Abstract
The support vector machine (SVM) is a popularly used classifier in applications such as pattern recognition, texture mining and image retrieval owing to its flexibility and interpretability. However, its performance deteriorates when the response classes are imbalanced. To enhance the performance of the support vector machine classifier in the imbalanced cases we investigate a new two stage method by adaptively scaling the kernel function. Based on the information obtained from the standard SVM in the first stage, we conformally rescale the kernel function in a data adaptive fashion in the second stage so that the separation between two classes can be effectively enlarged with incorporation of observation imbalance. The proposed method takes into account the location of the support vectors in the feature space, therefore is especially appealing when the response classes are imbalanced. The resulting algorithm can efficiently improve the classification accuracy, which is confirmed by intensive numerical studies as well as a real prostate cancer imaging data application.
Keywords: Classification, data-adaptive kernel, imaging data, imbalanced data, separating hyperplane, support vector machine
1. Introduction
Classification is perhaps one of the most important objectives for data mining. It has been widely used in a variety of research areas such as genomics studies, imaging analysis and finance. The Support Vector Machine (SVM), proposed by Vapnik and co-authors [7], is a useful statistical learning method for classification. Its excellent performance has been demonstrated in various applications including handwriting pattern recognition [9], text classification [14] and image retrieval [20]. Recently, applications of the SVM have been extended to artificial intelligence, such as knowledge transformation [21], environmental protection [1] and natural language process [16]. The core idea of the SVM is to map the current input space into a feature space so that the linear separability of the classes becomes as much as possible in the feature space [19], while in the input space it is not linearly separable. The feature space has a higher dimension than the input space and it is formed on the basis of a kernel function [13]. Hence, it is the kernel that is crucial to determine the performance of the SVM classifier. Often, the optimal kernel function is driven by the prior knowledge of the data, and the optimization process during training the SVM is typically limited by the specific form of the kernel function that is being used, especially when the number of training patterns is small. In applications such as image analysis and cancer detection [10], the number of the training instances of the target class is dramatically smaller than other classes, therefore the separating boundary by the SVM can be severely skewed towards the target class. In such instances, the false-negative rate can be significantly high in order to identify the target objects [24]. Hence, to improve the performance of an SVM when the real data are imbalanced, how to choose a suitable kernel and how to deal with imbalance issue are both considerations of this paper.
To determine a suitable kernel function appears to be somewhat difficult. A typical way to achieve the goal is to pre-specify the parametric form of several popularly used kernel functions, tune the optimal parameters with every given kernel function, and determine the most appropriate kernel function by comparing the classification performance of the estimated SVMs with all possible kernel functions. This procedure may be intensely time-consuming on tuning the parameters for different kernel function; further, the performance of an SVM may even be sensitive to the specific form of the kernel function. An alternative idea to construct a suitable kernel is, instead of attempting different kernels, to simply update any form of kernel by some transformation driven by real data [6]. Such kernel functions are called data-driven or data-adaptive kernels. The classification procedure benefits in the sense of saving time for tuning parameters for different kernel functions, and the performance of the SVM becomes less dependent on different forms of the kernels, therefore probably more suitable for a given data set.
Motivated by this idea, [3] proposed a two-stage training process for choosing an optimal kernel function in the support vector machine. Their idea is that the mapping with a kernel function introduces the Riemannian metric in the feature space, and a good kernel should have the property of enlarging the metric in the feature space, consequently broadening the spatial separation between two classes. Although the performance of their classifier improves over the usual one-stage classifier in many cases, this two-stage method is vulnerable to the choice of the location of the support vectors that depends on the density of the data points in the region. A modified method was proposed by [25], but the new algorithm still suffers a certain level of susceptibility, therefore can not be applied in high dimensional cases. Following Amari and Wu's idea, [23] proposed a kernel scaling technique, describing a more direct way to achieve the useful magnification ability. In their proposed method, the initial kernel function is transformed so that the magnification effect will decay from the separating surface along with the squared distance to the separating boundary. This ensures the magnification effect to be maximized along the separating surface and decays smoothly to a positive constant at the separating boundary. However, magnification effect decays too fast for those data points not far away from the separating boundary. Alternatively, modifying the kernel functions directly to deal with imbalance is attempted. The core idea is to modify the kernel function to enlarge more of the class boundary around the minority class than that around the majority class. In particular, [24] proposed a two-stage method, where the first stage of their method is to roughly find a separating boundary with a primary kernel function, and the kernel function is then rescaled in the second stage using a transformation that can amplify the Riemannian metric around the separating boundary found in the first stage. [18] applied asymmetric kernel scaling method to imbalanced binary classification. Their basic idea is to differently enlarge areas on each side of the separating surface so that the skewness toward minority will be compensated. However, the performance does not seem to be improved much.
Although these methods somewhat improve the performance of an SVM, none of these takes into account the possibility of imbalanced phenomenon. Imbalance of data tends to drag the separating hyperplane constructed by an SVM towards the minority class so that a test object is more likely to be classified into the majority class. Extreme imbalance in real data may even completely ruin the classifier in the sense that all minority objects are likely to be classified into the majority class if the imbalance is ignored, whatever kernel function is considered, such as the cancer data we have. This greatly impacts the classification performance.
