models. The training data used for the initial versions of the Petri dish and plaque detection models are available under the Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0) license on the Roboflow Universe platform: https://universe.roboflow.com/onepetri/onepetri. The benchmarking data set, analysis scripts, and raw data are available at the following link: https://github.com/mshamash/onepetri-benchmark.
Results
Trained Petri dish and bacteriophage plaque object detection models are accurate and precise
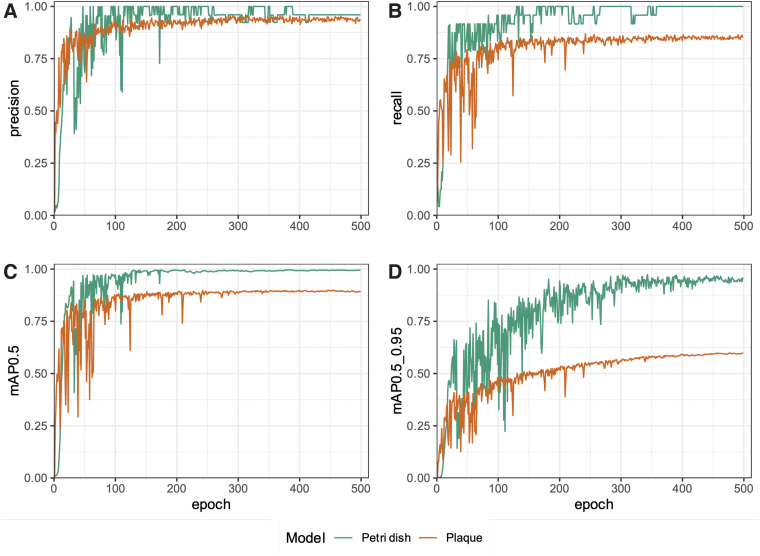
Due to the lack of publicly available trained object detection models able to identify Petri dishes and phage plaques, we first set out to train two such models. Using the YOLOv5 training scripts, we developed a model to detect common circular Petri dishes in a laboratory environment. The training and validation data sets comprised 75 and 23 images, respectively. The models trained using these images performed well and as such no additional images were added to the training data sets. After 500 epochs, the model achieved 96% precision (Fig. 1A) and 100% recall (Fig. 1B). In computer vision, the intersection over union (IoU), also known as the Jaccard index, is a measure to evaluate how well the detected object boundaries (obtained from testing the trained model) overlap with the actual object boundaries specified before training.20
FIG. 1.
The trained Petri dish and plaque object detection models have high recall and precision after 500 training epochs. Petri dish and plaque object detection model performance metrics were recorded throughout model training using the validation data sets to test the models after each round of training, over the 500 training epochs. The following metrics were recorded and included in the figure above: (A) precision, (B) recall, (C) mean average precision for an IoU of 0.50 (mAP0.5), and (D) mAP for IoU ranging from 0.50 to 0.95 (step size 0.05; mAP0.5_0.95). mAP, mean average precision; IoU, intersection over union.
The mAP@[0.5:0.95] (range of IoUs from 0.50 to 0.95, step size 0.05) and mAP@0.5 metrics are a measure of the model's mean average precision (mAP) at the indicated IoU threshold (or range of thresholds), where detections below the threshold are not counted. Models with high mAP@[0.5:0.95] values (typically >50%) are thus preferred, as this would suggest that the models' precision remains high despite increasingly stringent cutoff values for what can be considered a true-positive detection. The mAP at an IoU of 0.50 (mAP@0.5) was 99.5% (Fig. 1C), while the mAP@[0.5:0.95] was 95.7% (Fig. 1D).
Next, to be able to detect a wide variety of plaque morphologies on diverse agar colors, we developed a model to detect phage plaques using our tiled and augmented initial data set. The training and validation data sets comprised 2175 and 225 tiles, respectively. After 500 epochs, the model achieved 93.6% precision (Fig. 1A) and 85.6% recall (Fig. 1B). The mAP@0.5 was 89.5% (Fig. 1C), while the mAP@[0.5:0.95] was 59.9% (Fig. 1D).
All performance metrics for both models plateaued after around 300 epochs, indicating that it may have been sufficient to stop training at this point.
