Abstract
The histogram has all its bin widths equal to some non-random number arbitrary set by an analyst (EBWH). In the result, particular bin counts are random variables. This paper presents also a histogram that is constructed in a converse manner. Bin counts are all equal to some non-random number arbitrary set by an analyst (EBCH). In the result, particular bin widths are random variables. The first goal of the paper is a choose of constant bin width (of bin numbers k) in the EBWH, which maximize the similarity measure in the Monte Carlo simulation. The second goal is a choose of constant bin count in the EBCH, which maximize the similarity measure in the Monte Carlo simulation. The third goal is to present similarity measures between empirical and theoretical data. The fourth goal is the comparative analysis of two histogram methods by means of the frequency formula. The first additional goal is a tip how to proceed in EBCH when modulo(n,k)≠0. The second additional goal is the software in the form of a Mathcad file with the implementation of EBWH and EBCH.
Subject classification codes: 62G30, 62G07
KEYWORDS: Empirical density function, histogram, random number generator, Monte Carlo simulation, similarity measure
1. Introduction
Histograms are used extensively as non-parametric density estimators both to visualize data and to obtain summary quantities such as the entropy of the underlying density. The histogram is completely determined by two parameters, the bin width and the bin origin.
An important parameter that needs to be specified when constructing a histogram is the bin width . A major issue with all classifying techniques is how to select the number of bins . There is no “best” number of bins. Different values can reveal different features of the data. Using wider bins, where the density of the underlying data points is low, reduces noise due to sampling randomness. Whereas using narrower bins, where the signal drowns the noise, gives greater precision to the density estimation. Thus varying the bin width within a histogram can be beneficial. Nonetheless, equal-width bins are widely used.
Several rules of thumb exist for determining the number of bins, such as the belief that between 5 and 20 bins is usually adequate. Mathematical software such as Matlab uses 10 bins as a default. Scott [24,26], Freedman and Diaconis [10], Sturges [31] and many other statisticians derived formulas for bin width and number of bins. The latest publication on this subject is the paper of Knuth [14]. A large set of formulas for calculating the number of bins is presented in [8]. For more information on bin width and number of bins, please see Sections 5 and 6.
Equal-bin-width histograms (EBWHs) are not statistically efficient for two reasons [30]. First, the bins are blindly allocated, not adapting to the data. Second, the normalized frequency may be zero for many bins and there is no guarantee for the consistency of the density estimates. In [18,21], consistency of estimated data-based histogram densities has been proven and general partitioning schemes have been proposed. [29] suggests a variable bin width histogram by inversely transforming an equal bin width histogram obtained on the transformed data. In [15], denoising method based on variable bin width histograms and minimum description length principle is proposed. In [11], a new projective image clustering algorithm by using variable bin width histogram is proposed.
Another equally important parameter when creating a histogram is the bin origin. The investigator must choose the position of the bin origin (very often by using convenient ‘round’ numbers). This subjectivity can result to misleading estimations because a change in the bin origin can change the number of modes in the density estimation [9,26,28]. Another approach to avoid the problem of choosing the bin origin in the histogram is the ‘moving histogram’ as introduced by Rosenblatt [23]. An improved version of the ‘moving histogram’ is the kernel density estimator (KDE) [22]. One drawback presented by the KDEs is the large number of calculations required to compute them. Scott [25] suggested an alternative procedure to eliminate the influence of the chosen origin and proposed the averaged shifted histogram (ASH). Smoothing makes histograms visually appealing and suitable for application, see e.g. [12,30].
An optimally calibrated EBWH with real data often appears rough in the tails due to the paucity of data. This phenomenon is one of several reasons for considering histograms with adaptive meshes. In practice, finding the adaptive mesh is difficult. Thus some authors have proposed (nonoptimal) adaptive meshes that are easier to implement. One of the more intuitively appealing meshes has an equal number (or equal fraction) of points in each bin. Other adaptive histogram algorithms have been proposed in [37,34].
The adaptive histogram with equal bincounts [26] will be called in the rest of the paper as the equal-bin-count histogram (EBCH). In the literature, there are also the terms: the equal area histogram [6] and the dynamic bin-width histogram [38]. According to [6], the EBWH oversmooths in regions of high density and is poor at identifying sharp peaks. The EBCH oversmooths in regions of low density and so does not identify outliers. We agree with [6] that the EBWH did not attract too much attention in statistical literature and therefore we intend to change it.
This paper puts into consideration histogram that has all bin counts equal to some non-random number arbitrary but reasonably set by an analyst. In the result, particular bin widths are random variables. Mainly it results from the will to improve estimation accuracy of skewed histograms.