To deal with imbalance, two frameworks of methods, namely external and internal methods are available. External methods, also referred to as preprocessing methods, aim to balance the data sets before training SVMs by randomly undersampling the data from majority class or oversampling the data from the minority class or both [4,5,8,17]. Ensemble techniques are also included in external methods to balanced the data, by separating the majority class data into multiple sub-data sets each of which has similar numbers of the minority data set [15,22]. This procedure is usually achieved by random sampling with or without replacement. Then a set of individual SVM classifiers, each of which is trained based on a relatively balanced data set, are developed, and a test object is classified into the specific class by majority voting from all individual classifiers. However, these methods may suffer from the accuracy problem in the sense that partial information from the originial data set may be dropped such as re-sampling methods, and that the classification process becomes more time-consuming with a suitable number of individual classifiers such as the ensemble learning methods, where the number of individual classifiers may be difficult to determine [6]. Alternatively, internal methods, also known as the algorithm methods, are developed to account for imbalance [2,26]. Instead of balancing the data sets before training an SVM, internal methods modify the objective function of an SVM, using different costs on majority and minority misclassification to make different punishment. In this way, the effect of the imbalance is alleviated.
To enhance the performance of the SVM, we propose a new two-stage method using kernel function scaling. Following the primary SVM procedure in the first stage, the proposed method employs a conformal transformation driven by the given data and locally adapts the kernel function to the data location based on the skewness of the classes, and hence enlarges the magnification effect directly on the Riemannian manifold in the feature space, instead of in the initial input space. With the distance measured in the feature space, the conformal transformation can make full use of the updated information in the second stage. Our method benefits the classification procedure by firstly enhancing the accuracy of the SVM with a suitable data-adaptive kernel function, secondly incorporating the imbalance effect and finally saving computational cost.
Our research is partially motivated by a prostate cancer image study, but our method has a broad scope of application. Traditionally, areas of cancer are determined visually by examining the images of the suspected cancer areas, therefore is not completely reliable. It is thereby desirable to develop a diagnosis process by corresponding the imaging data to the histological imaging data. In this study, the images of the prostate gland are taken when the gland is in the body (in-vivo imaging data), and the gland is then surgically removed from the body and imaging data of the gland are taken again when it is outside the body (ex-vivo imaging data). Pathologists examine the sliced gland using high resolution microscope to identify the exact position of the cancer in the gland to form histological image. A co-registration process is employed to build the correspondence between the in-vivo imaging data and ex-vivo histological imaging data for the defined voxels, and a prediction model is constructed to predict cancer status for each of the voxels using the in-vivo imaging data. The predictive model is expected to be utilized for diagnosis, targeted biopsy and targeted treatment in the future. In this study, the raw intensity measurements of the prostate image are obtained using imaging techniques such as various types of Magnetic Resonance (MR) or Computed Tomography (CT). Usually there are around 170,000 to 200,000 voxels for each patient, with only 5% to 10% cancer voxels, which makes the classes of cancer and non-cancer to be extremely imbalanced. A classifier that can perform well for extremely imbalanced data is urgently needed, which promotes our consideration of support vector machine for dealing with imbalanced data.
The remainder of the paper is organized as follows. In Section 2, we present the framework and explain the geometric relation between an SVM and the kernel function. In Section 3, we describe the proposed method and discuss the benefits of the constructed classifier to show how the local separability is enhanced. In Section 4, we conduct extensive simulation studies to show the excellent performance of the proposed, and apply the proposed method to the real prostate cancer imaging data. Concluding remarks and discussion are included in Section 5.
2. Notation and framework
Consider a binary classification problem where a hyperplane is expected to separate the two classes of the response y, given sample data for Here for , is a vector in the input space , denoted as I, and represents the class index which takes values or . However, in many cases, a hyperplane does not exist in the input space to separate completely the two classes [13]. To get around this, an SVM method maps the input data into a higher dimensional feature space , denoted as F, using a nonlinear mapping function , and then searches a linear discriminant function or a hyperplane in the feature space F
| (1) |
where is vectorized parameter vector with l dimensions, is the dimension vector in the feature space, b is a scalar bias term, and denote the transpose operator. Hence, an individual point with observation can be classified by the sign of as long as the parameters and b are determined. The separating surface of the nonlinear classifier is determined by in the input space I. Theoretically, the solution to the SVM can be obtained by maximizing the aggregated margin between the separating boundaries [7]. This can be justified from the Structure Risk Minimization principle as minimizing an upper bound of the generalization error [13].
Mathematically, an SVM is the solution of minimizing
| (2) |
with respect to and b, which are subject to the constraints for where C is the so-called soft margin parameter that determines the trade-off between the optimal combinatorial choice of the margin and the classification error, is the vector of non-negative slack variables, and represents the norm. Equivalently, this optimization problem can be represented in the Lagrangian dual function with the form as
subject to the constraints for , where 's are the dual variables (the Lagrange Multiplers) by Lagrange Multiple Methods when solving the minimization problem in (2). Generally a scalar function , which is called a kernel function, is adopted to replace the inner product of the two vectors and in the dual function,
Let SV be the set . Then the corresponding 's where i belongs to SV are called support vectors, where the cardinality of SV is l, the dimension of the feature space F. Thus, to determine the hyperplane (1) in the feature space, we may not need to find explicitly. We need only to specify the inner products of and , which is available from the giving kernel . Then the kernel form of SVM can be written as
and the estimated bias term obtained by using the jth support vector is defined as [13] have proved that for different j in the support vectors set SV, the s are the same. In practice, we can take the average of all the estimated as the estimate of the bias term b.
