Abstract
Background: Enterobacter spp. are opportunistic pathogens that cause nosocomial infections. Bacteriophages could be used to treat antibiotic-resistant Enterobacter infections.
Materials and Methods: We used 10 genetically diverse clinical Enterobacter spp. isolates to identify lytic bacteriophages in hospital and municipal wastewater. Comparative genomics was performed on host bacterial isolates and isolated phages. Activity of each phage against all 10 host isolates was determined. We also tested phage activity against paired isolates from two patients who developed ceftazidime–avibactam resistance.
Results: Bacteria belonged to three Enterobacter species and Klebsiella aerogenes. We isolated 12 bacteriophages, most of which belonged to the Myoviridae and Autographiviridae families. Most phages were able to lyse multiple bacterial isolates, and many lysed isolates of different species. Ceftazidime–avibactam-resistant isolates were still phage susceptible, and one isolate showed increased susceptibility compared with the parent isolate.
Conclusion: The phages we isolated expand the diversity of Enterobacter-targeting phages, and could be useful for treating antibiotic-resistant Enterobacter infections.
Keywords: bacteriophage, Enterobacter spp, antibiotic resistance, phage therapy
Background
Enterobacter spp. are Gram-negative opportunistic pathogens that cause hospital-acquired infections. Although Enterobacter spp. make up only ∼5–7% of nosocomial bacterial infections, they are more frequently present in the ICU, having the third highest pathogen prevalence in respiratory tract infections (11.1%) and the fourth highest prevalence of surgical wound infections (10.3%).1 Enterobacter spp. also frequently cause antibiotic-resistant infections, and are named among the ESKAPE pathogens.2
The Enterobacter species E. cloacae and E. aerogenes (which has recently been redesignated Klebsiella aerogenes) are especially prone to developing antibiotic resistance, and carbapenem resistance has been previously noted among these strains.3 Carbapenem resistance has developed across Enterobacter through mutations that cause overexpression of the ampC gene,4,5 and through transmission of mobile genetic elements that carry carbapenemases.6 As carbapenems are typically used as last-resort antibiotics to treat infections,4,6 finding alternative ways to treat antibiotic-resistant Enterobacter spp. infections is important to prevent further morbidity and mortality.
One potential solution to antibiotic resistance across Enterobacter spp. is the use of bacteriophage therapy. Bacteriophages can be used either as an alternative to, or in combination with, antibiotics.7 Previous studies have shown that bacteriophages can be used to effectively treat a wide variety of bacterial infections, including those caused by enteric pathogens such as Klebsiella spp. and Escherichia coli.8,9 Compared with other members of the Enterobacteriaceae, Enterobacter phage therapy is less studied and much less attention has focused on the potential utility of Enterobacter-targeting phages; however, previous research suggests that bacteriophages can effectively infect and kill Enterobacter spp.10 Because of this, bacteriophages may be a promising therapeutic approach to treat antibiotic-resistant Enterobacter infections.
In this study, we isolated and characterized 12 bacteriophages that are capable of lysing genetically diverse clinical Enterobacter spp. isolates. We used whole genome sequencing and comparative genomics to study bacterial host and phage diversity, and tested the susceptibility of each host to each phage. We also investigated how phage susceptibility changed when Enterobacter spp. isolates evolved resistance to ceftazidime–avibactam in vivo during human infection. Our findings suggest that phages may be a viable approach for treating antibiotic-resistant Enterobacter spp. infections, and that the preemptive isolation of Enterobacter-targeting phages may be an effective mitigation strategy when Enterobacter spp. infections are likely to become highly antibiotic resistant.
Materials and Methods
Bacterial isolates
Nine different Enterobacter spp. isolates and one K. aerogenes isolate used for phage screening were collected from patients at the University of Pittsburgh Medical Center. For two patients, additional isolates with evolved ceftazidime–avibactam resistance were also collected.11 Because K. aerogenes has only recently been reclassified from E. aerogenes,3 we primarily refer to all isolates as Enterobacter isolates for clarity. Clinical isolates were collected from patients as part of routine clinical care and were stored as deidentified pure bacterial cultures.
Isolate collection was approved by the University of Pittsburgh Institutional Review Board (Protocols nos. PRO14080060 and PRO19110005). All isolates were cryopreserved in brain heart infusion (BHI) medium with 16.7% glycerol, and were stored at −80°C. Antimicrobial susceptibility testing against meropenem and ceftazidime–avibactam was performed using Sensititre Gram-Negative Susceptibility Testing Plates (Thermo Fisher Scientific, Waltham, MA) following the manufacturer's protocol. Susceptibility calls (sensitive/intermediate/resistant) were determined following CLSI guidelines.12
Wastewater collection and processing
Hospital wastewater was collected from the main sewer outflow of a Pittsburgh area hospital. Municipal wastewater was collected from two different municipal wastewater treatment facilities located in the Pittsburgh metropolitan area. Each wastewater sample was centrifuged at 3200 g for 20 min, and the supernatant was filtered using a 0.22 μm filter and concentrated by centrifuging in an Amicon filter unit (MilliporeSigma, Burlington, MA) at 3200 g for 15 min.