Scott [26] examines EBWH vs. EBCH for normal data (well Beta(5,5)) with and using the exact MISE as a function of for the EBWH and for the EBCH. He shows the poor performance of the EBCH when compared with EBWH, with the gap increasing with sample size .
In relation to Scott [26], this paper concerns not only the normal distribution but also the other three distributions divided into five groups according to the skewness value. Monte Carlo simulations are carried out for sample sizes using an appropriately defined similarity measure.
The first goal of the paper is a choose of bin numbers (of constant bin width) in the EBWH for analyzed distributions and sample sizes , which maximize the similarity measure in the Monte Carlo simulation. The second goal is a choose of constant bin count (of bin widths) in the EBCH for analyzed distributions and sample sizes, which maximize the similarity measure in the Monte Carlo simulation. The third goal is to present similarity measures between empirical and theoretical data. The fourth goal is the comparative analysis of two histogram methods by means of the frequency formula. The first additional goal is a tip how to proceed in EBCH when . The second additional goal is the software in the form of a Mathcad file (version 14) with the implementation of EBWH and EBCH.
This paper is arranged as follows. Section 2 is devoted to the preliminaries. Here is a tip how to proceed in EBCH when . Section 3 presents distributions selected to the Monte Carlo study. Section 4 presents distance and similarity measures between empirical and theoretical data. Section 5 is devoted to the EBWH whereas Section 6 is devoted to the EBCH. Section 7 compares analyzed histograms. Section 8 presents simulation and real data examples. The paper ends with conclusions. Tables 4–21, due to the size of the paper, are saved in a separate file as additional materials.
2. Preliminaries
The artistry of building the histograms consists in balancing between rough-hewn histogram and waving one. It is exemplified in Figure 1. The sample of n = 30 items on the basis of which the histograms were built was drawn from the Normal N(0,1) general population. The numerator in a histogram formula is the probability for the sample item to fall into particular bin.
Figure 1.
The EBWH, , bin count (left), (right), .
Let us employ the coefficient of variation (CoV) denoted of the binomial distribution as a measure of confidence of ith histogram segment
In the case of the Binomial distribution, we have
The coefficient of entire variation (CoEV) is
Replacing unknown with we get the sample CoV:
and the sample CoEV
The CoEV enables comparing confidences of two or more histograms.
Let us consider the Normal distribution censored from both sides at at (two areas of cut tiles are negligible) and build non-random histogram of five bins. In histogram of such name particular bin count is a product of sample size and a difference of the normal cumulative distribution functions calculated at bin borders. The results are presented in Table 1.
Table 1. The EBWH, bin width .
| Bin No. | Bin borders | Bin probabilities | ||
|---|---|---|---|---|
| 1 | −3 | −1.8 | 0.035 | 0.965 |
| 2 | −1.8 | −0.6 | 0.238 | 0.326 |
| 3 | −0.6 | 0.6 | 0.451 | 0.201 |
| 4 | 0.6 | 1.8 | 0.238 | 0.326 |
| 5 | 1.8 | 3 | 0.035 | 0.965 |
| 2.783 | ||||
What relates to the histogram of equal bin counts, all bin probabilities are, of course, equal to . So, all and . The histogram proposed in this paper has the accuracy of particular bins stabilized and is intended for use mainly on skewed histograms.
Let us consider another example. A sample of items is drawn from general population in which the feature of our interest denoted follows the lognormal distribution with PDF . Figure 2 shows the EBWH (left) and the EBCH (right).
Figure 2.
EBWH (left) and EBCH (right), , the lognormal distribution.
It is important that the EBCH reveals the mode when the EBWH does not!
At the end of this section, there is a tip how to proceed in EBCH when .
Create a matrix.
Locate sample elements in the first column of .
Sort the first column of in ascending order.
Set and .
Generate uniformly distributed random numbers .
Set .
Order according to values of the second column.
Calculate .
From the whole ordered sample, select a subsample .
Sort the subsample in ascending order.
On the basis of the above subsample, determine bin widths.
Add remaining sample elements to bins they belong to.
Calculate bin ordinates.