Typical kernels in the literature include the following forms [13]. The radial kernel has the form
| (3) |
with being a positive scalar function. A popularly used radial kernel is the Gaussian Radial Basis kernel where σ is the bandwidth parameter. Another popularly used kernel has the form of a polynomial function of the inner product of two vectors, where is the polynomial function. A popular polynomial kernel with degree d has the form
3. Methodology
3.1. Adaptive scaling on the kernel
The fact that the kernel function determines the performance of the SVM motivates us to consider to modify the kernel function to accommodate imbalanced observations so that the separation may be enlarged near the separating margins and surface differently for different classes, and further the overall classification may perform better. To achieve this, we propose a new two-stage adaptive scaling on the kernel with two goals. That is, in the first stage, we roughly obtain the temporary separating boundary and margins, and then refine them in the second stage. One goal is that the spatial resolution near the margin area needs to be increased so that the separability is enhanced, while keeping the decision boundary unchanged. The other goal is that the scaling process should only depend on the local support vectors, not those ones farther apart, and the scaling effect should decrease robustly and slowly with the distance approaching the boundary.
To be specific, we construct a standard SVM with a primary kernel K in the first stage to identify the possible support vectors and the separating surface. The kernel is then updated in the second stage as follows. Let be a positive scalar function such that
| (4) |
where and are vectors from the input space, and is a positive univariate scalar function. The kernel function K is updated as
| (5) |
where corresponds to the mapping that satisfies the transformation where
The above process is referred to as the adaptive scaling, and can be easily shown to satisfy the Mercer positivity condition, which is the sufficient condition for a real function to be a real kernel function. When has larger values at the support vectors than those at other data points, the updated mapping can increase the separation when a positive function is properly chosen. This modification of the kernel function can keep the spatial resolution stable within the feature space so that the spatial relationship between the sample points would not be changed, with properly chosen. Also, the computational cost turns out to be reasonable.
Inspired by the above idea, we propose to adaptively scale the primary kernel function K by constructing with the norm radial basis function
| (6) |
and
where is given by (1), AVG denotes the average operator, y is the class label associated with , and M is a tuning parameter controlling how much location information is included.
The proposed form of has many benefits. reflects the spatial information of the local support vectors in the feature space F, instead of the input space I. is the average distance between and the support vectors of different class within a radius of M. The average can comprise all the support vectors with different labels in the neighborhood of within the radius of M. When the data are imbalanced, the magnification tends to be biased towards the higher density support vectors [23]. With a proper M, the level of local balance is controlled. The setting in the proposed method can make the global minority class have higher density in a small neighborhood, making it balanced locally. As a result, the magnification tends to be comparable for the two classes locally instead of being extremely biased to the majority class. The data points that are misclassified by the original separating hyperplane are likely to be correctly classified by the updated hyperplane, and hence the performance of separation for two classes will be enhanced.
The magnification effect roughly achieves its largest near the separating boundary. It is easy to check that takes its maximum value on the separating hyperplane surface of , and decays slowly to at the boundary where . Thus, the resolution is amplified along the boundary surface, and the rate of decay based on -norm is moderate compared with the choice of -norm. In addition, by making use of the tuning parameter M to control the number of support vectors to be included in the kernel scaling process, the classifier can be adopted without much complexity.
3.2. Geometric interpretation of SVM kernels
From the geometry point of view, when the input space I is the Euclidean space, the Riemannian metric is then induced in the feature space F. Let be the mapped result of in F, i.e. . A small change in in the input space, , will be mapped into the vector in the feature space such that
where
| (7) |
Thus, the squared length of can be written in the quadratic form as
where .
Consequently, the matrix that is defined on the Riemannian metric can be derived from the kernel K, and is positive definite [3]. Theoretically, Lemma 3.1 shows the connection between a kernel function K and a mapping :
Lemma 3.1 [25] —
Suppose is a kernel function, and is the corresponding mapping in the support vector machine, then we have
(8)
where and
| (9) |
Let with . The factor indicates the magnification level on the local area in F under the mapping , thus is called the magnification factor. To increase the margin of separability between two classes, the spatial resolution around the boundary surface in F needs to be enlarged. This motivates us to increase the magnification factor around the boundary area between and . Therefore, the mapping , or equivalently, the related kernel K, is modified so that can be enlarged around the boundary. For the radial kernel with the form in (3), it is easy to check that However, it is not easy to control the magnification factor by simply changing , the entries of . Take the Gaussian kernel as an example, we have and . To increase the spatial resolution at a support vector x, we cannot simply increase σ. We need to accommodate the location information around the neighborhood of the support vector x, thus a universal parameter σ may not be used for all the support vector points since it cannot accommodate the neighborhood information; if different σ's for every single support vector are used, however, the number of parameters is too large to be specified. Furthermore, it is not a universal practice to change local resolution by only changing the scale parameter σ, since not all radial kernels have the scale parameter σ. The locations of the support vectors, which determine the separating boundary, are also needed to be included in consideration. Therefore, to increase the spatial resolution locally, we attempt to use the adaptive scaling on the kernel function. With the adaptive scaling, the metric , introduced from , is related to the original by the following proposition:
Proposition 3.2
Given a primary kernel function and a scalar function as in (5), the modified magnification factor is linked with the adaptive kernel function by
where is the ith element of the gradient of , is determined by (8), and . Particularly when i = j,
(10) where
Detailed proof is provided in the appendix. As the adaptive scaling is properly chosen, the goal of enlarging the separating margins and boundary can be achieved.
3.3. Adaptively scaled Gaussian RBF kernel
In real applications, the primary kernel K usually takes the Gaussian radial basis function kernel
| (11) |
We have the following results for Gaussian kernel.
Proposition 3.3
When a Gaussian radial basis kernel is used and a scalar function takes an arbitrary form, the modified magnification factor is given by
where , and is the indicator function.