Bacteriophage screening, passaging, and high-titer stock creation
To isolate bacteriophages present in filtered and concentrated wastewater, we used a soft agar overlay screening approach.2 For bottom agar plates, BHI with 1.5% agar was used, to which 1 mM CaCl2 and 1 mM MgCl2 were added. Bacterial isolates were inoculated into BHI medium and grown overnight at 37°C with shaking. In total, 100 μL of each bacterial isolate was mixed with 100 μL of wastewater, incubated at room temperature for 5 min, mixed with 5 mL of molten top agarose (BHI with 0.5% agarose, 1 mM CaCl2, and 1 mM MgCl2), and plated onto a bottom agar plate. The plates were incubated overnight at 37°C and examined the next day to identify clear lytic bacteriophage plaques.
Bacteriophages were passaged by picking an individual plaque with a sterile pipette tip and incubating with 100 μL of SM buffer (50 mM TrisCl pH 7.5, 100 mM NaCl, 8 mM MgSO4) overnight at 37°C. After incubation, serial 10-fold dilutions were made in SM buffer, and 2 μL of each dilution was spotted onto a plate containing 5 mL top agarose mixed with 100 μL bacterial culture, layered on a bottom agar plate. After overnight incubation at 37°C, an individual plaque was picked and passaged again. Each phage was passaged at least four times before the generation of high-titer stocks.
To create a high-titer stock, a single plaque was picked and incubated in 100 μL of SM buffer overnight. The next day, 100 μL of an overnight host bacteria culture was added to the phage/SM buffer mixture, incubated for 10 min at room temperature, mixed with 10 mL of top agarose, and 5 mL each was plated onto two bottom agar plates. These plates were incubated overnight at 37°C, checked for complete bacterial lysis, then flooded with 10 mL of SM buffer and incubated for 1 h at 37°C. SM buffer was removed from each plate, pooled together, spun down at 3200 g for 20 min, and filtered through a 0.22 μm filter. Phage-containing lysates were then extracted once with 0.1 volumes chloroform followed by extraction three times with 0.4 volumes of 1-octanol. Purified phages were then stored at 4°C.
Genome sequencing and analysis
Bacterial genomic DNA was extracted from 1 mL overnight cultures of each isolate grown in BHI medium using a Qiagen DNeasy Blood and Tissue Kit (Qiagen, Germantown, MD) following the manufacturer's protocol. Genome sequencing was performed at the Microbial Genome Sequencing Center (MiGS) at the University of Pittsburgh. Sequencing libraries were prepared with a Nextera kit (Illumina, San Diego, CA), and libraries were sequenced on a NextSeq 550 using 150-bp paired-end reads. Genomes were assembled with SPAdes v3.12.013 using default settings. Bacterial species were determined by examining the 16S rRNA locus as well as nucleotide BLAST of other genomic regions.
After assembly, genomes were annotated with Prokka v1.14.514 and compared with Roary v3.11.2.15 A core genome phylogenetic tree was generated using RAxML16 with the GTRCAT substitution model and 1000 iterations, and iTOL was used to visualize the phylogenetic tree.17 PHASTER was used to identify prophages,18 and those predicted to be intact and questionable were included, whereas incomplete prophages were excluded from analysis. Clustered regularly interspaced short palindromic repeats (CRISPR) presence was determined using CRISIPRCasFinder,19 and genomes were identified as CRISPR positive if they had CRISPR loci flanked by annotated Cas genes.
Mutations were identified between paired clinical isolates from the same patient with CLC Genomics Workbench v11 (Qiagen), by mapping reads from the resistant isolate to the annotated genome of the parent isolate. Variants were identified with a read depth cutoff of 10 and a variant frequency cutoff of 90%. Bacterial genome sequence data have been deposited in NCBI under BioProjects PRJNA577956 and PRJNA777034.
To extract bacteriophage genomic DNA, 500 μL of phage lysate was mixed with 500 μL of phenol–chloroform–isoamyl alcohol (25:24:1), then the mixture was vortexed and centrifuged at 16,000 g for 1 min. A 1 × volume of chloroform was added to the upper aqueous phase, and the sample was again vortexed and centrifuged for 1 min at 16,000 g. To the new upper aqueous phase, 1 μL glycogen, 0.1 × volume 3 M sodium acetate, and 2.5 × volume 100% ethanol were added, and the sample was incubated overnight at −20°C. The next day, samples were centrifuged at 4°C for 30 min at 16,000 g. The supernatant was removed, washed with 150 μL 70% ethanol, spun for 2 min at 16,000 g, the new supernatant was discarded, and the pellet was resuspended in 100 μL nuclease-free water.
DNA was then quantified with a Qubit fluorimeter (Thermo Fisher Scientific). Bacteriophage genomes were sequenced at MiGS using the same protocol already described. Genomes were assembled with SPAdes v3.12.013 using default settings, and phage contigs were extracted from each assembly and separated from contaminating host bacterial sequences by examining the differential read coverage of each contig, and with nucleotide BLAST.
Phage genomes were compared with one another and with publicly available genomes using nucleotide BLAST, Mauve,20 and PHASTER.18 Phage genomes were annotated with RAST,21 and were queried for the presence of antibiotic resistance and virulence genes using ResFinder,22 VirulenceFinder,23 and CARD.24 Assembled phage genomes were deposited in GenBank with accession nos. OL355123–OL355134.
Bacteriophage host range
To determine the host range of each isolated phage, every phage was tested on every bacterial isolate to determine its infectivity profile. Serial 10-fold dilutions were created from each bacteriophage stock in SM buffer, and 2 μL of each dilution was spotted onto a plate containing 5 mL top agarose mixed with 100 μL bacterial culture and layered on top of a bottom agar plate. This plate was incubated overnight at 37°C, and the next day plaques were counted, and infectivity was determined by calculating the plaque-forming units per mL of each phage lysate against each bacterial isolate.