An image processing pipeline for rapid Petri dish detection and bacteriophage plaque enumeration
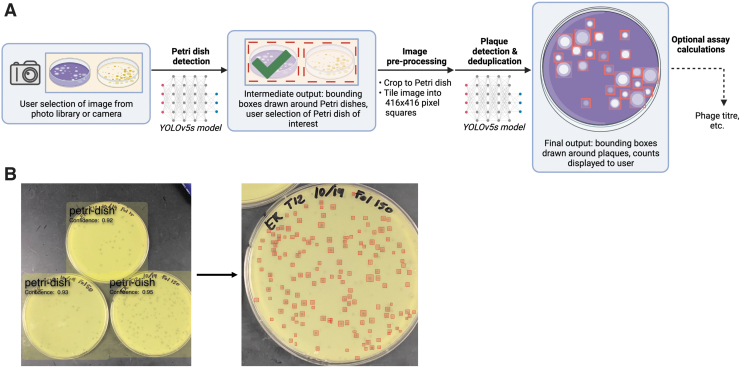
We then set out to create a mobile application wrapper for the trained models above, to allow for rapid phage plaque enumeration and assay calculations with a user-friendly interface while in the laboratory environment, without having to transfer images from a mobile phone or camera to a computer. The OnePetri mobile application was developed to fulfill this purpose and is currently available for download on the Apple App Store for free. Briefly, upon launching OnePetri, the user is first prompted to select an image for analysis (from photo library or to be taken with camera). The Petri dishes are then identified, and the user selects the Petri dish of interest to proceed with plaque enumeration analysis.
This approach allows users to serially analyze multiple Petri dishes from a single image, increasing throughput. The image is cropped to the Petri dish boundaries and tiled into overlapping tiles of 416 × 416 pixels in size, where plaques are then identified serially on each tile. Finally, plaque deduplication occurs to account for plaques that may have been identified twice on overlapping tiles, and the final counts are returned to the user (Fig. 2). In addition, OnePetri for iOS can automatically perform the necessary calculations to obtain phage titer from Petri dishes of multiple phage dilutions as needed by the user.
FIG. 2.
Overview of the OnePetri mobile application image processing pipeline for Petri dish detection and plaque enumeration. (A) Upon selecting an image for analysis, all circular Petri dishes are detected using the trained Petri dish detection model. The user selects the Petri dish they wish to analyze, and the image is cropped to fit that Petri dish of interest, tiled into overlapping 416 × 416 pixel squares, and resulting tiles are fed serially to the trained plaque detection model. The detected plaques are deduplicated to account for the overlapping tiles, which may have resulted in some plaques being detected twice, and the final annotated image is presented to the user. Optionally, the user may proceed with assay calculations within the application directly (e.g., to determine phage titer) using the obtained plaque counts and dilution volumes. Figure created with BioRender. (B) Example image processed in the OnePetri mobile application. Three Petri dishes are detected with high confidence scores. Upon selecting a Petri dish, 155 phage plaques are enumerated and highlighted with a red box. Image provided by the Howard Hughes Medical Institute's Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program.
OnePetri rapidly and precisely enumerates bacteriophage plaques on a mobile device
To compare OnePetri's accuracy with other currently available tools with a similar purpose, we benchmarked OnePetri against manual counts, as well as PST and CFU.AI, using a collection of 50 test images that the trained models would be seeing for the first time. One image had too many plaques to count manually (>1000) and was excluded from further analysis, despite OnePetri returning a value of 1641 plaques, a seemingly accurate value.
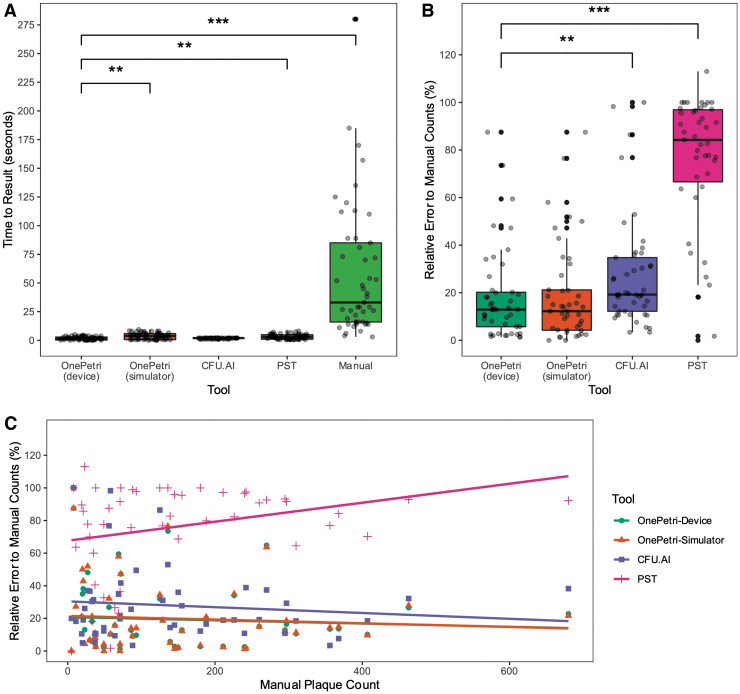
OnePetri was benchmarked directly on-device and using the iOS development simulator included with the Xcode IDE on macOS. The time to result was significantly shorter when using OnePetri on-device versus PST (p = 0.0029, nonparametric one-way analysis of variance [ANOVA] with Wilcoxon rank-sum test and Benjamini/Hochberg correction for multiple comparisons), manual counts (p < 0.001), and OnePetri in the iOS simulator (p = 0.0041) (Fig. 3A).
