Abstract
Indole-3-acetic acid (IAA) plays an important role in the growth and development of plants. In this study, a series of predominant strains were isolated and identified as Enterobacter sp. with remarkable IAA-producing capabilities. The IAA-producing strains are mainly tryptophan-dependent and have significantly high yields of IAA (3477 μg mL−1 and 3378 μg mL−1). The ipdC gene encoding indole-3-pyruvate decarboxylase was identified by genomic analysis and RT-qPCR analysis, indicating the involvement of the indole-3-pyruvic acid (IPyA) pathway of IAA biosynthesis. The IPyA pathway was also confirmed by the intermediate assay. The IAA product of microbial metabolites was isolated, purified and characterized. These microbes exhibiting IAA production significantly promoted the growth of maize, increasing root length, plant height, fresh weight and dry weight. Thus, Enterobacter sp. with high IAA production has great prospects in agricultural and industrial applications.
The strains have remarkable IAA-producing capabilities. Genomic analysis and intermediate assay indicated the involvement of the indole-3-pyruvic acid pathway of IAA biosynthesis. These microbes significantly promoted the growth of maize.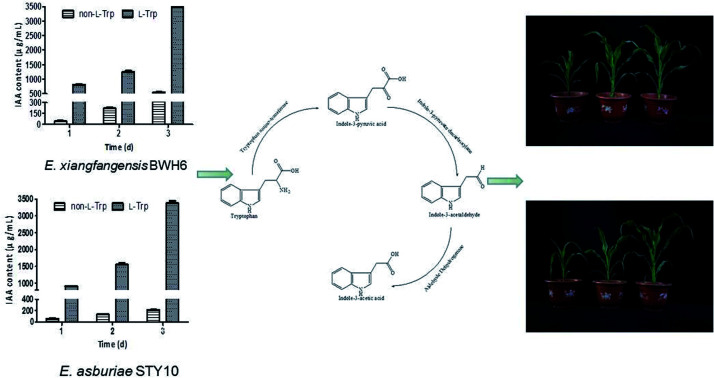
Introduction
Phytohormones, such as auxins and cytokinins, abscisic acids (ABA) and gibberellins (GAs), participate in many biological processes in plants. Indole-3-acetic acid (IAA), as one of the most abundant auxins, plays a key role in the regulation of various physiological processes such as cell division and elongation, vascular differentiation, gravitropism and phototropism.1 IAA can be synthesized by both plants and microbes,2–4 and IAA produced by microbes could promote plant growth.2
Numerous bacteria are capable of producing IAA, such as Bacillus, Enterobacter, Pseudomonas, Azospirillum, Agrobacterium and Rhizobium.3,4 Generally, IAA production has been obtained in a range of 10–200 μg mL−1 concentrations with or without the addition of 0.2–2 μg mL−1 of l-tryptophan (l-Trp).5–8 Recently, remarkable IAA production was achieved at concentrations of 200–250 μg mL−1 with Rheinheimera sp., Rhizobium sp. and Bacillus subtilis.9
IAA biosynthesis in microbes is accomplished by both tryptophan-dependent and tryptophan-independent pathways. In particular, l-Trp is an efficient precursor for IAA biosynthesis. In general, the following pathways are the main pathways involved in the conversion of tryptophan into IAA.10 The indole-3-pyruvate acid (IPyA) pathway (l-Trp → IPyA → indole-3-acetaldehyde (IAAld) → IAA) has been studied in detail in Enterobacter cloacae and Bacillus amyloliquefaciens FZB42.8 The indole-3-acetamide (IAM) pathway (l-Trp → IAM → IAA) has been discovered in Streptomyces spp.11 The tryptamine (TAM) pathway (l-Trp → TAM → IAAld → IAA) has been reported in Bacillus cereus.12 The tryptophan side chain oxidase (TSO) pathway (l-Trp → IAAld → IAA) has been demonstrated in Pseudomonas fluorescens CHA0.13 The indole-3-acetonitrile (IAN) pathway (l-Trp → IAN → IAA) has been confirmed in several strains of Agrobacterium and Rhizobium.14
In this study, E. xiangfangensis BWH6 and E. asburiae STY10 with high IAA production were isolated and identified. IAA-producing strains were mainly l-Trp-dependent. Genome analysis was conducted to predict the crucial gene participating in IAA biosynthesis, followed by RT-qPCR to identify gene expression associated with the key enzymes. An intermediate assay was performed to investigate the IAA synthetic pathway. Column chromatography was employed for the isolation, purification and characterization of IAA products of microbial metabolites. IAA-producing strains were further investigated for their effect on the growth of maize.
