Abstract
The 5′ and 3′ untranslated regions (UTRs) of coxsackievirus B3 (CVB3) RNA form highly ordered secondary structures that have been confirmed to play important regulatory roles in viral cap-independent internal translation initiation and RNA replication. We previously demonstrated that deletions in different regions of the 5′ UTR significantly reduced viral RNA translation and infectivity. Such observations suggested strongly that viral RNA translation and replication could be blocked if highly specific antisense oligodeoxynucleotides (AS-ODNs) were applied to target crucial sites within the 5′ and 3′ UTRs. In this study, seven phosphorothioate AS-ODNs were synthesized, and the antiviral activity was evaluated by Lipofectin transfection of HeLa cells with AS-ODNs followed by infection of CVB3. Analysis by Western blotting, reverse transcription-PCR, and viral plaque assay demonstrated that viral protein synthesis, genome replication, and infectivity of CVB3 were strongly inhibited by the AS-ODNs complementary to different regions of the 5′ and 3′ UTRs. The most effective sites are located at the proximate terminus of the 5′ UTR (AS-1), the proximate terminus of the 3′ UTR (AS-7), the core sequence of the internal ribosome entry site (AS-2), and the translation initiation codon region (AS-4). These AS-ODNs showed highly sequence-specific and dose-dependent inhibitory effects on both viral protein synthesis and RNA replication. It is noteworthy that the highest inhibitory activities were obtained with AS-1 and AS-7 targeting the termini of the 5′ and 3′ UTRs. The percent inhibition values of AS-1 and AS-7 for CVB3 protein VP1 synthesis and RNA replication were 70.6 and 79.6 for AS-1 and 73.7 and 79.7 for AS-7, respectively. These data suggest that CVB3 infectivity can be inhibited effectively by AS-ODNs.
Coxsackievirus B3 (CVB3) is a member of the genus Enterovirus of the family Picornaviridae (3). This virus is the most important viral myocarditis pathogen in humans and animals (34). Such importance is reflected in data from the World Health Organization global surveillance of viral diseases, where the coxsackie B viruses were ranked the number one cause of clinically evident cardiovascular diseases (14). In addition, there is considerable clinical and experimental evidence indicating that dilated cardiomyopathy, another common heart disease, may be a late consequence of viral myocarditis (19, 23, 34).
CVB3 is a single-stranded positive-polarity RNA virus. Like other picornaviruses, the 5′ untranslated region (UTR) of the CVB3 genome is unusually long (741 nucleotides [nt]) but, unlike eukaryotic mRNAs, is not capped with a 7-methylguanosine triphosphate group. Instead, it is covalently linked to a virus-encoded oligopeptide (VPg) (31). The viral genome is approximately 7.4 kb long with a polyadenyl tail at the 3′ end. The primary sequence of the genomic RNA serves as mRNA to direct synthesis of viral proteins using host protein translational machinery. Picornavirus RNA encodes a single long polyprotein, which is processed initially into three precursor polyproteins (P1, P2, and P3). Further processing of these precursors by three virus-encoded proteases, 2A, 3C, and 3CD, gives rise to mature structural and nonstructural proteins including four capsid proteins and the RNA-dependent RNA polymerase essential for viral replication (27).
It has long been known that the majority of cellular mRNAs in eukaryotic organisms initiate translation via a cap-dependent ribosomal scanning mechanism (18, 26). However, the initiation of protein translation in picornaviruses occurs by an unusual mechanism involving direct internal binding of the ribosome to a sequence element of the 5′ UTR of viral RNA (20), termed an internal ribosomal entry site (IRES) (9, 21, 22). The IRES directs binding of the small ribosomal subunit to viral RNA near the 3′ border of the IRES, independent of a cap structure at the 5′ terminus of the RNA. Recently, our work has confirmed the presence of an IRES within the 5′ UTR of CVB3 RNA by mutational analysis using both bicistronic plasmids and full-length CVB3 mutants (33, 54). Further mapping of various mutations demonstrated that the crucial sequence of the IRES of CVB3 is located roughly at stem-loops G, H, and I, spanning nt 439 to 639. This critical sequence was further analyzed by site-directed mutagenesis and demonstrated that the critical nucleotides of the IRES span the pyrimidine-rich tract between stem-loops G and H. A 46-nt deletion in this region abolished viral translation and infectivity (33). Therefore, the IRES plays an important role in the translation initiation of viral proteins. In addition, our recent work also found that the 5′ proximate terminus of 5′ UTR is critical for translation initiation of CVB3. Deletion of nt 1 to 63 of 5′ UTR greatly inhibited CVB3 translation (54). Similar findings have been reported in other picornaviruses, suggesting that the 5′ cloverleaf structure of the 5′ UTR may be responsible for viral replication (16, 53). In addition, recent reports have suggested that the 3′ UTRs of several picornaviruses are involved in viral RNA replication (35, 37, 41). Thus, by blocking crucial sites within the 5′ and 3′ UTRs of CVB3 through sequence-specific hybridization, viral protein translation and RNA replication will be inhibited.
Antisense (AS) RNA or DNA oligonucleotides have been considered promising agents for inhibiting viral replication due in part to their high specificity for viral RNA sequences. These agents have been successfully employed to inhibit human immunodeficiency virus (32), hepatitis B and C virus (5, 38, 39, 42), influenzavirus (2, 17), coronavirus (1), and respiratory syncytial virus (40). Recently, the Food and Drug Administration approved the first AS-based therapeutic product for the treatment of retinitis caused by cytomegalovirus infections in patients with AIDS (11). Although viral replication could be inhibited by unmodified oligonucleotides, their vulnerability to nuclease attack, combined with their intracellular distribution and uptake properties, have limited their therapeutic potential (4, 24). To avoid this problem and to improve the cellular uptake of oligonucleotides, phosphorothioate oligodeoxynucleotides (ODNs) encapsulated in liposomes were used in this study. Because the mode of AS action is highly specific, it is essential to carefully select appropriate viral target sequences. Based on our previous mutational mapping and other reports on CVB3 and other picornaviruses (15, 33, 35, 37, 50, 54). The 5′ and 3′ UTRs of CVB3 RNA were chosen as the major targets for designing As ODNs. In this report, seven AS-ODNs were synthesized and evaluated in HeLa cells infected with CVB3. Four of the seven showed specific and dose-dependent inhibition of CVB3 gene expression. Two of the four targeting the 5′ and 3′ proximate termini of CVB3 genomic RNA demonstrated the strongest antiviral activity.
MATERIALS AND METHODS
Design and synthesis of AS-ODNs.