FIG. 3.
The OnePetri mobile application rapidly detects plaques with minimal error compared with other tools. OnePetri (on-device and in the iOS simulator) was benchmarked against CFU.AI, PST, and manual counts. The (A) total time to obtain plaque counts (in seconds) and (B) the relative error rate of each tool (%), comparing counts from the tool to gold standard manual counts, were calculated and compared (n = 49 images in the benchmarking data set analyzed using each tool, **p ≤ 0.01, ***p ≤ 0.001, nonparametric one-way ANOVA using Wilcoxon rank-sum test with Benjamini/Hochberg correction for multiple comparisons). (C) The relative error rate of each tool (%) was compared with the manual plaque count value. The resulting correlations are overlaid. Note that the OnePetri-Device (green circle) and OnePetri-Simulator (orange triangle) correlation lines mostly overlap. ANOVA, analysis of variance; CFU, colony forming units; PST, Plaque Size Tool.
No significant difference in time to result was seen when comparing OnePetri (on-device) with CFU.AI (p = 0.4833). The mean time to result for OnePetri on-device was 1.91 s, OnePetri in simulator was 3.76 s, CFU.AI was 1.80 s, PST was 3.21 s, and manual counts were 57.98 s (Fig. 3A). OnePetri on-device was ∼2 × faster than the iOS development simulator, 1.7 × faster than PST, and 30 × faster than manual counts, on average.
We compared the relative percent error of each approach with manual counts to get a sense of the overall accuracy of each tool. This value was calculated by taking the absolute value of the difference between actual and expected plaque counts, dividing by the expected plaque count, and multiplying by 100%. OnePetri on-device had the lowest median relative error of 12.90%, with a rate of 12.26% in the simulator, while CFU.AI and PST had median relative errors of 19.23% and 85.71%, respectively, with these differences remaining significant after correcting for multiple comparisons (Fig. 3B, nonparametric one-way ANOVA with Wilcoxon rank-sum test and Benjamini/Hochberg correction for multiple comparisons).
Finally, we investigated whether the relative error rates of each tool correlated with the true plaque counts of the images (Fig. 3C). All four of the calculated Pearson correlations were very weak, indicating no strong relationship between any tool's error rate and the number of plaques on the Petri dish: OnePetri on-device (ρ = −0.07, R2 = 0.01), OnePetri in iOS simulator (ρ = −0.08, R2 = 0.01), CFU.AI (ρ = −0.11, R2 = 0.01), PST (ρ = 0.04, R2 = 0.001).
Discussion
Phage enumeration through manual plaque counting has long been the gold standard in the field, despite the time-intensive nature of this approach. Over the years, several tools have been developed to help automate this approach, with varying levels of user intuitiveness and accuracy. However, most tools have been developed for desktop computers, requiring Petri dish images to be uploaded to the computer for analysis, removing the user from their workflow in the laboratory. To this end, we developed OnePetri, a set of object detection models, and a mobile application, which can perform rapid plaque counting in a high-throughput manner, directly in a laboratory environment, and which will improve regularly with ongoing model updates.
Using a diverse training data set of Petri dish and plaque assay images from the HHMI SEA-PHAGES program, we were able to train YOLOv5s object detection models that could detect Petri dishes and phage plaques with high precision and recall (Fig. 1). When benchmarked on a set of 50 images that the models have not been previously exposed to, OnePetri running on a physical iOS device was significantly faster for plaque counting than PST and manual counting (Fig. 3A).