Results
Isolation of bacteriophages
To determine whether hospital and/or municipal wastewater contained Enterobacter-targeting bacteriophages, we conducted a bacteriophage screen using 10 clinical Enterobacter spp. isolates (Table 1). For each water sample, bacteria were incubated with filtered wastewater, then top agar was added and overlaid on top of a bottom agar plate (see Materials and Methods for details). This method successfully yielded clear lytic plaques from multiple different samples, and phages were isolated and propagated by picking and passaging individual plaques. In total, 12 bacteriophages were isolated and studied further.
Table 1.
Bacterial Isolates Used in This Study
| Isolate | Species | Genome length (bp) | %GC | MEM | CZA | Used for phage screening | CRISPR ± | Prophages | References |
|---|---|---|---|---|---|---|---|---|---|
| 607 | Enterobacter cloacae | 4,580,599 | 55.6 | S | S | Yes | − | 2 | This study |
| 608 | E. cloacae | 4,862,673 | 55.5 | S | S | Yes | + | 4 | This study |
| 609 | Enterobacter dissolvens | 5,184,790 | 54.8 | S | S | Yes | − | 4 | This study |
| 649 | E. cloacae | 4,909,029 | 55.0 | S | S | Yes | + | 6 | This study |
| 955 | Klebsiella aerogenes | 5,152,230 | 55.1 | R | S | Yes | + | 3 | This study |
| 971 | E. cloacae | 4,887,763 | 55.3 | S | S | Yes | − | 5 | This study |
| 973 | E. cloacae | 4,733,519 | 55.5 | S | S | Yes | + | 3 | This study |
| 1665 | Enterobacter kobei | 5,399,822 | 54.7 | S | S | Yes | − | 12 | This study |
| Surv186 | E. cloacae | 4,568,799 | 55.7 | I | S | Yes | − | 1 | 33 |
| Surv196 | E. cloacae | 4,623,312 | 55.7 | R | R | No | − | 1 | 33 |
| Ent634 | E. cloacae | 4,957,562 | 55.4 | S | S | Yes | + | 5 | 33 |
| Ent630 | E. cloacae | 4,958,359 | 55.4 | R | R | No | + | 5 | 33 |
CRISPR, clustered regularly interspaced short palindromic repeats; CZA, ceftazidime–avibactam; MEM, meropenem; S/I/R, sensitive/intermediate/resistant according to CLSI criteria.12
Bacterial genomic analysis
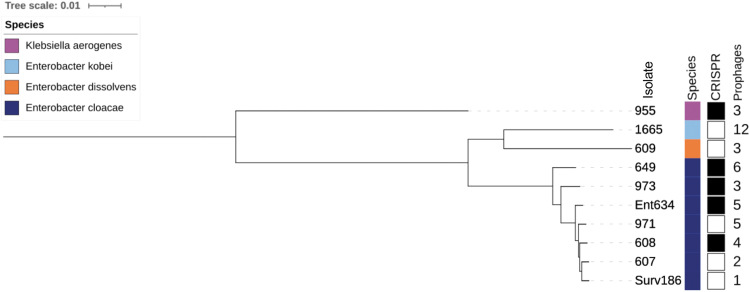
To determine the genetic diversity of the 10 Enterobacter spp. isolates used for phage screening, the genome of each isolate was sequenced on the Illumina platform (Table 1). We determined the species, the presence of CRISPR loci, and the number of prophages present in each genome using online bioinformatics tools (Supplementary Table S1).18,19 We also made a core genome phylogeny of all 10 isolates (Fig. 1). Seven of the isolates used for screening belonged to the E. cloacae complex, whereas the remaining isolates were found to belong to Enterobacter dissolvens, Enterobacter kobei, and K. aerogenes (Table 1 and Fig. 1).
FIG. 1.
Genome phylogeny of 10 clinical Enterobacter spp. isolates used for bacteriophage screening. The core genome phylogeny was built with RAxML and is annotated with isolate name, species, presence of CRISPR-Cas genes (black, present; white, absent), and the number of predicted prophages in each isolate genome. CRISPR, clustered regularly interspaced short palindromic repeats.
Half of the 10 isolates contained CRISPR-Cas genes, including 4 of the 7 E. cloacae isolates and the single K. aerogenes isolate. Bacterial isolate genomes contained varying numbers of prophages, ranging from 1 prophage in isolate Surv186 to 12 prophages in isolate 1665 (Table 1, Fig. 1, and Supplementary Table S1). We found no significant difference between the presence or absence of CRISPR-Cas genes and the number of prophages present in an isolate's genome (two sample t-test, [log-transformed] p = 0.72); however this may be due to a small sample size.
Bacteriophage genomic analysis
To determine the genetic diversity of the isolated bacteriophages, genomic DNA was extracted from each phage and was sequenced on the Illumina platform. We analyzed the genome length, GC content, predicted family, and predicted subfamily of each isolated phage (Table 2). Overall, we isolated 12 bacteriophages from three different families (Myoviridae, Autographiviridae, and Siphoviridae) and three different subfamilies (Table 2). When broken up by water source, there were subtle differences among the diversity of phages isolated.
Table 2.