ODNs targeting the 5′ UTR were designed based on our previous mutational mapping of the cis- and trans-acting translational sequence elements (33, 54), such as the putative translation initiation factor binding site, the IRES, and the surrounding sequence of the initiation codon. The ODNs located at the 3′ UTR were designed according to the tertiary structure of the 3′ UTR of CVB3 RNA (35, 50). These three ODNs targeting the 3′ UTR were designed to disrupt the kissing interaction of two predominant hairpin loops. To avoid using too many control oligomers in the first round of the evaluation, an ODN (AS-S) with the same length and an average GC content but with a random sequence was designed as a general control of all seven ODNs. After the highest inhibitory activities were obtained with AS-1 and AS-7, these two AS-ODNs were chosen for further evaluation of their specificity using two new controls for each, which were designed with scrambled and reverse sequences, respectively. The controls were analyzed with DNA Strider software to avoid any sequence complementation with CVB3 genomic RNA.
The AS-ODNs were synthesized by the standard phosphoramidite chemistry method using an Applied Biosystems DNA/RNA synthesizer on a 1-μM scale at the Biotechnology Laboratory, University of British Columbia. In order to replace the phosphodiester bonds within the oligonucleotides with phosphorothioates, the oxidation step was substituted with a sulfurization procedure using Beaucage's reagents. The oligonucleotide derivatives were purified by reverse-phase high-pressure liquid chromatography and lyophilized, and the powder was dissolved in distilled water. All oligonucleotides were 20-mers. Figure 1 shows the location of each AS-ODN within the 5′ and 3′ UTRs of CVB3 RNA (25) used in this investigation. The AS-ODN sequences are shown in Table 1.
FIG. 1.
Targets of the AS-ODNs within the proposed secondary structures of the 5′ and 3′ UTRs of CVB3 RNA (50, 54). The 5′ UTR (nt 1 to 741) contains three AS-ODN blocking sites. AS-1 blocks the proximal terminus of the 5′ UTR at nt 1 to 20. AS-2 (nt 557 to 576) and AS-3 (nt 583 to 602) target the IRES region. AS-2 is complementary to the polypyrimidine tract of the IRES core sequence. AS-3 targets the downstream region of the AS-2 near the 3′ boundary of the IRES. AS-4 (nt 733 to 752) blocks the translation initiation codon AUG region including 9 nt of the 5′ UTR. Within the 3′ UTR (nt 7300 to 7399), three AS-ODN targets were selected. AS-5 (nt 7301 to 7320) and AS-6 (nt 7340 to 7359) target stem-loops L and M, respectively. AS-7 (nt 7380 to 7399) blocks the 3′ proximal terminus of the 3′ UTR. The general scrambled oligomer control, AS-S, was synthesized at the same length and random sequence with no annealing target in the CVB3 genomic RNA. Additional oligomer controls for AS-1 and AS-7 are listed with all other oligomers together in Table 1.
TABLE 1.
AS phosphorothioate ODNs used in anti-CVB3 evaluation
| ODNa | Sequence (5′ → 3′) | Target in CVB3 RNAb |
|---|---|---|
| AS-1 | CAACCCACAGGCTGTTTTAA | nt 1–20, 5′ end of the 5′ UTR |
| AS-1-s | CGTAGTACACTTACTAACGC | Scrambled sequence of AS-1 |
| AS-1-r | AATTTTGTCGGACACCCAAC | Reverse sequence of AS-1 |
| AS-2 | AGGAATAAAATGAAACACGG | nt 557–576, IRES core sequence |
| AS-3 | CAATTGTCACCATAAGCAGC | nt 583–602, IRES, downstream of AS-2 |
| AS-4 | TGAGCTCCCATTTTGCTGTA | nt 733–752, AUG start codon region |
| AS-5 | TTATTTCAAATTGTCTCTAA | nt 7301–7320, 3′ UTR, stem-loop L |
| AS-6 | TATCTGGTTCGGTTAGCACA | nt 7340–7359, 3′ UTR, stem-loop M |
| AS-7 | CCGCACCGAATGCGGAGAAT | nt 7380–7399, 3′ end of the 3′ UTR |
| AS-7-s | ACGACGTCGATCGAACGACG | Scrambled sequence of AS-7 |
| AS-7-r | TAAGAGGCGTAAGCCACGCC | Reverse sequence of AS-7 |
| AS-S | ACGTTGCAACGTCGTATCAT | General scrambled sequence |
All oligomers are 20 nt long. AS-S is scrambled AS as a general control.
The nucleotide positions within CVB3 genomic RNA are shown (25).
Virus, cell culture, and transfection.
Stock CVB3 was generously provided by Reinhard Kandolf and was stored at −80°C. Virus was grown in HeLa cells (American Type Culture Collection), and titers were routinely redetermined at the beginning of all individual experiments.
Transfection of HeLa cells with ODNs was conducted in 24-well plates. HeLa cells were seeded in plates (1.2 × 105 cells/well) and grown in minimum essential medium (MEM) containing 10% fetal calf serum at 37°C. After incubation for 20 h, the cells reached about 85% confluency and were washed with phosphate-buffered saline (PBS). A transfection mixture containing AS-ODN (final concentration of 1.0 or 10 μM) and 4 μg of Lipofectin (GIBCO-BRL) in 200 μl of Opti-medium (GIBCO-BRL) was added to each well. The transfection mixture was prepared as follows. In a sterile tube, 16 μl of Lipofectin was added to 384 μl of Opti-medium, mixed well gently, and kept at room temperature for 60 min. In another sterile tube, 4 μl of the oligonucleotide at appropriate concentration was added to 400 μl of Opti-medium, and the sample was mixed well gently and kept at room temperature for 60 min. The two tubes were combined, mixed well, and kept at room temperature for another 30 min. HeLa cells were overlaid with this final transfection mixture of Lipofectin-ODN (200 μl/well) and incubated for 6 h at 37°C. Then the cells were washed with PBS and infected with 200 μl of CVB3 supernatant at a multiplicity of infection (MOI) of 0.01 for 60 min. After infection, the cells were washed with PBS, overlaid with 200 μl of complete MEM, and incubated for 24 h at 37°C in a humidified 5% CO2 incubator. Finally, the supernatants from each treatment were collected by centrifugation at 4,000 × g for 5 min. The resulting supernatant was aliquoted and kept at −80°C until use.
Western blot detection of CVB3 structural protein VP1.