Results
Determination of phytohormones
IAA plays a central role in modulating plant growth and development. A number of microorganisms in the rhizosphere, such as Agrobacterium, Bacillus, Pseudomonas, and Rhizobium, are able to produce IAA.15
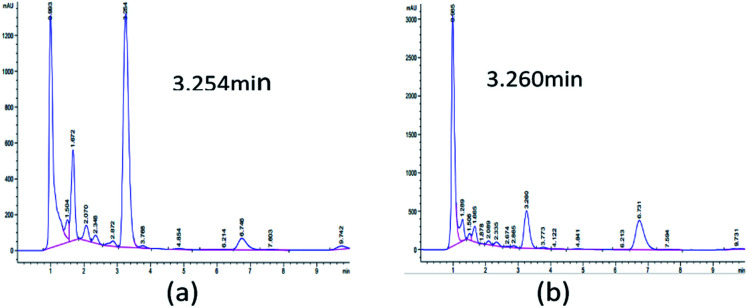
Initially, 10 strains were identified by Salkowski's reagent colorimetry to be able to produce IAA.16 The strain STY10 was isolated from maize root endophytes, and the strain BWH6 was isolated from maize rhizosphere soil.17 IAA in microbial metabolites was analysed by HPLC to ensure sensitive and accurate measurement. The retention times of ABA, GA3, IAA and ZA were 5.451, 1.719, 3.260 and 1.226 min, respectively. When the bacterial strains were incubated in LB liquid medium at 37 °C for 3 days, 553 μg mL−1 and 214 μg mL−1 concentrations of IAA were obtained from the strains BWH6 and STY10, respectively (Fig. 1a and b). Kumla et al. described the highest IAA level observed for Astraeus odoratus (54.56 ± 2.21 μg mL−1), followed by Pisolithus orientalis (42.27 ± 3.18 μg mL−1), Phlebopus portentosus (40.72 ± 2.87 μg mL−1), Pisolithus albus (32.65 ± 3.25 μg mL−1) and Gyrodon suthepensis (22.56 ± 2.32 μg mL−1).5 Compared with the current studies, remarkable IAA production was achieved from BWH6 and STY10 in the absence of l-Trp.
Fig. 1. HPLC analysis of phytohormones. Strains BWH6 (a) and STY10 (b) were incubated in LB liquid medium at 37 °C for 3 days.
Molecular identification
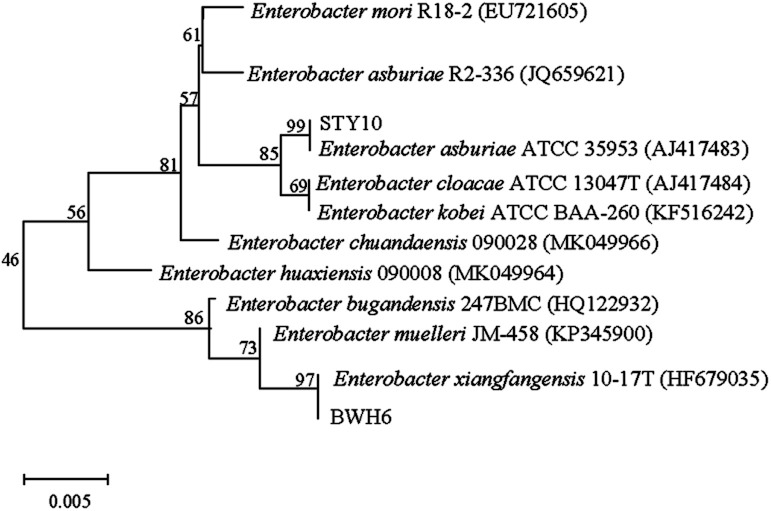
In this study, bacterial strains were identified based on 16S rRNA gene sequencing (Fig. 2). Strain BWH6 was identified as Enterobacter xiangfangensis BWH6, as it showed 99.94% similarity with E. xiangfangensis 72231 (MN304301). Strain STY10 was identified as Enterobacter asburiae STY10, as it showed 99.94% similarity with E. asburiae R2-336 (JQ659621). The obtained nucleotide sequences were submitted to NCBI GeneBank under accession no. MN696244 for E. xiangfangensis BWH6 and MZ303737 for E. asburiae STY10.
Fig. 2. Phylogenetic relationships of strains. The accession numbers for the sequences used are shown in parentheses after the strain designation. Bootstrap values are shown for each node that had >50% support in a bootstrap analysis of 1000 replicates. The scale bar indicates 0.005 substitutions per site.
IAA production based on l-tryptophan
Generally, l-Trp is considered a precursor of IAA, and it can be obtained from root exudates or released from degrading roots and microbial cells, which are the natural sources of l-Trp for the rhizosphere microbes that may enhance auxin biosynthesis in the rhizosphere.18 As reported, 1.5 μg mL−1 IAA was obtained from the Rubrivivax benzoatilyticus JA2 in the absence of l-Trp, which was enhanced to 12–14 μg mL−1 by 0.5–2 μg mL−1 concentrations of l-Trp.6 The highest concentration of IAA (220 μg mL−1) was observed for Enterobacter sp. I-3 in l-Trp-supplemented medium.7 Xu et al. screened twelve IAA-producing strains from watermelon rhizosphere soil.19 The highest yield of IAA was recorded for Streptomyces CL05, which produced 117.3 mg L−1 IAA in the medium supplemented with 0.2 g L−1l-Trp.19
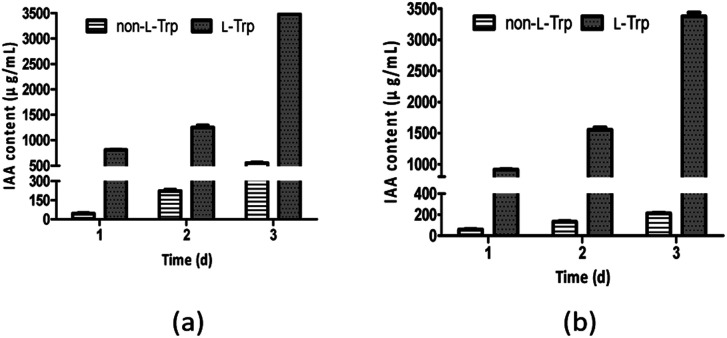
As shown in Fig. 3, IAA production improved as l-Trp concentration increased. The IAA yields of both strains increased from 1 day to 3 days. The maximum IAA concentrations (3477 μg mL−1 and 3378 μg mL−1) were observed with the addition of 2 mg mL−1l-Trp, which were 6.29-fold and 15.79-fold higher than those in the absence of l-Trp for E. xiangfangensis BWH6 (553 μg mL−1) and E. asburiae STY10 (214 μg mL−1), respectively. The maximum IAA concentrations were observed with bacterial growth at 2.97 and 2.91 of OD600 values for E. xiangfangensis BWH6 and E. asburiae STY10, respectively.