Genomic Diversity of 12 Enterobacter-Targeting Bacteriophages
| Phage | Source | Host isolate | Host species | Genome length (bp) | %GC | Predicted family | Predicted subfamily | Predicted genus |
|---|---|---|---|---|---|---|---|---|
| ENC20 | Municipal wastewater | 973 | Enterobacter cloacae | 181927 | 44.5 | Myoviridae | Tevenvirinae | — |
| ENC19 | Municipal wastewater | 973 | E. cloacae | 179729 | 44.6 | Myoviridae | Tevenvirinae | — |
| ENC7 | Hospital wastewater | Surv186 | E. cloacae | 178940 | 44.9 | Myoviridae | Tevenvirinae | — |
| ENC22 | Hospital wastewater | 1665 | Enterobacter kobei | 178942 | 44.9 | Myoviridae | Tevenvirinae | — |
| ENC25 | Hospital wastewater | 607 | E. cloacae | 178842 | 44.9 | Myoviridae | Tevenvirinae | — |
| ENC9 | Hospital wastewater | Surv186 | E. cloacae | 173639 | 40.1 | Myoviridae | Tevenvirinae | — |
| ENC16 | Municipal wastewater | 971 | E. cloacae | 43486 | 51.9 | Autographiviridae | Slopekvirinae | Koutsourovirus |
| ENC31 | Hospital wastewater | 649 | E. cloacae | 40200 | 52.1 | Autographiviridae | Studiervirinae | Kayfunavirus |
| ENC2–2 | Hospital wastewater | 649 | E. cloacae | 40193 | 52.1 | Autographiviridae | Studiervirinae | Kayfunavirus |
| ENC14 | Municipal wastewater | 955 | Klebsiella aerogenes | 39996 | 52.6 | Autographiviridae | Studiervirinae | Przondovirus |
| ENC18 | Municipal wastewater | 973 | E. cloacae | 38849 | 50.4 | Autographiviridae | Studiervirinae | Teetrevirus |
| ENC13 | Hospital wastewater | Ent634 | E. cloacae | 46484 | 51.8 | Siphoviridae | — | — |
Of the seven bacteriophages isolated from hospital wastewater, four belonged to the Myoviridae family and the Tevenvirinae subfamily (Table 2). Two Autographiviridae and one Siphoviridae phage were also isolated from hospital wastewater. Among the five phages isolated from municipal wastewater, two Myoviridae phages were isolated that were also predicted to belong to the Tevenvirinae subfamily, as well as three Autographiviridae phages predicted to belong to the Slopekvirinae and Studiervirinae subfamilies (Table 2).
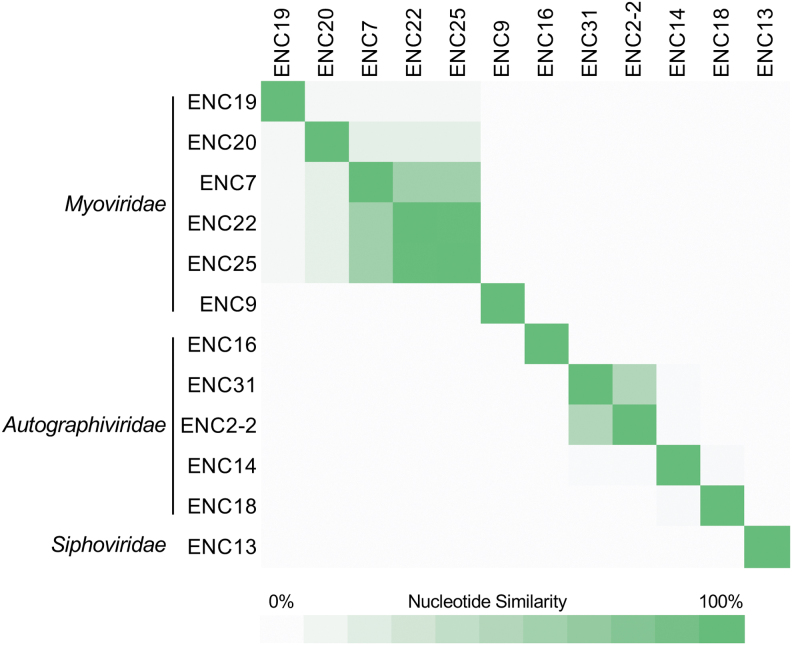
We compared the genomes of all isolated phages to one another, and found that although overall genetic diversity was high between different phages, some phages were genetically similar to one another (Fig. 2). The Myoviridae phages ENC22 and ENC25 were very similar, despite being isolated on different bacterial host species (Table 2). Both phages also showed moderate similarity to ENC7 and weak similarity to ENC19 and ENC20.
FIG. 2.
Nucleotide similarity between 12 isolated Enterobacter-targeting phages. Phages are organized by predicted family, which are labeled to the left. Phage genomes were compared with one another using nucleotide BLAST to determine sequence coverage and nucleotide identity for each pairwise comparison. Coverage and identity values were multiplied to calculate the nucleotide similarity for each comparison. Similarity values range from 0% to 100%, and are shown with green shading (darker shading indicates higher similarity).
The final Myoviridae phage, ENC9, showed no nucleotide similarity to the other Myoviridae phages, despite belonging to the same predicted subfamily. Among the Autographiviridae phages we isolated, ENC2-2 and ENC31 were only moderately similar to one another, even though they were isolated on the same host isolate and both were isolated from hospital wastewater. None of the other Autographiviridae phages showed nucleotide similarity with one another.