Thirty microliters of the resulting supernatant from each ODN treatment was denatured at 95°C for 3 min and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The sample was then transferred to a nitrocellulose membrane at 75 V for 60 min. The membrane was blocked with PBS containing Tween 20 and 5% milk at room temperature for 2 h and then incubated with primary antibody (rabbit immunoglobulin G to CVB3 VP1) (Denka Seiken Co., Ltd.) diluted 1:2,000 at 4°C overnight. The membrane was washed by PBS containing Tween 20 for 30 min and incubated with secondary antibody (anti-rabbit immunoglobulin G conjugated to horseradish peroxidase) (Transduction Ltd.). The membrane was washed again for 30 min. The signal detection was conducted by the enhanced chemiluminescence method per the manufacturer's instructions (Amersham Pharmacia Biotech, Inc.). Films were analyzed by densitometric scanning of the bands, and the density values of CVB3 VP1 were represented as means ± standard deviations (SD). The means ± SD of controls were normalized to a value of 100. Mean ± SD values of the treatment groups were calculated with respect to the control values. All experiments were performed four times.
Detection of CVB3 RNA by RT-PCR.
One hundred microliters of supernatant containing CVB3 particles from each treatment was added to 1 ml of TRIZOL Reagent (GIBCO-BRL). Viral RNA was extracted following the procedure described in the instructional manual. At the final step, the pellets were dissolved in 30 μl of diethyl pyrocarbonate-treated water and than kept at −80°C. Reverse transcription (RT) was conducted according to the manufacturer's instructions (GIBCO-BRL) using 5 μl of extracted CVB3 RNA and 1.2 μl of 20 μM RT primer (GCATTCAGCCTGGTCTCA, nt 780 to 797 of CVB3 RNA genome). After incubation at 42°C for 60 min, the samples were heated at 99°C for 5 min to stop the reaction. In order to amplify CVB3 cDNA, a PCR was carried out by following the standard method in a volume of 100 μl containing 5 μl of RT product and 2 μl of 20 μM upstream primer (AGCCTGTGGGTTGATCCCAC, nt 8 to 27) and 2 μl of 20 μM downstream primer (AATTGTCACCATAAGCAGCCA, nt 581 to 601). The reaction was run for 20 cycles with the following parameters: denaturation at 94°C for 30 s, annealing at 58°C for 40 s, and extension at 72°C for 45 s. For the negative control, water was substituted for cDNA. Twenty microliters of PCR product from each sample was analyzed by 0.8% agarose gel electrophoresis. The bands were scanned using a densitometer, and the mean density for each band was calculated as described above. Each experiment was repeated three times.
Plaque assay.
Viral plaque assay was carried out using supernatants collected from the HeLa cell monolayers treated with AS-ODN at a final concentration of 10 μM. HeLa cells were seeded into 6-well plates (8 × 105 cells/well) and incubated at 37°C for 20 h. When cell confluency reached approximately 90%, cells were washed with PBS to remove fetal bovine serum and then overlaid with 1 ml of supernatant diluted 1:10. The cells were incubated at 37°C for 60 min, the supernatants were removed, and the cell were washed with PBS. Finally, cells were overlaid with 2 ml of sterilized soft Bacto Agar-MEM (1.5% Bacto Agar–2× MEM [1:1]). The cells were incubated at 37°C for 72 h, fixed with Carnoy's fixative (75% ethanol–25% acetic acid) for 30 min and then stained with 1% crystal violet. The plaques were counted, and the viral PFU per milliliter was calculated. Supernatants from HeLa cell monolayer treated with control AS-ODNs were used as controls. Each experiment was repeated three times. The inhibitory activity of each AS-ODN was calculated with respect to the value for the corresponding control.
Statistical analysis.
All values are expressed as means ± SD. Statistical significance was evaluated using the Student t test for paired comparison. A P value of <0.01 was considered statistically significant.
RESULTS
Establishment of in vitro cell system.
To establish an optimal in vitro evaluation system using HeLa cells, three parameters were tested. The first was cell confluency. The optimal cell confluency for transfection should usually be 30 to 50%. However, at this cell confluency, it is hard to determine the effects of AS-ODNs on viral multiplication because most transfected cells died quickly after only 12 h with CVB3 infection. This observation was attributed to the high transfection efficiency leading to nonspecific toxicity to the cells, especially at a high oligonucleotide concentration. Through a series of experiments, we found that the optimal cell confluency for AS-ODN transfection followed by CVB3 infection is 85%, even though the transfection efficiency would be somewhat affected. The second consideration was the MOI. Unlike some viruses, CVB3 replicates and assembles very fast in HeLa cells. Therefore, it is important to choose a suitable MOI for CVB3 to evaluate the antiviral effects of AS-ODNs effectively. MOIs of 0.1, 0.01, and 0.001 were tested with the AS-ODNs at a final concentration of 10 μM. We found that an MOI of 0.01 was the best choice for this experiment because the most meaningful inhibitory effects were obtained under this condition. The third parameter was the dosage of AS-ODNs. ODNs at two different final concentrations (1 and 10 μM) were evaluated under the optimal conditions mentioned above. Ten micromolar appeared to be the best dosage for this in vitro evaluation system (see Fig. 2). Under these conditions, additional experiments were conducted by transfection of control ODN (AS-S) to determine the toxic effects on HeLa cell growth with the treatment using Lipofectin only as AS-S's negative control. The results demonstrated no significant differences between the AS-S and Lipofectin groups in terms of cell growth, CVB3 VP1 synthesis, and RNA replication, indicating that AS-S is a reliable control AS-ODN (data not shown). Last, the active AS1, AS-7, and their corresponding scrambled and reverse ODN controls (10 μM) were simply added to cell culture without transfection for 6 h, and no cellular toxicity on HeLa cells in terms of morphology and cell count was found.
FIG. 2.
Inhibitory effects of AS-ODNs, at a final concentration of 10 μM, on CVB3 structural protein VP1 synthesis. (A) Western blot analysis. HeLa cells were transfected with 10 μM AS-ODNs by a Lipofectin method for 6 h, infected with CVB3 at an MOI of 0.01 for 1 h, and then incubated in complete MEM for 24 h. The cell-free supernatants containing viral proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The viral protein VP1 was detected by Western blotting and enhanced chemiluminescence analysis using rabbit antiserum to VP1. Seven AS-ODNs and a control oligomer (AS-S) are indicated above the lanes and correspond to the bars in panel B. (B) Quantitation of VP1 protein in the inhibition assay using the AS-ODNs indicated in panel A. The VP1 bands were measured by scanning X-ray film using a densitometer. The mean density of each product was calculated with respect to the control values as described in Materials and Methods. Each experiment was repeated four times. The SD values of data are shown as error bars in the figures. The differences between the values for AS-S and ODNs AS-1, AS-2, AS-3, AS-4, and AS-7 were statistically significant (P < 0.01).
