Abstract
Cortico-striatal projections are critical components of forebrain circuitry that regulate motivated behaviors. To enable the study of the human cortico-striatal pathway and how its dysfunction leads to neuropsychiatric disease, we developed a method to convert human pluripotent stem cells into region-specific brain organoids that resemble the developing human striatum and include electrically active medium spiny neurons. We then assembled these organoids with cerebral cortical organoids in three-dimensional cultures to form cortico-striatal assembloids. Using viral tracing and functional assays in intact or sliced assembloids, we show that cortical neurons send axonal projections into striatal organoids and form synaptic connections. Medium spiny neurons mature electrophysiologically following assembly and display calcium activity after optogenetic stimulation of cortical neurons. Moreover, we derive cortico-striatal assembloids from patients with a neurodevelopmental disorder caused by a deletion on chromosome 22q13.3 and capture disease-associated defects in calcium activity, showing that this approach will allow investigation of the development and functional assembly of cortico-striatal connectivity using patient-derived cells.
INTRODUCTION
Neural activity in cortico-striatal circuits of the forebrain coordinates motivated behaviors and movement1, 2. In this pathway, glutamatergic neurons in the cerebral cortex project to the striatum where they connect primarily to GABAergic medium spiny neurons that subsequently connect to downstream circuits in the basal ganglia (Fig. 1a). Dysfunctions in neural circuits of cortico-striatal pathway are thought to contribute to neurodevelopmental disorders such as autism spectrum disorder (ASD), schizophrenia and Obsessive-Compulsive Disorder2–5. However, how these circuits are assembled in human development and how functional defects arise in disease in human patients are still poorly understood. Therefore, new in vitro approaches to image and functionally manipulate human cortico-striatal circuits will facilitate research on their roles in neuropsychiatric disorders6.
FIGURE 1. Generation of 3D human striatal spheroids (hStrS).
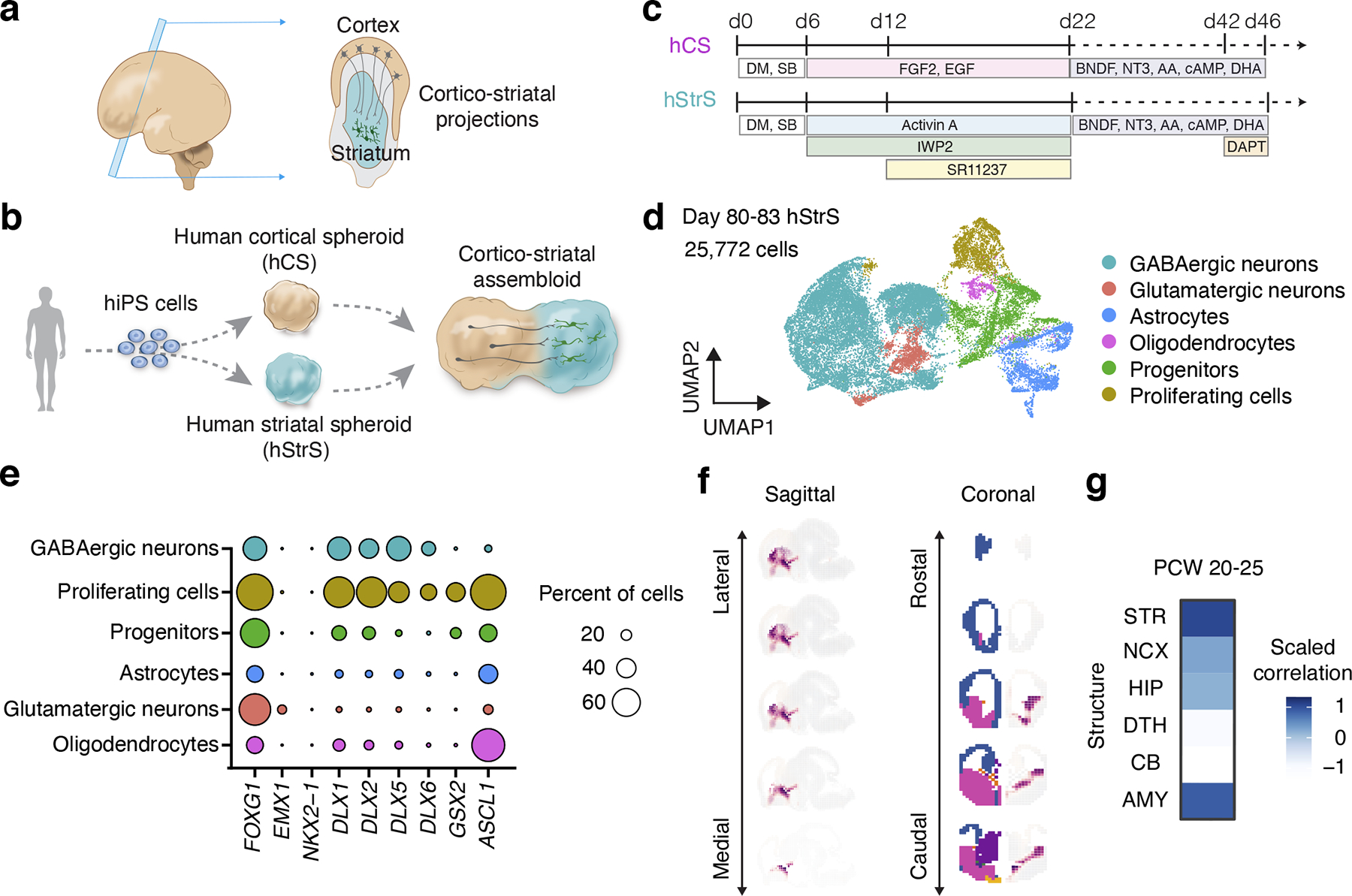
(a) Schematic diagram describing cortico-striatal projection in the developing human forebrain at mid-gestation, and (b) in vitro modeling of the cortico-striatal projections using assembloid derived from hiPS cells. (c) Differentiation condition for human cortical spheroids (hCS) and human striatum spheroids (hStrS). (d) UMAP visualization of single cell RNA expression in hStrS at day 80–83 of in vitro differentiation (n = 25,772 cells from 3 hiPS cell lines). (e) Dot plot shows percentage of selected markers for each cell cluster. (f) VoxHunt spatial brain mapping of the GABAergic neuron cluster in hStrS onto E13.5 mouse brain data from the Allen Brain Institute, and (g) correlation to the BrainSpan dataset of the human developing brain (PCW 20–25).
Human pluripotent stem (hPS) cells, including human induced pluripotent stem (hiPS) cells, have the ability to differentiate into any of the germ layers and, with the advent of three-dimensional (3D) culture methods7, to self-organize in brain organoids8 and generate diverse cell types in the central nervous system9–13. These cultures can be, in principle, derived from any individual and can be employed to study cell specification, model cell-cell interactions14 and to investigate disease15–18. We previously developed an approach to model cell-cell interactions during human brain development by generating region-specific brain organoids, also known as spheroids, and combining them in vitro to generate 3D cellular structures named assembloids19. Using this approach, we modeled human interneuron migration into the cerebral cortex and identified phenotypes associated with genetic neurodevelopmental disease19. Although hPS cells have been differentiated into striatal neurons in 2D cultures20–22, to our knowledge they have not been used to generate striatal 3D organoids or cortico-striatal assembloids.
Here, we first developed a protocol to generate human 3D brain organoids resembling the lateral ganglionic eminence (LGE), which gives rise to the striatum in development23, 24. We leveraged transcriptomic trajectories of the developing human striatum and identified a small molecule that modulates the retinoic acid receptor pathway and that, in combination with Wnt and activin, generates 3D human striatal spheroids (hStrS). As illustrated by single cell RNA-seq analyses and morphological analysis of cells, this approach yielded a diverse group of striatal cells, including medium spiny neurons that develop dendritic spines without a need for in vivo transplantation. Next, we assembled hStrS with organoids resembling the cerebral cortex to form cortico-striatal assembloids (Fig. 1b). Using morphological analyses, retrograde viral tracing, live calcium imaging and patch-clamp, we showed that cortical neurons in the assembloids send axonal projections into the striatum and functionally connect to medium spiny neurons. Lastly, we identified functional defects in striatal neurons in assembloids derived from a cohort of 22q13.3 deletion syndrome (22q13.3DS)25 patients.
RESULTS
Generation of 3D human striatal spheroids (hStrS)
To identify differentiation conditions that favor an LGE fate, we used a two-step HDR-based genome editing to derive a reporter hiPS cell line that expresses a fluorescent protein under the promoter of the LGE-related transcription factor GSX226 (GSX2::mCherry; Extended Data Fig. 1, Source Data 1). Similar to an approach we previously used for deriving 3D spheroids resembling the medial ganglionic eminence (MGE)19, we applied several small molecules at early stages of 3D neural differentiation and used the GSX2 reporter cell line in combination with immunostaining for the early striatal marker CTIP2 to verify the presence of LGE cells. We generated uniform 3D spheroids by aggregating ~10,000 dissociated hiPS cells in AggreWell-800™ plates. To induce neural differentiation, we applied modulators of the SMAD and Wnt pathways as well as Activin A, which has been shown to promote striatal differentiation22 (Extended Data Fig. 2a). To identify other pathways that we could modulate for LGE patterning, we inspected the BrainSpan transcriptome dataset of the developing human striatum27. The retinoid X receptor gamma (RXRG) is one of top differentially expressed genes in the early developing human striatum (Extended Data Fig. 2e), and adding SR11237– a selective agonist of retinoid X receptors– in combination with IWP-2 and Activin A significantly increased the proportion of GSX2+ and CTIP2+ cells at day 15 of differentiation as compared to human cortical spheroids (hCS) or applying Activin A only (*P = 0.01 for hCS versus AcISr and *P = 0.01 for A versus AcISr in Extended Data Fig. 2c, **P = 0.009 for Extended Data Fig. 2d; Extended Data Fig. 2b–d). We tested the effect of multiple concentration of SR11237 (25 nM, 50 nM, and 100 nM) on the expression of selected LGE markers by RT-qPCR (Extended Data Fig. 2f,g). We found a dose-dependent increase in DLX5, BCL11B (CTIP2) and MEIS2. We also tested an earlier exposure to SR11237 (from day 6), but this did not result in higher expression of the LGE markers. Therefore, we proceeded with using 100 nM of SR11237 from day 12. At day 22 of in vitro differentiation, neural spheroids derived in the presence of SR11237 also showed high gene expression levels of the forebrain marker FOXG1, the LGE markers DLX5, GSX2, BCL11B (CTIP2) and MEIS2 as compared to hCS, but did not express the hypothalamic marker RAX or spinal cord marker HOXB4 (n = 4–9 spheroids from 2–4 hiPS cell lines; Extended Data Fig. 2h). Cultures were subsequently maintained in medium containing BDNF, NT3, AA, cAMP, DHA and DAPT (Fig. 1c). We referred to neural spheroids derived by exposure to Activin A + IWP-2 + SR11237 (AcISr) as human striatal spheroids (hStrS). A comparison to previous approaches to derive striatal neurons in 2D is included in Supplementary Table 5.
To comprehensively study cell diversity in hStrS, we performed droplet-based single cell RNA-seq (scRNA-seq) analysis at day 80–83 of in vitro differentiation (n = 25,772 cells from 3 hiPS cell lines). UMAP dimensionality reduction identified 6 major cell clusters (Fig. 1d), including a major group of GABAergic neurons (STMN2+, SYT1+) expressing the GABA synthesizing enzyme genes GAD1 and GAD2 (56.99%), a cluster of dividing progenitors expressing TOP2A (8.67%) and one of ventral forebrain progenitors expressing GSX2, ASCL1, HOPX (13.82%), as well astrocytes expressing AQP4 and GFAP (11.58%). We also observed a small group of glutamatergic neurons expressing the glutamate transporters SLC17A7 and SLC17A6 (6.79%) and a cluster of oligodendrocytes expressing SOX10 and OLIG2 (2.12%) (Fig. 1d, e and Extended Data Fig. 3a–e, Supplementary Table 6,7). Overall, the forebrain marker FOXG1 and the ventral forebrain markers DLX1, DLX2, DLX5 and DLX6 were broadly expressed, while the MGE marker NKX2–1 was only present in 0.002% of cells (Fig. 1e and Extended Data Fig. 3c), which suggests that hStrS mostly include LGE-related cells. Cell proportions were similar across the three hiPS cell lines analyzed (with GABAergic neurons ranging from 50.12 to 66.25 %) and gene expression correlations between pairs of lines were high (2242–1 versus 1205–4: R = 0.97, ***P < 0.001; 1205–4 versus 8858–3: R = 0.98, ***P < 0.001; 2242–1 versus 8858–3: R = 0.98, ***P < 0.001) (Extended Data Fig. 3f–h). To more unbiasedly verify the regional identity of cells in hStrS, we also mapped the scRNA-seq data onto three-dimensional in situ hybridization data from the Allen Brain Atlas using the VoxHunt algorithm28. We found that the GABAergic neuron cluster in hStrS mapped onto the ventral forebrain of the E13.5 mouse brain (Fig. 1f). In addition, we mapped hStrS onto the BrainSpan human transcriptomic dataset27. We found that the GABAergic neuronal cluster in hStrS showed the highest scaled correlation with GE (ganglionic eminences) when compared to primary brain samples at early stages (PCW 5–10) and with the striatum when compared to samples at PCW 10–25 (Fig. 1g and Extended Data Fig. 4a). We also found a correlation with the developing amygdala, which is consistent with previous reports of an LGE origin for some neurons in the amygdala29. In contrast, the glutamatergic neurons cluster in hStrS was highly correlated with the neocortex (NCX) and the amygdala (AMY) in BrainSpan, and these cells appeared to be either dorsal forebrain- (EMX1+) or amygdala- related (expressing the early developing amygdala marker, TFAP2D) (Extended Data Fig. 3a,c, 4b,c). We next further analyzed the GABAergic cluster in hStrS and found neurons expressing SST, CALB1, CALB2, TH, NOS1 and NPY and only very few PVALB and CHAT expressing cells (Extended Data Fig. 4d,e). Immunostaining in hStrS confirmed expression of CALB1 and CALB2 (Extended Data Fig. 4f). We also compared hStrS to our previously derived human subpallium spheroid (hSS)19 (Extended Data Fig. 5a–e). UMAP visualization showed that hSS and hStrS share several clusters including cluster 2 (enriched for cell proliferation markers, including TOP2A), cluster 4 (enriched for progenitors including HOPX+ cells), cluster 6, 8 and 16 (enriched for glial lineage-related markers and expressing S100B and MBP). Cluster 10 and 13 were mostly hSS specific and expressed LHX6 and SST, which are suggestive of an MGE fate30, 31. These results suggest that hSS (MGE-like) and hStrS (LGE-like) display some cluster similarities, but they resemble overall different domains of the ventral forebrain. Lastly, immunostaining of hStrS at day 80–85 confirmed the presence of MAP2+ neurons, GFAP+ astrocytes, MBP+ oligodendrocytes (Extended Data Fig. 5f) and ASCL1+ progenitors (Extended Data Fig. 6b) and showed cells co-expressing FOXP2 and GAD65 (Extended Data Fig. 6c), GAD67 and CTIP2, DARPP32 and CTIP2 (Fig. 2a, Extended Data Fig. 6d), which are indicative of striatal medium spiny neurons. Overall, we found that ~50% of cells expressed CTIP2 and 8.5% expressed DARPP32 in plated hStrS cultures at day 65 (Extended Data Fig. 6e–i). Up to 30%of NeuN+ cells expressed DARPP32 at day 80–90 hStrS (Fig. 2b,c; the patterns of expression of CTIP2 and DARPP32 in the E18.5 mouse striatum is shown in Extended Data Fig. 6a). We also found expression of both DRD1 and DRD2 in hStrS, as well as expression of the striosome marker TAC1 and the matrix marker PENK (Extended Data Fig. 6j). In addition, immunocytochemistry confirmed expression of the D2 receptors in hStrS at day 124 (Extended Data Fig. 6k).
FIGURE 2. Characterization of 3D human striatal spheroids (hStrS).