We also analyzed the gene contents of the isolated phages to evaluate their potential utility for the treatment of clinical Enterobacter spp. infections. The single Siphoviridae phage we isolated (ENC13) encoded a predicted phage integrase as well as a predicted phage repressor protein, suggesting that this phage is capable of undergoing lysogeny. No other phages were found to encode genetic markers of lysogeny. We also searched for predicted antibiotic resistance, virulence, and toxin–antitoxin system genes in all phage genomes, and did not find any such genes in the genomes of any phages we isolated. These data suggest that with the exception of phage ENC13, the phages we isolated could be considered for potential use in phage therapy.
Bacteriophage host range
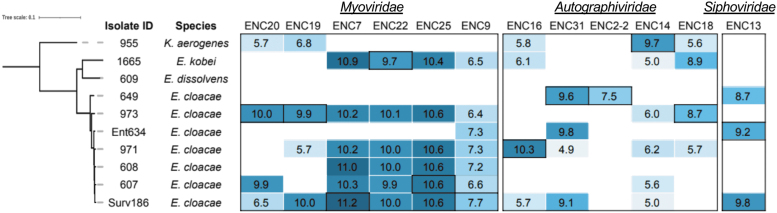
Since bacteriophages often have unique ranges of bacterial hosts that they can infect, we determined the host range of each isolated phage by calculating the titer of each phage against each Enterobacter isolate (Fig. 3). All but one phage (ENC2-2) could lyse multiple isolates, and five phages were able to lyse five or more isolates (Fig. 3). Nine phages were able to infect isolates belonging to different species, and five phages infected Enterobacter spp. as well as K. aerogenes (Fig. 3). In general, the Myoviridae phages we isolated were able to infect more clinical isolates compared with the Autographiviridae and Siphoviridae phages.
FIG. 3.
Host range of isolated Enterobacter-targeting bacteriophages. Log10 rates of infectivity (PFU/mL) of each isolated bacteriophage against each Enterobacter spp. isolate are shown. Larger numbers and darker shading indicate a greater ability to infect an isolate. Blank entries indicate no lytic activity. Outlined values indicate the initial host isolate of each bacteriophage. PFU/mL, plaque-forming units per mL.
Myoviridae phages isolated from hospital wastewater (ENC7, ENC9, ENC22, and ENC25) were able to infect more isolates compared with Myoviridae phages isolated from municipal wastewater. One bacterial isolate (E. dissolvens 609) was unable to be infected with any of the isolated phages. Without other E. dissolvens isolates available for comparison, it is unclear whether the phage resistance in this isolate is strain or species specific. We compared phage infectivity between isolates with and without CRISPR loci, and found no significant difference in the number of bacteriophages able to infect the isolates in either group (two sample t-test, p = 0.39). Likewise, no correlation was found between the number of prophages in an isolate's genome and the number of bacteriophages able to infect that isolate (Spearman correlation test, p = 0.45).
Bacteriophage infectivity of ceftazidime–avibactam-resistant isolates
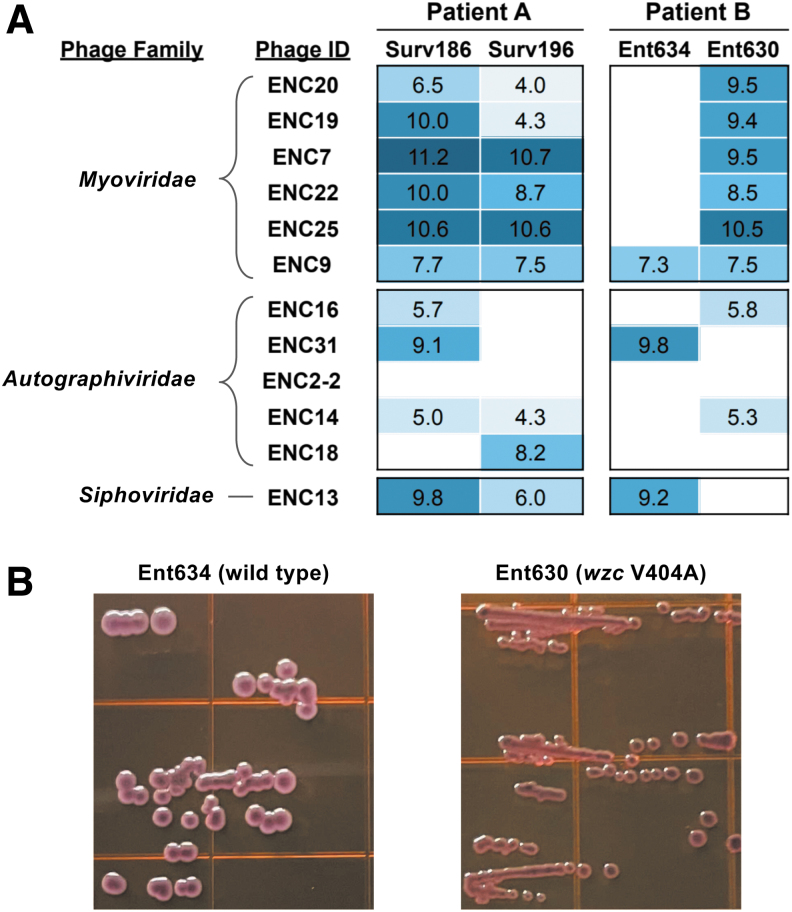
Two of the E. cloacae isolates used for phage isolation (Surv186 and Ent634) were collected from patients that subsequently developed ceftazidime–avibactam-resistant infections (Table 1).11 In both cases, resistance was due to mutations in the R2 loop of the AmpC beta-lactamase. The phage susceptibilities of the ceftazidime–avibactam-resistant isolates (Surv196 and Ent630, respectively) were determined and were compared with the susceptibilities of the corresponding parent isolates (Fig. 4A).