Inhibitory effects of AS-ODNs on CVB3 VP1 protein synthesis.
To evaluate the effects of AS-ODNs on CVB3 translation, viral structural protein VP1 was detected by Western blotting after transfection. Since CVB3 RNA encodes a single long polyprotein including structural and nonstructural proteins, the synthesis of structural protein VP1 fully represents CVB3 translation efficiency. In order to compare the antiviral action of each AS-ODN with the negative-control oligomer AS-S, the statistically analyzed means and SD were normalized by a value which converted the VP1 mean of control group AS-S to 100. Two different doses of ODNs were used to evaluate their antiviral activities. When a 10 μM concentration ODNs was used (Fig. 2), several oligomers showed potent antiviral activity. Of these oligomers, AS1 and AS7 blocking the termini of 5′ and 3′ UTRs showed the strongest inhibitory effects on VP1 synthesis. Compared with the negative controls (AS-S), the percent inhibition values of CVB3 VP1 synthesis were 84.6, 75.2, 40.6, 56.3, and 88.5 for AS-1, AS-2, AS-3, AS-4, and AS-7, respectively. However, no marked inhibitory activities were observed in groups AS-5 and AS-6. On the contrary, ODN AS-5 slightly stimulated the synthesis of CVB3 VP1, which may be due to experimental variation, since similar results were not obtained from other evaluations of the same ODN.
The evaluation was also performed at a final AS-ODN concentration of 1 μM. Similar inhibitory patterns were observed but at a lower degree of inhibition (Fig. 3) compared with treatment at the 10 μM dosage. The percentages of inhibition for CVB3 VP1 synthesis were 67.9, 55.7, 33.1, 55.3, and 72.8 for AS-1, AS-2, AS-3, AS-4, and AS-7, respectively. Again, no significant inhibitory activities for AS-5 and AS-6 were observed.
FIG. 3.
Inhibitory effects of AS-ODNs, at a final concentration of 1 μM, on CVB3 structural protein VP1 synthesis. HeLa cells were transfected with 1 μM AS-ODN by a Lipofectin method for 6 h, infected with CVB3 at an MOI of 0.01 for 1 h, and then incubated in complete MEM for 24 h. The cell-free supernatants were collected by centrifugation. The viral protein VP1 in supernatants was detected and quantitated by the methods described in the legend to Fig. 2. Each experiment was repeated four times. The differences between the values for AS-S and ODNs AS-1, AS-2, AS-3, AS-4, and AS-7 were statistically significant (P < 0.01).
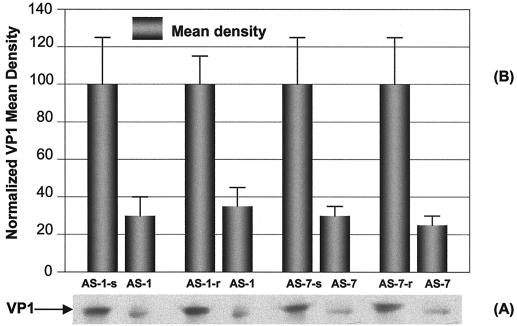
Based on the first round of evaluation, the two most active oligomers, AS-1 and AS-7, were chosen for further evaluation. The specificities of these two oligomers were confirmed by using additional corresponding scrambled and reverse sequences as negative controls. At 10 μM dosage, the percent inhibition values of CVB3 VP1 synthesis were 70.6 for AS-1 and 73.7 for AS-7 compared with the scrambled control value (AS-1-s or AS-7-s). There were significant differences between AS and scrambled oligomers. Similar inhibitory results were obtained with reverse control groups, namely, 66.5 for AS-1 and 76.7 for AS-7 (Fig. 4).
FIG. 4.
Further controlled evaluation of specific inhibition of AS-1 and AS-7, at a final concentration of 10 μM, on CVB3 structural protein VP1 synthesis. HeLa cells were transfected with 10 μM AS-1, AS-1-s, AS-1-r, AS-7, AS-7-s, or AS-7-r by a Lipofectin method and then infected with CVB3 at an MOI of 0.01. After incubation, the cell-free supernatants were collected, and the viral protein VP1 for each sample was detected (A) and quantitated by densitometry (B). The means ± SD of the controls were normalized to a value of 100. Mean ± SD values of ODN treatments were calculated with respect to the control values. The data from four independent experiments were analyzed. The difference between each control and its respective ODN was statistically significant (P < 0.01).
Inhibitory effects of AS-ODNs on CVB3 RNA replication.
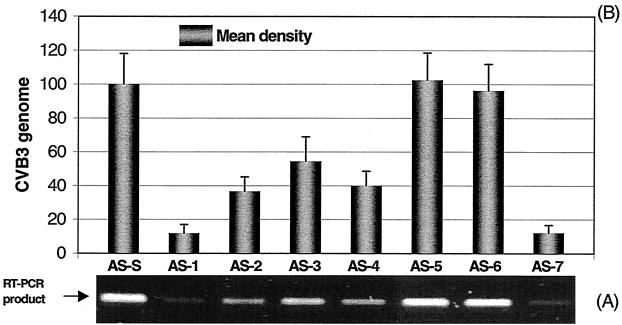
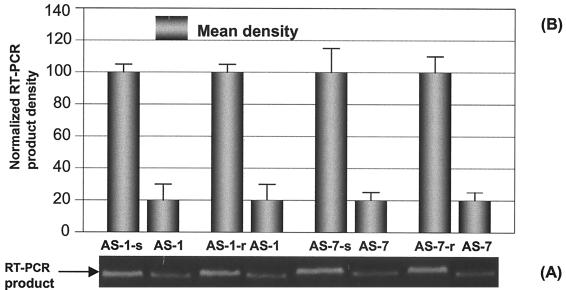
To evaluate the effects of AS-ODNs on CVB3 replication, viral genomic RNA was amplified and quantitatively analyzed by RT-PCR and densitometry. We performed PCR with different numbers of cycles and found that 20 cycles was optimal for demonstrating the most significant differences among samples treated with different AS-ODNs. In order to compare the antiviral action of each AS-ODN with the negative-control oligomer, the statistically analyzed means and SD were normalized by a value, which converted the CVB3 cDNA mean of the control group AS-S to 100. Two different doses of AS-ODNs were used to evaluate their antiviral activities. When 10 μM ODN was applied (Fig. 5), the inhibitory trend on CVB3 RNA synthesis was quite similar to that seen for VP1 synthesis. Of the seven oligomers, AS-1 and AS-7 blocking the termini of 5′ and 3′ UTRs, respectively, again showed the strongest antiviral activity. Compared with the control oligomer AS-S, the percentages of inhibition were 88.2, 63.3, 45.5, 59.9, and 87.9 for AS-1, AS-2, AS-3, AS-4, and AS-7, respectively. There were no remarkable inhibitory effects on CVB3 RNA synthesis for AS-5 and AS-6. When 1 μM AS-ODN was employed, similar data were obtained (Fig. 6). The percentages of inhibition for CVB3 RNA synthesis were 81.2, 58.1, 26.1, 50.9, and 79.1 for AS-1, AS-2, AS-3, AS-4, and AS-7, respectively. No significant inhibitory effects were observed in groups AS-5 and AS-6. Again, the evaluation was further conducted to confirm the specificity of the most potent oligomers AS-1 and AS-7 by using their corresponding scrambled and reverse sequence as negative controls. At 10 μM dosage, compared with the scrambled control AS-1-s and AS-7-s, the percentages of inhibition of CVB3 RNA synthesis were 79.6 for AS-1 and 79.7 for AS-7. There were significant differences of antiviral activity between AS and scrambled oligomers. Similar data were obtained with reverse control oligomers (80.3 for AS-1 and 78.7 for AS-7) (Fig. 7).