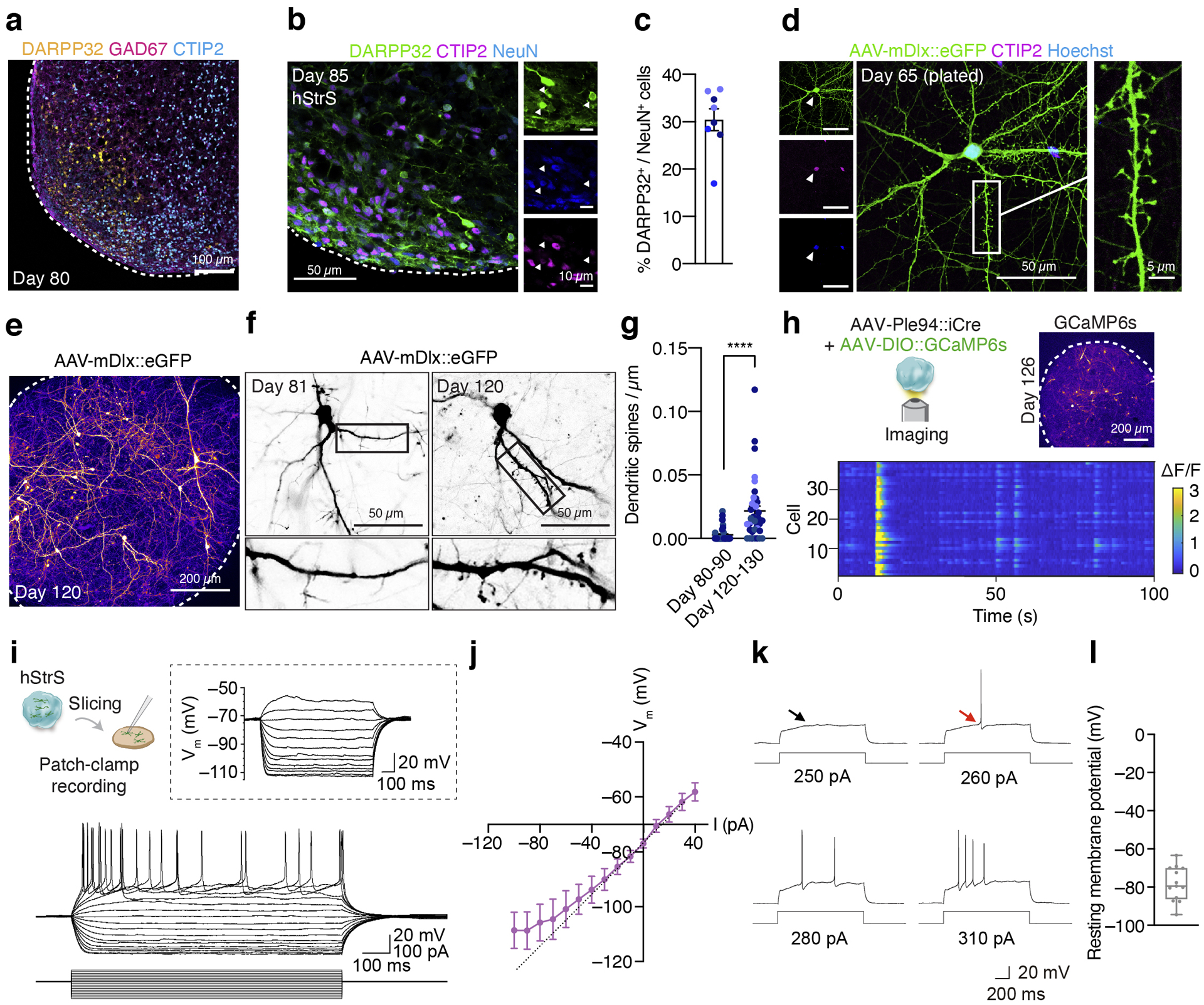
(a) Immunostaining for DARPP32 (yellow), GAD67 (magenta) and CTIP2 (cyan) in hStrS at day 80. n = 5 hiPS cell lines. Scale bar: 100 μm. (b,c) Immunostaining for DARPP32 (green), CTIP2 (magenta), and NeuN (blue), and quantification of CTIP2+ and DARPP32+ cells in hStrS at day 85. n = 8 neural differentiation experiments of 3 hiPS cell lines. (d) Representative image showing dissociated hStrS cells labelled with AAV-mDlx::eGFP reporter, and. eGFP (green), CTIP2 (magenta), and Hoechst (blue). Scale bar: 50 μm (left) and 5 μm (right). Immunostainings were repeated in dissociated hStrS cultures from 4 independent differentiation experiments with similar results. (e–g) Dendritic spine morphology in the hStrS neurons labelled with AAV-mDlx::eGFP and quantification of number of dendritic spines (day 80–90: n = 40 neurons from 3 neural differentiation experiments of 2 hiPS cell lines; day 120–130: n = 38 neurons from 4 differentiation experiments of 3 iPS cell lines; two-tailed, Mann-Whitney test, ****P < 0.0001). (h) Calcium imaging of hStrS neurons expressing the genetically encoded calcium indicator GCaMP6s at day 126 of differentiation. GCaMP6s is induced by iCre expression under a minipromoter including regulatory region of striatal gene GPR88 (Ple94). Heatmap showing ΔF/F of GCaMP6s signal from 39 cells. Imaging was repeated in hStrS from 3 independent differentiation experiments with similar results. Scale bar: 200 μm. (i) Whole cell patch-clamp recording and representative membrane response of hSyn1::eYFP labeled hStrS neurons following intracellular current pulse injection. (j) Current-frequency (I-V) curve showing inward rectification in hSyn1::eYFP+ hStrS neurons (n= 17 cells from hStrS at day 110–120). (k) Representative traces showing slow-ramp depolarization of hSyn1::eYFP+ hStrS neurons at day 160. The black arrow indicates slow-ramp depolarization, the red arrow indicates delayed first spike. (l) Resting membrane potential (n = 13 cells from hStrS at day 160–170). Data show mean ± s.e.m. Box plots show maximum, third quartile, median, first quartile, and minimum values.
Medium spiny neurons in the striatum form abundant dendritic spines during the development32. To inspect the morphology of hStrS neurons, we labeled GABAergic cells with an AAV driving eGFP expression from a Dlx5/6 enhancer (AAV-mDlx::eGFP33). In 2D plated hStrS, we found dendritic spines with thick spine heads as early as day 65 of differentiation (Fig. 2d, Extended Data Fig. 6e,i). In intact 3D hStrS, the number of spines increased significantly from day 80–90 to day 120–130 (****P < 0.0001; Fig. 2e–g; Extended Data Fig. 6l,m). The number of dendritic spines did not appear to be related to differences in the proportion of glutamatergic neurons in hStrS derived from two hiPS cell lines (Extended Data Fig. 6n).
We next examined neural activity in hStrS using live imaging of the genetically encoded calcium indicator GCaMP634. To restrict expression to GABAergic medium spiny neurons in hStrS, we used an AAV expressing improved Cre (iCre) under a mini-promoter for the striatal gene GPR88 (AAV-Ple94::iCre35) (Extended Data Fig. 6o). The majority of hStrS cells labelled by AAV-Ple94::iCre and AAV-EF1a::DIO-eYFP were GABAergic cells (85% GAD65+, 23% GAD67+) (Extended Data Fig. 6 p–s). Co-infection with AAV-Ple94::iCre and AAV-EF1a::DIO-GCaMP6s followed by live imaging of intact hStrS, showed spontaneous calcium events that were often synchronized in an imaging field (n = 39 cells; Fig. 2h and Supplementary Video 1). The spontaneous and synchronized calcium signals were blocked by application of the glutamate receptors antagonists NBQX and APV, but not the GABAA receptor antagonist bicuculine (Extended Data Fig. 7a–e). This suggests that spontaneous activity may be related to intrinsic glutamatergic transmission in hStrS. We also verified the gene expression of NKCC1 and KCC2, which encode cation-chloride co-transporters that regulate the developmental transition in GABAergic transmission36. Single cell RNA-seq analysis at day 80–83 showed that the majority of GABAergic neurons express KCC2, although NKCC1 was also expressed in some of cells (Extended Data Fig. 7f). RT-qPCR experiments also showed that NKCC1 expression decreased slightly from day 15 to day 170 (Extended Data Fig. 7g). In contrast, KCC2 expression was relatively low at early stages and increased by day 93 (Extended Data Fig. 7h).
To further examine if hStrS neurons display characteristic electrophysiological features of striatal medium spiny neurons, we analyzed the intrinsic membrane properties of hStrS neurons. We found that 70% of hStrS cells at day 110–120 (12 out of 17) showed inward rectification (Fig. 2i,j). At later stages of differentiation (day 160–170), but not at early stages (day 110–120), we found that 15% of hStrS neurons (2 out of 13 cells) displayed slow-ramp depolarization with delayed first spike (Fig. 2k) and hyperpolarization of the resting membrane potential (RMP) (–78.2 ± 2.4 mV; Fig. 2l). These properties are reminiscent of observations in postnatal medium spiny neurons in rodents37.
Generation of cortico-striatal assembloids
To assemble hStrS and hCS, we first labeled hCS with an AAV-hSyn1::eYFP and, separately, labeled hStrS with an AAV-hSyn1::mCherry. We then placed hCS and hStrS in contact to each other in a conical tube, as we have previously described for generating forebrain assembloids. After 72 hours, the two region-specific spheroids were fused to form a cortico-striatal assembloid, which was subsequently transferred to an ultra-low attachment plate. Over the next 21 days, we observed a progressive increase in YFP+ projections from hCS into hStrS (**P = 0.001 for Fig. 3c and P = 0.17 for Fig. 3d; Fig. 3a–d, Extended Data Fig. 8a, Supplementary Video 2). Projections in hCS-hStrS were unilateral from hCS into hStrS (Fig. 3c) but not vice versa (Fig. 3d), a feature consistent with cortico-striatal circuits in vivo2. Live imaging of intact, fluorescently labeled cortico-striatal assembloids also revealed dynamic movement in hStrS of growth cones from hCS neurons (Fig. 3e and Supplementary Video 3). Moreover, hCS eYFP+ terminals within hStrS showed expression of the glutamate transporter VGLUT1 at 21 days after fusion (daf) (Fig. 3f). Furthermore, we found projecting hCS neurons coming in close proximity to PSD95+ puncta on dendrites of hStrS neurons (Extended Data Fig. 8b).
FIGURE 3. Generation of cortico-striatal assembloids.
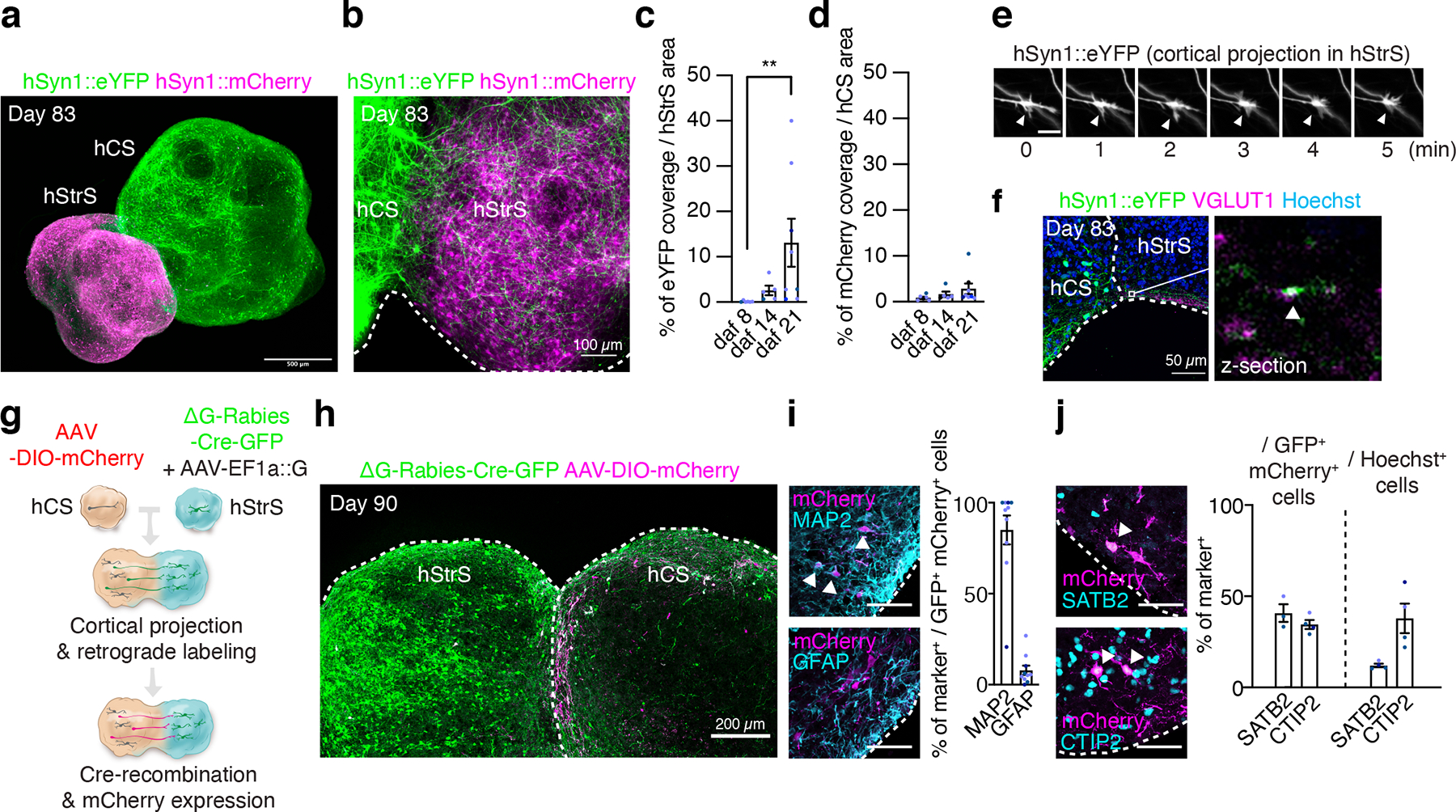
(a) Three-dimensional immunostaining of CUBIC-cleared cortico-striatal assembloids expressing AAV-hSyn1::eYFP in hCS and AAV-hSyn1::mCherry in hStrS (day 83). Scale bar: 500 μm. (b) Axon projections (YFP+) from hCS into hStrS in cortico-striatal assembloids at 21 days after fusion (daf) (day 83 of differentiation). Scale bar 100 μm. (c) Quantification of percentage of eYFP fluorescence coverage per hStrS area, and (d) of mCherry fluorescence over hCS area. n = 6 assembloids for daf 8, n = 5 assembloids for daf 14, and n= 8 assembloids for daf 21; Kruskal-Wallis test, ***P = 0.0002, **P = 0.001 with Dunnett’s multiple comparison test for daf 21 versus daf 8 in c, and Kruskal-Wallis test, P = 0.17 for d. (e) Time lapse imaging in the hStrS side of cortico-striatal assembloids showing growth cones of hCS neurons labeled with eYFP. Arrow heads indicate the growth cone. Scale bar: 10 μm. Imaging was repeated in assembloids from 3 independent differentiation experiments with similar results. (f) Representative image showing glutamatergic terminal labeled with VGLUT1 in the hStrS side of cortico-striatal assembloids. Immunostainings were repeated in assembloids from 2 independent differentiation experiments with similar results. (g, h) Experimental design schematic and image showing retrograde viral labeling of projection neurons from hCS into hStrS using ΔG-Rabies-Cre-GFP virus. Cells labeled with AAV-DIO-mCherry in hCS that project to hStrS and receive the ΔG-Rabies-Cre-GFP retrogradely co-express GFP+ and mCherry+. Immunostainings were repeated in assembloids from 4 independent differentiation experiments with similar results. (i) Representative image and quantitative results showing percentage of neurons (MAP2+) and glial cells (GFAP+) that are retrogradely labeled (GFP+/mCherry+); n = 10 assembloids from 4 differentiation experiments of 3 hiPS cell lines. Scale bar: 50 μm. (j) Percentage of SATB2+ or CTIP2+ that are retrogradely labeled in the hCS side of assembloids (GFP+/mCherry+) (left), as compared to percentages of SATB2+ and CTIP2+ cells out of all cells in the same slice (right); n = 3 assembloids for SATB2+ / GFP+ mCherry+ cells, 4 assembloids for CTIP2+ / GFP+ mCherry+ cells, and 4 assembloids for SATB2+ and CTIP2+ / Hoechst+ cells from 2 differentiation experiments of 2–3 hiPS cell lines. Scale bar: 50 μm. Data show mean ± s.e.m.
To identify what cortical cell types project from hCS into hStrS, we implemented retrograde and trans-neuronal labelling with ΔG-Rabiesvirus-Cre-GFP38 in assembloids (Fig. 3g). Specifically, we infected hStrS with a ΔG-Rabiesvirus-Cre-GFP and a AAV-EF1a::rabies glycoprotein (G), which is required for trans-synaptic spreading of the rabies virus39. Separately, we infected hCS with an AAV-DIO-mCherry, which drives mCherry expression following Cre recombination. Two days after viral delivery, we assembled hCS and hStrS and cultured them as assembloids for another 28 days. We observed extensive expression of GFP in hStrS and cells co-expressing GFP and mCherry on the cortical side of the assembloid (Fig. 3h), as well as mCherry+ projection from hCS into hStrS (Extended Data Fig. 8b). We found that the majority of GFP+/mCherry+ cells in hCS co-expressed the neuronal marker MAP2 (85.0%), and only 7.92% expressed the glial lineage marker GFAP (Fig. 3i). A major population of neurons projecting into the striatum is the cortical intra-telencephalic projection neurons expressing the transcription factor SATB2. Deep layer CTIP2+ cortical neurons of the pyramidal tract also send important collaterals into the striatum2, 40, 41. We found a 3.4× enrichment in the proportion of SATB2+ in the GFP+/mCherry+ population compared to all other cells in the same hCS section (*P = 0.01) and, overall, ~41% of the retrogradely labeled hCS neurons were SATB2+ (Fig. 3j). CTIP2+ cells accounted for ~34% of GFP+/mCherry+ cells. We note that some cells in hCS expressed GFP in the absence of mCherry, and this may be due to differences in the viral transduction in hCS (AAV-based) and hStrS (rabies-based). Although there are still technical challenges with tracing connectivity42, including ascertaining trans-synaptic tracing with rabies virus, these data suggest that projection neurons in assembloids are biased towards certain neuronal populations.