The proof is outlined in the appendix. The result is neat in the sense that under the Gaussian RBF kernel assumption the updated magnification factor depends only on the information from the adaptive scaling function . To make bigger, we need to make the positive scalar and its first order derivative bigger, which is satisfied using the proposed method. When the Euclidean metric is used, the magnification factor is a constant By Proposition 3.3 and the proposed in (6), the ratio of the new to the old magnification factors is given by
| (12) |
The ratio from (12) indicates that the magnification does not change along the separating surface ; the magnification is the largest when the contours are tight with large ; the magnification is the smallest at the separating margin of , taking values . Therefore, we can adaptively tune according to the local allocation of support vectors.
There are several attempts on choosing c(x) in the literature that can be viewed as the special cases of our proposed method. [3] considered the function where k is a positive constant. Using this function, the support vectors need normally located near the boundary, so that the magnifying effect in the vicinity of support vectors can be large around the boundary. This function can be quite sensitive to the spatial locations of the support vectors found in the first step, and thus the magnification becomes larger at higher density regions of support vectors but drops dramatically at lower density regions. A modified function was proposed in [25] by setting different for different support vectors, so that the local density of support vectors can be accommodated. Though improvement of the performance is achieved using this modified function, the computational cost becomes huge and the performance in high dimensions is uncertain. Those kernel modifications are based on the distance of the observed support vectors in the input space, while we are searching for hyperplane in the feature space. Therefore the effect of the scaling may not be intuitive and effective. [23] proposed a different with distance considered in the feature space to achieve the magnifying effect. They suggest where is given in (1) and k is a positive constant for all support vectors. This function takes its maximal value on the separating surface and decay to at the margin where . However, the tuning parameter k is fixed throughout the whole region, thus the local information cannot be accommodated. When the density of local support vectors is high, the separation can be inaccurate and inefficient. The norm is used in the function, which decays the resolution too fast from the separating surface to the constant at the margin , making the separation performance unstable in high dimensional cases.
4. Numerical assessments
We conduct extensive simulation studies to access the performance of the proposed method, and compare the proposed method with other methods in the literature, including traditional one-stage SVM, the two-stage SVM of [25] and the two-stage SVM of [23]. The proposed adaptive kernel scaling support vector machine is also applied to a real prostate cancer image study to assess the classification performance using cross validation, and compare with the popular used classifier in the literature.
4.1. Simulation studies
We evaluate the proposed method under situations of balanced and imbalanced cases. For all the two-stage algorithms, a Gaussian RBF kernel K is used in the first-stage standard SVM to find the locations of the support vectors. Based on these support vectors, the kernel function is adaptively rescaled to with the corresponding from each method, and second-stage SVM is conducted by using the modified kernel . The separating hyperplane is then obtained, and sample points are classified based on the hyperplane. We will assess the performance of different methods by two popular measures. One is the misclassification rates, defined as We also adopt popularly used F-score, defined as
| (13) |
where and ; TP, FP and FN are true positive, false positive and false negative numbers, respectively.
For values of the tuning parameters, namely the cost parameter C and the Gaussian RBF kernel bandwidth parameter σ in the kernel function, we consider the same setting as in [23]. The sample size n is fixed as 200 for all scenarios. The cost parameter C in (2) is chosen from the set and σ takes value from the set . As M is the threshold controlling the size of the local neighborhood, it is chosen by grid search from the set times the maximal Euclidean distance between all pairs of data points in the sample. Five fold cross validation is employed to obtain the misclassification rate for each simulated data set, and the whole process is repeated 100 times. We compare the classification performance by considering different scenarios and report selected results in Tables 1 to 2 due to page limit.
Table 1.
Misclassification rates (in %) for Settings 1 to 3. I: One-stage SVM; II: [25]; III: [23]; IV: Our proposed method.
| Setting 1 (50% vs 50%) | Setting 2 (25% vs 75%) | Setting 3 (10% vs 90%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | σ | I | II | III | IV | I | II | III | IV | I | II | III | IV |
| 8 | 0.1 | 16.3 | 13.2 | 12.5 | 11.1 | 19.3 | 15.4 | 12.1 | 10.5 | 22.3 | 18.6 | 16.2 | 13.3 |
| 0.5 | 9.6 | 7.7 | 6.9 | 5.7 | 14.2 | 12.3 | 10 | 9.5 | 19.2 | 15.4 | 14.8 | 11.6 | |
| 5 | 7.2 | 5.3 | 4.7 | 4.9 | 12.3 | 9.6 | 9.1 | 7.6 | 16.7 | 13.2 | 10.3 | 8.2 | |
| 40 | 0.1 | 18.7 | 17.7 | 14.1 | 11.9 | 21.2 | 15.6 | 15.8 | 13.1 | 23.9 | 19.6 | 17.1 | 14.0 |
| 0.5 | 15.5 | 12.3 | 11.1 | 10.6 | 16.6 | 14.6 | 12.7 | 11.9 | 18.5 | 18.5 | 18.5 | 18.5 | |
| 5 | 9.3 | 7.1 | 6.2 | 5.9 | 14.5 | 11.2 | 10.1 | 8.2 | 15.4 | 12.9 | 11.8 | 10.1 | |
| 100 | 0.1 | 23.0 | 18.6 | 16.3 | 13.2 | 21.1 | 17.2 | 15.6 | 13.9 | 25.7 | 19.9 | 18.6 | 15.1 |
| 0.5 | 14.0 | 9.9 | 8.3 | 7.3 | 18.0 | 16.1 | 13.3 | 12.1 | 21.2 | 17.8 | 16.3 | 13.5 | |
| 5 | 12.2 | 7.1 | 7.8 | 6.2 | 17.2 | 13.6 | 12.8 | 8.9 | 18.2 | 15.2 | 14.2 | 11.2 | |
Table 2.