FIG. 5.
Inhibitory effects of AS-ODNs, at a final concentration of 10 μM, on CVB3 RNA replication. HeLa cells were transfected with 10 μM AS-ODN by a Lipofectin method for 6 h, infected with CVB3 at an MOI of 0.01 for 1 h, and then incubated in complete MEM for 24 h. The cell-free supernatants were collected by centrifugation. Viral RNA was prepared from 100 μl of the resulting supernatant of each treatment. RT-PCR was conducted for 20 cycles under standard conditions. (A) Agarose gel electrophoresis of RT-PCR products. Twenty microliters of PCR product from each treatment was run on a 0.8% agarose gel. The AS-ODNs and the control (AS-S) are marked above the lanes. (B) Quantitation of the viral RNA in the inhibition assays using AS-ODNs corresponding to those in panel A. The bands of RT-PCR products were scanned using a densitometer. The mean density of each band was calculated with respect to that of the control as described in Materials and Methods. Each experiment was repeated three times. The differences between the values for AS-S and ODNs AS-1, AS-2, AS-3, AS-4, and AS-7 were statistically significant (P < 0.01).
FIG. 6.
Inhibitory effects of AS-ODNs, at a final concentration of 1 μM, on CVB3 RNA replication. HeLa cells were transfected with 1 μM AS-ODN by a Lipofectin method for 6 h, infected with CVB3 at an MOI of 0.01 for 1 h, and then incubated in complete MEM for 24 h. Viral RNA was detected by RT-PCR (A) and quantitated by densitometry (B) as described in the legend to Fig. 5. Each experiment was repeated three times. The differences between the values for AS-S and ODNs AS-1, AS-2, AS-3, AS-4, and AS-7 were statistically significant (P < 0.01).
FIG. 7.
Further controlled evaluation of specific inhibition of AS-1 and AS-7, at a final concentration of 10 μM, on CVB3 RNA replication. HeLa cells were transfected with 10 μM AS-1, AS-1-s, AS-1-r, AS-7, AS-7-s, or AS-7-r by a Lipofectin method and followed by infection with CVB3 at an MOI of 0.01. After incubation, the cell-free supernatants were collected and the viral RNA was detected by RT-PCR (A) and quantitated by densitometry (B) as described in the legend to Fig. 4. Each experiment was repeated three times. The difference between each control and its respective ODN was statistically significant (P < 0.01).
Inhibitory effect of AS-ODN on CVB3 infectivity.
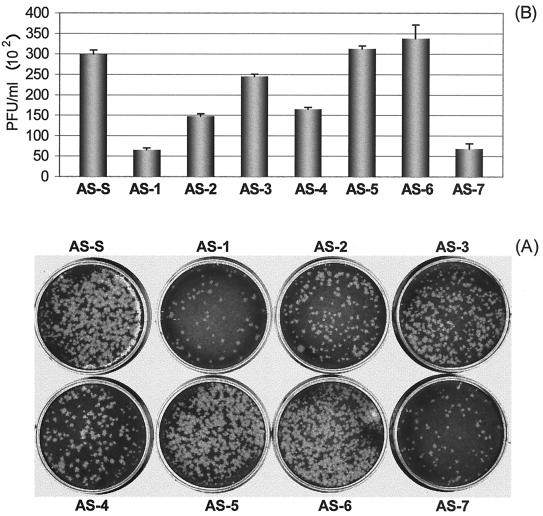
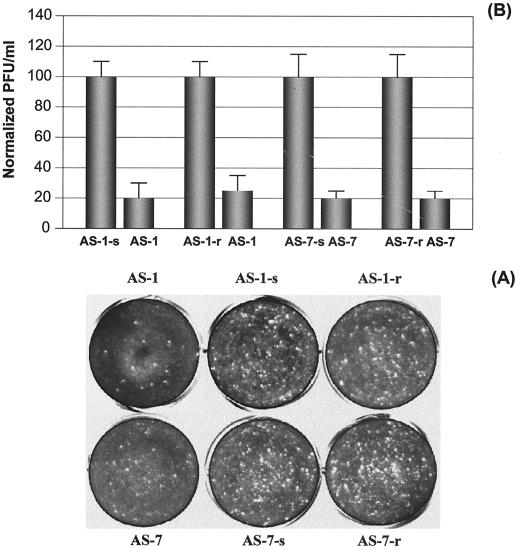
A viral plaque assay was employed to measure the antiviral activity of AS-ODNs at the optimal dosage of 10 μM determined above. The viral plaques were counted 3 days after overlaying soft agar on a HeLa cell monolayer (Fig. 8), and the numbers of PFU per milliliter were calculated. By normalizing the data with that of the negative-control AS-S, we found that AS-ODNs AS-1 and AS-7 showed the strongest antiviral activities (78.4 and 77.4%), followed by AS-2 (50.7%) and AS-4 (45%). AS-3 showed only slight inhibition (18%) of plaque-forming ability. However, AS-5 and AS-6 did not demonstrate noticeable inhibition of CVB3 infectivity. These results correlated very well with those obtained by measuring inhibitory effects on CVB3 VP1 production and RNA replication. These observations were further confirmed by plaque assays with AS-1 and AS-7 and their corresponding controls mentioned above. The percent inhibition values of CVB3 infectivity normalized against scrambled and reverse controls are 80.5 and 76.7 for AS-1 and 79.9 and 80.1 for AS-7, respectively (Fig. 9).