Functional neural circuits in cortico-striatal assembloids
To determine if neurons projecting from hCS can form functional synaptic connection with hStrS neurons, we implemented optogenetics with simultaneous calcium imaging in cortico-striatal assembloids. To detect optically-evoked calcium responses, we first virally expressed hSyn1::ChrimsonR-tdTomato in hCS, an opsin that excites neurons in response to red-shifted light43, and separately in hStrS, we delivered Ple94::iCre and DIO-GCaMP6s (Fig. 4a). After 2 days of viral infection, we assembled hCS and hStrS and imaged them around day 90 of differentiation (Fig. 4b). We found that application of 625 nm light reliably elicited calcium responses in hStrS cells (Fig. 4c, d). The median ΔF/F of GCaMP6 signals was significantly higher following light stimulation as compared to the median ΔF/F at randomly selected time-points (n = 180 cells, ***P = 0.0002, Extended Data Fig. 8d,e). Responding cells were also able to respond to different frequency of light simulation in sequential stimulation experiments (Extended Data Fig. 8f). We also observed, however, non-responding cells or cells that fired spontaneously during optogenetic stimulation (Fig. 4d, Extended Data Fig. 8e). Lastly, light-induced calcium responses were blocked by application of the AMPA receptor antagonist NBQX (20 μM) and the NMDA receptor antagonist APV (50 μM) (****P < 0.0001; Fig. 4d, e), and this effect was reversible (Extended Data Fig. 8g), suggesting that some of these calcium signals are mediated by glutamatergic transmission from hCS into hStrS. In addition, LED exposure, in the absence of an opsin in hCS, did not induce calcium response in cortico-striatal assembloids (Extended Data Fig. 8h,i). We also quantified the median amplitude of ΔF/F GCaMP6 signals over time in hStrS and found an increase in the amplitude of optically-evoked GCaMP6 signals at day 100–120 and at day 140–150 as compared to hStrS before day 100 (Extended Data Fig. 8j). This result suggests that there may be a time-dependent increase in strength or frequency of synaptic connection in cortico-striatal assembloids.
FIGURE 4. Functional neural circuits in cortico-striatal assembloids.
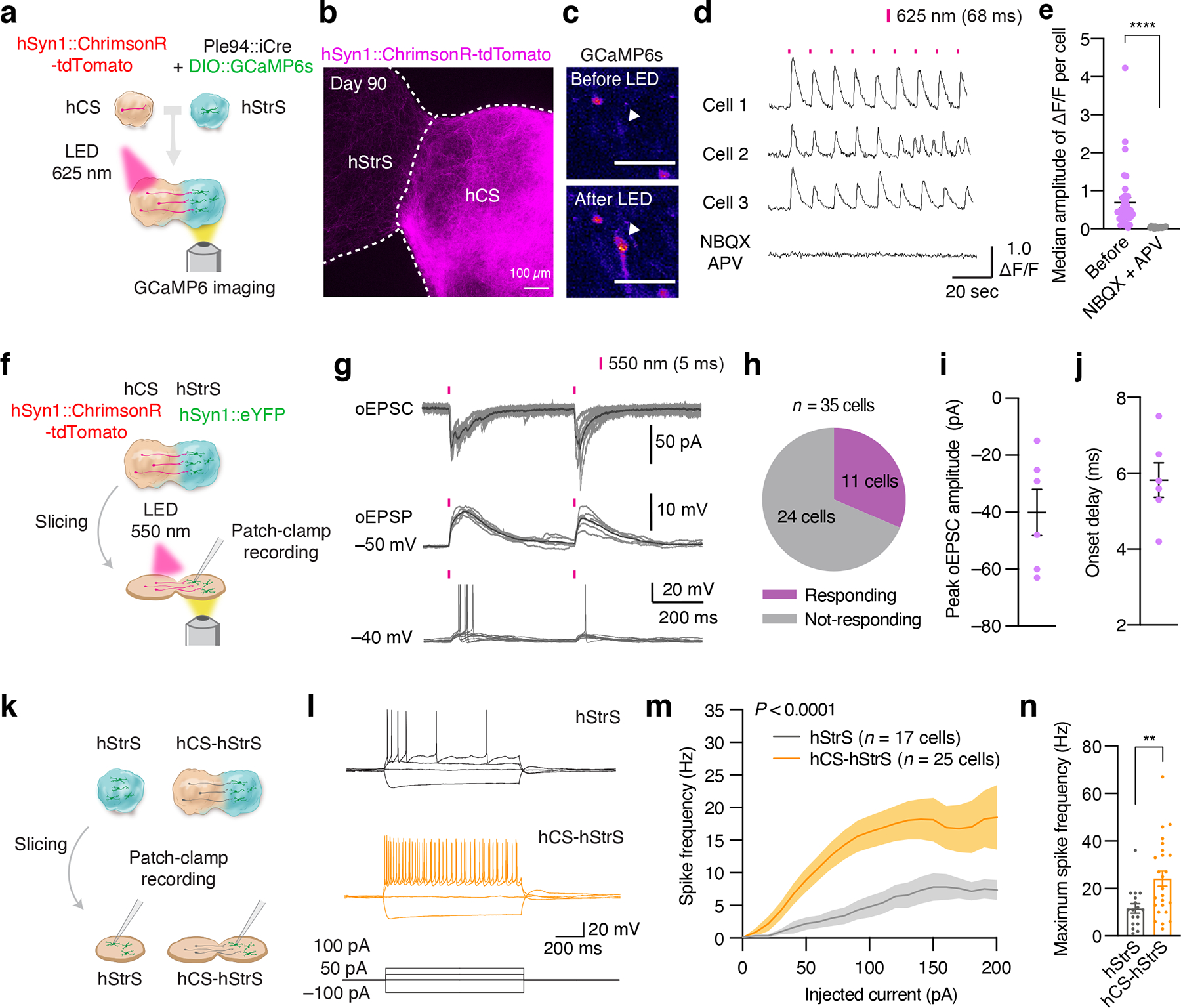
(a) Optogenetics coupled with calcium imaging of the cortico-striatal assembloid. hCS expressing AAV-ChrimsonR-tdTomato and hStrS expressing AAV-Ple94::iCre and AAV-EF1a::DIO-GCaMP6s are assembled. GCaMP6 expressing neurons in hStrS were imaged with 625 nm LED light stimulation (68 ms) at day 90–145 of differentiation. (b) Axon projection of ChrimsonR-tdTomato expressing neurons in hCS to hStrS of cortico-striatal assembloids at day 90. Scale bar: 100 μm. Imaging was repeated in assembloids from one differentiation experiment. (c) Cells in hStrS responding to 625 nm LED stimulation. Scale bar: 50 μm. Imaging was repeated in assembloids from 5 independent differentiation experiments with similar results. (d) Representative traces of GCaMP6s imaging, and (e) median amplitude of ΔF/F per cell of before and during NBQX (20 μM) and APV (50 μM) treatment. n = 36 cells for Before NBQX + APV, and 17 cells for during NBQX + APV; two-tailed Mann-Whitney test ****P < 0.0001. (f) Schematic diagram illustrating method for whole cell patch-clamp recording with optogenetic activation in cortico-striatal assembloids. (g) Representative traces of optically-evoked EPSC (oEPSC), optically-evoked EPSP (oEPSP) and neuronal firing of hStrS neurons by LED light stimulation (5-ms duration of 550 nm whole field LED illumination). (h) Percentage of responding cells (11/35 cells), (i) Peak oEPSC amplitude and (j) onset delay (n = 6 cells) of hStrS neurons in cortico-striatal assembloids. (k) Schematic showing whole-cell patch-clamp recording in hStrS or cortico-striatal assembloids in slices. (l) Representative electrophysiology traces of neurons in hStrS and hCS-hStrS neurons. (m) Frequency-current (F-I) curve showing spike frequency versus current injected in hStrS and hCS-hStrS neurons (n = 17 cells in hStrS, n = 25 cells in hCS-hStrS from 3 hiPS cell lines; two-way ANOVA, F1,840 = 131.6, ****P < 0.0001 for current injection), and (n) Maximum spike frequency (n = 17 cells in hStrS, n = 25 cells in hCS-hStrS from 3 hiPS cell lines; two-tailed unpaired t-test **P = 0.005). Data show mean ± s.e.m.
To further characterize connectivity in cortico-striatal assembloids, we combined optogenetic stimulation with electrophysiological whole-cell recording in slices (Fig. 4f). We found that delivery of 550 nm light stimulation can induce optically-evoked excitatory postsynaptic currents (oEPSCs), optically-evoked excitatory postsynaptic potentials (oEPSPs), as well as firing of neurons in hStrS (Fig. 4g). The success rate was moderately high, with 11 cells out of 35 recorded cells (Fig. 4h) responding with approximately 40 pA of average oEPSC amplitude (Fig. 4i) and 6 ms of onset delay (Fig. 4j). This indicates that neurons in hStrS can synaptically connect with hCS projecting neurons in the cortico-striatal assembloids.
Glutamatergic projections from the cerebral cortex into the striatum are thought to play a key role in the developmental maturation of medium spiny neurons32. To examine whether the assembly of hCS and hStrS impacts the electrophysiological properties of neurons in hStrS, we performed whole-cell patch clamp recordings in sliced cortico-striatal assembloids (Fig. 4k). We found increased intrinsic excitability of hSyn1::eYFP labelled neurons on the hStrS side of hCS-hStrS as compared to neurons in hStrS at the same in vitro stage (P < 0.0001, **P = 0.005; Fig. 4l–n). We also analyzed individual action potential and found shorter spike half width in hCS-hStrS without a change in the amplitude or spike threshold (Extended Data Fig. 9a–h), which is consistent with findings in the mouse developing striatum44. Neither capacitance nor input resistance was changed (P = 0.68 for Extended Data Fig. 9i, P = 0.82 for Extended Data Fig. 9j; Extended Data Fig. 9i, j) suggesting that the physiological change was not due to differences in the cell size or surface area of patched cells. Moreover, the resting membrane potentials was also not changed before and after assembly (P = 0.87; Extended Data Fig. 9k). We also observed a higher frequency of spontaneous EPSC (sEPSC) in hStrS neurons following assembly (***P = 0.0006; Extended Data Fig. 9l,m), while the number of dendritic spines in hStrS neurons was not changed following assembly (Extended Data Fig. 9n), suggesting that assembly with hCS may not affect the morphology of hStrS neurons. Taken together, these results indicate that functional connectivity can be assessed in cortico-striatal assembloids derived from hiPS cell in vitro and that this assembly results in changes of intrinsic firing properties in hStrS.
Modeling disease-related cellular phenotypes in cortico-striatal assembloids
Lastly, we studied whether cortico-striatal assembloids can be used to model cellular defects associated with genetic neurodevelopmental disease. Phelan-McDermid syndrome also known as 22q13.3 deletion syndrome (22q13.3DS) is a severe disorder characterized by global developmental delay, severe intellectual impairment, delayed speech and autism spectrum disorder. The syndrome is caused by a deletion on chromosome 22q13.3 that includes the SHANK3 gene encoding a key postsynaptic scaffold protein25. SHANK3 is highly expressed in the striatum and considered to be essential for cortico-striatal circuit development and function, and its loss is considered to be a key player in the pathogenesis of 22q13.3DS4, 37, 45–48. Defects in cortico-striatal connectivity have been previously suggested in Shank3 knockout mice37. However, whether cellular abnormalities are present in heterozygous human cells in cortico-striatal circuits from patients is not known. To investigate if we can model cortico-striatal defects in 22q13.3DS, we first differentiated hiPS cell lines from three 22q13.3DS patients into hCS and, separately, into hStrS (Fig. 5a; characterization of hiPS cells including morphology, pluripotency marker expression and genome integrity and microdeletion ascertainment by SNP arrays are shown in Fig. 5b, Extended Data Fig. 10a–c). We found that patient-derived hiPS cells can be efficiently aggregated and differentiated in 3D cultures to derive hStrS (Fig. 5b, Extended Data Fig. 10d) and, as expected, expressed the forebrain marker FOXG1 and the LGE markers GSX2, MEIS2, BCL11B (CTIP2), but not spinal cord maker HOXB4 (Extended Data Fig. 10e). The area of 22q13.3DS hCS and hStrS was not different from controls (Fig. 5c) and all 22q13.3DS lines could efficiently generate DARPP32+/NeuN+ neurons (Fig. 5d, Extended Data Fig. 10f). We next generated cortico-striatal assembloids from 3 patients with 22q13.3DS and 3 control subjects and performed calcium imaging (Fig. 5e–g). We found that neurons labeled with AAV-mDlx::GCaMP6f in the hStrS side of cortico-striatal assembloids displayed increased number of calcium spike events in 22q13.3 (*P = 0.01; Fig. 5e–g). Interestingly, when tested at the same stage of in vitro differentiation, this defect was not present in hStrS derived from 22q13.3DS (P = 0.54; Fig. 5h–j). To follow up on these changes in global calcium events in 22q13.3DS hStrS and cortico-striatal assembloids, we used GCaMP6 signals to calculate mean correlation coefficients between cells. We found reduced network synchronization in both 22q13.3DS hStrS and cortico-striatal assembloids (****P < 0.0001 in l, *P < 0.01 in m; Fig. 5k–m). Taken together, these results indicate that cortico-striatal assembloids can be used to recapitulate altered neural activity of human cells in the disease.
FIGURE 5. Modeling altered neural activity in cortico-striatal assembloids derived from 22q13.3 deletion syndrome patients.
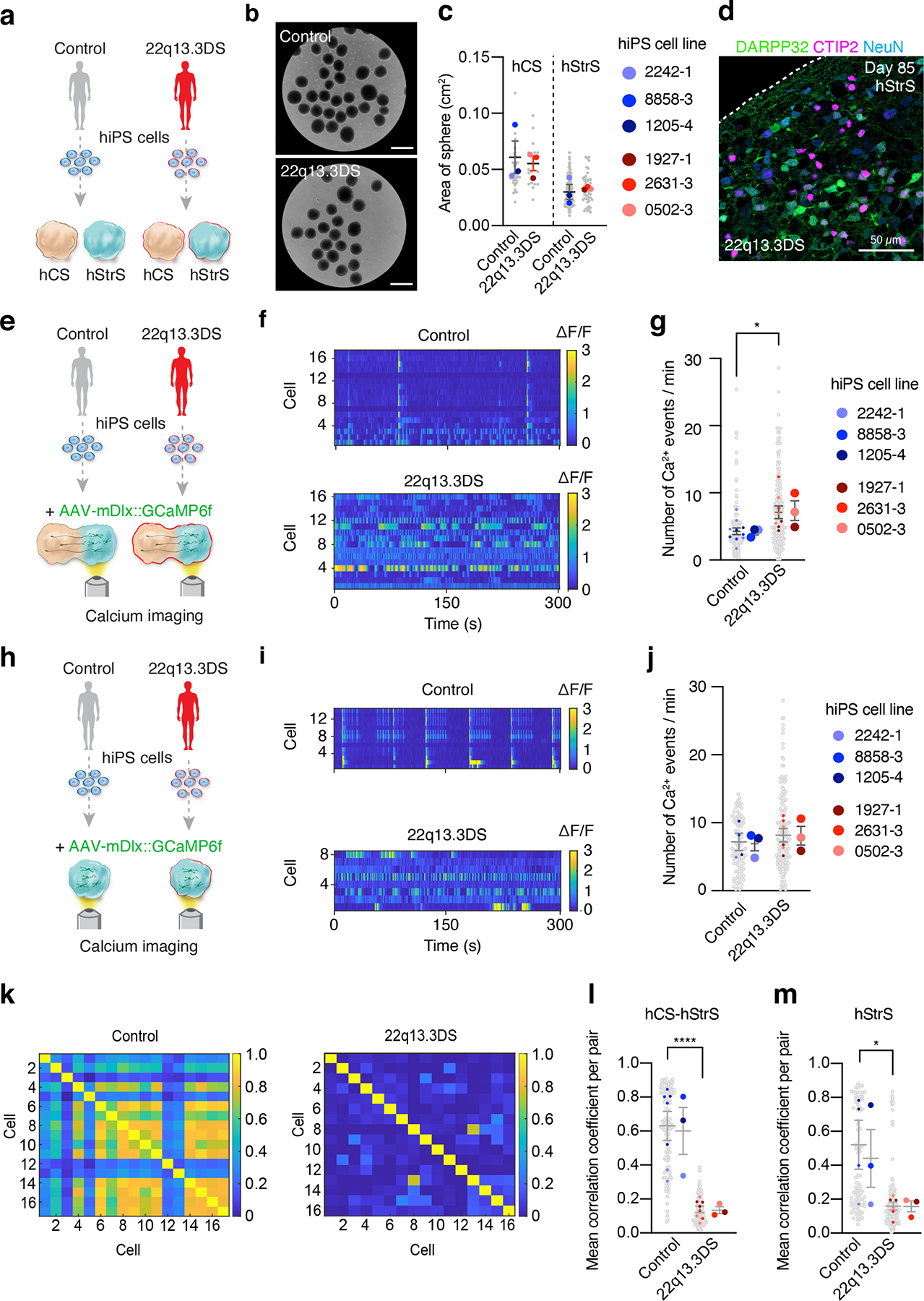
(a) Schematic illustrating the generation of hCS and hStrS derived from control and 22q13.3 deletion (22q13.3DS) hiPS cells. (b) Representative images of 3D neural spheroids (at day 5) derived from control and 22q13.3DS hiPS cell lines. Scale bars: 1 mm. (c) Quantification of the area of hCS and hStrS derived from control and 22q13.3DS at day 87–145; n = 40 spheroids for control hCS, n = 25 spheroids for 22q13.3DS hCS, n = 94 spheroids for control hStrS, n = 51 spheroids for 22q13.3DS from 4 differentiation experiments of 3 hiPS cell lines for control and 3 hiPS cell lines for 22q13.3DS; one-way ANOVA, F3,8 = 3.2, P = 0.07. (d) Immunostaining for DARPP32 (green), CTIP2 (magenta) and NeuN (blue) in day 85 hStrS derived from 22q13.3DS; n = 3 hiPS cell lines. Scale bar: 50 μm. (e) Schematic showing the functional characterization of cortico-striatal assembloids derived from 22q13.3 deletion patients. (f,g) Calcium imaging of hStrS neurons labeled with AAV-mDlx::GCaMP6f in hCS-hStrS (day 80, 18 daf) derived from control and 22q13.3DS patients, and quantitative result showing number of Ca2+ events per minute (n = 142 control cells from 10 assembloids with 3 hiPS cell lines, n = 164 22q13.3DS cells from 8 assembloids with 3 hiPS cell lines; two-tailed unpaired t-test *P = 0.01). (h) Schematic illustrating the calcium imaging of hStrS derived from 22q13.3DS patients. (i,j) Calcium imaging of hStrS neurons labeled with AAV-mDlx::GCaMP6f (day 80, 18 daf) derived from control and 22q13DS patients, and quantitative result showing number of Ca2+ events per minute (n = 91 control cells from 4 assembloids with 3 hiPS cell lines, n = 143 22q13.3DS cells from 6 assembloids with 3 hiPS cell lines; two-tailed unpaired t-test P = 0.54. (k) Heatmap showing mean correlation coefficient of ΔF/F of GCaMP6 per pair of cells in cortico-striatal assembloids from control (left) and 22q13.3DS (right), and (l,m) quantification in cortico-striatal assembloids (l) and spheroids (m). n = 127 cells for control from 7 assembloids and n = 173 cells for 22q13.3DS from 8 assembloids; two-tailed, unpaired t-test ****P < 0.0001 in l, n = 86 cells for control from 4 spheroids and n = 168 cells for 22q13.3DS from 6 spheroids; two-tailed, unpaired t-test *P = 0.01 in m. Data shown are mean ± s.e.m. Gray dots indicates cells, small colored dots show spheroids/assembloids and large colored dots show different hiPS cell line in c, g, j, l, m.