3. Distributions selected to the Monte Carlo study
The lognormal distribution is a very good distribution for skewness modeling. The second distribution selected for skewness modeling is the generalized gamma distribution . This distribution includes as special cases a number of well-known distributions in statistics and theory of reliability [32]. Table 2 presents the family of these distributions. A key advantage of is not that already existing distributions are special cases of , but that it fills the “space” between them. In other words, allows the parametric description of empirical distributions that are indescribable by the traditional distributions. Parameter values of were selected to obtain the same levels of skewness as
Table 2. Groups of distributions selected to the Monte Carlo study.
| Group name | Distribution | Skewness value |
|---|---|---|
| Zero skewness | ||
| Constant skewness | ||
| Weak skewness | ||
| Medium skewness | ||
| Strong skewness |
Monte Carlo study is carried out based on distributions as follows: standard normal , exponential , . These distributions are divided into five groups (see Table 2).
Figure 3 presents PDF of and for selected parameter values and skewness values (in parenthesis).
Figure 3.
PDF of and with parameter values and skewness values (in parenthesis).
4. Distance and similarity measures between empirical and theoretical data
In the literature, there are many definitions of distance and similarly measures between empirical and theoretical PDFs. These measures are defined for discrete, finite values of and by the integral for all in the domain. Discrete measure are reviewed and categorized in both syntactic and semantic relationships [4]. Below we define four integral measures: distance measures and similarity measures . measure the distance between PDFs and compare the area under PDFs.
The first popular distance measure is the mean square error (MSE) defined as
If and are identical, then . No maximum exists when PDFs are not identical. According to [6], relying on asymptotics of the IMSE leads to inappropriate recommendations.
The second distance measure similar to the Pearson's chi-square statistic is also noteworthy [6]
If and are identical, then . No maximum exists when PDFs are not identical.
The third measure is the Matusita distance [19] given by
If and are identical, then . No maximum exists when PDFs are not identical.
The next measure is the Bhattacharyya similarity measure [1] defined as
Measure takes values in interval . If and are identical, then . If for any , we have and , then . The same value of integral function we obtain for and .
We propose a new similarity measure between the actual PDF and the empirical PDF defined as
The similarity measure takes values in interval . If and are identical, then . Figure 4 presents the graphical representation of .
Figure 4.
The graphical interpretation of the new similarity measure. The shaded area reflects the measure .
Why is the measure better than ? Figure 5 shows PDFs of and . The measure is more reliable than and will be used in the simulation study.
Figure 5.
N(0,1) versus N(3,1). Similarity study, .
5. Equal-bin-width histogram (EBWH)
Many statisticians have attempted to determine an optimal value, but these methods generally make strong assumptions about the shape of the distribution. Depending on the actual data distribution and the goals of the analysis, different values may be appropriate, so experimentation is usually needed to determine an appropriate width. There are, however, various useful guidelines and rules of thumb.
Let us define the set of ordered sample elements as . Sturges [30] proposed the rule of determining numbers of bins defined as . In practice, according to Scott [24], Sturges’ rule is applied by dividing the sample range into . Technically, Sturges’ rule is a number-of-bins rule rather than a bin-width rule. Much simpler is to adopt the convention that all histograms have an infinite number of bins, only a finite number of which are nonempty. Further, the equal-bin-count histogram (EBCH), which are considered in next section, does not use equal-width bins. Thus focusing on bin width rather than number of bins seems appropriate.
In this section, we compare 17 rules for determining bin width .
The Scott's normal rule is defined as [24]
where is the sample standard deviation, is optimal for random samples of normally distributed data as it minimizes the IMSE of the density estimate [23].
The Freedman–Diaconis normal rule, based on the percentiles, is defined by [10]
If the data are in fact normal, the rule is about 77% of the rule as the in this case.
The third rule, which refers to minimizing cross-validation estimated squared error, is an approach of minimizing IMSE from Scott's normal rule. Using leave-one-out cross validation, it can be generalized beyond normal distribution. The third rule has a form [36]
where is the number of data points in the th bin, and choosing the value of (which minimizes ) will minimize IMSE.
The fourth rule is as follows: the optimal bin width is obtained by minimizing the cost function [27]
where
The Doane rule attempts to improve its performance in the case of non-normal data and is given by [7]
where is the estimated third-moment-skewness.
The Mosteller–Tukey rule [20], used inter alia in Microsoft Excel, is given by
The Sturges rule has the form [31]
Rules comes from a binomial distribution and implicitly assumes an approximately normal distribution. This formula can perform poorly for and also in case of data that are not normally distributed.
The Rice rule [8], being a simple alternative to , is given by
Others rules are as follows [8]:
Cochran rule [5]: ,
Cencov rule [3]: ,
Bendat and Piersol rule [2]: ,
Larson rule [17]: ,
Velleman rule [35]: ,
Terrel and Scott rule [33]: ,
Ishikawa rule [13]: ,
Anonymous 1 rule [8]: ,
Anonymous 2 rule [8]: .