Fig. 3. IAA content in the presence of l-tryptophan (l-Trp). Strains BWH6 (a) and STY10 (b) were incubated in LB liquid medium supplemented with or without 2 mg mL−1 of l-Trp at 37 °C for 1 day to 3 days.
Bacterial growth l-tryptophan for IAA production
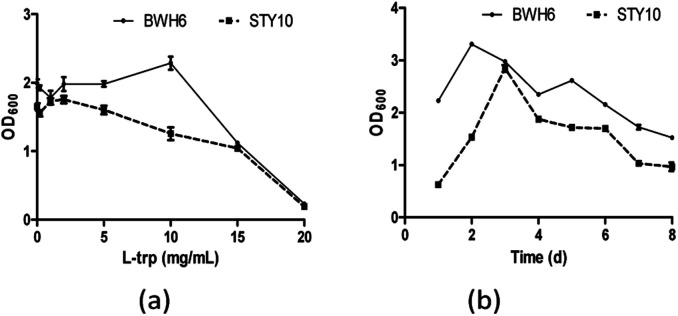
When l-Trp was increased, bacterial growth was improved for E. xiangfangensis BWH6, with strong activity under 10 μg mL−1l-Trp (Fig. 4a). Nevertheless, bacterial growth was reduced notably as l-Trp increased for E. asburiae STY10. In addition, the growth of the strain was reduced remarkably after 3 days of cultivation with a significantly reduced OD600 value (Fig. 4b). As culture time increased, the nutrients in the medium were gradually depleted, which resulted in reduced growth of the strain. Swain et al. described that IAA production of B. subtilis increased linearly from 2 days to 8 days, which was accompanied with the growth of the strain.20
Fig. 4. Bacterial growth in the presence of l-tryptophan (l-Trp). (a) The strains were incubated in LB liquid medium at 37 °C for 3 days. (b) The strains were incubated in LB liquid medium at 37 °C with the addition of 2 mg mL−1l-Trp.
IAA biosynthesis
Modern sequencing technologies make it possible to obtain genome information. Clusters of orthologous genes (COGs) analysis indicated that E. xiangfangensis BWH6 genes are mainly involved in carbohydrate transport and metabolism (8.34%), transcription (7.21%), organic ion transport and metabolism (7.13%), amino acid transport and metabolism (6.65%) and energy production and conversion (5.02%) (Fig. S1†). Specifically, 68 genes were involved in the biosynthesis, transport and metabolism of secondary metabolites. However, 24.08% of the sequences were functionally unknown. Gene Ontology (GO) analysis revealed 3710 genes were annotated into GO terms, accounting for 77.52% of the total gene sequence. The gene functions were mainly involved in biological processes and molecular functions, of which nine genes were involved in secondary metabolism (Fig. S2†). According to Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, 2939 genes were annotated, accounting for about 61.41% of the total gene sequence (Fig. S3†). KEGG analysis of E. xiangfangensis BWH6 identified the ipdC gene, which encodes indole-3-pyruvate decarboxylase, an enzyme that can catalyse the conversion of indole-3-pyruvic acid (IPyA) to indole-3-acetaldehyde (IAAld) in the IPyA pathway (Fig. S3 and S4†).
The length of ipdC is 1659 bp, encoding 559 amino acids (Fig. 5). The similarity of the amino acid sequence is 99.82% with indole-3-pyruvate decarboxylase from Enterobacter cloacae (KGY930.98.1). The ipdC gene has been identified in the IPyA pathway of microbes such as Azospirillum brasilense and Pseudomonas putida.10,21,22 However, other genes encoding tryptophan aminotransferase [EC:2.6.1.27] and aldehyde dehydrogenase [EC:1.2.1.3] were not found by genome analysis, probably owing to the low comparison rate between the strain and the previously known strains or the low comparison rate between the observed enzymes and those from known sources, which failed to pass the comparison threshold of KEGG analysis. The indole-3-pyruvate acid (IPyA) pathway is the most important pathway for IAA biosynthesis, and it has been described in various bacteria such as Enterobacter cloacae, Azospirillum brasilense, and Arthrobacter pascens.21,25
Fig. 5. Nucleotide sequence and amino acid sequence of ipdC.