Using mean values for comparison, OnePetri on-device was ∼30 × faster than manual counts, representing significant time savings for the user, especially when analyzing multiple Petri dishes. Notably, OnePetri on-device was also significantly faster than using the iOS simulator included in the Xcode development suite, highlighting the need to benchmark iOS applications on-device rather than in simulators for accurate real-world values. No significant difference was seen in inference times between OnePetri on-device and CFU.AI.
Despite having quicker detection times, OnePetri significantly outperformed CFU.AI and PST in terms of relative error rates when comparing plaque counts from each tool to the true manual counts using the benchmarking image data set (Fig. 3B). The median error rate for OnePetri on-device (12.90%) was ∼1.5 × and 6.6 × lower than CFU.AI (19.23%) and PST (85.71%), respectively. No significant difference was observed between OnePetri error rates on-device versus in the iOS simulator.
While a median error rate of 12.90% is quite low relative to the other tools, there remains room for improvement. Given the current error rate, we recommend that this version of OnePetri be used only when this level of error is acceptable for the assay at hand, and users should first evaluate how OnePetri performs with the phage/host pairings before considering replacing manual counts entirely. There was essentially no correlation between the relative error rates of each tool and the number of plaques per Petri dish (Fig. 3C).
During benchmarking, we remarked that the Petri dish's background surface (e.g., on a dark laboratory bench, or held up against room light) can affect results, with more accurate counts being obtained when the Petri dish was against a dark surface. Furthermore, certain plaque morphologies, such as the “bull's eye,” were sometimes incorrectly detected with the current version of OnePetri's plaque detection model. CFU.AI also often struggled with this plaque morphology, while PST was able to detect about half of the “bull's eye” plaques on the images tested. OnePetri does require that images be of sufficient resolution for individual plaques to be distinguishable by the machine learning models, although we did not test this directly.
However, all supported devices (modern smartphones from past 5 to 7 years) have camera resolutions that are well beyond what would likely be the minimum image size for reliable results.
The user-friendly approach we developed for image analysis on a mobile device allows users to serially analyze multiple Petri dishes within a single image, increasing throughput and reducing the time to results (Fig. 2). Multiple detection parameters (object detection confidence thresholds and plaque deduplication overlap thresholds) can be easily changed within the application itself, allowing users to fine-tune the application's performance to their unique setup, should the default values not be ideal. The recently released PST also allows for fine-tuning of detection parameters; however, it is not as user-friendly, and requires the user to be comfortable with installing and running applications from the command line on a computer, and is only able to process one Petri dish per image.
Our unique machine learning approach for Petri dish and plaque detection allows for improved accuracy over traditional image processing approaches, such as those used in PST. In addition, the object detection models we developed can be improved upon further using user-submitted data due to the inherent trained nature of machine learning model.
The phage titration assay is the only assay currently supported within the mobile application. Upon entering the volume of sample plated and the corresponding plate dilutions, the initial phage titer is calculated based on the number of plaques present on serially diluted plates. Support for additional phage and bacterial assays is planned for late-2021/early-2022. OnePetri does not currently support spot assays for approximating phage titer and requires that each Petri dish contain phage of a single dilution, as all plaques on a given dish are counted assuming they are from the same diluted sample.
Unlike PST, OnePetri does not currently directly measure or infer individual plaque size. This may be added in a future version of OnePetri, along with support for exporting a summary report of all Petri dishes analyzed in a given session. A version of the OnePetri mobile application that supports Android devices is currently under development and should be released early-2022.
Conclusion
We present a pair of trained object detection machine learning models for the identification of Petri dishes and phage plaques, as well as OnePetri, a mobile application for iOS that leverages these models for the rapid and reproducible enumeration of phage plaques. OnePetri is now freely available to download from the Apple App Store on iOS. The application source code, trained models, training data, benchmarking data set, and analysis scripts are all available for download under open-source licenses. When compared with the manual counting gold standard, as well as CFU.AI and PST, OnePetri had minimal relative error with significantly lower time to results.
Acknowledgments
We are especially grateful to the Howard Hughes Medical Institute's Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program for proving the large collection of Petri dish and plaque assay images to be used for model training and testing—this project would not have been possible without their generous contribution. We would also like to thank Hendrik Kueck (Pocket Pixels, Inc.) for his guidance on topics in computer vision and machine learning object detection model training, as well as Mohamed Traore (Roboflow, Inc.) and the entire Roboflow team for their support and for providing access to their image annotation and preprocessing platform. We would like to acknowledge those who helped test and provided feedback on OnePetri for iOS during the beta testing period, before its official release.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC), the Fonds de recherche du Québec—Nature et technologies (FRQNT; #282402), and AbbVie Canada to M.S.