DISCUSSION
In this study, we demonstrate the generation of human 3D striatal cultures that include morphologically and functionally mature striatal neurons that can be assembled with cortical glutamatergic projection neurons to model human cortico-striatal circuits in vitro. Classical ex vivo explants experiments49 and, more recently, in vivo functional studies in behaving animals50 serve as critical experimental models to functionally interrogate cortico-striatal pathways and to model disease in knockout animals. To date, however, there are no cellular models that allow these cell-cell interactions to be proved in a functional human preparation while also taking into account the complex genetic architecture of basal ganglia disorders. Previous methods to derive striatal neurons from hPS cells have yielded relatively immature cells that have limited their application to disease modeling21, 22. Although the proportion of neurons in hStrS is not as high as in conventional striatal 2D cultures21, 22, we found that neurons in hStrS form abundant dendritic spines and recapitulate intrinsic electrophysiological properties described in rodent medium spiny neurons. Moreover, assembly of hStrS with cortical neurons accelerates the intrinsic functional maturation of striatal neurons, which could be due to glutamate release following assembly and mediated through NMDA receptor activation and subsequent modulation of potassium channels functions51. Future studies could investigate if assembly with midbrain organoids producing dopamine in three-part assembloids would even further advance their electrophysiological properties52.
The main advantage of our approach is its modularity. Cortical neurons, but not neurons in striatal 3D cultures, project into the counterpart spheroid, which is reminiscent of the directionality in this pathway in vivo2. To probe the functionality of this 3D human cellular model, we implemented rabies virus tracing and optogenetic stimulation coupled with live imaging of genetically encoded calcium indicators delivered to specific cell types. Notably, we found that projecting cortical neurons were biased to express a marker for intra-telencephalic projecting neurons, although a considerable fraction expressed a marker of corticofugal neurons, which send collaterals towards the basal ganglia2. Moreover, optogenetic stimulation of cortical neurons can reliably trigger calcium activity or even action potentials in medium spiny neurons in cortico-striatal assembloids and these are mediated by glutamate transmission. Although more studies using orthogonal mapping methods are required, these experiments suggest that connectivity with some level of specificity is possible in vitro. Future experiments should also explore whether selective cortical connectivity through dopamine receptor D1 or D2 pathways can be intrinsically achieved in vitro or if activity through the cortico-striatal-thalamic loop is required for this selectivity.
Modeling cytoarchitectural features of specific brain regions in vitro is a central goal of brain organoid research. Although we did not directly observe striosome and other matrix structures that are present in the striatum53, it remains to be seen if these features can organize at later stages of maturation in vitro, following midbrain assembly or, perhaps, as a consequence of broader circuit input after in vivo transplantation in rodents. Alternatively, it could be that embedding organizer-like structures, such as organoids secreting sonic hedgehog (SHH)54, may induce spatial organization in hStrS.
Our system has a number of potential applications for the study of human striatal development and the pathophysiology of diseases of the cortico-striatal circuits. Here we used it to capture cellular defects associated with 22q13.3DS in which the postsynaptic, striatum-enriched SHANK3 gene is hemizygously deleted. Previous work in homozygous knockout rodent models suggested potential functional defects in the cortico-striatal pathway. Indeed, we found that while hStrS displayed similar spontaneous calcium activity in 22q13.3DS as compared to control, following assembly with hCS, medium spiny neurons become hyperactive in patient-derived cultures. This increase in intrinsic neuronal activity is in line with previously reported defects in mouse and human cortical neurons37, 55, although a functional cellular defect in human striatal cells in 22q13.3DS has not been reported. Future studies should use more subjects to test these defects in the context of various 22q13.3 chromosomal deletions and identify what the role of SHANK3 is, establish if this phenotype is cell intrinsic (using hybrid assembloids as we have previously shown for forebrain assembloids19) and discover the specific contribution of cortical glutamatergic neurons to dysfunction in disease assembloids. We found network synchronization changes in 22q13.3DS in assembloids as well as in hStrS. Future studies should investigate how this phenotype is related to cell composition in hStrS. Interestingly, Shank3–/–mice display a reduction in the density of dendritic spines in the striatum as well as changes in glutamate receptor expression, which are related to an increase in dendritic length4. Therefore, it’s possible that changes in cell morphology and synaptic function in patient-derived medium spiny neurons could also contribute to network alterations in 22q13.3DS.
In addition to uncovering the molecular mechanism underlying the assembly-dependent cellular defect following loss of SHANK3, our system should be useful for modeling other neuropsychiatric disorders of the cortico-striatal circuit, including Tourette syndrome, autism spectrum disorder and obsessive-compulsive disorder3. Moreover, it may allow the investigation of trans-neuronal cortico-striatal spreading of huntingtin in Huntington’s disease56. Ultimately, circuits built in assembloids generated by combining various region-specific brain organoids may advance our understanding of neuronal connectivity in disease and accelerate the search for novel therapeutics for neuropsychiatric disorders.
MATERIALS & METHODS
Characterization and maintenance of hiPS cells
Human induced pluripotent stem (hiPS) cell lines used in this study were validated using standardized methods as previously described57. Cultures were frequently tested for and maintained Mycoplasma free. A total of 5 control hiPS cell lines derived from fibroblasts collected from 5 healthy subjects and 3 hiPS cell lines derived from fibroblasts collected from 3 patients with 22q13.3DS were used for experiments (Supplementary Table 3). Approval for this study was obtained from the Stanford IRB panel and informed consent was obtained from all subjects.
Genome-wide SNP genotyping of hiPS cells
Genome-wide SNP genotyping was performed using Illumina genome-wide GSAMD-24v2–0 SNP microarray at the Children’s Hospital of Philadelphia (CHOP). B-allele frequency (BAF) and probe level log R ratio (LRR) were generated using the Illumina GenomeStudio software (version 2.0.4).
Generation of GSX2::mCherry hiPS cells using CRISPR/Cas9 genome editing
The GSX2::mCherry hiPS cell reporter line was generated using scarless genome editing method as we previously described58. Briefly, guide RNA/Cas9 expression vectors were constructed by ligation of BbsI-digested px330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene, Plasmid #42230) with annealed oligos using T4 ligase (NEB). Editing design (position of homology arms, guide RNA targets and primer for genotyping) and sequence of donor vectors are included in Extended Data Figure 1 and Supplementary Table 1 and 2. For gRNA design, no off-target candidates of all guide RNAs used in this study were found by COSMID tool59 under the default search condition. Donor DNA plasmids were constructed using NEBuilder HiFi DNA Assembly (NEB, E2621S). The left and right homology arm for donor plasmids were amplified by nested PCR using genomic DNA extract from K562 cells (ATCC, ATCC® CCL-243™), and other fragments were obtained by PCR amplification from previously reported vectors. 2 × 106 hiPS cells were electroporated using the P3 Primary Cell 4D-Nucleofector™ X L kit (V4XP-3024) and the 4D-Nucleofector system (Lonza, AAF-1002B) with 5 μg of pX330 plasmid and 5 μg of donor plasmids, according to manufacturer’s protocol (program:CA-137). Cells were then cultured for 3 days into one well of a Matrigel™-coated 6-well plate in mTeSR™1 media (STEMCELL technologies, 85850) supplemented with 10 μM Y27632 (Tocris, 1254) and then in mTeSR™1 media without Y27632. At 7 days post-nucleofection and following a 1 hour pre-treatment with 10 μM Y27632, cells were sorted (twice) using magnetic beads selection on MS and LD columns (Miltenyi Biotec, 130-042-201, 130-042-901) for 1st and 2nd editing, respectively, and then transferred into column with a wash buffer (1% human albumin, 0.5 μM EDTA and 10 μM Y27632 in PBS). CD8+ cells were selected with CD8 MicroBeads, magnetic beads conjugated with anti-human CD8 (Militenyi Biotec, 130-045-201, 1:5 dilution). Seven days later, single hiPS colonies were picked up based on expression of GFP and genotyped by PCR with the following primers:
FW: CTTCTATGTCGACTCGCTCATCATC
RV: GGGCTGCTTTGAAAAAGTGAGATTA
Image Lab software (v5.2.1) was used for gel image acquisition.
Generation of hStrS and hCS from hiPS cells
For neural differentiation, hiPS cells were cultured on vitronectin-coated plates (5 μg ml–1, Thermo Fisher Scientific, A14700) in Essential 8 medium (Thermo Fisher Scientific, A1517001). Cells were passaged every 4–5 days with UltraPure™ 0.5 mM EDTA, pH 8.0 (Thermo Fisher Scientific, 15575). For the generation of 3D neural spheroids, hiPS cells were incubated with Accutase® (Innovative Cell Technologies, AT104) at 37°C for 7 min and dissociated into single cells. One to two days before spheroid formation, hiPS cells were exposed to 1% dimethylsulfoxide (DMSO) (Sigma-Aldrich, 472301) in Essential 8 medium. For aggregation into spheroids, approximately 3 × 106 single cells were added per AggreWell-800 well in Essential 8 medium supplemented with the ROCK inhibitor Y27632 (10 μM, Selleckchem, S1049), centrifuged at 100 g for 3 min, and then incubated at 37°C in 5 % CO2. After 24 hours, spheroids consisting of approximately 10,000 cells were collected from each microwell by pipetting medium in the well up and down with a cut P1000 pipet tip and transferred into ultra-low attachment plastic dishes (Corning, 3262) in Essential 6 medium (Thermo Fisher Scientific, A1516401) supplemented with the SMAD pathway inhibitors dorsomorphin (2.5 μM, Sigma-Aldrich, P5499) and SB-431542 (10 μM, R&D Systems, 1614). For the first 5 days, Essential 6 medium was changed every day and supplemented with dorsomorphin and SB-431542.
To generate hStrS, day 6 spheroids were transferred to neural medium containing Neurobasal™-A medium (Thermo Fisher Scientific, 10888022), B-27™ without vitamin A (Thermo Fisher Scientific, 12587010), GlutaMAX™ (1:100, Thermo Fisher Scientific, 35050079), Penicillin-Streptomycin (10,000 U/mL) (1:100, Thermo Fisher Scientific, 15140122) and supplemented with the Wnt pathway inhibitor IWP-2 (2.5 μM, Selleckchem, S7085) and recombinant Human/Murine/Rat activin A (50 ng ml−1, PeproTech, 120–14P). On day 11, neural medium was supplemented with the retinoid X receptor (RXR) agonist, SR11237 (100 nM, Tocris, 3411) in addition to the compounds described above. From day 22, to promote differentiation of the neural progenitors into neurons, neural medium was supplemented with brain-derived neurotrophic factor (BDNF; 20 ng ml–1, PeproTech, 450–02), NT3 (20 ng ml–1, PeproTech, 450–03), L-Ascorbic Acid 2-phosphate Trisodium Salt (AA; 200 μM, Wako, 323–44822), N6, 2’-O-Dibutyryladenosine 3’, 5’ -cyclic monophosphate sodium salt (cAMP; 50 μM, Millipore Sigma, D0627) and cis-4, 7, 10, 13, 16, 19-Docosahexaenoic acid (DHA; Millipore Sigma, D2534). From day 42, neural spheroids were supplemented with the DAPT (2.5 μM, STEMCELL Technologies, 72082) in addition to BDNF, NT3, AA, cAMP and DHA. From day 46, only neural medium containing B-27™ Plus Supplement (Thermo Fisher Scientific, A3582801) was used for media changes every 4 days.
hCS were generated as previously described12, 57, 60. From day 22, the neural medium was supplemented with BDNF NT3, AA, cAMP and DHA. From day 46, only neural medium containing B-27™ Plus Supplement was used for media changes every 4 days.
Dissociation of neural spheroids for 2D culture
Three to four randomly selected hStrS were collected into a 1.5 mL Eppendorf tube in a solution containing 10 unit/mL papain (Worthington Biochemical, LS003119), 1 mM EDTA, 10 mM HEPES (pH 7.4), 100 μg/mL BSA, 5 mM L-Cys and 500 μg/mL DNase I (Roche, 10104159001). Cells were incubated at 37°C for 15 min and gently shaken every 5 min. Papain was inactivated with fetal bovine serum (FBS) and hStrS were then gently triturated with a P1000 pipette. Samples were centrifuged twice at 1,300 rpm for 7 min, filtered with a 70 μm Flowmi cell strainer (Bel-Art, H13680–0070) and then suspended in Neurobasal media. Approximately 1 × 105 cells were seeded on 15 mm round coverslips (Warber Instruments, 64–0713) that were coated with approximately 0.001875 % poly-ethylenimine (PEI, Sigma-Aldrich, 03880). Neurons were cultured in neural basal medium containing B-27™ without vitamin A for 7 days (with half media changes every other day). From day 7, cells were cultured in Neurobasal media containing B-27™ Plus (with half media changes every week). At day 54, 100 or 104, neurons were fixed in 4% PFA, 4% sucrose dissolved in PBS for 20 min at 37°C.
Cryoprotection and immunocytochemistry
hCS, hStrS, assembloids and E18.5 mouse brains were fixed in 4% paraformaldehyde (PFA)/phosphate buffered saline (PBS) overnight at 4°C. They were washed in PBS and transferred to 30% sucrose/PBS for 2–3 days until the 3D cultures or tissue sank in the solution. Subsequently, they were rinsed in optimal cutting temperature (OCT) compound (Tissue-Tek OCT Compound 4583, Sakura Finetek) and 30% sucrose/PBS (1:1), and embedded. For immunofluorescence staining, 10–18 μm thick sections were cut using a Leica Cryostat (Leica, CM1850). Cryosections were washed with PBS to remove excess OCT and blocked in 10% normal donkey serum (NDS, Abcam, ab7475), 0.3% Triton X-100 (Millipore Sigma, T9284–100ML), 1% BSA diluted in PBS for 1 hour at room temperature. The sections were then incubated overnight at 4°C with primary antibodies diluted in PBS containing 2% NDS and 0.1% Triton X-100. PBS was used to wash the primary antibodies and the cryosections were incubated with secondary antibodies in PBS containing 2% NDS, 0.1% Triton X-100 for 1 hour.