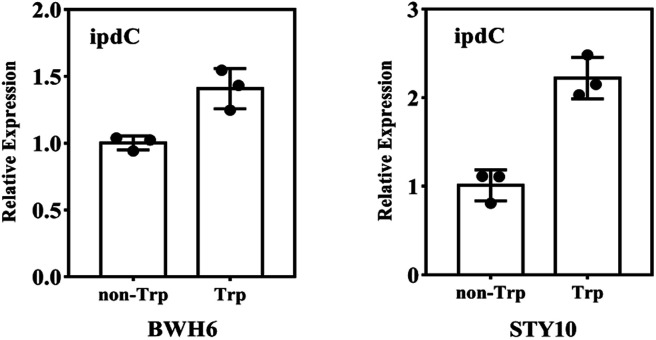
The ipdC gene expression was analysed by quantitative PCR analysis. The expression of ipdC under l-Trp treatment was 1.4 times and 2.1 times higher than that under non-l-Trp treatment for E. xiangfangensis BWH6 and E. asburiae STY10, respectively (Fig. 6). This up-regulation under l-Trp treatment indicates that the ipdC was induced by l-Trp.
Fig. 6. RT-qPCR analysis of ipdC. The transcript level of 16s rRNA was used as an internal control. The experiment was conducted with three replicates, and the results shown are the mean values ± SD.
Intermediate assay
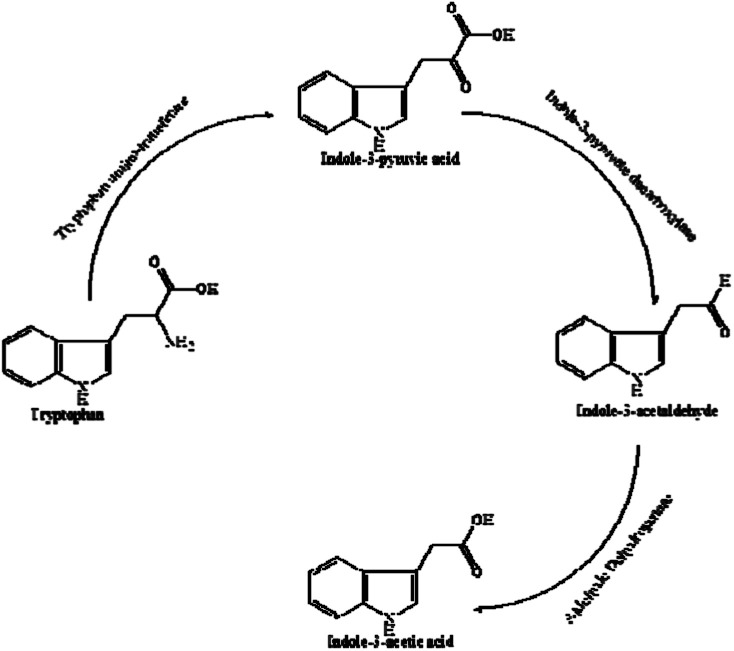
IAA biosynthesis was further investigated by an intermediate assay. HPLC analysis was used for the quantification of the intermediates. The retention times of indole-3-acetamide (IAM), indole-3-acetonitrile (IAN), tryptamine (TAM), indole-3-pyruvate acid (IPyA), l-Trp and IAA were 1.865, 3.829, 0.979, 1.290 and 3.280 min, respectively. As shown in Table 1, IPyA, TAM and IAA were detected when the medium was supplemented with l-Trp, which is the precursor of IPyA and TAM. As the fermentation time was increased, the amount of IPyA and TAM were improved. Meanwhile, l-Trp, IPyA, TAM and IAA were also detected without the addition of l-Trp indicating that both of the strains are able to synthesize l-Trp on their own. In the IPyA pathway, l-Trp is converted into IPyA by an aminotransferase. IPyA is then decarboxylated to indole-3-acetaldehyde (IAAld) by indole-3-pyruvate decarboxylase, which is the rate-limiting step. After that, IAAld is oxidized into IAA (Fig. 7). The IPyA pathway was confirmed for both strains by the intermediate assay.
The content of indole compounds.
| Time | E. xiangfangensis BWH6 | E. asburiae STY10 | |||
|---|---|---|---|---|---|
| Non-l-Trp | l-Trp | Non-l-Trp | l-Trp | ||
| l-Trp | 1 d | 0 | 925.23 ± 114.44 | 33.84 ± 35.1 | 97.05 ± 18.11 |
| 2 d | 42.14 ± 3.45 | 857.12 ± 135.62 | 14.21 ± 3.25 | 429.60 ± 35.32 | |
| 3 d | 72.64 ± 42.92 | 1727.57 ± 245.52 | 185.8 ± 42.22 | 1510.88 ± 166.22 | |
| IAA | 1 d | 47.19 ± 16.34 | 831.91 ± 143.51 | 62.11 ± 21.22 | 914.03 ± 121.11 |
| 2 d | 207.89 ± 51.51 | 1121.47 ± 96.11 | 134.50 ± 31.21 | 1571.49 ± 99.26 | |
| 3 d | 573.269 ± 61.72 | 3410.22 ± 99.12 | 215.25 ± 41.12 | 3399.14 ± 133.25 | |
| IPyA | 1 d | 0 | 93.48 ± 8.99 | 0 | 116.0 ± 1.65 |
| 2 d | 0 | 75.67 ± 6.65 | 0 | 67.2 ± 3.88 | |
| 3 d | 36.37 ± 7.58 | 107.31 ± 9.48 | 63.34 ± 2.18 | 138.5 ± 3.94 | |
| TAM | 1 d | 297.23 ± 33.34 | 405.89 ± 46.56 | 292.41 ± 39.22 | 256.392 ± 36.66 |
| 2 d | 668.93 ± 62.18 | 457.13 ± 68.65 | 853.40 ± 68.78 | 314.91 ± 48.65 | |
| 3 d | 927.98 ± 117.69 | 1181.81 ± 158.43 | 1501.72 ± 87.69 | 1150.92 ± 148.23 | |
Fig. 7. IAA biosynthetic pathway.