FIG. 8.
Inhibition of plaque formation by AS-ODNs at a final concentration of 10 μM. (A) Viral plaque assay. HeLa cell monolayers at ∼90% confluency were infected for 1 h with supernatants containing viral particles and diluted 1:10, and then were overlaid with 2 ml of 0.75% Bacto Agar–MEM. Seventy-two hours postinfection, cells were fixed with Carnoy's fixative and stained with 1% crystal violet. The plaques were counted, and the viral titer (PFU per milliliter) was calculated. The bars in panel B represent the values (PFU per milliliter) obtained from four experiments. The differences in values (PFU per milliliter) between AS-S and ODNs AS-1, AS-2, AS-3, AS-4, and AS-7 were statistically significant (P < 0.01).
FIG. 9.
Further controlled evaluation of specific inhibition of plaque formation by AS-1 and AS-7 at a final concentration of 10 μM. All the procedures are the same as described in the legend to Fig. 8 except that two more controlled oligomers, scrambled and reverse sequences, for each AS-ODN were used. (A) Viral plaque assay. (B) Values (PFU per milliliter) for the control and treatment in each group. Each experiment was repeated three times. The means ± SD of controls were normalized to 100, and the values of treatments were calculated accordingly. The difference between each control and its respective ODN treatment was statistically significant (P < 0.01).
DISCUSSION
The most important factor in determining effectiveness and specificity of AS-ODNs is the selection of target sites within CVB3 RNA. The unusually long 5′ UTR of CVB3 RNA forms a highly ordered secondary structure and plays an important role in controlling viral translation, replication, and cardiovirulence (10, 33, 54). Our recent mutational mapping of the IRES has confirmed that ribosomes initiate translation in CVB3 by binding to the polypyrimidine stretch in the IRES of 5′ UTR and then migrating to the AUG initiation codon (54). In this situation, AS-ODNs presumably block translation through the inhibition of binding of ribosomes and/or other RNA-binding proteins that are essential in forming the ribosomal complex and in the migration of ribosomal subunits along the mRNA. Therefore, it is possible that the AS-ODN (AS-2) functions in translation arrest by blocking the landing of the ribosome and/or RNA-binding proteins at the IRES (Fig. 1). In addition, it should be noted that the polypyrimidine tract located between stem-loops G and H (5′UUCAUUUU3′, nt 562 to 569) appears as an open single-stranded structure based on the computer-predicted secondary structure model (54). This implies that AS-ODN can easily form stable hybridization at this region. From this point of view, it is obvious why this AS-ODN showed such potent inhibitory effects, while another AS-ODN (AS-3) also binding to IRES region did not show the same potency in inhibiting the synthesis of CVB3 VP1. Because the target site of AS-3 is in a hairpin-like secondary structure, it is difficult for AS-3 to form a stable duplex with the viral sequence. Similar situations occurred for AS-5 and AS-6, targeting two hairpin-like structures of 3′ UTR. As for AS-4 covering the translation start codon, the strong inhibitory effect is expected, since AS-ODNs blocking the translation initiation codon should theoretically show a certain degree of inhibitory effect on a mRNA's translation initiation. Similar observations have been reported by other investigators (2, 5, 39, 49).
Interestingly, two of the AS-ODNs with the strongest inhibitory effects are located at both ends of CVB3 RNA. It is known that the 5′ and 3′ UTRs of the CVB3 RNA form highly ordered tertiary structures serving as recognition features for the RNA-binding domain of host proteins and have important cis-acting functions in the replication and translation of the RNAs (8, 35, 37, 41, 53, 54). Evidence to date suggests that a cloverleaf structure of poliovirus formed by the 5′-terminal 88 nt of the RNA binds viral proteins 3AB and 3CD and a host protein of 36 kDa (16). A cellular factor that specifically binds to the 3′ UTR of poliovirus, coxsackievirus, and rhinovirus was detected. Mutations within the 3′ UTR, which decrease the affinity of the RNA for the cellular factor, decrease RNA replication and virus viability (36). Furthermore, our concurrent UV cross-linking experiments suggest that certain host proteins are able to interact with both the 5′ UTR (nt 1 to 249) and 3′ UTR (nt 7299 to 7399) terminal regions of CVB3 genome (P. K. M. Cheung, C. M. Carthy, L. Bohunek, A. K. Wang, J. E. Wilson, B. M. McManus, and D. C. Yang, unpublished data). The extensive stem-loops and secondary structures within the 5′ and 3′ UTRs are likely the recognition sites of certain common binding proteins connecting both ends of the viral genome (28–30). The interaction between the 5′ and 3′ ends of the viral genome via common binding protein(s) has been suggested to play a crucial role in viral replication and possibly in translation initiation (28–30, 52). In addition, there has been evidence of the viral 5′ and 3′ UTRs interacting to enhance viral translation and/or transcription (42, 52); hence, a closed-loop translation model of mRNA has been proposed (6, 12, 46, 51). Thus, blocking the terminal region of CVB3 RNA either in the 5′ UTR or 3′ UTR can possibly inhibit binding of cellular proteins which serve as translation or replication initiation factors and in turn, block the formation of a circular replication unit of viral RNA. Such a mechanism may explain why two AS-ODNs (AS-1 and AS-7) blocking termini of CVB3 RNA showed such powerful inhibitory effects. In addition, CVB3 translation and replication may affect each other and thereby result in a negative-feedback effect. When translation is inhibited, the synthesis of structural proteins (such as VP1) and the nonstructural proteins (including RNA-dependent RNA polymerase) essential for virus replication is reduced, which will make it difficult for viral RNA transcription and particle assembly to occur. Decreased viral RNA synthesis will in turn reduce translation efficiency. More importantly, some of the AS-ODNs may block not only the landing of translational machinery but also the binding of the protein factors required within viral RNA for gene expression. Therefore, the AS-ODNs can inhibit CVB3 gene expression at both the translational and transcriptional levels.