Rules are defined using only the sample size and therefore Table 3 lists number of bins for these rules and sample sizes used in the simulation study. It can be observed that there is a great diversity of k values obtained.
Table 3. Number of bins for selected sample size and rules .
| n | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 3 | 4 | 4 | 1 | 2 | 4 | 3 | 6 | 2 | 6 | 4 | 3 |
| 15 | 3 | 4 | 4 | 1 | 2 | 5 | 3 | 7 | 3 | 6 | 4 | 3 |
| 20 | 4 | 5 | 5 | 2 | 2 | 6 | 3 | 8 | 3 | 6 | 5 | 4 |
| 25 | 5 | 5 | 5 | 2 | 2 | 6 | 4 | 10 | 3 | 6 | 5 | 4 |
| 30 | 5 | 5 | 6 | 2 | 3 | 7 | 4 | 10 | 3 | 6 | 5 | 4 |
| 40 | 6 | 6 | 6 | 2 | 3 | 8 | 4 | 12 | 4 | 6 | 6 | 5 |
| 50 | 7 | 6 | 7 | 3 | 3 | 8 | 4 | 14 | 4 | 7 | 6 | 5 |
| 60 | 7 | 6 | 7 | 3 | 3 | 9 | 4 | 15 | 4 | 7 | 6 | 5 |
| 80 | 8 | 7 | 8 | 4 | 4 | 10 | 5 | 17 | 5 | 7 | 7 | 6 |
| 100 | 10 | 7 | 9 | 4 | 4 | 11 | 5 | 20 | 5 | 8 | 7 | 6 |
The rules determining bin width are compared in the Monte Carlo simulations using the similarity measure. The algorithm calculating values of the similarity measure
| (1) |
is as follows:
Set sample size n.
- Repeat the following steps times:
- Generate random sample based on distribution with PDF .
- Present obtained values in an increasing order, i.e. .
- Calculate bin width according to an appropriate formula.
- Calculate number of bins .
- Set bins .
- Calculate bin counts .
- Calculate empirical density function .
- Calculate value of the similarity measure according to (1)
Calculate value of the similarity measure .
Monte Carlo simulations are carried out for five groups of distributions. The similarity measure is calculated for a given sample size and rules based on the same data. Values do not significantly affect values. The rules and are calculated, according to the definition, for the SN distribution only. The rules and give bad results for skewness groups of distribution, e.g. for and we have ( ), ( ). Therefore and are calculated for the zero skewness group only. Tables 4–12 (see additional materials) show values for analyzed distributions. The highest values for a given sample size are in bold. Rules , which maximizes for a given sample size, will be used in Section 7 (see Tables 22–25).
Table 26. The measure for rules in the EBWH and for bin count in the EBCH, , .
| 0.479 | 0.575 | 0.437 | 0.531 | 0.275 | 0.275 | 0.615 | 0.531 | 0.719 | 0.332 |
| 0.485 | 0.437 | 0.386 | 0.731 | 0.760 | 0.764 | 0.742 | 0.652 | 0.572 | 0.404 |
Table 22. Values of the frequency formula . Zero and constant skewness group.
| Zero skewness group | Constant skewness group | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.203 | 0.193 | 0.499 | |||||||
| 0.386 | 0.305 | 0.507 | |||||||
| 0.292 | 0.267 | 0.433 | |||||||
| 0.231 | 0.219 | 0.417 | |||||||
| 0.242 | 0.220 | 0.385 | |||||||
| 0.203 | 0.173 | 0.356 | |||||||
| 0.142 | 0.125 | 0.306 | |||||||
| 0.148 | 0.124 | 0.293 | |||||||
| 0.094 | 0.087 | 0.244 | |||||||
| 0.049 | 0.058 | 0.181 | |||||||
Table 23. Values of the frequency formula . Weak skewness group.
| 0.284 | 0.303 | |||||
| 0.263 | 0.210 | |||||
| 0.283 | 0.150 | |||||
| 0.277 | 0.166 | |||||
| 0.272 | 0.192 | |||||
| 0.247 | 0.154 | |||||
| 0.175 | 0.140 | |||||
| 0.201 | 0.151 | |||||
| 0.134 | 0.091 | |||||
| 0.092 | 0.092 | |||||
Table 24. Values of the frequency formula . Medium skewness group.