Dissociated cultures on glass coverslips were fixed in 4% PFA and 4% sucrose in PBS for 20 min at 37°C and then rinsed for 5 min with PBS twice. hiPS cell cultures on glass coverslips were fixed in 4% PFA in PBS for 20 min at room temperature, and rinsed with PBS twice for 5 min. Coverslips were blocked in 10% NDS (Abcam, ab7475), 0.3% Triton X-100 (Millipore Sigma, T9284–100ML), 1% BSA diluted in PBS for 1 hour at room temperature, and then incubated overnight at 4°C with primary antibodies diluted in PBS containing 2% NDS and 0.1% Triton X-100. PBS was used to wash the primary antibodies and the coverslips were incubated with secondary antibodies in PBS with 2% NDS, 0.1% Triton X-100. The following primary antibodies were used for staining: anti-Calbindin (Synaptic Systems, #214 011, 1:200, Lot# 214011/5), anti-Calretinin (rabbit, SWANT, CR7697, 1:200), anti-MAP2 (guinea pig, Synaptic Systems 188 004, 1:200 dilution, Lot# 2–26), anti-GFAP (rabbit, DAKO, Z0334, 1:1000 dilution, Lot#20035993), anti-MBP (rat, Millipore, MAB386, 1:300 dilution), anti-ASCL1 (mouse, BD, 556604, 1:200 dilution, Lot# 6063825), anti-FOXP2 (mouse, Santa Cruz Biotechnology, sc-517261, 1:100 dilution, Lot# 1517), anti-GAD65 (goat, R&D systems, AF2247, 1:100 dilution, Lot# UCX0218101), anti-CTIP2 (rat, Abcam, ab18465 1:300 dilution, Lot# GR322373–6, Lot# GR3272266–4), anti-DARPP32 (Cell Signaling, 2306, 1:200 dilution, Lot# 7), anti-DARPP32 (rabbit, abcam, #ab40801, 1:200, Lot#GR3213231–3), anti-NeuN (mouse, abcam, ab104224, 1:200, Lot#GR3341933–1), anti-GAD67 (mouse, Millipore Sigma, MAB5406, 1:200 dilution, Lot# 2676521), anti-GFP (chicken, GeneTex, GTX13970, 1:1000 dilution, Lot# 821905323, Lot# 821704840, Lot# 821805508), anti-DRD2 (rabbit, MBL, #MC-1405, 1:200, Lot#107522), anti-mCherry (rabbit, GeneTex, GTX128508, 1:500 dilution, Lot# 42025), anti-PSD95 (guinea pig, Invitrogen, #MA1–045, 1:200, Lot#SC249676), anti-SATB2 (mouse, Abcam, ab51502, 1:50 dilution, Lot# GR178264–4), anti-OCT4 (C30A3) (rabbit, Cell Signaling Technology, #2840, 1:200 dilution, Lot# 9), anti-SSEA4 (MC813) (mouse, Cell Signaling Technology, #4755, 1:200 dilution, Lot# 4). Alexa Fluor dyes, donkey anti-rabbit IgG (H+L) highly cross-adsorbed secondary antibody, Alexa Fluor 488 (Thermo Fisher SCIENTIFIC, A-21206, 1:1000 dilution), donkey anti-mouse IgG (H+L) highly cross-adsorbed secondary antibody, Alexa Fluor 568 (Thermo Fisher SCIENTIFIC, A10037, 1:1000 dilution, Lot#2110843), donkey anti-tabbit IgG (H+L) h highly cross-adsorbed secondary antibody, Alexa Fluor 568 (Thermo Fisher SCIENTIFIC, A10042, 1:1000 dilution, Lot#2136776), donkey anti-mouse IgG (H+L) H highly cross-adsorbed secondary antibody, Alexa Fluor 647 (Thermo Fisher SCIENTIFIC, A-31571, 1:1000 dilution, Lot#1839633), donkey anti-goat IgG (H+L) highly cross-adsorbed secondary antibody, Alexa Fluor 647 (Thermo Fisher SCIENTIFIC, A-21447, 1:1000 dilution, Lot#1661244), Alexa Fluor® 488 AffiniPure, donkey anti-chicken IgY (IgG) (H+L) (Jackson ImmunoResearch Inc, 703-545-155, 1:1000 dilution), Alexa Fluor® 488 AffiniPure donkey anti-guinea pig IgG (H+L) (Jackson ImmunoResearch Inc, 706-545-148, 1:1000 dilution), Alexa Fluor® 647 AffiniPure donkey anti-rat IgG (H+L) (Jackson ImmunoResearch Inc, 712-605-153, 1:1000 dilution), Alexa Fluor® 647 AffiniPure donkey anti-rabbit IgG (H+L) (Jackson ImmunoResearch Inc, 711-605-152, 1:1000 dilution), Alexa Fluor® 647 AffiniPure donkey anti-rat IgG (H+L) (Jackson ImmunoResearch Inc, 712-605-153, 1:1000 dilution), Alexa Fluor® 647 AffiniPure donkey anti-guinea pig IgG (H+L) (Jackson ImmunoResearch Inc, 706-605-148, 1:1000 dilution) were used at 1:1,000 dilution, and nuclei were visualized with Hoechst 33258 (Thermo Fisher Science, H3569, 1:10,000 dilution, Lot#1829927). Cryosections and coverslips were mounted for microscopy on glass slides using Aquamount (Polysciences, 18606), and imaged on a Keyence BZ-X710, a Zeiss M1 Axioscope or a Leica TCS SP8 confocal microscopes. Images were processed with the Fiji (ImageJ, v2.1.0, NIH).
Real-time qPCR
mRNA from hCS and hStrS at day 15 and 22 were isolated using the RNeasy Mini kit (Qiagen, 74106) with DNase I, Amplification Grade (Thermo Fisher Scientific, 18068–015). Template cDNA was prepared by reverse transcription using the SuperScript™ III First-Strand Synthesis SuperMix for qRT-PCR (Thermo Fisher Scientific, 11752250). qPCR was performed using the SYBR™ Green PCR Master Mix (Thermo Fisher Scientific, 4312704) on a ViiA7 Real-Time PCR System (Thermo Fisher Scientific, 4453545). Primers used in this study are listed on Supplementary Table 4.
Single cell RNA-seq library preparation and data analysis
To obtain a single cell suspension, randomly selected 3–4 hStrS from each hiPS cell line at day 80 or 83 were collected into a 1.5 mL Eppendorf tube containing 10 unit/mL papain (Worthington Biochemical, LS003119) and 500 μg/mL DNase I (Roche, 10104159001). The samples were then incubated at 37°C incubator for 15 min (with gentle shaking every 5 min). Papain was inactivated with 10% FBS in neurobasal media, and hStrS were gently triturated using a P1000 pippette. Samples were centrifuged 1,300 rpm for 7 min, filtered with a 70 μm Flowmi cell strainer (Bel-Art, H13680–0070), and suspended in 0.04% BSA in PBS (Millipore-Sigma, B6917–25MG) at a concentration of 1,000 cells/μL. Approximately 16,000 cells were loaded onto a Chromium Single cell 3’ chip (Chromium™ Chip B Single Cell Kit, 10x Genomics, PN-1000154) with Chromium™ Single Cell 3’ Library & Gel Bead kit v3 (10x Genomics, PN-1000075) and cDNA library was generated with the Chromium™ Single Cell 3’ Library Construction kit v3 (10x Genomics, PN-1000078) according to manufacturer’s instructions. Each library was sequenced using the Illumina NovaSeq S4 2 × 150 bp by Admera Health. Quality control, UMI counting of Ensemble genes and aggregation of samples were performed by count and aggr functions in the Cell Ranger software (version 3.1.0). Further downstream analyses were performed using the R package Seurat (version 3.1.4)61. Genes on the X and Y chromosome were removed from the count matrix to avoid biases in clustering due to the sex of the hiPS cell line donors. Cells with more than 10,000 or less than 100 detected genes or with mitochondrial content higher than 15% were also excluded. Genes that were not expressed in at least three cells were not included in the analysis. Gene expression was normalized using a global-scaling normalization method (normalization.method = “LogNormalize”, scale.factor = 10000), and the 2,000 most variable genes were selected (selection.method =“vst”) and scaled (mean=0 and variance=1, for each gene) before principal component analysis (PCA). The top 15 principal components were used to cluster (resolution of 0.5) using the FindNeighbors and FindClusters function and to visualize with Uniform Manifold Approximation and Projection (UMAP). Clusters were grouped based on expression of known marker genes and differentially expressed genes as identified with the FindAllMarkers function (Supplementary Table 6), and more resolved clusters and top 10 genes are shown in Extended Data Fig. 3d,e and Supplementary Table 7. Unbiased spatial mapping of hStrS GABAergic neurons cluster was performed using VoxHunt28. Briefly, 100 most variable features from the E13.5 mouse brain ISH Allen Brain Atlas data were selected and similarity maps were calculated. These maps were then plotted in the sagittal and coronal views. Comparison of hStrS GABAergic neurons cluster data to BrainSpan transcriptomics data of microdissected human brain tissue was also performed using VoxHunt with the default setting. Integration of hSS19 (GSE93811, BD Resolve) and hStrS scRNAseq data set was performed using standard Seurat v3 integration workflow62. Prior to finding anchors between the individual data set, the gene expression of two samples was normalized with the NormalizeData function, and the 2,000 most variable genes were selected (Selection.method =“vst”) using the FindVariableFeatures function. Anchors were identified using the FindIntegrationAnchors function with dimensionality 5, and the standard workflow for visualization with UMAP and clustering were then used. Whether clusters are shared or not shared between hSS and hStrS was determined based on a bias for one of the conditions in the proportion of cells in each cluster. For instance, cluster 10 includes 21.5% of all hSS, but only 1.07% of hStrS; therefore, we considered this to be a hSS biased cluster. On the other hand, an almost equal percentage of cells in both hSS (5.38%) and hStrS (6.04%) contributed to cluster 4; therefore, we considered this a shared clustered. These percentages for each condition can be seen in Extended Data Figure 5c.
Viral labeling and live cell imaging
Viral infection of 3D neural spheroids was performed as previously described19, 60. Briefly, hCS or hStrS were transferred into a 1.5 mL Eppendorf tube containing 200 μL of neural medium and incubated with the virus overnight at 37°C, 5% CO2. Next day, fresh 800 μL of culture media was added. The following day, neural spheroids were transferred into fresh culture media in ultra-low attachment plates (Corning, 3471, 3261). For live cell imaging, labeled hCS or hStrS or assembloids were transferred into a well of Corning™ 96-Well microplate (Corning, 4580) in 150 μL of neural media or on a 20 mm glass coverslip in a 35 mm glass bottom well (Cellvis, D35-20-0-N), and incubated in an environmentally controlled chamber for 15–30 min before imaging on a Leica TCS SP8 confocal microscope.
The viruses used were: AAV-DJ-mDlx-GFP-Fishell-133 (Addgene, #83900), AAV-DJ-mDlx-GCaMP6f-Fishell-233 (Addgene, #83899), AAV-DJ-hSyn1::eYFP (Stanford University Neuroscience Gene Vector and Virus Core, GVVC-AAV-16), AAV-DJ-CaMKIIa::eYFP (Stanford University Neuroscience Gene Vector and Virus Core, GVVC-AAV-8), AAV-DJ-hSyn1::mCherry (Stanford University Neuroscience Gene Vector and Virus Core, GVVC-AAV-17), Rabies-ΔG-Cre-eGFP (Salk Institute Viral Vector Core), AAV-DJ-EF1a-CVS-G-WPRE-pGHpA63 (Addgene, #67528), AAV-DJ-EF1-DIO-mCherry (Stanford University Neuroscience Gene Vector and Virus Core, GVVC-AAV-14), AAV1-Syn::ChrimsonR-tdT43 (Addgene, #59171-AAV1), AAV-DJ-Ple94 (GPR88)-iCre35 (pEMS1995, Addgene, #49125), AAV-DJ-EF1-DIO-eYFP (Stanford University Neuroscience Gene Vector and Virus Core, GVVC-AAV-13), AAV-DJ-EF1a-DIO-GCaMP6s (Stanford University Neuroscience Gene Vector and Virus Core, GVVC-AAV-91).
For imaging and quantification of dendritic spines in hStrS, mDlx-eGFP+ cells were imaged using a 10x and 20x objective on a Leica TCS SP8 confocal microscope, and images were analyzed with Fiji (ImageJ, v2.1.0, NIH). The number of dendritic spines were counted on a primary and secondary dendrite within 200 μm from the soma.
For calcium imaging, hStrS or assembloids were labeled with DJ-Ple94::iCre & AAV-DJ-EF1a::DIO-GCaMP6s or mDlx-GCaMP6f-Fishell-2 and placed on a well of Corning™ 96-Well microplate (Corning, 4580) or a 20 mm coverslip glass in a 35 mm glass bottom plate in neural media, and imaged using a 10x objective on a Leica TCS SP8 confocal microscope. GCaMP6 was imaged at a frame rate of 14.7 frames/sec, and results were analyzed with Fiji (ImageJ, v2.1.0, NIH) and MATLAB (vR2018a, 9.4.0, MathWorks).
Generation of cortico-striatal assembloids
To generate cortico-striatal assembloids, hCS and hStrS were derived separately and fused by placing them in close proximity in a 1.5 ml Eppendorf tubes for 3 days in an incubator. Media was carefully changed on day 2, and on day 3 assembloids were placed in 6-well or 60 mm ultra-low attachment plate (Corning, 3471, 3261) in neural medium described. Assembly was performed between days 60 to 76 of differentiation.
Clearing and 3D staining of cortico-striatal assembloids
To optically clear cortico-striatal assembloids, we applied the hydrophilic chemical cocktails-based CUBIC (clear, unobstructed brain/body imaging cocktails and computational analysis) protocol64, 65. Briefly, cortico-striatal assembloids at day 83 were fixed with 4% PFA/4% sucrose/PBS at 37°C for 20 min and incubated at 4°C overnight. Next day, the assembloids were washed twice with PBS and incubated in Tissue-Clearing Reagent CUBIC-L (TCI, T3740) at 37°C for 2 days. Assembloids were washed three times with PBS for 2 hours, then stained with anti–GFP (1:1000 dilution) and anti–mCherry (1:1000 dilution) antibodies in PBS containing 0.2% Triton X-100 (Millipore Sigma, T9284–100ML) and 3% NDS (Abcam, ab7475) at 37°C for 2 days. Assembloids were subsequently washed three times with PBS for 2 hours, then incubated with secondary antibodies (Alexa, 1:1000 dilution) in PBS containing 0.2% Triton X-100 and 3% NDS at 37°C for 2 days. For refractive index (RI) matching, assembloids were incubated with Tissue-Clearing Reagent CUBIC-R+ (TCI, T3741) at room temperature for 2 days. CUBIC-cleared assembloids were then transferred into a well of Corning™ 96-Well microplate (Corning, 4580) in 150 μL of CUBIC-R+ solution and imaged using a 10x objective on a Leica TCS SP8 confocal microscope.
Axon projection imaging in cortico-striatal assembloids
Projections into hStrS of AAV-DJ-hSyn1::eYFP+ cells from hCS or into hCS of AAV-DJ-hSyn1::mCherry+ cells from hStrS were imaged under environmentally controlled conditions in cortico-striatal assembloids using the Leica TCS SP8 confocal microscope with a motorized stage. Assembloids were transferred to a well in a Corning™ 96-Well microplate (Corning, 4580) in 150 μL of culture media and incubated in an environmentally controlled chamber 15–30 min before imaging. Images were taken using a 10x objective lens at a depth of 0–500 μm. The percentage of eYFP coverage in hStrS and of mCherry coverage in hCS were quantified with Fiji (ImageJ, v2.1.0, NIH).
Retrograde rabies tracing in cortico-striatal assembloids
For retrograde neural tracing experiments in cortico-striatal assembloids, hCS were labeled with AAV-DJ-EF1-DIO-mCherry and hStrS were labeled with ΔG-Rabies-eGFP-Cre and AAV-DJ-EF1a-CVS-G-WPRE-pGHpA. After viral infection, hCS and hStrS were assembled and maintained in culture with media changes every 4 days. At 28 days after assembly, cortico-striatal assembloids were fixed overnight in 4% PFA in PBS at 4°C and processed for immunostaining. Images were collected with a Leica TCS SP8 confocal microscope and analyzed in Fiji (ImageJ, v2.1.0, NIH).
Optogenetic stimulation and calcium imaging
Cortico-striatal assembloids expressing AAV1-Syn::ChrimsonR-tdTomato in hCS, and AAV-DJ-Ple94::iCre & AAV-DJ-EF1a::DIO-GCaMP6s in hStrS were placed on a 20 mm coverslip glass in a 35 mm glass bottom plate in neural media and imaged using a 10x objective on a Leica TCS SP8 confocal microscope. For optogenetic stimulation, ChrimsonR-tdTomato+ cells in hCS were activated with 625 nm light using an optical fiber-coupled LED (400 μm-diameter, 13.2 mW, Thorlabs). GCaMP6s was imaged at a frame rate of 14.7 frames/sec. Stimulation experiments included 1500 frames, and one 625 nm of LED (68 ms) were applied every 150 frames or 300 frames. The pulse was generated by a Cyclops LED connected to the Leica TCS SP8. Results were analyzed with Fiji (ImageJ, v2.1.0, NIH) and MATLAB (version R2018a, 9.4.0, MathWorks). After ROI registration, raw time-series were transformed to relative changes in fluorescence: ΔF/F(t)= (F(t)-F0)/F0. To verify if responses were time-locked to the LED stimulation, ΔF/F responses were compared to ΔF/F values at randomly selected time points. For time-locked ΔF/F, the amplitudes of ΔF/F from each cell were calculated as the maximum ΔF/F value within a 20 to 30 frames (1,360 to 2,040 ms) window following LED stimulation (minus the mean of the baseline 1 second before the stimulation). The median was used to exclude shape-dependent artifacts; the median amplitude of ΔF/F was compared before and after treatment with the AMPA receptor antagonist NBQX (20 μM, Tocris, 0373), the NMDA receptor antagonist D-APV (100 μM, Tocris, 0106), and the GABAA receptor antagonist (+)– bicuculine (50 μM, Tocris, 0130). To analyze synchronization of calcium signals, mean correlation coefficient per pair of cells of ΔF/F of GCaMP6 signal in hStrS and cortico-striatal assembloids were analyzed in MATLAB.