TAM pathway analysis
HPLC analysis was used for the quantification of the metabolites. A large amount of tryptamine (TAM) was found in the fermentation broth of E. xiangfangensis BWH6 and E. asburiae STY10. TAM is also the precursor of IAA synthesis in the TAM pathway; thus, both of the strains were investigated under the addition of TAM (2 mg mL−1). As shown in Table 2, IAA content was not obviously improved when the culture medium was supplemented with TAM. Meanwhile, genes encoding key enzymes in the TAM pathway were not found by KEGG annotation analysis. Thus, the TAM pathway of IAA biosynthesis does not appear to be active for either of the strains. A quantity of TAM could be produced by E. xiangfangensis BWH6 and E. asburiae STY10.
IAA content of strains with TAM.
| Time | E. xiangfangensis BWH6 | E. asburiae STY10 | |
|---|---|---|---|
| Non-TAM | 1 d | 47.19 ± 16.34 | 62.11 ± 21.22 |
| 2 d | 207.89 ± 51.51 | 134.50 ± 31.21 | |
| 3 d | 573.26 ± 61.72 | 215.25 ± 41.12 | |
| TAM | 1 d | 65.10 ± 18.98 | 68.03 ± 21.12 |
| 2 d | 139.18 ± 23.45 | 215.23 ± 22.88 | |
| 3 d | 172.01 ± 22.89 | 297.22 ± 45.32 |
Purification and characterization of IAA
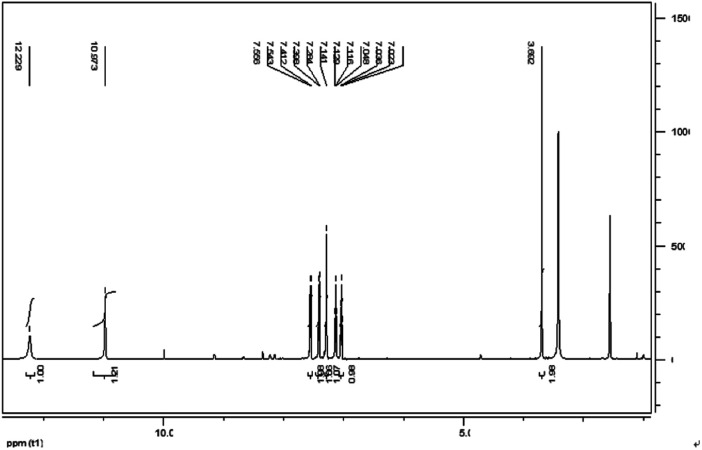
IAA was subjected to column chromatography by eluting with hexane/ethyl acetate (1/2, v/v). The chemical structure of the IAA product was characterized as shown in Fig. 8. The 1H NMR spectrum results (400 MHz, DMSO-d6) (ppm) were as follows: 12.23 (s, 1H), 10.97 (d, 1H), 7.55 (d, 1H), 7.41 (s, 1H), 7.28 (s, 1H), 7.13 (t, 1H), 7.03 (t, 1H), 3.69 (s, 1H). As E. xiangfangensis BWH6 and E. asburiae STY10 produced IAA at high levels, the resulting IAA was readily isolated at levels that could be useful for industrial applications.
Fig. 8. 1H NMR spectrum of IAA products of microbial metabolites.