| 0.364 | 0.57 | |||||
| 0.340 | 0.679 | |||||
| 0.330 | 0.713 | |||||
| 0.264 | 0.725 | |||||
| 0.254 | 0.722 | |||||
| 0.230 | 0.680 | |||||
| 0.16 | 0.676 | |||||
| 0.172 | 0.719 | |||||
| 0.230 | 0.710 | |||||
| 0.187 | 0.696 | |||||
Table 25. Values of the frequency formula . Strong skewness group.
| 0.433 | 0.941 | |||||
| 0.495 | 0.982 | |||||
| 0.45 | 4 | 0.991 | ||||
| 0.414 | 0.995 | |||||
| 0.408 | 5 | 0.997 | ||||
| 0.325 | 5 | 1 | ||||
| 0.265 | 5 | 1 | ||||
| 0.229 | 1 | |||||
| 0.28 | 1 | |||||
| 0.24 | 1 | |||||
6. Equal-bin-count histogram(EBCH)
In the EBWH, all the bins are of equal width that is arbitrary prescribed. Bin counts are random numbers. In the EBCH, the bin counts are arbitrary prescribed to be equal in all the bins. The width of each particular bin is adjusted so that bin counts to be equal. Bin borders are set precisely in the middle between two neighboring observations that have fallen into previous and next bin.
Let us define the set of ordered sample elements as . If denotes a bin count, then the number of bins is given by . The bin count is divisor ( ). The rules in the EBWH are marked as , so the bin widths in EBCH are marked as .
The bin width can be calculated as
| (2a) |
| (2b) |
A set of bins can be written as
| (2c) |
Empirical density function is defined as
| (3) |
where and obviously.
The similarity measure related to the EBCH is given by
| (4) |
The Monte Carlo simulation is carried out for the optimal constant bin count which maximizes . Simulation results for a given sample size and constant bin counts are obtained based on the same data.
The algorithm calculating values of the similarity measure (4) is as follows:
Set a sample size n.
Set a bin count , where is divisor ( ).
Calculate the number of bins .
- Repeat the following steps times:
- Generate random sample distributed according to distribution.
- Present obtained values in an increasing order, i.e. .
- Calculate bin width according to (2a)–(2b).
- Set bins .
- Calculate empirical density function (3).
- Calculate value of the similarity measure (4).
Calculate value of the similarity measure
Tables 13–21 (see additional materials) show the optimally selected bin count determined by the highest value of for a given distribution and a sample size. The value increases with a sample size . The optimally selected bin count will be used in Section 7 (Tables 22–25).
7. EBWH vs. EBCH – comparative study
This section is devoted to the comparative study of the EBWH and EBCH by means of the frequency formula. The Monte Carlo study uses the results presented in Sections 5 and 6. These results were calculated to obtain optimal bin widths in the EBWH and optimal constant bin counts in the EBCH.
The rules (r) with the highest value of (Tables 4–12) and the bin counts with the highest value of (Tables 13–21) are selected for the comparative study. The algorithm describing this study is as follows:
Set a counter .
- Repeat the following steps times:
- Calculate the similarity measure related to the EBWH with optimally selected rules in Section 5.
- Calculate the similarity measure related to the EBCH with optimally selected η values in Section 6.
- If , then .
Calculate a frequency formula .
Tables 22–25 present the values of the frequency formula for five distribution groups. The cases with for a given sample size are in bold.
The EBWH is recommended to the zero skewness group (Table 22), the constant skewness group (Table 22) and the weak skewness group of unimodal distributions (Table 23) (see Figure 3).
An advantage of the EBCH over the EBWH (C greater than 0.5) becomes apparent when population distributions are both modeless (see Figure 3) and of medium or great skewness (Tables 24 and 25). The greater the skewness, the greater an advantage of the EBCH over the EBWH. Examples 1–5 confirm this fact.
8. Examples
8.1. Simulation examples
Example 1. Figure 6 compares the EBWH and EBCH for the SN distribution and sample . Optimally selected rule is (Table 22), optimally selected (Table 22). Similarity measure for the EBWH is higher than for the EBCH.
Figure 6.
EBWH (left, ) versus EBCH (right, ) for SN distribution and .
To show an advantage of the EBCH over the EBWH, we will use three distributions. The first one is the generalized gamma distribution (Example 2), the second one is the compound normal distribution (Example 3) and the third one is the lognormal distribution (Examples 4 and 5).
Example 2. Figure 7 compares histograms for the and . Optimally selected rule is (Table 24), optimally selected (Table 24). Similarity measure for the EBCH is higher than for the EBWH.
Figure 7.