The patterns of inhibition observed in this study were considered sequence specific regarding AS-1 and AS-7 because strict ODN controls (scrambled and reverse) for AS-1 and AS-7 were used. However, it still can be argued that the antiviral effects may be due in part to the so-called sequence-dependent but nonantisense effect in which sequence-specific interactions between ODNs and cellular proteins occur (7, 45, 47, 48). Since our AS-ODNs did not produce notable negative effects on HeLa cell growth in the tests of toxicity, this possibility is unlikely. Additionally, several reports indicated the nonsequence-specific effects of ODNs, particularly with the chemically modified molecules. For example, ODNs can bind in a sequence-independent manner the gp120 protein of human immunodeficiency virus type 1, viral polymerases, RNase H, bovine serum albumin, the receptor for platelet-derived growth factor, and other cellular proteins (7, 13, 43–45). Whether this similar event can happen in CVB3 needs to be explored in the future to determine the detailed mechanisms of antiviral action of our AS oligomers.
In conclusion, AS phosphorothioate ODNs targeting the termini of 5′ and 3′ UTRs, the core sequence of the IRES, and the translation initiation codon region possess specific and strong anti-CVB3 activity. This is the first report to demonstrate that translation and replication of CVB3 in tissue culture cells can be specifically inhibited by AS-ODNs. Our observations suggest that these oligomers have great potential for further development as effective, ameliorative therapeutic agents for the control of coxsackievirus-induced heart, pancreas, brain, liver, and muscle diseases.
ACKNOWLEDGMENTS
We thank Reinhard Kandolf, University of Tubingen, Germany for providing us with CVB3.
This work was supported in part by a grant from the Medical Research Council of Canada (D. C. Yang and B. M. McManus), studentships from the Heart and Stroke Foundation of British Columbia and Yukon (P. Cheung and C. Carthy).
REFERENCES
- 1.Abdou S, Collomb J, Sallas F, Marsura A, Finance C. Beta-cyclodextrin derivatives as carriers to enhance the antiviral activity of an antisense oligonucleotide directed toward a coronavirus intergenic consensus sequence. Arch Virol. 1997;142:1585–1602. doi: 10.1007/s007050050182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abe T, Hatta T, Takai K, Nakashima H, Yokota T, Takaku H. Inhibition of influenza virus replication by phosphorothioate and liposomally encapsulated oligonucleotides. Nucleosides Nucleotides. 1998;17:472–478. doi: 10.1080/07328319808005191. [DOI] [PubMed] [Google Scholar]
- 3.Abelmann W H. Viral myocarditis and its sequelae. N Engl J Med. 1973;24:145–152. doi: 10.1146/annurev.me.24.020173.001045. [DOI] [PubMed] [Google Scholar]
- 4.Agrawal S. Importance of nucleotide sequence and chemical modifications of antisense oligonucleotides. Biochim Biophys Acta. 1999;1489:53–68. doi: 10.1016/s0167-4781(99)00141-4. [DOI] [PubMed] [Google Scholar]
- 5.Alt M, Renz R, Hofschneider P H, Paumgartner G, Caselmann W H. Specific inhibition of hepatitis C viral gene expression by antisense phosphorothioate oligodeoxynucleotides. Hepatology. 1995;22:707–717. [PubMed] [Google Scholar]
- 6.Borman A M, Deliat F G, Kean K M. Sequences within the poliovirus internal ribosome entry segment control viral RNA synthesis. EMBO J. 1994;13:3149–3157. doi: 10.1002/j.1460-2075.1994.tb06613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crooke S T, Bennet C F. Progress in antisense oligonucleotide therapeutics. Annu Rev Pharmacol Toxicol. 1996;36:107–129. doi: 10.1146/annurev.pa.36.040196.000543. [DOI] [PubMed] [Google Scholar]
- 8.Currey K M, Shapiro B A. Higher structures of coxsackievirus B3 5′ nontranslated region RNA. Curr Top Microbiol Immunol. 1997;223:169–190. doi: 10.1007/978-3-642-60687-8_8. [DOI] [PubMed] [Google Scholar]
- 9.Dorner A I, Semler B L, Jackson R J, Hanecak R, Duprey E, Wimmer E. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J Virol. 1984;50:507–514. doi: 10.1128/jvi.50.2.507-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn J J, Chapman N M, Tracy S, Romero J R. Genomic determinants of cardiovirulence in coxsackievirus B3 clinical isolates: location to the 5′ nontranslated region. J Virol. 2000;74:4787–4797. doi: 10.1128/jvi.74.10.4787-4794.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox J L. FDA approves first antisense drug for CMV retinitis. ASM News. 1998;64:678–679. [Google Scholar]
- 12.Gallie D R. The cap and poly (A) tail function synergistically to regulate messenger RNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 13.Gao W Y, Han F S, Storm C, Egan W, Cheng Y C. Phosphorothioate oligonucleotides are inhibitors of human DNA polymerases and RNase H: implications for antisense technology. Mol Pharmacol. 1992;41:223–229. [PubMed] [Google Scholar]
- 14.Grist N R, Reid D. Epidemiology of viral infections of the heart. In: Banatvala J E, editor. Viral infections of the heart. London, England: Edward Arnold; 1993. pp. 23–31. [Google Scholar]
- 15.Haller A A, Nguyen J C, Semler B L. Minimum internal ribosomal entry site required for poliovirus infectivity. J Virol. 1995;67:7461–7471. doi: 10.1128/jvi.67.12.7461-7471.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris K S, Xiang W, Alexander L, Lane W S, Paul A V, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 17.Hatta T, Nakagawa Y, Takai K, Nakada S, Yokota T, Takaku H. Inhibition of influenza virus RNA polymerase and nucleoprotein gene expression by unmodified, phosphorothioated and liposomally encapsulated oligonucleotides. Biochem Biophys Res Commun. 1996;223:341–346. doi: 10.1006/bbrc.1996.0896. [DOI] [PubMed] [Google Scholar]
- 18.Hershey J W B. Translation control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 19.Hosenpud J D, Novick R J, Breen T J, Daily O P. The registry of the International Society for Heart and Lung Transplantation: eleventh official report—1994. J Heart Lung Transplant. 1994;13:561–570. [PubMed] [Google Scholar]
- 20.Jackson R J, Howell M T, Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990;15:477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- 21.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang S K, Kräusslich H-G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson R A, Palacios I. Dilated cardiomyopathy in the adult. N Engl J Med. 1982;307:119–126. [Google Scholar]