| Setting 1 (50% vs 50%) | Setting 2 (25% vs 75%) | Setting 3 (10% vs 90%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | σ | I | II | III | IV | I | II | III | IV | I | II | III | IV |
| 8 | 0.1 | 0.676 | 0.712 | 0.723 | 0.711 | 0.648 | 0.689 | 0.677 | 0.732 | 0.482 | 0.523 | 0.619 | 0.679 |
| 0.5 | 0.741 | 0.755 | 0.772 | 0.785 | 0.661 | 0.708 | 0.711 | 0.752 | 0.507 | 0.551 | 0.632 | 0.698 | |
| 5 | 0.783 | 0.808 | 0.813 | 0.815 | 0.672 | 0.723 | 0.730 | 0.765 | 0.523 | 0.588 | 0.686 | 0.754 | |
| 40 | 0.1 | 0.623 | 0.679 | 0.691 | 0.688 | 0.610 | 0.635 | 0.636 | 0.687 | 0.462 | 0.500 | 0.576 | 0.651 |
| 0.5 | 0.653 | 0.701 | 0.726 | 0.755 | 0.635 | 0.677 | 0.683 | 0.722 | 0.491 | 0.523 | 0.601 | 0.601 | |
| 5 | 0.719 | 0.754 | 0.771 | 0.783 | 0.656 | 0.699 | 0.703 | 0.737 | 0.513 | 0.552 | 0.636 | 0.710 | |
| 100 | 0.1 | 0.578 | 0.626 | 0.658 | 0.654 | 0.554 | 0.599 | 0.601 | 0.662 | 0.433 | 0.476 | 0.522 | 0.601 |
| 0.5 | 0.629 | 0.683 | 0.711 | 0.732 | 0.577 | 0.611 | 0.631 | 0.698 | 0.483 | 0.520 | 0.569 | 0.623 | |
| 5 | 0.641 | 0.718 | 0.726 | 0.730 | 0.592 | 0.634 | 0.655 | 0.721 | 0.498 | 0.539 | 0.601 | 0.677 | |
Scenario 1: Balanced Data.
In this case, the proportion of the two different classes is around 50%, denoted as Setting 1. Two-dimensional input data are considered as which are independently generated from the uniform distribution in the area of , . Two classes are separated by the function , where
for all i from 1 to n in the sample.
It is evident from Tables 1 and 2 that the proposed method outperforms the considered competitors. When σ gets larger with a fixed C, the misclassification rates yielded from all the methods tend to decrease, while the F-scores tend to increase. When σ is relatively small, the proposed method performs better than the methods of [25] and [23]. If σ is relatively large, all methods produce nearly the same results. This is because when σ is large, the feasible solution set is large, and all the methods are capable of finding the optimal solution. Correspondingly, when C increases, the budget for misclassification gets bigger, which means more tolerance is permitted so that the two classes can be separated. In this scenario, that is found to be roughly the same, and approximately equals to the reciprocal of . This makes sense because in the balanced-data case, the density of the support vectors is roughly the same for both classes, and the averages of the distance to the separating boundary in the feature space for each data point are fairly close.
Scenario 2: Imbalanced Data.
For the case of imbalanced data, we consider that the proportions of two different labels are significantly different, in two settings representing moderate imbalance and extreme imbalance. We first take the proportions of the two classes as 25% versus 75%, denoted as Setting 2. We still choose a two-dimensional input as uniformly distributed in the area . The two classes are labeled by the function :
where C and σ are chosen in the same way as in Scenario 1. We also apply the same procedure to a more extreme case with proportions of versus , denoted as Setting 3. Similar to Scenario 1, we classify the data with the primary kernel K to find the locations of the support vectors of the standard SVM process, and repeat the classification with the proposed adaptive scaling kernel . Different combinations of the cost C and σ are utilized and five fold cross validation is applied to obtained misclassification rate. 1000 runs are taken and the outcomes are summarized in Tables 1 and 2. Note that in the imbalanced scenarios, we have adopted a different data generation procedure due to the consideration of the reliability of our proposed algorithm with different structure. In fact, we have obtained a similar performance of different classifiers with the synthetic data sets created in the way of creating the balanced data in Scenario 1. However, it is not reported due to page limit.
It is seen that the performance of all the methods decays for imbalanced data due to non-uniformly distributed support vectors. The trends of misclassification rates in all scenarios are similar to those in the balanced data case. The proposed method still works the best compared with all other methods considered here. tends to change in an opposite way along with the density changes of the support vectors around a specific point. When the ratio of proportions become more extreme, changes sharply. The spatial location of the support vectors in the feature space F is therefore seen to be taken into consideration by the proposed method.
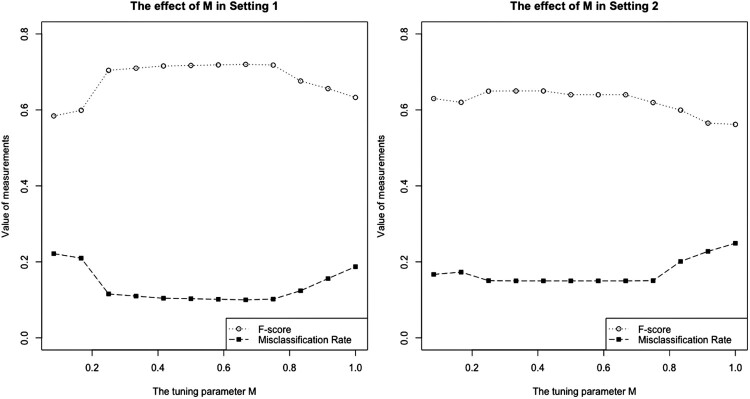
Specifically for our proposed method, the effect of the tuning parameter M in (6) is illustrated from Figure 1. When M is relatively small, the local neighborhood contains roughly very few data points that satisfy the requirement, which leads to an almost constant scaling effect. As M is getting large, the performance of both the F-score and the misclassification rate become better. When M gets closer to the maximum, almost all of the data points in the sample tend to be included into the neighborhood as if little adaptive scaling were impose. Besides, the proposed method performs quite closely and stably when M is around half to two thirds of the maximal distance. In this case, the performance gets poorer.