EBWH (left, ) versus EBCH (right, ) for distribution and .
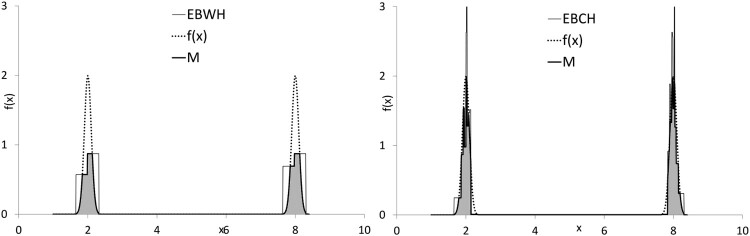
Example 3. Figure 8 compares histograms for the and . Simulation study shows (see Table 26) that optimally selected rule is and optimally selected . Similarity measure for the EBCH is higher than for the EBWH.
Figure 8.
EBWH (left, ) versus EBCH (right, ) for distribution and .
8.2. Resampling from real data examples
Determining histograms is a form of statistical reasoning. Therefore, this paper becomes devoted to assessing a power of statistical reasoning. In order to act properly “assessors” have to know a matter of facts to state whether reasoning is false or true. Assessors’ task causes examples presented in the paper differ from real data examples. The conclusions are drawn on the basis of results of the Monte Carlo experiment that serves to carry out resampling. For this reason, the following requirements must be met before applying the reasoning scheme proposed in this paper:
Analytical form of a distributions that “holds” in considered general populations has to be known with accuracy to parameter values! Otherwise, actuality remains unknown and results of statistical reasoning are not assessable.
Data used to determine histograms have to be exact data, i.e. not grouped and not censored. Otherwise, histograms cannot be determined.
The whole histogram competition is performed into eight steps:
Dig up a set of real data that seem to come from evidently abnormal population.
On the of-chance perform by-eye goodness-of-fit test plotting data on the Gaussian probability paper.
Having abnormality confirmed choose such a theoretical probability distribution that fits data in question. Further it will be treated as and called the actual distribution. Like in Step 2 perform appropriate by-eye goodness-of-test.
Having correctness of the choice of step 3 confirmed estimate parameters of the actual distribution.
Get a sample with the Monte Carlo resampling method from the actual distribution.
Determine EBWH and EBCH for optimally selected rule and bin count.
Calculate similarity measures of both histograms to the actual distribution.
Compare histograms with respect to similarity measure and point out a winner.
Example 4. Lai and Xie [16] present data from a lung cancer study for the treatment of veterans. Data represent survival days for 97 patients with lung cancer from therapy (see Table 27), skewness equals 1.944.
Figure 9.
Gaussian probability paper for survival days from Example 4 (left, ) and repair times from Example 5 (right, ).
Figure 10.
Lognormal probability paper for survival days from Example 4 (left, ) and repair times from Example 5 (right, ).
Figure 11.
Empirical and theoretical lognormal quantile function for survival days from Example 4 (left, ) and repair times from Example 5 (right, ).
Figure 12.
EBWH (left, ) versus EBCH (right, ) for survival days from Example 4, .
Figure 13.
EBWH (left, ) versus EBCH (right, ) for repair times from Example 5, , .
Table 28. The measure for rules in the EBWH and for bin count in the EBCH, , .
| 0.766 | 0.749 | 0.694 | 0.735 | 0.587 | 0.587 | 0.761 | 0.735 | 0.812 | 0.631 |
| 0.715 | 0.694 | 0.666 | 0.756 | 0.801 | 0.813 | 0.831 | 0.798 | 0.773 | 0.630 |
Table 29. The measure for rules in the EBWH and for bin count in the EBCH, , .
| 0.790 | 0.763 | 0.746 | 0.763 | 0.631 | 0.631 | 0.775 | 0.763 | 0.795 | 0.684 |
| 0.763 | 0.746 | 0.720 | 0.744 | 0.794 | 0.789 | 0.666 |
Table 27. Computer implementation of Examples 4 and 5.
| Step of above algorithm | Example 4 | Example 5 |
|---|---|---|
| 1 | Survival days | Repair times |
| 2 | A population the data come from is definitely abnormal (Figure 9, left) | A population the data come from is definitely abnormal (Figure 9, right) |
| 3 | The data come from the lognormal population (Figure 10, left) | The data come from the lognormal population (Figure 10, right) |
| 4 | The actual distribution that holds in the population in question is the (Figure 11, left) | The actual distribution that holds in the population in question is the (Figure 11, right) |
| 5 | ||
| 6 | Optimally selected rules is and bin count (Table 28). Comparison of histograms for analyzed data (Figure 12) | Optimally selected rules is and bin count (Table 29). Comparison of histograms for analyzed data (Figure 13) |
| 7 | Similarity measure for the EBCH is higher than for the EBWH | Similarity measure for the EBCH is higher than for the EBWH |
Example 5. Lai and Xie [16] present 46 repair times (in hours) for an airborne communication transceiver (see Table 27), skewness equals 2.987.