Whole cell recordings
Whole-cell recordings in neural spheroid slices were performed as previously described19, 57. Briefly, assembloid or spheroid slices were embedded in 4% agarose and transferred into an artificial cerebrospinal fluid (aCSF) containing: 126 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 1 mM MgSO4, 2mM CaCl2, 26 mM NaHCO3 and 10 mM D-(+)-glucose. Slices were cut at 200 μm at room temperature using a Leica VT1200 vibratome and maintained in aCSF at room temperature.
Whole-cell patch-clamp recordings from hStrS slices were performed under an upright slicescope microscope (Scientifica). Slices were perfused with aCSF (bubbled with 95% O2 and 5% CO2) and cells were recorded at 30°C. hSyn1::eYFP+ neurons were patched with a borosilicate glass pipette filled with an internal solution containing 127 mM potassium gluconate, 8 mM NaCl, 4 mM MgATP, 0.3 mM Na2GTP, 10 mM HEPES, 0.6 mM EGTA, pH 7.2 adjusted with KOH (290 mOsm). Data were acquired with a MultiClamp 700B amplifier (Molecular Devices) and Digidata 1550B digitizer (Molecular Devices), low-pass filtered at 2 kHz and digitized at 20 kHz, analyzed with the pClamp software (version 10.6, Molecular Devices). The liquid junction potential was calculated using JPCalc66, and recordings were corrected with an estimated −15 mV liquid junction potential67–69.
Spontaneous EPSCs were recorded at −70 mV in a voltage-clamp mode and analyzed with the MiniAnalysis software (v6.0.3, Synaptosoft). For F-I curves, cells were current-clamped at −60 mV, current steps (1 s duration) were given with an increment of 10 pA. Data were analyzed with in-house programs in MATLAB (MathWorks). For LED stimulation, 5 ms long whole-field illumination at 550 nm (maximal power, CoolLed) was applied through the 40x objective and optically-evoked EPSC (oEPSC) were recorded at −70 mV. AAV-hSyn1::eYFP+ neurons close to the hCS side of cortico-striatal assembloids were randomly selected for recordings.
Animals
Pregnant ICR/CD1 mice were obtained from Charles River. Whole brains from E18.5 mouse embryo were fixed with cold 4% PFA/PBS, and samples were prepared following the staining method described above. Approval for mouse experiments was obtained from the Stanford University, Administrative Panel on Laboratory Animal Care (APLAC).
Statistics
Data are presented as mean ± s.e.m. or box plots showing maximum, third quartile, median, first quartile and minimum values. Raw data was tested for normality of distribution, and statistical analyses were performed using paired or unpaired t-test (two-tailed), Mann-Whitney test, Kruskal-Wallis test, Wilcoxon test, or one-way ANOVA test with multiple-comparison, and two-way ANOVA. Sample sizes were estimated empirically. GraphPad Prism version 8.4.2 was used for statistical analyses.
Reporting Summary
Further information on research design and reagents are shown in the Life Sciences Reporting Summary, which is published alongside the paper and linked to this article.
Extended Data
Extended Data Fig. 1. Generation of the GSX2 gene reporter hiPS cell line.
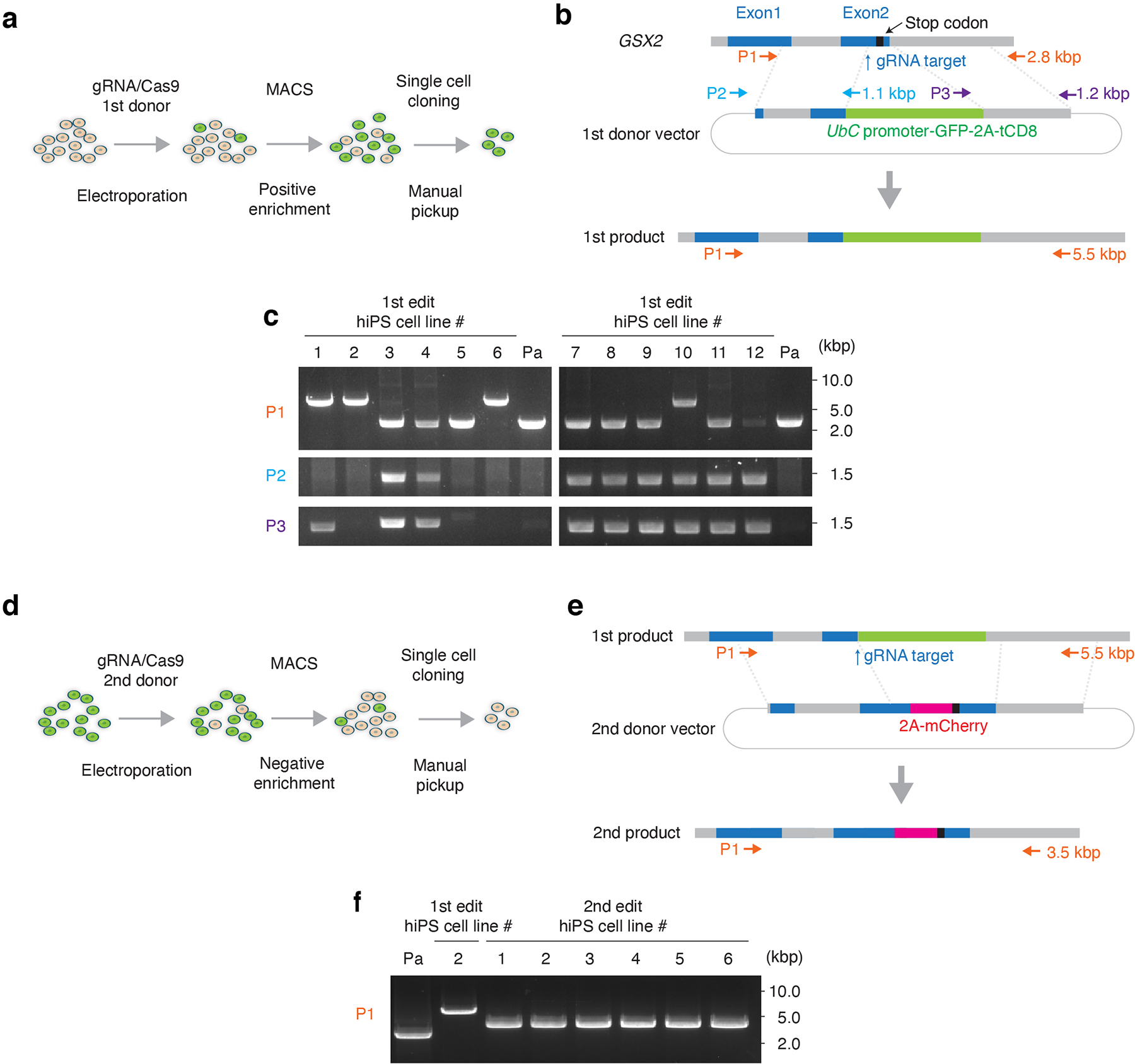
(a) Schematic showing the 1st step of the HDR-based genome editing and (b) the design of the 1st donor plasmids. The genomic sequence around the stop codon of GSX2 gene was replaced by EGFP and the truncated CD8 (tCD8) expression cassette. Arrows indicate the position of primer sets used for the genotyping PCR. Details on genome editing design and the sequence of 1st donor vector are available in Supplementary Table 1. (c) Genotyping and detection of random integration. Primer set 1 (P1) shows targeted integration of the selection marker expression cassette in the GSX2 locus. Primer set 2 (P2) and set 3 (P3) show random integration. For hiPS cell lines #2 and #6, a single 5.5 kb band was detected by P1 and no band was detected by P2 or P3. Subsequent ddPCR showed that 2 copies of exogenous UbC promoter were integrated in #2 and #6. Line #2 was used in the next step of genome engineering. Pa: parental hiPS cell line. (d) Schematic showing the 2nd editing step, and (e) the design of the 2nd donor plasmid. The selection marker expression cassette was replaced by mCherry. Arrows show the primer sets used in the genotyping PCR. The size of amplicons is indicated by arrows. Information on the 2nd donor is available in Supplementary Table 2. (f) Genotyping of the negatively selected lines by PCR. The single 3.5 kbp band indicate biallelic editing. Line #1 from the 2nd editing step was used for further experiments. Full-length, unprocessed gel images for c and f are included in Source Data 1.
Extended Data Fig. 2. Generation of hStrS.
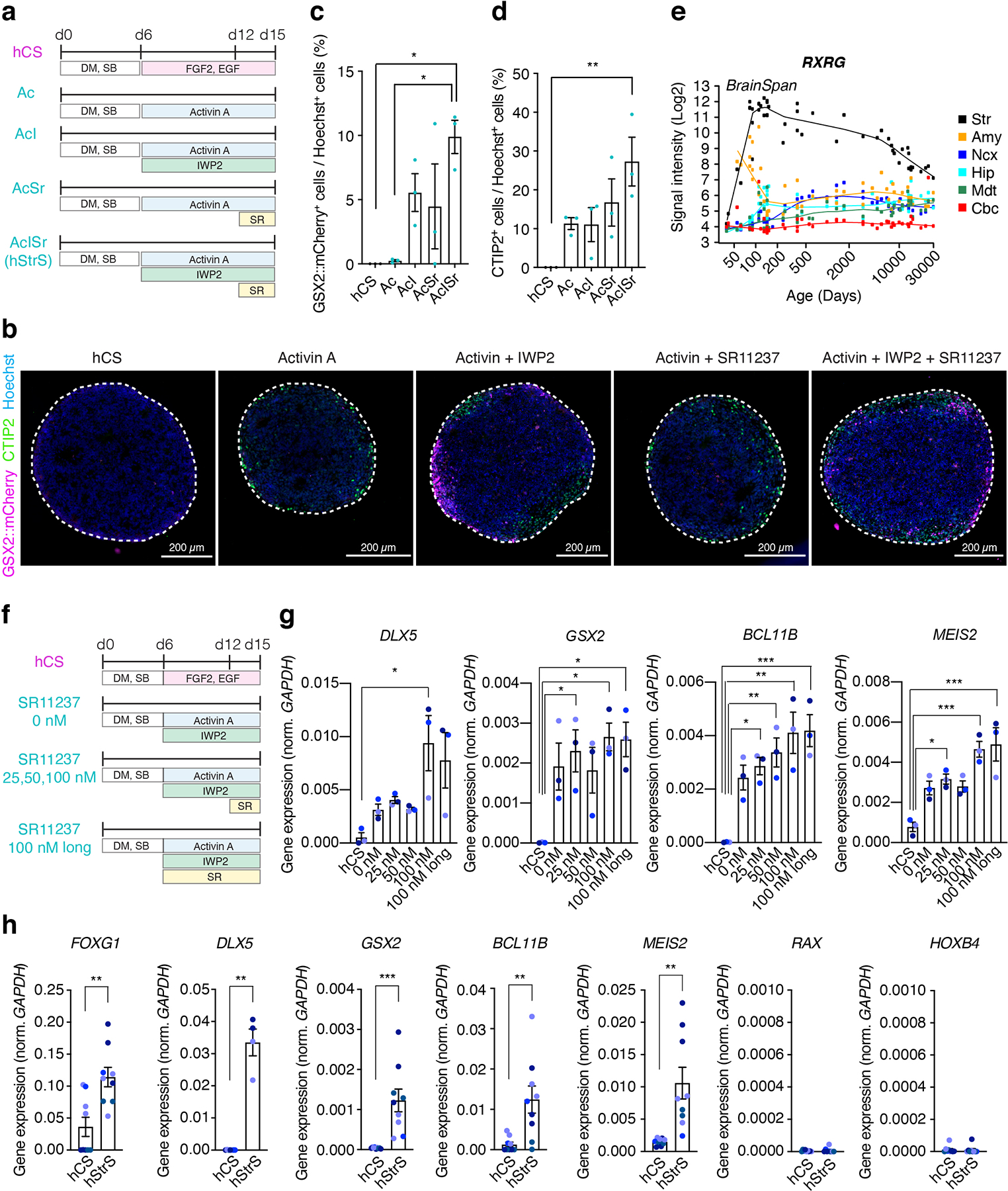
(a) Schematic illustrating the differentiation conditions for striatal differentiation as compared to the protocol for generating hCS. Ac (Activin), AcI (Activin + IWP-2), AcSr (Activin + SR11237), AcISr (Activin + IWP-2 + SR11237) (b) Immunostaining of neural spheroids at day 15 differentiation in the reporter GSX2::mCherry line. Scale bars: 200 μm. (c–d) Percentage of (c) GSX2::mCherry+ cells and (d) CTIP2+ cells in neural spheroids at day 15 (n = 3 differentiation experiments with GSX2::mCherry line; one-way ANOVA, F4,10 = 5.7, P = 0.01, following Tukey’s multiple comparison test: *P = 0.01 for hCS vs AcISr and *P = 0.01 for A vs AcISr in c, one-way ANOVA F4,10= 5.03, P = 0.01, following Tukey’s multiple comparison test for d; **P = 0.009 for hCS vs AcISr). (e) Gene expression pattern of RXRG in developing striatum (Str), amygdala (Amy), neocortex (Ncx), hippocampus (Hip), mediodorsal nucleus thalamus (Mdt) and cerebellar cortex (Cbc) in the BrainSpan transcriptome dataset (https://hbatlas.org/). (f) Schematic illustrating differentiation conditions and different SR11237 concentrations as compared to the protocol for generating hCS, and (g) level of gene expression (RT-qPCR) for DLX5, GSX2, BCL11B (CTIP2), and MEIS2. n = 3 neural spheroids from 3 hiPS cell lines; one-way ANOVA, F5,12 = 4.6, P = 0.01, following Tukey’s multiple comparison test: *P = 0.01 for hCS vs 100 nM for DLX5, one-way ANOVA, F5,12 = 4.6, P = 0.01, following Tukey’s multiple comparison test: *P = 0.03 for hCS vs 25 nM, *P = 0.01 for hCS vs 100 nM, *P = 0.01 for hCS vs long 100 nM for GSX2, one-way ANOVA, F5,12 = 9.07, P = 0.0009, following Tukey’s multiple comparison test: *P = 0.01 for hCS vs 25 nM, **P = 0.005 for hCS vs 50 nM, **P = 0.001 for hCS vs 100 nM, ***P = 0.001 for hCS vs long 100 nM for BCL11B, one-way ANOVA, F5,12 = 11.69, P = 0.0003, following Tukey’s multiple comparison test: *P = 0.02 for hCS vs 25 nM, ***P = 0.0005 for hCS vs 100 nM, ***P = 0.0003 for hCS vs long 100 nM for MEIS2. (h) Gene expression (by RT-qPCR) of FOXG1, DLX5, GSX2, BCL11B (CTIP2), MEIS2, RAX, and HOXB4. For FOXG1, GSX2, BCL11B (CTIP2), MEIS2, RAX and HOXB4: n = 9 neural spheroids (hCS or hStrS) from 3 differentiation experiments of 4 hiPS cell lines. For DLX5: n = 7 hCS from 2 differentiation experiments of 4 hiPS cell lines, n = 4 hStrS from 2 differentiation experiments of 2 hiPS cell lines. Two-tailed unpaired t-test **P = 0.002 for FOXG1, two-tailed Mann-Whitney test **P = 0.006 for DLX5, two-tailed unpaired t-test **P = 0.004 for GSX2, two-tailed unpaired t-test ***P = 0.0007 for BCL11B, two-tailed unpaired t-test **P = 0.002 for MEIS2, two-tailed Mann-Whitney test P = 0.67 for RAX, two-tailed unpaired t-test P = 0.88 for HOXB4. The 2242–1 hiPS cell line is shown in orchid blue, 8858–3 in blueberry blue, 1205–4 in midnight blue, and 0524–1 in ocean blue. Data shown are mean ± s.e.m.
Extended Data Fig. 3. Characterization of hStrS.