FIG. 4.
Differences in bacteriophage infectivity and colony morphology between pairs of ceftazidime–avibactam-susceptible and resistant Enterobacter cloacae isolates from two different patients. (A) Phage infectivity against Surv186 versus Surv196 (Patient A) and Ent634 versus Ent630 (Patient B). Spearman correlation test of phage infectivity for Patient A (log-transformed) p = 0.041; Patient B (log-transformed) p = 0.130. (B) Colony morphology of isolates from Patient B grown on EMB agar. The genome of isolate Ent630 encodes a V404A mutation in the capsular polysaccharide biosynthetic operon tyrosine kinase wzc. EMB, eosin methylene blue.
Between Surv186 and Surv196, we found that overall phage infectivity was correlated between the parent and ceftazidime–avibactam-resistant isolates, as most bacteriophages had comparable infectivity against both isolates (Fig. 4A; Spearman correlation test of log-transformed titers p = 0.041). Conversely, we observed different phage infectivity profiles between Ent634 and Ent630, with the ceftazidime–avibactam-resistant Ent630 isolate being overall more susceptible to bacteriophage infection than Ent634 (Fig. 4A). Phage infectivity of Ent634 was not significantly correlated with infectivity of Ent630 (Spearman correlation test of log-transformed titers p = 0.13), further suggesting that there was a difference between the phage infectivity in these two isolates.
Examination of mutations in the genome of Ent630 compared with Ent634 revealed a mutation (Val404Ala) in the tyrosine kinase wzc, which is part of the predicted capsular polysaccharide biosynthetic operon.25,26 To look for a difference in capsular polysaccharide production between Ent634 and Ent630, both isolates were grown on eosin methylene blue agar, which revealed differences in overall colony size and color between the isolates (Fig. 4B). These data suggest that the wzc mutation identified in the ceftazidime–avibactam-resistant Ent630 isolate may have caused alterations in capsular polysaccharide production, which likely impacted phage susceptibility. In this case, the capsular polysaccharide alterations appear to have increased the phage susceptibility of the ceftazidime–avibactam-resistant derivative isolate.
Discussion
In this study, we isolated 12 bacteriophages that could target a panel of 10 genetically distinct Enterobacter spp. clinical isolates in vitro. These phages constitute a promising start toward Enterobacter phage therapy efforts. Although the current status of Enterobacter spp. bacteriophage research is rather limited, our findings are in line with other studies of Enterobacteriaceae phages, which suggest that phages have high potential as an effective therapy to treat antibiotic-resistant infections.27–30 Furthermore, our findings are also consistent with prior reports of phages working in concert with antibiotics to improve bacterial killing.31,32
Because we used a diverse panel of bacterial host isolates, we identified a variety of genetically distinct phages that could infect these isolates. Out of the 12 phages we isolated, most were predicted to belong to either the Myoviridae or Autographiviridae families. Although previous research on Enterobacter phages is limited, a prior study also found Enterobacter phages belonging to the Myoviridae family.33 The same study also found Myoviridae phages to have a broad host range across Enterobacter spp.33
Autographiviridae phages targeting Enterobacter spp. have not been previously described in the literature; however, the genomes of several Autographiviridae phages isolated on Enterobacter spp. have been deposited in the NCBI database, and they have also been described for other pathogens.34,35 One prior study found Enterobacter phages belonging to the Siphoviridae family,36 which is in agreement with our results.
We found that many phages could infect Enterobacter spp. isolates belonging to different species. Many of the phages we isolated against E. cloacae were also able to target E. kobei, and a few phages were also active against K. aerogenes, although with lower titers. Similarly, ENC14 was isolated against K. aerogenes, and although it also showed activity against E. kobei and E. cloacae, titers were greatly reduced against these other species. The single E. dissolvens isolate tested was not susceptible to any of the phages that were isolated. No literature exists on E. dissolvens bacteriophages, so it is unclear whether or not the phage resistance we observed is a general property of E. dissolvens, or is specific to the single isolate that we tested.
Because the CRISPR-Cas system is one way that bacteria can defend themselves from bacteriophage infection,37 we expected to find a correlation between the presence of CRISPR genes and the number of bacteriophages able to infect a given isolate. Likewise, because prophages are integrated into their hosts genome, they can limit bacteriophage infection by altering host mechanisms to prevent other phages from integrating into the same host.38,39 However, we found no correlation between CRISPR presence and prophage abundance, nor between either of these measures and bacteriophage infectivity in our study.
Although these results did not necessarily align with our expectations, the small number of isolates studied here likely limited our ability to detect such associations. In addition, phage susceptibility may be determined by the gene content of prophages, not just their abundance.39 Further studies would be needed to determine whether CRISPR presence and prophage abundance are correlated, or not, with phage susceptibility in Enterobacter spp.