What causes that in many cases the EBCH more precisely reflects the actual density function than the EBWH? Because EBWH is insensitive to contents of data set. Notice that EBWH is determined mainly by extreme values of data set. The content is mechanically sliced. In contrast to this in case of EBCH the content is sliced intelligently for equilibrated accuracy of local density estimates.
The software in the form of a Mathcad file (version 14) implementing EBWH and EBCH for sample size n is available at https://sulewski.apsl.edu.pl/index.php/publikacje. This file also contains a tip how to proceed in EBCH when . If you want to compare both histograms, use the theoretical distribution.
9. Conclusion
The EBWH is recommended to symmetric distributions and distributions with constant nonzero skewness (e.g. exponential distribution). The EBCH is recommended to asymmetric distributions, especially in situations when skewness is not weak and empirical PDF is modeless. It has been proved that the greater skewness the greater the advantage of the EBCH over the EBWH. The EBCH should be used for bimodal distributions (see Examples 3 and 4). The greater distance between modes the greater the advantage of the EBCH over the EBWH.
Supplementary Material
Acknowledgements
The author is grateful to the unknown Referees and the Associate Editor for their valuable comments that contributed to the improvement of the original version of the paper.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Aherne F.J., Thacker N.A., and Rockett P.I., The Bhattacharyya metric as an absolute similarity measure for frequency coded data. Kybernetika 34(4) (1998), pp. 363–368. [Google Scholar]
- 2.Bendat S.J., and Piersol A.G., Measurements and Analysis of Random Data, John Wiley & Sons, New York, 1966. [Google Scholar]
- 3.Cencov N.N., Estimation of an unknown distribution density from observations. Soviet Math 3 (1962), pp. 1559–1566. [Google Scholar]
- 4.Cha S.H., Comprehensive survey on distance/similarity measures between probability density functions. City 1(2) (2007), pp. 1. [Google Scholar]
- 5.Cochran W.G., Some methods for strengthening the common χ 2 tests. Biometrics 10(4) (1954), pp. 417–451. doi: 10.2307/3001616 [DOI] [Google Scholar]
- 6.Denby L., and Mallows C., Variations on the histogram. J. Comput. Graph. Stat. 18(1) (2009), pp. 21–31. doi: 10.1198/jcgs.2009.0002 [DOI] [Google Scholar]
- 7.Doane D.P., Aesthetic frequency classifications. Am. Stat. 30(4) (1976), pp. 181–183. [Google Scholar]
- 8.Doğan İ, and Doğan N., Determination of the number of bins/classes used in histograms and frequency tables: a short bibliography. TurkStat. J. Stat. Res. 7(2) (2010), pp. 77–86. [Google Scholar]
- 9.Fox J., Describing univariate distributions, in Modern Methods of Data Analysis, Fox J., Long J.S., eds., Sage, London, 1990. pp. 58–125. [Google Scholar]
- 10.Freedman D., and Diaconis P., On the histogram as a density estimator: L 2 theory. Zeitschr. Wahrsch. Theor. Verwan. Gebiete 57(4) (1981), pp. 453–476. doi: 10.1007/BF01025868 [DOI] [Google Scholar]
- 11.Gao S., Zhang C., and Chen W.B., A variable bin width histogram based image clustering algorithm. In 2010 IEEE Fourth International Conference on Semantic Computing (1990), pp. 166–171.
- 12.Hardle W., and Scott D.W., Smoothing by weighted averaging of rounded points. Comp. Stat 7 (1992), pp. 97–128. [Google Scholar]
- 13.Ishikawa K., Guide to Quality Control, Unipub, Kraus International, White Plains, New York, 1986. [Google Scholar]
- 14.Knuth K.H., Optimal data-based binning for histograms and histogram-based probability density models. Digit. Signal. Process. 95 (2019), pp. 102581. 10.1016/j.dsp.2019.102581 [DOI] [Google Scholar]
- 15.Kumar V., and Heikkonen J., Denoising with flexible histogram models on minimum description length principle. arXiv preprint arXiv:1601.04388.