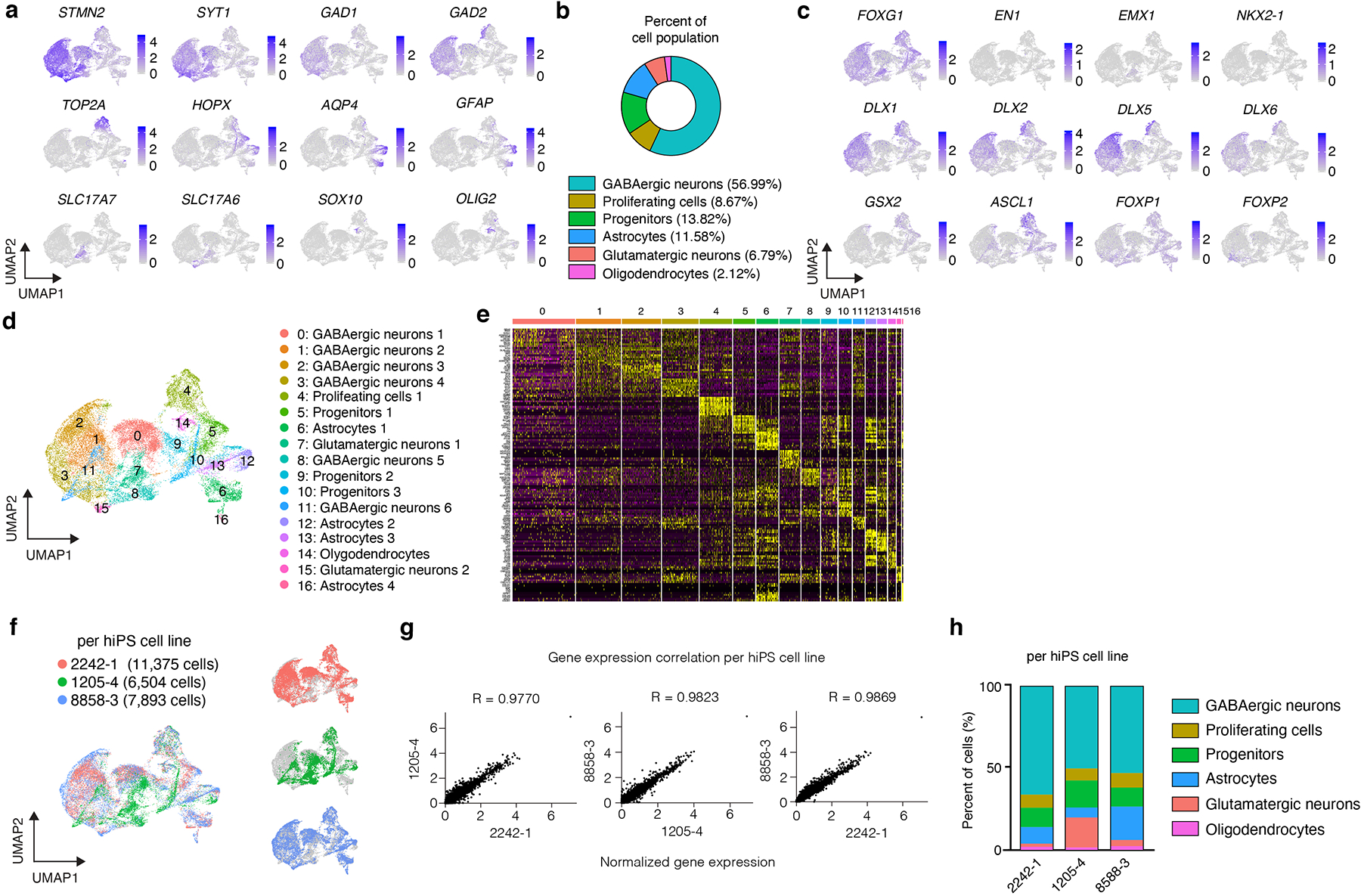
(a) UMAP visualization of expression of selected genes in the hStrS single cell RNA-seq data at day 80–83 of in vitro differentiation (n = 25,772 cells from 3 hiPS cell lines). (b) Percentage of major cell type clusters in hStrS. (c) Expression of forebrain (FOXG1), midbrain, hindbrain (EN1), dorsal forebrain (EMX1), LGE and MGE markers in hStrS. (d) UMAP visualization of the resolved single cell RNA-seq data of hStrS, and (d) heat map for the top 10 genes in each cluster. (f) UMAP visualization of the single cell RNA-seq data color coded by the hiPS cell lines: 2242–1 (red), 1205–4 (green), 8858–3 (blue). (g) Plots showing the Pearson correlation of the normalized average gene expression between each of the three hiPS cell lines used. (h) Graph showing the percentage of cells in each of the three hiPS cell lines belonging to each cluster in hStrS.
Extended Data Fig. 4. Characterization of hStrS.
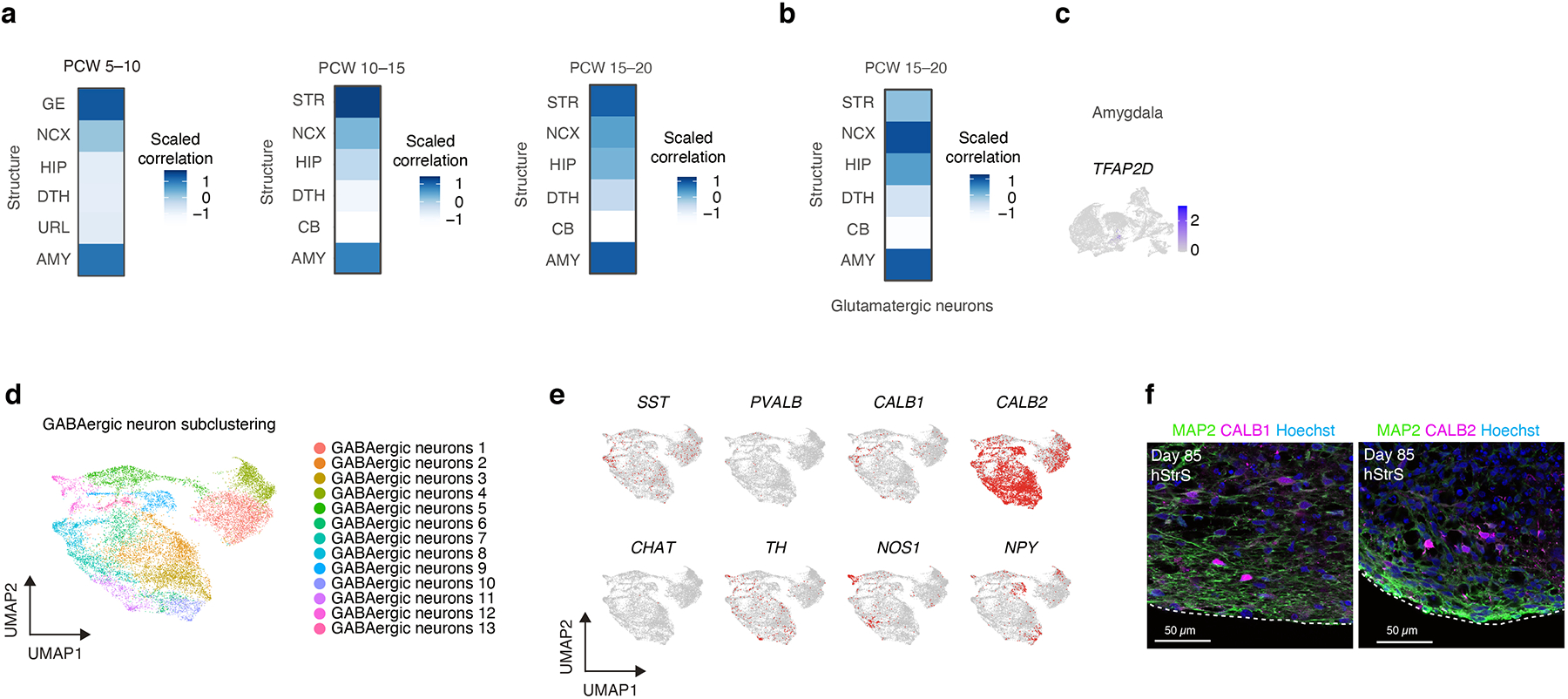
(a) VoxHunt mapping of the hStrS GABAergic neuron cluster to the BrainSpan human brain dataset (PCW 5–10, 10–15, and 15–20). (b) VoxHunt mapping of the glutamatergic neuronal cluster in hStrS. (c) Expression of early amygdala marker TFAP2D in hStrS. (d) UMAP visualization of single cell RNA expression in GABAergic neurons subcluster of hStrS at day 80–83 of in vitro differentiation. (e) Expression of SST, PVALB, CALB1, CALB2, CHAT, TH, NOS1, NPY in the hStrS GABAergic subcluster. (f) Left, immunostaining for MAP2 (green), CALB1 (magenta), Hoechst (blue) in hStrS at day 85. Right, immunostaining for MAP2 (green), CALB2 (magenta), Hoechst (blue) in hStrS at day 85. n = 4 hiPS cell lines. Scale bar: 50 μm.
Extended Data Fig. 5. Comparison of hStrS to hSS.
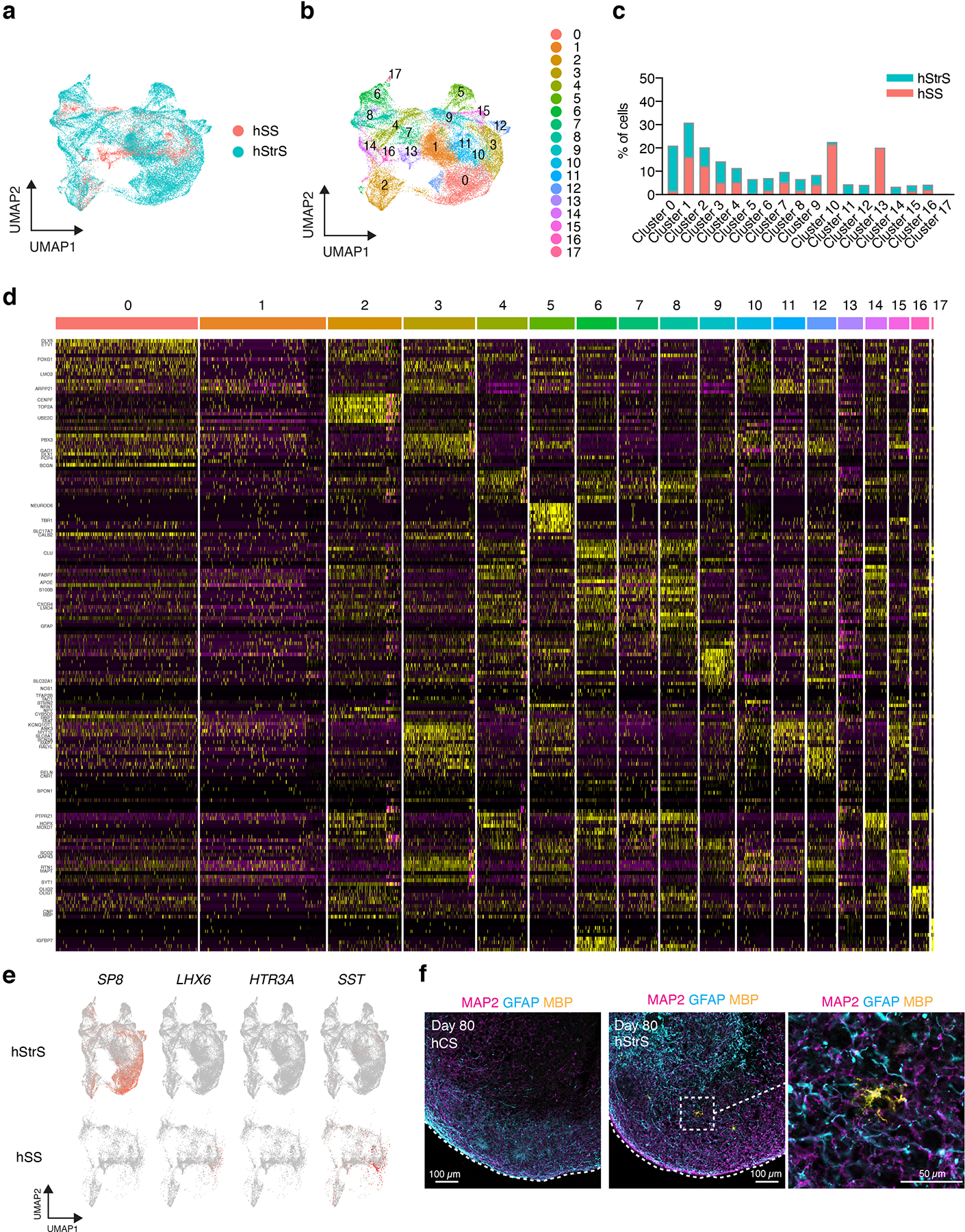
(a,b) UMAP visualization of scRNA-seq data from hStrS and hSS. (c) Percentage of cells from hStrS and hSS in each cluster. (d) Heat map for the top 10 genes in each cluster. (e) Expression of SP8, LHX6, HTR3A, SST in hStrS (top) and hSS (bottom). (f) Representative immunocytochemistry images of hCS and hStrS (day 80) for the neuron marker MAP2 (magenta), astrocyte marker GFAP (cyan) and oligodendrocyte marker MBP (yellow). Scale bar: 100 μm for left and middle images, and 50 μm for right image. Immunostainings were repeated in spheroids from 2 independent differentiation experiments with similar results.
Extended Data Fig. 6. Characterization of hStrS neurons.
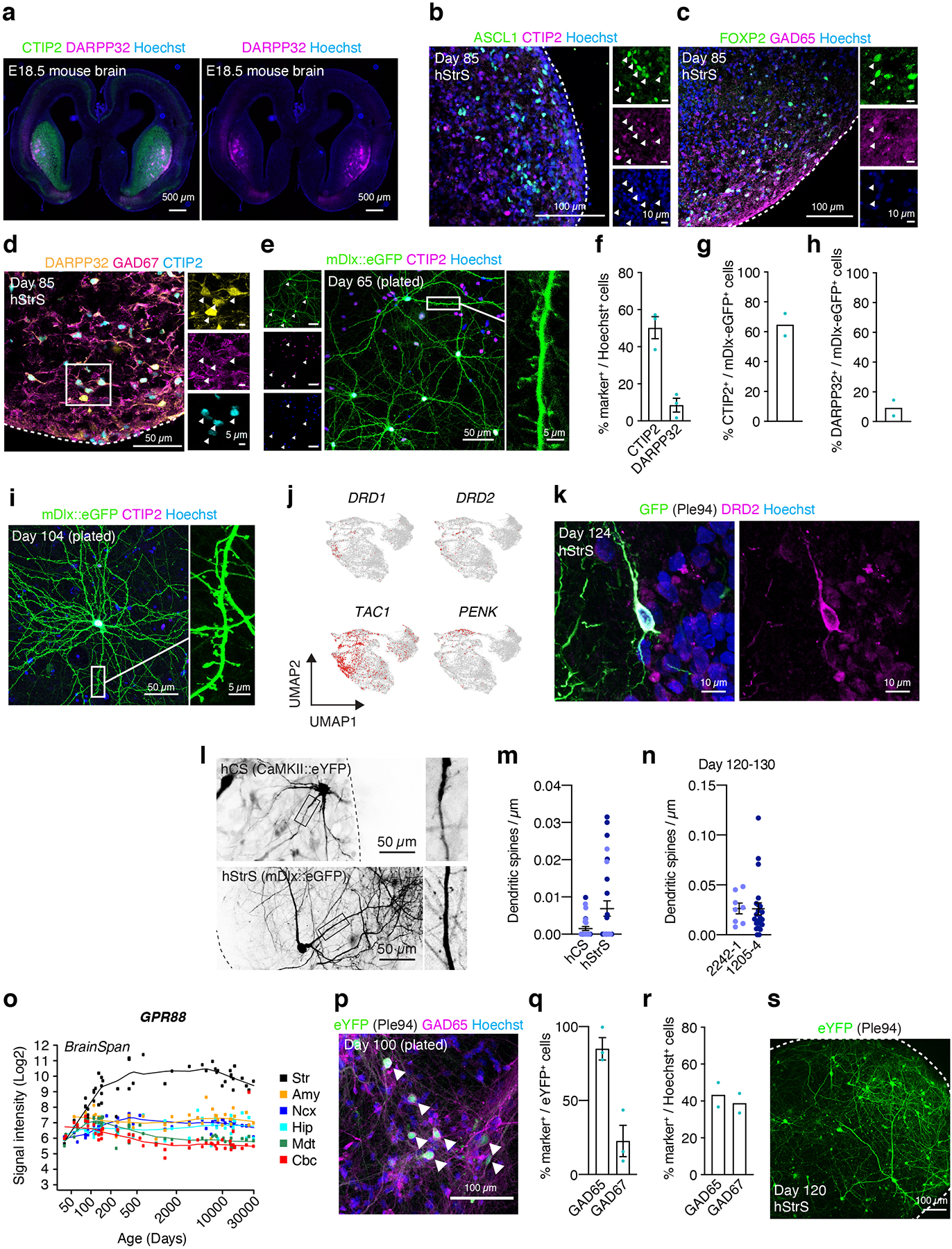
(a) Representative image of an E18.5 mouse brain section immunostained with antibodies against CTIP2 (BCL11B) (green), DARPP32 (magenta) and Hoechst (blue). Scale bar: 500 μm. (b–d) Immunostaining for ASCL1 (green), CTIP2 (magenta), Hoechst (blue) in b, FOXP2 (green), GAD65 (magenta) and Hoechst (blue) in c, DARPP32 (yellow), GAD67 (magenta) and CTIP2 (cyan) in d, hStrS at day 85. n = 3 hiPS cell lines. Scale bar: 100 μm in b,c, and 50 μm in d. (e–i) Representative immunocytochemistry images of dissociated hStrS neurons that have been labeled with an AAV-mDlx::eGFP at day 65 or at day 104. Scale bar: 50 μm for the left and middle image, and 5 μm for the right image (e). 50 μm for left, and 5 μm for right image (i). Quantification of CTIP2+ and DARPP32+ cells in mDlx::eGFP infected and 2D-plated hStrS cells. n = 3 neural spheroids from 2 hiPS cell lines in f, n = 2 neural spheroids from 2 hiPS cell lines in h. (j) Expression of DRD1, DRD2, TAC1, PENK in hStrS. (k) Immunostaining for GFP (green), DRD2 (magenta), Hoechst (blue), hStrS at day 124. Scale bar: 10 μm. Immunostainings were repeated in spheroids from 3 independent differentiation experiments with similar results. (l,m) Dendritic spine morphology of hCS neurons labelled with CaMKII::eYFP and of hStrS neurons labelled with AAV-mDlx::eGFP. Quantification of the number of dendritic spines (day 100–110: n = 26 neurons for hCS, n = 27 neurons for hStrS; from 2 hiPS cell lines; two-tailed, Mann-Whitney test, P = 0.17). (n) Quantification of number of dendritic spines (day 120–130: n = 8 neurons from the 2242–1 line, n = 22 neurons from the 1205–4 line; two-tailed, Mann-Whitney test, P = 0.44). (o) Gene expression of GPR88 in developing human brain in the BrainSpan transcriptome dataset (https://hbatlas.org/). (p) Representative image of dissociated hStrS neurons labeled with Ple94::iCre and DIO-eYFP at day 100. Expression of eYFP is induced by iCre expression under a mini-promoter including regulatory region of striatal gene GPR88 (Ple94). Arrow heads indicate eYFP+/GAD65+ cells. Scale bar: 100 μm. (q) Quantitative results showing percentage of GAD65+ and GAD67+ cells out of eYFP+ cells following recombination with the Ple94 reporter. n = 3 hiPS cell lines. (r) Percentage of GAD65+ and GAD67+ cells out of Hoechst+ cells (n = 2 differentiation with 2 iPS cell lines. (s) Representative confocal live image of hStrS labeled with Ple94::iCre and DIO-eYFP at day 120. Scale bar: 100 μm. Data shown are mean ± s.e.m. Imaging were repeated in spheroids from 2 independent differentiation experiments with similar results.