One clinically relevant question that we asked in this study was whether the evolution of ceftazidime–avibactam resistance in Enterobacter spp. also resulted in changes in phage susceptibility. In two separate cases where E. cloacae isolates evolved ceftazidime–avibactam resistance in vivo, we found either no difference or an increase in phage susceptibility among ceftazidime–avibactam-resistant isolates compared with their susceptible parent isolates. In the case of increased phage susceptibility in the resistant isolate, we identified a mutation in the tyrosine kinase wzc, which is part of the capsular polysaccharide biosynthetic operon.11,40 Specifically, wzc serves as a master regulator for both polymerization and translocation of the capsular polysaccharide across the outer membrane.26
A recent study of Klebsiella pneumoniae-targeting phages found that in vitro evolved phage resistance was often associated with wzc disruption and a resulting loss of capsular polysaccharide,41 whereas here we found that wzc mutation was associated with increased phage susceptibility. The wzc mutation we identified in Ent630 is unique and has not been previously observed in sequenced Enterobacter spp. genomes. When we compared the colony morphology of this isolate with that of its parent isolate, we saw that colonies of the resistant Ent630 isolate were smaller and differently colored, suggesting differences in capsule production in this isolate. Because bacterial capsule content can impact phage susceptibility,42 we suspect that altered capsule production in Ent630 likely enables its increased phage susceptibility. Confirming this link will be a focus of our future study in this area.
There were several limitations in this study. Our study was limited by a small sample size, and we were unable to identify associations between CRISPR or prophage presence and phage susceptibility. We isolated phages from wastewater sampled in only two locations, and it is likely that additional testing of wastewater from other locations as well as other types of samples would yield different phages than the those we describe here. In addition, although our finding that the E. dissolvens isolate we tested was completely phage resistant may be noteworthy, we do not have an explanation for why this might be.
Finally, we only tested two ceftazidime–avibactam-resistant isolates, thus it is unclear how applicable our results are to the treatment of multidrug-resistant Enterobacter spp. more generally. Nonetheless, our findings are consistent with other studies that have shown evidence for phage–antibiotic synergy,43–45 where increased drug resistance sometimes comes at a fitness cost to the bacteria, making them more susceptible to phage.
Conclusion
Overall, we find that lytic Enterobacter-targeting bacteriophages have promise as potential therapeutics for antibiotic-resistant Enterobacter spp. infections. We believe that bacteriophages constitute a potentially useful tool to combat the growing morbidity and mortality due to these infections. Furthermore, the preemptive isolation of bacteriophages that target Enterobacter spp. could be a viable approach for treating infections that are likely to develop antibiotic resistance.
Supplementary Material
Acknowledgments
We gratefully acknowledge Drs. Lee Harrison and Jane Marsh, who provided some of the Enterobacter isolates used for phage isolation as part of the EDS-HAT project (National Institutes of Health grant R01AI127472). Alecia Rokes also aided in Enterobacter spp. isolate collection and sequencing. Finally, we thank all members of the Van Tyne laboratory, in particular Hayley Nordstrom, Shu-Ting Cho, and Emily Dembinski, for helpful input and assistance throughout the study.
Author's Contributions
A.G.F. and D.V.T. designed the experiments. A.G.F, J.M.P., D.R.E., K.J.W., and C.L.M. performed the experiments. A.G.F and D.V.T analyzed the data. A.G.F and D.V.T. wrote the article with input from all authors.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
Research reported in this publication was funded, in part, by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award no. UM1AI104681. We also gratefully acknowledge research support from the University of Pittsburgh Department of Medicine. The funders of this study had no role in study design, data collection and analysis, decision to publish, or in preparation of the article.
Supplementary Material
References
- 1. Sanders WE Jr., Sanders CC. Enterobacter spp.: Pathogens poised to flourish at the turn of the century. Clin Microbiol Rev. 1997;10(2):220–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed Res Int. 2016;2016:2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davin-Regli A, Lavigne JP, Pagès JM. Enterobacter spp.: Update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin Microbiol Rev. 2019;32(4):e00002-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Doumith M, Ellington MJ, Livermore DM, et al. . Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother. 2009;63(4):659–667. [DOI] [PubMed] [Google Scholar]
- 5. Woodford N, Dallow JW, Hill RL, et al. . Ertapenem resistance among Klebsiella and Enterobacter submitted in the UK to a reference laboratory. Int J Antimicrob Agents. 2007;29(4):456–459. [DOI] [PubMed] [Google Scholar]
- 6. Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J Infect Dis. 2017;215(suppl_1):S28–S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaudhry WN, Concepción-Acevedo J, Park T, et al. . Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS One. 2017;12(1):e0168615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Herridge WP, Shibu P, O'Shea J, et al. . Bacteriophages of Klebsiella spp., their diversity and potential therapeutic uses. J Med Microbiol. 2020;69(2):176–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bolocan AS, Callanan J, Forde A, et al. . Phage therapy targeting Escherichia coli—A story with no end? FEMS Microbiol Lett. 2016;363(22):fnw256. [DOI] [PubMed] [Google Scholar]