Figure 1.
Changes of F-score and misclassification rate against different values of M. The left panel shows the result for the balanced case in Setting 1, and the right panel shows that for the imbalanced case in Setting 2. Similar results in Setting 3 have been found as Setting 2.
4.2. KEEL data sets
Our proposed method is applied to two real imbalanced data sets to test the performance, namely Ecoli [12], Yeast1 [11] and Yeast2 [11] from KEEL date set repository, compared with the performance from state-of-art classifiers, including the standard SVM, the random forest and the logistic regression. Preliminary descriptions of the two data sets are presented in Table 3. In particular, Ecoli is moderately imbalanced, while both Yeast1 and Yeast2 are extremely imbalanced. All covariates are standardized before different classifiers are applied. To compare the performance, the proposed algorithm and all other classifiers are applied to each of the three data sets with the corresponding parameters tuned using the 5-fold cross validation from the same sets as used on the synthetic data sets, respectively. The whole process is repeated 100 times so that no random split bias will affect the results.
Table 3.
Description of three real KEEL data sets. Ecoli is moderately imbalanced. Both Yeast1 and Yeast2 are extremely imbalanced.
| Name | Number of Covariates | Type of Covariates | Sample Size | % of Positive | Imbalance Ratio | Missing |
|---|---|---|---|---|---|---|
| Ecoli | 7 | Continuous | 336 | 22.94 | 3.36 | No |
| Yeast1 | 7 | Continuous | 459 | 6.54 | 14.30 | No |
| Yeast2 | 8 | Continuous | 693 | 4.33 | 22.10 | No |
Results of classification on three data sets are reported as the average of the hundred repetitions in Table 4. The proposed method has the smallest test errors among all classifiers on all three data sets, with similar training errors compared with other classifiers. The F-score from our proposed method is greater than other classifiers, though not that much. All the methods tend to estimate the recall rate very close to 1, while the proposed method shows the greatest precision rate. The standard errors of the estimated F-score and the test error rate from the proposed method and other classifiers are similar respectively (so not reported), mush smaller those from the logistic regression. The results on the real KEEL data sets have confirmed the superb performance of the proposed method.
Table 4.
Performances of different classifiers on the three KEEL data sets. F is the F-score, the Test is the test error rate and Prec is the precision.
| E. coli | Yeast1 | Yeast2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methods | F | Test | Prec | Recall | F | Test | Prec | Recall | F | Test | Prec | Recall |
| Proposed | 0.997 | 0.005 | 1.000 | 1.000 | 0.996 | 0.007 | 1.000 | 0.996 | 0.983 | 0.012 | 0.985 | 0.969 |
| SVM | 0.976 | 0.045 | 0.952 | 1.000 | 0.962 | 0.050 | 0.966 | 0.992 | 0.936 | 0.065 | 0.935 | 0.921 |
| RF | 0.991 | 0.026 | 0.977 | 0.998 | 0.981 | 0.033 | 0.964 | 0.998 | 0.942 | 0.047 | 0.948 | 0.970 |
| [25] | 0.982 | 0.033 | 0.981 | 0.992 | 0.972 | 0.044 | 0.951 | 0.985 | 0.928 | 0.056 | 0.950 | 0.964 |
| [23] | 0.973 | 0.041 | 0.957 | 0.973 | 0.972 | 0.056 | 0.943 | 0.985 | 0.931 | 0.061 | 0.935 | 0.959 |
| Logit | 0.932 | 0.081 | 0.939 | 0.951 | 0.952 | 0.103 | 0.911 | 0.926 | 0.920 | 0.127 | 0.889 | 0.864 |
4.3. Ontario prostate cancer MRI data
We apply the proposed method to a prostate cancer imaging study conducted by the Prostate Cancer Imaging Study Team at the University of Western Ontario. The statistical objective is to classify cancer/non-cancer status by examining the in-vivo imaging data such as MRI, CT and ultrasound. The obtained data have correspondence of in-vivo imaging data and ex-vivo histological cancer/non-cancer for each defined voxel in a whole prostate. Twenty-one patients are involved in the first phase of the study. For each patient, three specific MR image intensity measurements on each voxel are used as input variables, and the cancer/non-cancer status of the voxel is used as the class label. All the input measurements are standardized as a usual step for the analysis.
The proposed data-adaptive scaling procedure is applied to the data to classify each voxel to have cancer or non-cancer status. Specifically, a first round of the standard SVM with the Gaussian kernel is conducted, and the support vectors are roughly obtained. The proposed adaptive scaling function is applied to update the kernel function, and a second-stage SVM is then conducted based on the updated kernel. The estimated hyperplane is employed as the rule for classification. To choose suitable tuning parameters M, C and σ for each method, we employ the 7-fold cross validation method, where the 21 patients are randomly grouped into 7 groups with equal sizes; 6 groups of patients are used as training data, and the remaining group is set aside as the test data. The whole process is repeated 100 times to remove possible random splitting bias for the cross validation.
We also analyze the data using the scaling methods of [25] and [23] apart from the traditional SVM, random forest and logistic regression. Training and testing error rates and F-scores for all classifiers are reported. For adaptive scaling SVMs, the number of support vectors and the misclassification rates are reported. ROC curves are included for the comparison of the proposed method with other classifiers.