Growth promoting assay
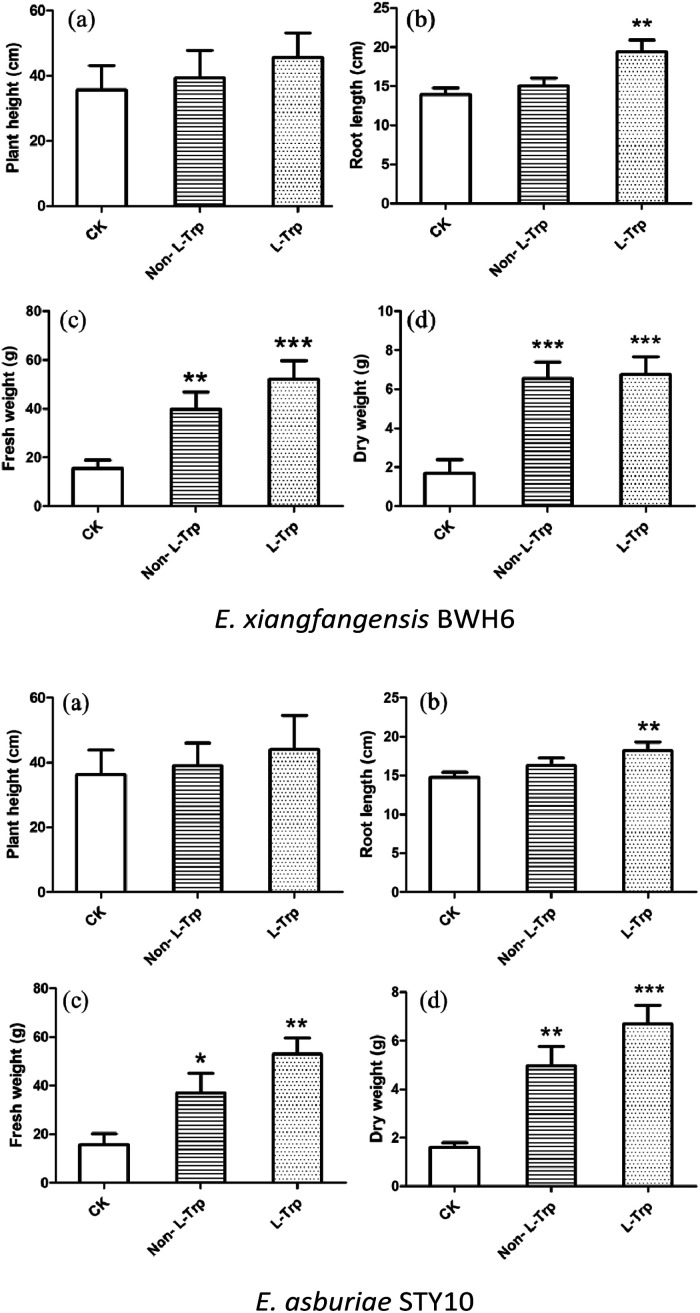
The effects of strains with IAA-producing activity were studied on the growth and development of maize (Fig. 9). The root length, plant height, fresh weight and dry weight of maize were significantly increased by these growth-promoting strains (Fig. 10). The plant height, root length, fresh weight and dry weight of maize seedlings were increased by 1.08, 1.08, 2.59 and 3.78 times by the addition of E. xiangfangensis BWH6 and 1.08, 1.10, 2.30, and 3.06 times by the addition of E. asburiae STY10 in the absence of l-Trp.
Fig. 9. Effects of strains on the growth of maize.
Fig. 10. Effects of strains on phenotypic trait vales of maize. The results are represented as the mean values of three replicates with standard deviations (SD) shown as error bars. Statistical significance as determined by Student's t-test is indicated by asterisks as follows: *p ≤ 0.05, **p < 0.01, ***p < 0.001.
As higher IAA production was achieved in the culture medium supplemented with of l-Trp, the growth of maize was improved obviously more than that under the non-l-Trp treatment. The plant height, root length, fresh weight and dry weight of maize seedlings were increased by 1.27, 1.38, 3.39 and 4.00 times for E. xiangfangensis BHW6 and 1.19, 1.23, 3.32 and 4.25 times for E. asburiae STY10, respectively. Thus, remarkable growth promotion was observed in maize.
Materials and methods
Sample collection
Soil samples and corn stalks collected from a local corn farm in Harbin, China were stored in sealed and sterile bags at 4 °C. Chemicals were purchased from Aladdin Reagent Co., Ltd. (Shanghai, China) and used as received without further purification.
Determination of IAA content
The adhering soil on root systems was washed with sterile water, and the surfaces of roots were treated with NaClO (5%, w/v) for 3 min, following by hydrogen peroxide solution (3%, w/v) for 3 min. Then, the roots were washed with sterile water. The suspension of the root power was incubated on LB medium for 48 h at 30 °C to screen for endophytic bacteria. The suspension of rhizosphere soil was incubated on LB medium for 48 h at 30 °C to isolate rhizosphere bacteria. The IAA-producing capability of microbes was initially screened according to the method of Gordon and Weber with some modifications.16 The obtained bacterial strains were incubated in LB liquid medium supplemented with or without l-tryptophan (l-Trp) at 37 °C and 30 °C for 3 days. Each experiment was conducted in triplicate. After that, the fermentation broth was centrifuged at 10 000 rpm for 10 min. Then, 2 mL of the supernatant was combined with 4 mL of Salkowski reagent (50 mL of 35% HClO4 and 1 mL of 0.5 M FeCl3). The solution was incubated in darkness for 30 min at 40 °C. The absorbance of IAA was measured at 530 nm using a UV-Vis spectrophotometer (UV-6100, Metash, Shanghai, China).
Molecular identification
DNA was extracted from each strain using the bacterial DNA kit according to the manufacturer's instructions (TIANGEN, Beijing, China). Primers are shown in Table 3. The polymerase chain reaction (PCR) was performed in a 50 μL system including template DNA (2 μL), forward primer (2 μL), reverse primer (2 μL), 2× master mix (25 μL), and ddH2O (19 μL) (TIANGEN). PCR products were sequenced at the Huada Gene Company (Beijing, China). The sequencing results were compared using the Basic Local Alignment Search Tool (BLAST) program on NCBI. A phylogenetic tree was constructed using the neighbor-joining model as implemented in MEGA 5.0.