References
- 1. Ács N, Gambino M, Brøndsted L. Bacteriophage enumeration and detection methods. Front Microbiol. 2020;11:594868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen F, Lu J, Binder BJ, et al. . Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR Gold. Appl Environ Microbiol. 2001;67(2):539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Noble RT, Fuhrman JA. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol. 1998;14:113–118. [Google Scholar]
- 4. Kropinski AM, Mazzocco A, Waddell TE, et al. . Enumeration of bacteriophages by double agar overlay plaque assay. In: Clokie MRJ, and Kropinski AM; eds. Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. Totowa, NJ: Humana Press; 2009: 69–76. [DOI] [PubMed] [Google Scholar]
- 5. Abedon ST, Yin J. Bacteriophage plaques: theory and analysis. In: Clokie MRJ, and Kropinski AM; eds. Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. Totowa, NJ: Humana Press; 2009: 161–174. [DOI] [PubMed] [Google Scholar]
- 6. Culley S, Towers GJ, Selwood DL, Henriques R, et al. . Infection counter: Automated quantification of in vitro virus replication by fluorescence microscopy. Viruses. 2016;8(7):201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Katzelnick LC, Coello Escoto A, McElvany BD, et al. . Viridot: An automated virus plaque (immunofocus) counter for the measurement of serological neutralizing responses with application to dengue virus. PLoS Negl Trop Dis. 2018;12(10):e0006862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trofimova E, Jaschke PR. Plaque Size Tool: An automated plaque analysis tool for simplifying and standardising bacteriophage plaque morphology measurements. Virology. 2021;561:1–5. [DOI] [PubMed] [Google Scholar]
- 9. Cacciabue M, Currá A, Gismondi MI. ViralPlaque: A Fiji macro for automated assessment of viral plaque statistics. PeerJ. 2019;7:e7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Russell DA, Hatfull GF. Phages DB: the actinobacteriophage database. Bioinformatics. 2017;33(5):784–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paszke A, Gross S, Massa F, et al. . PyTorch: An imperative style, high-performance deep learning library. In: Wallach H, Larochelle H, Beygelzimer A, d'Alché-Buc F, and Fox EGarnett R; eds. Advances in Neural Information Processing Systems. Vancouver, BC, Canada: Curran Associates, Inc., 2019. [Google Scholar]
- 12. Jocher G, Stoken A, Borovec J, et al. . ultralytics/yolov5: v5.0—YOLOv5-P6 1280 models, AWS, Supervise.ly and YouTube integrations. Geneva, Switzerland: Zenodo; 2021. [Google Scholar]
- 13. Redmon J, Divvala S, Girshick R, et al. . You Only Look Once: Unified, Real-Time Object Detection. Ithaca, NY, USA: arXiv; 2016. [Google Scholar]
- 14. Wang C-Y, Liao H-YM, Yeh I-H, et al. . CSPNet: A New Backbone that can Enhance Learning Capability of CNN. Ithaca, NY, USA: arXiv; 2019. [Google Scholar]
- 15. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2021. [Google Scholar]
- 16. Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- 17. Constantin A-E, Patil I. ggsignif: R Package for Displaying Significance Brackets for ‘ggplot2’. PsyArxiv; 2021.
- 18. Wickham H. Reshaping data with the reshape Package. J Stat Softw. 2007;21(12):1–20. [Google Scholar]
- 19. Wickham H, Averick M, Bryan J, et al. . Welcome to the tidyverse. J Open Source Softw. 2019;4(43):1686. [Google Scholar]
- 20.Rezatofighi H, Tsoi N, Gwak J, et al. Generalized intersection over union: A metric and a loss for bounding box regression. 2019. IEEECVF Conference on Computer Vision and Pattern Recognition. New York, NY, USA: CVPR. 2019. pp 658–666.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Swift source code and Xcode project for OnePetri for iOS is available under the GNU General Public License v3.0 (GPL-3.0) at the following link: https://github.com/mshamash/OnePetri. The trained machine learning models (PyTorch and Apple MLModel formats) are available at the following link: https://github.com/mshamash/onepetri-models. The training data used for the initial versions of the Petri dish and plaque detection models are available under the Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0) license on the Roboflow Universe platform: https://universe.roboflow.com/onepetri/onepetri. The benchmarking data set, analysis scripts, and raw data are available at the following link: https://github.com/mshamash/onepetri-benchmark.