The result of classifiers with different SVMs are displayed in Table 5. By comparing the training and testing accuracy of the proposed method and other methods, we find that the proposed method generally works better than the competitors. The number of the support vectors obtained from the proposed method is smaller than those yielded from other methods. This can reduce the complexity in high dimensional input space, since the estimated decision surface is the summation of a linear combination of the values of the kernel functions K over all the support vectors found in the first stage; when the number of the support vectors is smaller, the values of the kernel that need to be calculated are much fewer. Compared with other conformal transformation methods that accommodate the location of support vectors, the introduction of the tuning for the location of support vectors can greatly reduce the number of parameters that are needed to be tuned in the validation procedure.
Table 5.
Performance of different classifiers on the prostate cancer image data.
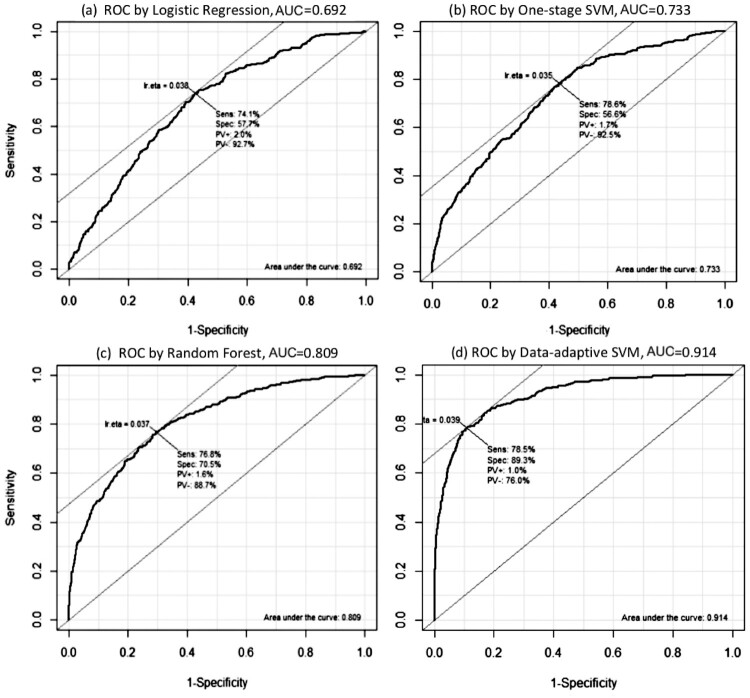
When comparing the proposed method to other classification methods, including Logistic Regression, Random Forest, and one-stage SVM, ROC curves are given in Figure 2. It is evident that the proposed method performs much better than the other classifiers. The prostate cancer image data set has extremely imbalanced cancer/non-cancer labels. It is not surprising the traditional methods perform poorly, while the proposed method can incorporate the imbalance situation and performs well.
Figure 2.
ROC curves produced by Logistic Regression, One-stage SVM, Random Forest and Data-Adaptive SVM with AUC values.
5. Discussion
In this paper, we propose a new method of data-adaptive scaling on the kernel function in the support vector machine. Our method picks the information of the relative position of the support vectors in order to obtain a more robust solution. The model adopts the idea that the Riemannian metric in the feature space introduced by mapping with a kernel can enlarge the spatial separation between two classes, and that the locally adaptive kernel function based on the skewness of the class boundary can enhance the kernel, and hence, increase the accuracy of the classification. Simulation studies and the real data application demonstrate that our method outperforms other classification methods in terms of both accuracy and robustness. Our method can be readily extended to multi-class cases.
To make our method more attractive, we may further pursue research in several aspects. The adaptive kernel involves two stages of the SVM procedure which are time-consuming with large data. If the spatial location of the support vectors can be approximately found and drawn with prior information, the first round of SVM may be removed, and those support vector candidates can be all used in the conformal transformation directly. It would be interesting to develop a more efficient algorithm based on this assumption. Another aspect worth exploration is pertinent to the variable selection. Choosing fewer variables simultaneously in the adaptive scaling transformation can significantly reduce the model complexity, thus reducing the computational cost. When the input variables are contaminated with measurement error, which is quite common in modern scientific studies such as gene expression study in cancer research, the performance of the classifier is likely to be affected. It would be interesting to investigate the measurement error effects on the proposed method and to develop more flexible classification methods for error-prone data.
Acknowledgments
This work has been supported by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) and a team grant from the Canadian Institute of Health Research. Liu's research is partially supported by the Fundamental Funds for Central Universities.
Appendices.
Appendix 1. Proof of Lemma 3.1.
By the definition of a reproducing kernel function with its values and the corresponding scalar eigenfunctions , we have
where . Then the kernel is represented as
By rescaling the function as the kernel function can be further present as
where and is the transpose operator. Thus, if we further define
| (A1) |
and
as in (7) and (9), it follows that
The lemma gives how a mapping is associated with the corresponding kernel function K. Thus, given a specific form of a kernel function and an adaptive scaling function , we have the Propositions 3.2 and 3.3.
Appendix 2. Proof of Proposition 3.2.
Assume the primary kernel function as and a scalar function as in (4). If we define
then by Lemma 3.1,
In particularly when i = j, it is easy to check that
and define them in short as , it follows that
Thus, given a specific form of the primary kernel function K, the adaptive scaling mapping can be calculated. When the Gaussian RBF kernel is applied, we have Proposition 3.3, as proved in the following.
Appendix 3. Proof of Proposition 3.3.