Primers used in the studya.
| Primer name | Sequence (5′ to 3′) | T m (°C) |
|---|---|---|
| 16S rRNA-F | TAACACCTGGCTTCTGCTAC | 55 |
| 16S rRNA-R | GCGCCAATAAACATCAGGATAAT | 55 |
| STY10 ipdC-F | CGACGGTGAGTTTCGTCATT | 55 |
| STY10 ipdC-R | GTCGTTAATACCCGGTCGATTT | 57 |
| BWH6 ipdC-F | CACGCCCGGATAGAGAATAAA | 56 |
| BWH6 ipdC-R | AGCCGTGGTGATACTCAAAG | 56 |
F and R indicate forward and reverse primer, respectively.
Genomic analysis and RT-qPCR analysis
Genome sequencing analysis was performed by Personalbio Company (Beijing, China). DNA was prepared using a genomic DNA extraction kit (TIANGEN). Whole genome shotgun (WGS) sequencing and next generation sequencing (NGS) were employed. The Illumina NovaSeq Sequencing platform (Illumina, San Diego, CA, USA) was used to sequence the paired-end (PE) reads. GeneMarkS software was used to predict the protein coding genes in the bacterial genome.
RNA was extracted according to the RNA extraction kit (TIANGEN). DNA removal and cDNA synthesis were performed using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen, Beijing, China). RT-qPCR was performed with fast SYBR mixture kit (TaKaRa, Dalian, China) on an ABI 7500 fast real-time PCR platform (Life Technologies, Foster City, CA, USA). The CT data of qPCR were analysed by comparative cycle threshold (CT) method (2−ΔΔCT) for relative quantification with the 16s rRNA gene as the internal reference gene for normalization.23,24 The experiments were performed in triplicate, and the results are represented by the mean values of three replicates with standard deviations (SD) shown as error bars. Statistical significance as determined by Student's t-test was assessed at a p ≤ 0.05 threshold.
Phytohormones and intermediate assay
The bacterial strains were inoculated into LB liquid medium at 37 °C and 180 rpm for 3 days. After that, the broth was adjusted to pH 2.0 with 5 mol L−1 HCl and centrifuged at 10 000 rpm for 8 min. The resulting supernatant was extracted using ethyl acetate (10 mL × 2). After ethyl acetate was removed by rotary evaporator at 40–50 °C, each product was dissolved in methanol, which was filtered through a 0.22 μm film. HPLC analysis was conducted on an Agilent 1260 platform (Agilent Technologies, Santa Clara, CA, USA) equipped with an Agilent XDB-C18 column (4.6 × 250 mm). The mobile phase was methanol/water/acetic acid (45/54.2/0.8, v/v/v), and the injection volume was 10 μL. The experiments were performed in triplicate. IAA, GA3, ABA and ZA were used as the standards for the determination of phytohormones. IAM, IPyA, IAN, l-Trp and TAM were used as standards for the intermediate assay.
Isolation and purification of IAA product
The crude IAA product was extracted by ethyl acetate and was subjected to column chromatography (1.5 × 30 cm) eluted with hexane/ethyl acetate (1/2, v/v). After the organic solvent was evaporated at 40–50 °C, the product was dried in a vacuum for 2 h. The 1H NMR spectrum of IAA was determined on a NMR spectrometer (Bruker AVI 400 MHz; Bruker, Bremen, Germany).
Plant growth promotion assay
Strains were incubated at 37 °C and 180 rpm for 3 days in LB medium (yeast extract, 5 g; tryptone, 10 g; sodium chloride, 10 g; agar powder 15–20 g; distilled water, 1000 mL) at pH 6.8–7.2 supplemented with or without 2 mg mL−1l-tryptophan. The soil was sterilized initially. When two true leaves of maize grew, one volume of the fermentation broth of the strain that was diluted into 50 times the volume of water, which was used to irrigate the maize seedling roots every 10 days (for six total irrigation times). The control group was treated with water. After 60 days, the maize seedlings were phenotyped for plant height, root length and fresh weight. The maize seedlings were then dried at 60 °C to determine the dry weight. The experiments were performed in triplicate, and the results are presented as mean values of three replicates with standard deviations (SD) shown as error bars. Statistical significance was determined by Student's t-test at significance levels indicated by asterisks as follows: *p ≤ 0.05, **p < 0.01, ***p < 0.001.
Conclusion
E. xiangfangensis BWH6 and E. asburiae STY10 were determined to secrete 3477 μg mL−1 and 3378 μg mL−1 concentrations of IAA in the presence of l-tryptophan. Genomic analysis and RT-qPCR indicated that the IAA biosynthesis of the strain was mainly occurring through the IPyA pathway, which was confirmed by an intermediate assay. The IAA product of microbial metabolites was isolated by column chromatography and characterized with NMR. The growth of maize was significantly promoted by these strains. Thus, these strains have a crucial economic value through their great prospects in agricultural and industrial applications.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflicts of interest
All authors declares that they have no conflict of interest.