- 10. Manohar P, Tamhankar AJ, Lundborg CS, et al. . Therapeutic characterization and efficacy of bacteriophage cocktails infecting Escherichia coli, Klebsiella pneumoniae, and Enterobacter species. Front Microbiol. 2019;10:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shields RK, Iovleva A, Kline EG, et al. . Clinical evolution of AmpC-mediated ceftazidime-avibactam and cefiderocol resistance in Enterobacter cloacae complex following exposure to cefepime. Clin Infect Dis. 2020;71(10):2713–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. CLSI. Performance Standards for Antimicrobial Susceptibilty Testing, 29th ed. CLSI Supplement M100. Malvern, Pennsylvania, USA: Clinical and Laboratory Standards Institute. 2019. [Google Scholar]
- 13. Bankevich A, Nurk S, Antipov D, et al. . SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012;19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. [DOI] [PubMed] [Google Scholar]
- 15. Page AJ, Cummins CA, Hunt M, et al. . Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ciccarelli FD, Doerks T, von Mering C, et al. . Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311(5765):1283–1287. [DOI] [PubMed] [Google Scholar]
- 18. Arndt D, Grant JR, Marcu A, et al. . PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44(W1):W16–W21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Couvin D, Bernheim A, Toffano-Nioche C, et al. . CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018;46(W1):W246–W251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Darling ACE, Mau B, Blattner FR, et al. . Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aziz RK, Bartels D, Best AA, et al. . The RAST server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bortolaia V, Kaas RS, Ruppe E, et al. . ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Malberg Tetzschner AM, Johnson JR, Johnston BD, et al. . In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J Clin Microbiol. 2020;58(10):e01269-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alcock BP, Raphenya AR, Lau TTY, et al. . CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–D525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cress BF, Englaender JA, He W, et al. . Masquerading microbial pathogens: Capsular polysaccharides mimic host-tissue molecules. FEMS Microbiol Rev. 2014;38(4):660–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang Y, Liu J, Clarke BR, et al. . The molecular basis of regulation of bacterial capsule assembly by Wzc. Nat Commun. 2021;12(1):4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mulani MS, Kamble EE, Kumkar SN, et al. . Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front Microbiol. 2019;10:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cano EJ, Caflisch KM, Bollyky PL, et al. . Phage therapy for limb-threatening prosthetic knee Klebsiella pneumoniae infection: Case report and in vitro characterization of anti-biofilm activity. Clin Infect Dis. 2021;73(1):e144–e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anand T, Virmani N, Kumar S, et al. . Phage therapy for treatment of virulent Klebsiella pneumoniae infection in a mouse model. J Glob Antimicrob Resist. 2020;21:34–41. [DOI] [PubMed] [Google Scholar]
- 30. Easwaran M, De Zoysa M, Shin HJ. Application of phage therapy: Synergistic effect of phage EcSw (PhiEcSw) and antibiotic combination towards antibiotic-resistant Escherichia coli. Transbound Emerg Dis. 2020;67(6):2809–2817. [DOI] [PubMed] [Google Scholar]
- 31. Gu Liu C, Green SI, Min L, et al. . Phage-antibiotic synergy is driven by a unique combination of antibacterial mechanism of action and stoichiometry. mBio. 2020;11(4):e01462-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chan BK, Sistrom M, Wertz JE, et al. . Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep. 2016;6:26717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manohar P, Nachimuthu R, Lopes BS. The therapeutic potential of bacteriophages targeting gram-negative bacteria using Galleria mellonella infection model. BMC Microbiol. 2018;18(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zurabov F, Zhilenkov E. Characterization of four virulent Klebsiella pneumoniae bacteriophages, and evaluation of their potential use in complex phage preparation. Virol J. 2021;18(1):9–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mizuno CM, Luong T, Cederstrom R, et al. . Isolation and characterization of bacteriophages that infect Citrobacter rodentium, a model pathogen for intestinal diseases. Viruses. 2020;12(7):737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li E, Wei X, Ma Y, et al. . Isolation and characterization of a bacteriophage phiEap-2 infecting multidrug resistant Enterobacter aerogenes. Sci Rep. 2016;6:28338–28338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Makarova KS, Haft DH, Barrangou R, et al. . Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9(6):467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dedrick RM, Jacobs-Sera D, Bustamante CA, et al. . Prophage-mediated defence against viral attack and viral counter-defence. Nat Microbiol. 2017;2:16251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bondy-Denomy J, Qian J, Westra ER, et al. . Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016;10(12):2854–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tiwari M, Panwar S, Kothidar A, et al. . Rational targeting of Wzb phosphatase and Wzc kinase interaction inhibits extracellular polysaccharides synthesis and biofilm formation in Acinetobacter baumannii. Carbohydr Res. 2020;492:108025. [DOI] [PubMed] [Google Scholar]
- 41. Hesse S, Rajaure M, Wall E, et al. . Phage resistance in multidrug-resistant Klebsiella pneumoniae ST258 evolves via diverse mutations that culminate in impaired adsorption. mBio. 2020;11(1):e02530-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hyman P, Abedon ST. Bacteriophage host range and bacterial resistance. Adv Appl Microbiol. 2010;70:217–248. [DOI] [PubMed] [Google Scholar]
- 43. Segall AM, Roach DR, Strathdee SA. Stronger together? Perspectives on phage-antibiotic synergy in clinical applications of phage therapy. Curr Opin Microbiol. 2019;51:46–50. [DOI] [PubMed] [Google Scholar]
- 44. Tagliaferri TL, Jansen M, Horz HP. Fighting pathogenic bacteria on two fronts: Phages and antibiotics as combined strategy. Front Cell Infect Microbiol. 2019;9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim M, Jo Y, Hwang YJ, et al. . Phage-antibiotic synergy via delayed lysis. Appl Environ Microbiol. 2018;84(22):e02085-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.