When we apply in Theorem 3.3 the Gaussian RBF kernel as in (11), it is found that
and
for any i and j, so the third term in the result of Proposition 3.2 is 0, and the second term of Proposition 3.2 is changed into . Further, when ,
while when i = j,
thus, the first term of Proposition 3.2 becomes
Combining all the above results, Proposition 3.3 is proved. ♯
Funding Statement
This work has been supported by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC), a team grant from the Canadian Institute of Health Research, and the SHUFE-2018110185 Startup Fund of Shanghai University of Finance and Economics
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Ahmad M., Rajapaksha A.U., Lim J.E., Zhang M., Bolan N., Mohan D., Vithanage M., Lee S.S., and Ok Y.S., Biochar as a sorbent for contaminant management in soil and water: A review, Chemosphere 99 (2014), pp. 19–33. [DOI] [PubMed] [Google Scholar]
- 2.Akbani R., Kwek S., and Japkowicz N.. Applying support vector machines to imbalanced datasets, European Conference on Machine Learning, Springer, 2004, pp. 39–50
- 3.Amari S.i. and Wu S., Improving support vector machine classifiers by modifying kernel functions, Neural. Netw. 12 (1999), pp. 783–789. [DOI] [PubMed] [Google Scholar]
- 4.Batuwita R. and Palade V., Micropred: effective classification of pre-mi s for human mi gene prediction, Bioinformatics 25 (2009), pp. 989–995. [DOI] [PubMed] [Google Scholar]
- 5.Batuwita R. and Palade V.. Efficient resampling methods for training support vector machines with imbalanced datasets, The 2010 International Joint Conference on Neural Networks (IJCNN). IEEE Publishing, 2010, pp. 1–8
- 6.Batuwita R. and Palade V.. Class Imbalance Learning Methods for Support Vector Machines, Imbalanced Learning, chap. 5, Wiley-Blackwell, 2013, pp. 83–99. Available at https://onlinelibrary.wiley.com/doi/abs/ 10.1002/9781118646106.ch5 [DOI]
- 7.Boser B., Guyon I., and Vapnik V.. A training algorithm for optimal margin classifiers, Proceedings of the Fifth Annual Workshop on Computational Learning Theory, ACM, 1992, pp. 144–152
- 8.Chawla N.V., Bowyer K.W., Hall L.O., and Kegelmeyer W.P., Smote: synthetic minority over-sampling technique, J. Artificial Intelligence Res. 16 (2002), pp. 321–357. [Google Scholar]
- 9.Cortes C. and Vapnik V., Support-vector networks, Mach. Learn. 20 (1995), pp. 273–297. [Google Scholar]
- 10.Fawcett T. and Provost F., Adaptive fraud detection, Data. Min. Knowl. Discov. 1 (1997), pp. 291–316. [Google Scholar]
- 11.Fernández A., del Jesus M.J., and Herrera F., Hierarchical fuzzy rule based classification systems with genetic rule selection for imbalanced data-sets, Int. J. Approx. Reason. 50 (2009), pp. 561–577. [Google Scholar]
- 12.Fernández A., García S., del Jesus M.J., and Herrera F., A study of the behaviour of linguistic fuzzy rule based classification systems in the framework of imbalanced data-sets, Fuzzy Sets and Syst. 159 (2008), pp. 2378–2398. [Google Scholar]
- 13.Hastie T., Tibshirani R., and Friedman J., The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed, Springer Series in Statistics, Springer New York, New York, NY, 2009. [Google Scholar]
- 14.Joachims T.. Text categorization with Support Vector Machines: Learning with many relevant features, European Conference on Machine Learning. Springer, Berlin Heidelberg, 1998, pp. 137–142
- 15.Kang P. and Cho S.. EUS SVMs: Ensemble of under-sampled SVMs for data imbalance problems, International Conference on Neural Information Processing, Springer, 2006, pp. 837–846
- 16.Kim Y.. Convolutional neural networks for sentence classification, Preprint (2014) arXiv:1408.5882
- 17.Lessmann S.. Solving Imbalanced Classification Problems with Support Vector Machines, International Conference on Artificial Intelligence, Vol. 4, 2004, pp. 214–220
- 18.Maratea A. and Petrosino A.. Asymmetric kernel scaling for imbalanced data classification, International Workshop on Fuzzy Logic and Applications, 2011. pp. 196–203.
- 19.Schölkopf B. and Smola A.J., Learning with Kernels: Support Vector Machines Regularization, Optimization, and Beyond, MIT press, Cambridge, MA, 2002. [Google Scholar]
- 20.Tong S. and Chang E.. Support vector machine active learning for image retrieval, Proceedings of the Ninth ACM International Conference on Multimedia, ACM, 2001, pp. 107–118
- 21.Vapnik V. and Izmailov R., Knowledge transfer in svm and neural networks, Ann. Math. Artif. Intell. 81 (2017), pp. 3–19. [Google Scholar]
- 22.Wang B.X. and Japkowicz N., Boosting support vector machines for imbalanced data sets, Knowl. Inf. Syst. 25 (2010), pp. 1–20. [Google Scholar]
- 23.Williams P., Li S., Feng J., and Wu S.. Scaling the kernel function to improve performance of the support vector machine, Advances in Neural Networks–ISNN 2005, Springer, 2005, pp. 831–836
- 24.Wu G. and Chang E.Y.. Adaptive feature-space conformal transformation for imbalanced-data learning, International Conference on Machine Learning, 2003, pp. 816–823
- 25.Wu S. and Amari S.I., Conformal transformation of kernel functions: A data-dependent way to improve support vector machine classifiers, Neural Process. Lett. 15 (2002), pp. 59–67. [Google Scholar]
- 26.Yang C.Y., Yang J.S., and Wang J.J., Margin calibration in svm class-imbalanced learning, Neurocomputing 73 (2009), pp. 397–411. [Google Scholar]