Supplementary Material
Acknowledgments
This work was supported by Leading Talent Echelon Backup Leaders Project of Heilongjiang Province, China (First-class Fund of Innovative R & D); Postdoctoral Scientific Research Start-up-Fund of Heilongjiang Province, China (LBH-Q19053) Key Program of Natural Science Foundation of Heilongjiang Province, China (JJ2020ZD0111); Major Science and Technology Program of “Millions” Project in Heilongjiang Province, China (2019ZX16B01); Young Scientist Training Project of Heilongjiang Academy of Agricultural Sciences (2021).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra05659j
Notes and references
- Boopathi T. Balamurugan V. Gopinath S. Sundararaman M. J. Plant Growth Regul. 2013;32(4):758–766. doi: 10.1007/s00344-013-9342-8. [DOI] [Google Scholar]
- Lin H. R. Shu H. Y. Lin G. H. Microbiol. Res. 2018;216:30–39. doi: 10.1016/j.micres.2018.08.004. [DOI] [PubMed] [Google Scholar]
- Lim J. H. Kim S. D. J. Korean Soc. Appl. Biol. Chem. 2009;52(5):531–538. doi: 10.3839/jksabc.2009.090. [DOI] [Google Scholar]
- Han R. Y. Li R. R. Yang F. Li H. Y. Qian Y. D. Zheng G. P. Guo Y. X. Jing R. Y. Microbiol. China. 2019;46(5):1030–1040. [Google Scholar]
- Kumla J. Suwannarach N. Matsui K. Lumyong S. PLoS One. 2020;15(1):e0227478. doi: 10.1371/journal.pone.0227478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujahid M. Sasikala C. Ramana C. V. Appl. Microbiol. Biotechnol. 2011;89:1001–1008. doi: 10.1007/s00253-010-2951-2. [DOI] [PubMed] [Google Scholar]
- Park J. M. Radhakrishnan R. Kang S. M. Lee I. J. Indian J. Microbiol. 2015;55(2):207–212. doi: 10.1007/s12088-015-0515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris E. E. Iglesias D. J. Talon M. Borriss R. Mol. Plant-Microbe Interact. 2007;20(6):619–626. doi: 10.1094/MPMI-20-6-0619. [DOI] [PubMed] [Google Scholar]
- Rupal K S. Raval V. H. Saraf M. Biocatal. Agric. Biotechnol. 2020;23:101435. doi: 10.1016/j.bcab.2019.101435. [DOI] [Google Scholar]
- Patten C. L. Glick B. R. Can. J. Microbiol. 1996;42(3):207–220. doi: 10.1139/m96-032. [DOI] [PubMed] [Google Scholar]
- Manulis S. Haviv-Chesner A. Brandl M. T. Lindow S. E. Barash I. Mol. Plant-Microbe Interact. 1998;11(7):634–642. doi: 10.1094/MPMI.1998.11.7.634. [DOI] [PubMed] [Google Scholar]
- Perley J. W. Stowe B. B. Plant Physiol. 1966;41(2):234–237. doi: 10.1104/pp.41.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhänsli T. Defago G. Haas D. J. Gen. Microbiol. 1991;137(10):2273–2279. doi: 10.1099/00221287-137-10-2273. [DOI] [PubMed] [Google Scholar]
- Kobayashi M. Izui H. Nagasawa T. Yamada H. Proc. Natl. Acad. Sci. U. S. A. 1993;90(1):247–251. doi: 10.1073/pnas.90.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohite B. J. Soil Sci. Plant Nutr. 2013;13(3):638–649. [Google Scholar]
- Gordon S. A. Weber R. P. Plant Physiol. 1951;26(1):192–195. doi: 10.1104/pp.26.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga J. Adachi T. Hidaka H. Mol. Genet. Genomics. 1991;226(1–2):10–16. doi: 10.1007/BF00273581. [DOI] [PubMed] [Google Scholar]
- Martens D. A. Frankenberger W. T. Plant Soil. 1994;166:281–290. doi: 10.1007/BF00008341. [DOI] [Google Scholar]
- Xu W. H. Lyu Z. H. Shi Y. R. Jiang J. Y. Hu Y. L. Wang Z. G. Acta Agric. Univ. Zhejiangensis. 2018;30(5):778–786. [Google Scholar]
- Swain M. R. Naskar S. K. Ray R. C. Pol. J. Microbiol. 2007;56(2):103–110. [PubMed] [Google Scholar]
- Zakharova E. A. Shcherbakov A. A. Brudnik V. V. kripko N. G. S. Bulkhin N. S. Ignatov V. V. Eur. J. Biochem. 1999;259(3):572–576. doi: 10.1046/j.1432-1327.1999.00033.x. [DOI] [PubMed] [Google Scholar]
- Costacurta A. Keijers V. Vanderleyden J. Mol. Genet. Genomics. 1994;243(4):463–472. doi: 10.1007/BF00280477. [DOI] [PubMed] [Google Scholar]
- Livak K. J. Schmittgen T. D. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ryu R. J. Patten C. L. J. Bacteriol. 2008;190(21):7200–7208. doi: 10.1128/JB.00804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. S. Guo R. Yu F. Chen X. Zhao H. Y. Li H. X. Wu J. Int. J. Mol. Sci. 2018;19(2):443–457. doi: 10.3390/ijms19020443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.