- 16.Lai C.D., and Xie M., Stochastic Ageing and Dependence for Reliability, Springer Science & Business Media, New York, 2006. [Google Scholar]
- 17.Larson H.J., Statistics: An Introduction, John Wiley & Sons, New York, 1975. [Google Scholar]
- 18.Lugosi G., and Noble A.B., Consistency of data-driven histogram methods for density estimation and classification. Ann. Stat. 24 (1996), pp. 687–706. doi: 10.1214/aos/1032894460 [DOI] [Google Scholar]
- 19.Matusita K., Decision rules based on distance for problems of fit, two samples and estimation. Ann. Math. Statist 26 (1955), pp. 631–641. doi: 10.1214/aoms/1177728422 [DOI] [Google Scholar]
- 20.Mosteller F., and Tukey J.W., Data Analysis and Regression, A Second Course in Statistics, Addison-Wesley, Reading, MA, 1977. [Google Scholar]
- 21.Nobel A.B., Histogram regression estimation using data dependent partitions. Ann. Stat. 24(3) (1996), pp. 1084–1105. doi: 10.1214/aos/1032526958 [DOI] [Google Scholar]
- 22.Parzen E., On estimation of a probability density function and mode. Ann. Math. Statist. 33(3) (1962), pp. 1065. doi: 10.1214/aoms/1177704472 [DOI] [Google Scholar]
- 23.Rosenblatt M., Remarks on some nonparametric estimates of a density function. Ann. Math. Statist. 27 (1956), pp. 832–837. doi: 10.1214/aoms/1177728190 [DOI] [Google Scholar]
- 24.Scott D.W., On Optimal and Data-Based Histograms, Biometrika 66(3) ( 1979), pp. 605-610. [Google Scholar]
- 25.Scott D.W., Averaged shifted histograms: effective nonparametric density estimators in several dimensions. Ann. Stat. 13 (1985), pp. 1024–1040. doi: 10.1214/aos/1176349654 [DOI] [Google Scholar]
- 26.Scott D.W., Multivariate Density Estimation: Theory, Practice, and Visualization, John Wiley and Sons, New York, 2015. [Google Scholar]
- 27.Shimazaki H., and Shinomoto S., A method for selecting the bin size of a time histogram. Neural Comput. 19(6) (2007), pp. 1503–1527. doi: 10.1162/neco.2007.19.6.1503 [DOI] [PubMed] [Google Scholar]
- 28.Silverman B.W., Density Estimation for Statistics and Data Analysis, Chapman & Hall, London, 1986. [Google Scholar]
- 29.Simonoff J.S., Smoothing Methods in Statistics, Springer-Verlag, New York, 1996. [Google Scholar]
- 30.Song M., and Haralick R.M., Optimally Quantized and Smoothed histograms, Proceedings of the Joint Conference of Information Sciences, Durham, NC, 21(153) 2002, pp. 894–897. [Google Scholar]
- 31.Sturges H.A., The choice of a class interval. J. Am. Stat. Assoc. (1926), pp. 65–66. doi: 10.1080/01621459.1926.10502161 [DOI] [Google Scholar]
- 32.Sulewski P., Uogólniony rozkład gamma w praktyce statystyka [Generalized gamma distribution in statistic practice], Akademia Pomorska, Słupsk, 2008, in Polish.
- 33.Terrel G.R., and Scott D.W., Oversmoothed nonparametric density estimates. J. Am. Stat. Assoc. 80(389) (1985), pp. 209–214. doi: 10.1080/01621459.1985.10477163 [DOI] [Google Scholar]
- 34.Van Ryzin J., A histogram method of density estimation. Comm. Statist 2 (1973), pp. 493–506. doi: 10.1080/03610927308827093 [DOI] [Google Scholar]
- 35.Velleman P.F., Interactive computing for exploratory data analysis I: Display algorithms. 1975 Proceedings of the Statistical Computing Section (1976), pp. 142–147, Washington, DC: American Statistical Association.
- 36.Wasserman L., All of Statistics, Springer, New York, 2004. [Google Scholar]
- 37.Wegman E.J., Maximum Likelihood estimation of a unimodal density function. Ann. Statist 41 (1970), pp. 457–471. doi: 10.1214/aoms/1177697085 [DOI] [Google Scholar]
- 38.Yang J., Li Y., Tian Y., Duan L., and Gao W., Multiple kernel active learning for image classification. In 2009 IEEE International Conference on Multimedia and Expo (2009), pp. 550–553.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.