- 24.Juliano R L, Akhtar S. Liposomes as a drug delivery system for antisense oligonucleotides. Antisense Res Dev. 1992;2:165–176. doi: 10.1089/ard.1992.2.165. [DOI] [PubMed] [Google Scholar]
- 25.Klump W M, Bergmann I, Muller B C, Ameis D, Kandolf R. Complete nucleotide sequence of infectious coxsackievirus B3 cDNA: two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J Virol. 1990;64:1573–1583. doi: 10.1128/jvi.64.4.1573-1583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krausslich H G, Wimmer E. Viral proteinases. Annu Rev Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- 28.Lahser F C, Marsh L E, Hall T C. Contributions of the brome mosaic virus RNA-3 3′-nontranslated region to replication and translation. J Virol. 1993;67:3295–3303. doi: 10.1128/jvi.67.6.3295-3303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai M M C. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 30.Leathers V, Tanguay R, Kobayashi M, Galli D R. A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol Cell Biol. 1993;13:5331–5347. doi: 10.1128/mcb.13.9.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y F, Nomoto A, Detjen B M, Wimmer E. The genome linked protein of picornaviruses: a protein covalently linked to poliovirus genome RNA. Proc Natl Acad Sci USA. 1977;74:59–63. doi: 10.1073/pnas.74.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lisziewicz J, Sun D, Klotman M, Agrawal S, Zamecnik P, Gallo R. Specific inhibition of human immunodeficiency virus type 1 replication by antisense oligonucleotides: an in vitro model for treatment. Proc Natl Acad Sci USA. 1992;89:11209–11213. doi: 10.1073/pnas.89.23.11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z W, Carthy C M, Cheung P K, Bohunek L, Wilson J E, McManus B M, Yang D C. Structural and functional analysis of the 5′ untranslated region of coxsackievirus B3 RNA: in vivo translational and infectivity studies of full-length mutants. Virology. 1999;265:206–217. doi: 10.1006/viro.1999.0048. [DOI] [PubMed] [Google Scholar]
- 34.McManus B M, Kandolf R. Myocarditis: evolving concepts of cause, consequence, and control. Curr Opin Cardiol. 1991;6:418–427. [Google Scholar]
- 35.Melchers W J G, Hoenderop J G J, Bruinsslot H J, Pleij C W A, Pilipenko E V, Agol V I, Galama J M D. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mellits K H, Meredith J M, Rohll J B, Evans D J, Almond J W. Binding of a cellular factor to the 3′ untranslated region of the RNA genomes of entero- and rhinoviruses plays a role in virus replication. J Gen Virol. 1998;79:1715–1723. doi: 10.1099/0022-1317-79-7-1715. [DOI] [PubMed] [Google Scholar]
- 37.Mirmomeni M H, Hughes P J, Stanway G. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J Virol. 1997;71:2363–2370. doi: 10.1128/jvi.71.3.2363-2370.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moriya K, Matsukura M, Kurokawa K, Koike K. In vivo inhibition of hepatitis B virus gene expression by antisense phosphorothioate oligonucleotides. Biochem Biophys Res Commun. 1996;218:217–223. doi: 10.1006/bbrc.1996.0038. [DOI] [PubMed] [Google Scholar]
- 39.Offensperger W B, Offensperger S, Walter E, Teubner K, Igloi G, Blum H E, Gerok W. In vivo inhibition of duck hepatitis B virus replication and gene expression by phosphorothioate modified antisense oligodeoxynucleotides. EMBO J. 1993;12:1257–1262. doi: 10.1002/j.1460-2075.1993.tb05767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Player M R, Barnard L, Torrence P F. Potent inhibition of respiratory syncytial virus replication using a 2-5A-antisense chimera targeted to signals within the virus genomic RNA. Proc Natl Acad Sci USA. 1998;95:8874–8879. doi: 10.1073/pnas.95.15.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohll J B, Moon D H, Evans D J, Almond J W. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz D E, Hardins C C, Lemon S M. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5′ nontranslated RNA of hepatitis A virus. J Biol Chem. 1996;271:14134–14142. doi: 10.1074/jbc.271.24.14134. [DOI] [PubMed] [Google Scholar]
- 43.Stein C A. Dose antisense exist? Nat Med. 1995;1:119–121. doi: 10.1038/nm1195-1119. [DOI] [PubMed] [Google Scholar]
- 44.Stein C A, Cheng Y C. Antisense oligonucleotides as therapeutic agents—is the bullet really magical? Science. 1993;261:1004–1012. doi: 10.1126/science.8351515. [DOI] [PubMed] [Google Scholar]
- 45.Stein C A, Krieg A M. Problems in interpretation of data derived from in vitro and in vivo use of antisense oligodeoxyribonucleotides. Antisense Res Dev. 1994;4:67–69. doi: 10.1089/ard.1994.4.67. [DOI] [PubMed] [Google Scholar]
- 46.Tarun S Z, Sachs A B. Association of the yeast poly (A) tail binding protein with translation initiation factor eIF4G. EMBO J. 1997;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 47.Vaerman J L, Lammineur C, Lewalle P, Deldime F, Blumenfeld M, Martiat P. BCR-ABL antisense oligodeoxyribonucleotides suppress the growth of leukemic and normal hematopoietic cells by a sequence specific but nonantisense mechanism. Blood. 1995;86:3891–3896. [PubMed] [Google Scholar]
- 48.Wagner R W. The state of art in antisense research. Nat Med. 1995;1:1116–1118. doi: 10.1038/nm1195-1116. [DOI] [PubMed] [Google Scholar]
- 49.Wakita T, Wands J R. Specific inhibition of hepatitis C virus expression by antisense oligodeoxynucleotides. J Biol Chem. 1994;269:14205–14210. [PubMed] [Google Scholar]
- 50.Wang J, Bakkers J M, Galama J M, Bruins Slot H J, Pilipenko E V, Agol V I, Melchers W J. Structural requirements of the higher order RNA kissing element in the enteroviral 3′ UTR. Nucleic Acids Res. 1999;27:485–490. doi: 10.1093/nar/27.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wells S E, Hillner P W, Vale R D, Sachs A B. Circulization of mRNA by eukaryotic translational initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 52.Witherell G W, Wimmer E. Encephalomyocarditis virus internal ribosomal entry site RNA-protein interactions. J Virol. 1994;68:3183–3192. doi: 10.1128/jvi.68.5.3183-3192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiang W, Harris K S, Alexander L, Wimmer E. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J Virol. 1995;69:3658–3667. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang D C, Wilson J E, Anderson D R, Bohunek L, Cordeiro C, Kandolf R, McManus B M. In vitro mutational and inhibitory analysis of the cis-acting translational elements within the 5′ untranslated region of coxsackievirus B3: potential targets for antiviral action of antisense oligomers. Virology. 1997;228:63–73. doi: 10.1006/viro.1996.8366. [DOI] [PubMed] [Google Scholar]