Extended Data Fig. 7. Functional characterization of hStrS.
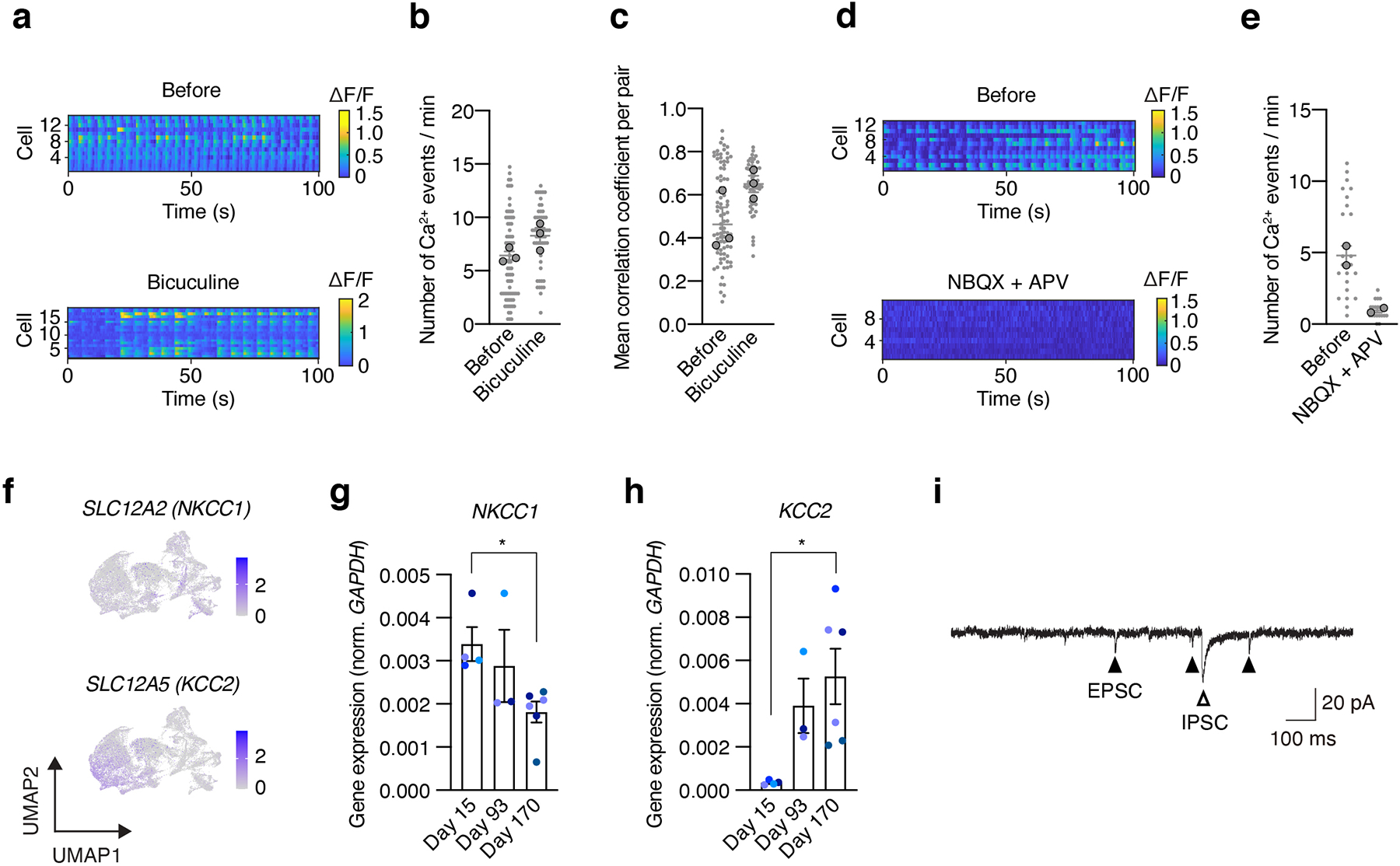
(a–e) Effect of bicuculine (50 μM) (a–c) and NBQX (20 μM) + APV (50 μM) (d,e) on calcium signals (GCaMP6s) in hStrS neurons at day 104. GCaMP6s was induced by iCre expression under a minipromoter that includes the regulatory region of the striatal gene GPR88 (Ple94). Heatmap showing ΔF/F of GCaMP6s signals. n = 74 cells before and n = 55 cells after bicuculine treatment in b; two-tailed, Mann-Whitney test, P = 0.09. n = 87 cells before and n = 56 cells after bicuculine exposure in c; two-tailed, unpaired t-test, P = 0.10. n = 25 cells before and n = 21 cells after NBQX + APV exposure in e. Data show mean ± s.e.m. (f) Expression of SLC12A2 and SLC12A5 in hStrS. (g,h) RT-qPCR for NKCC1 and KCC2. n = 4 neural spheroids from 4 hiPS cell lines at day 15, n = 3 neural spheroids from 3 hiPS cell lines at day 93, n = 6 neural spheroids from 3 hiPS cell lines at day 170; Kruskal-Wallis test, *P = 0.01, Dunn’s multiple comparisons test: *P = 0.02 for day 15 vs day 170 in NKCC1, one-way ANOVA, F3,12 = 4.38, P = 0.02, following Tukey’s multiple comparison test: *P = 0.04 for day 15 vs day 170 in KCC2. (i) Representative recording of spontaneous IPSC in hSyn1::eYFP expressing hStrS neurons at day 160. The 2242–1 hiPS cell line is shown in orchid blue, 8858–3 in blueberry blue, 1205–4 in midnight blue, 0524–1 in ocean blue, 0410–1 in aqua blue. Data show mean ± s.e.m.
Extended Data Fig. 8. Characterization of cortico-striatal assembloids.
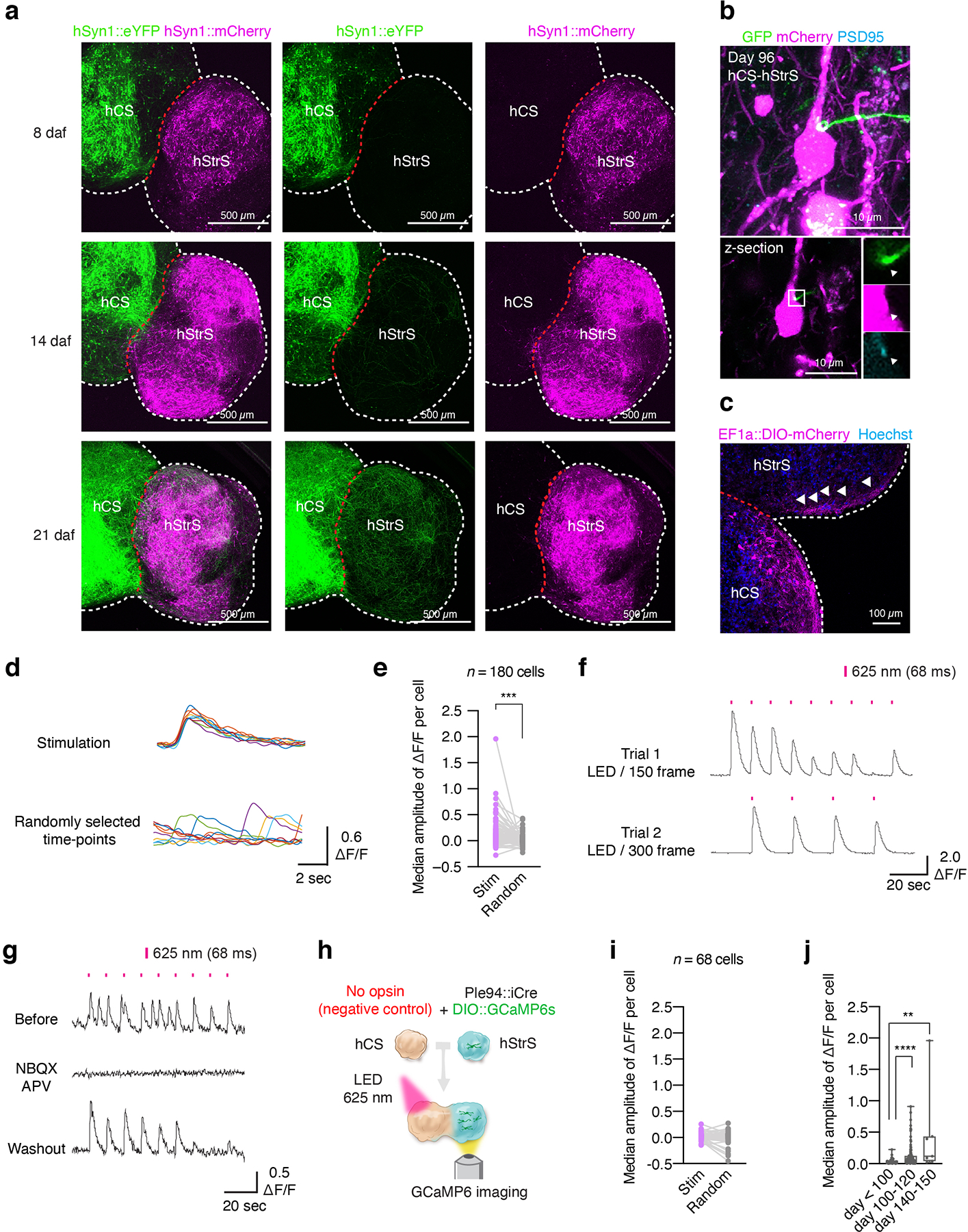
(a) Representative images of cortico-striatal assembloids at 8, 14, 21 days after assembly (daf). Scale bar: 500 μm. Imaging was repeated in assembloids from 2 independent differentiation experiments with similar results. (b) hCS neurons (Syn1::YFP+) projecting towards PSD95+ puncta on dendrites of mCherry+ hStrS neurons at day 96. Immunostainings were repeated in assembloids from 2 independent differentiation experiments with similar results. (c) mCherry+ projections from hCS in cortico-striatal assembloids (hCS was infected with a AAV-EF1a::DIO-mCherry; hStrS was infected with ΔG-Rabies virus-Cre-GFP and a AAV-EF1a::G). Scale bar: 100 μm. (d,e) Comparison of stimulus-triggered change in amplitude of ΔF/F of GCaMP6 signals to random time-locked ΔF/F in the same cell; (d) Representative traces of ΔF/F from real stimulation (top) and randomly selected time-points (bottom), and (e) quantitative results. n = 180 cells from 10 assembloids with 3 hiPS cell lines; two-tailed Wilcoxon test ***P = 0.0002. (f) Representative trace of ΔF/F of GCaMP6 signal at LED / 150 frame (top) and at LED / 300 frame (bottom). (g) Representative traces of GCaMP6s imaging and median amplitude of ΔF/F per cell before, during NBQX (20 μM) and APV (50 μM) treatment and after wash. Data show mean ± s.e.m. (h,i) Schematics of a control optogenetics coupled with calcium imaging experiment in cortico-striatal assembloids. Quantitative results of ΔF/F from a real stimulation and a randomly selected time-point at day 108. n = 68 cells from 3 assembloids derived from 2 hiPS cell lines; two-tailed Wilcoxon test, P = 0.33. (j) Quantitative results of ΔF/F from day 90 to day 145 cortico-striatal assembloids. n = 68 cells from 3 assembloids derived from 2 hiPS cell lines; Kruskal-Wallis test, ****P < 0.0001, Dunn’s multiple comparisons test: ****P < 0.0001 for day < 100 vs day 100–120, **P = 0.004 for day < 100 vs day 140–150. Data show mean ± s.e.m. Box plots show maximum, third quartile, median, first quartile, and minimum values.
Extended Data Fig. 9. Electrophysiological characterization of cortico-striatal assembloids.
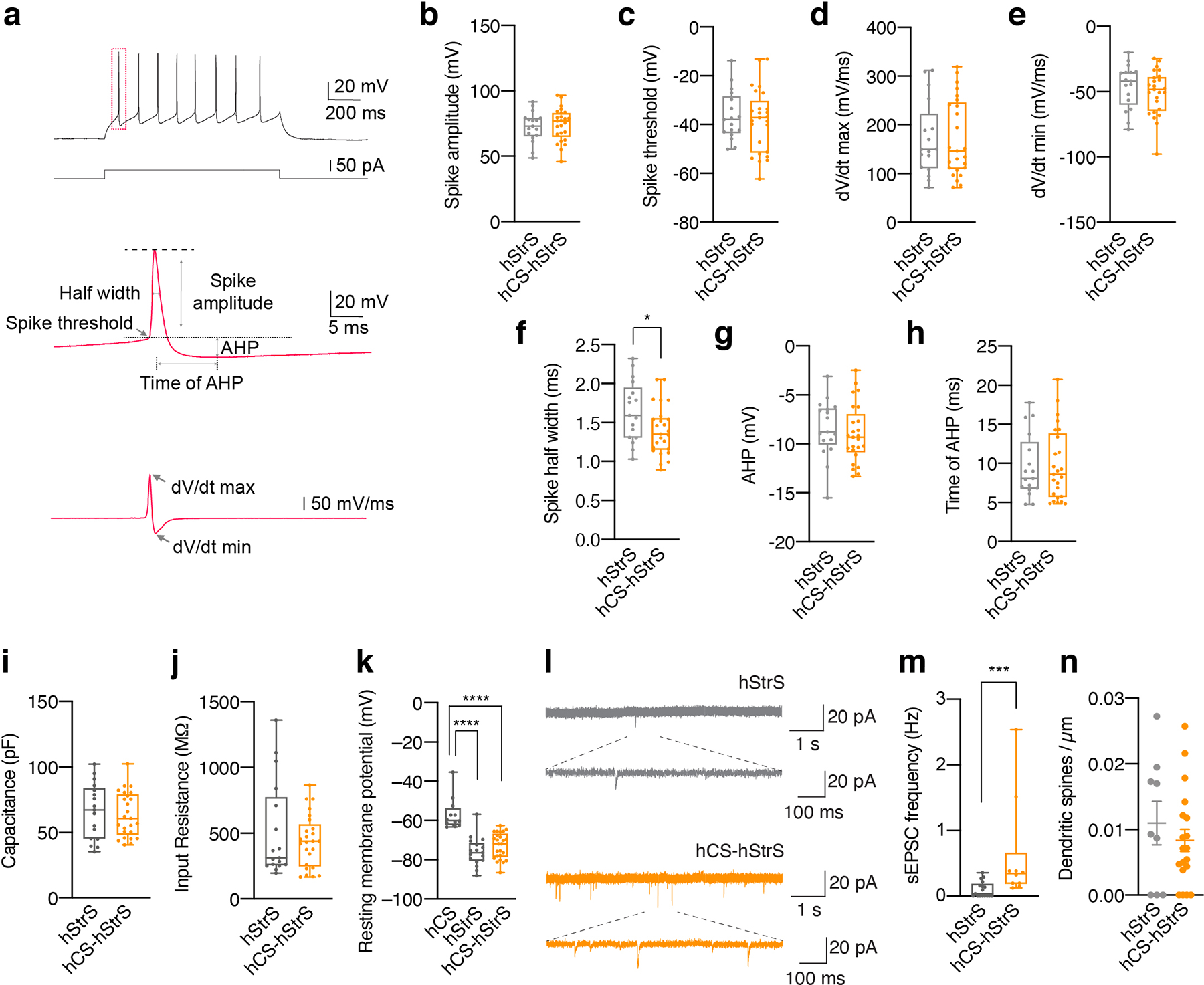
(a) Analyses on individual action potential traces in hStrS and hCS-hStrS neurons. (b) Quantification of spike amplitude, (c) spike threshold, (d) dV/dt max, (e) dV/dt min, (f) spike half width, (g) AHP, (h) time of AHP, (i) capacitance, (j) input resistance and (k) resting membrane potential in hCS, hStrS and hCS-hStrS neurons; n = 17 cells from hStrS, n = 25 cells from hCS-hStrS derived from 3 hiPS cell lines; two-tailed unpaired t-test P = 0.64 for b, two-tailed unpaired t-test P = 0.60 for c, two-tailed unpaired t-test P = 0.94 for d, two-tailed unpaired t-test P = 0.37 for e, two-tailed unpaired t-test *P = 0.02 for f, two-tailed unpaired t-test P = 0.53 for g, two-tailed unpaired t-test P = 0.87 for h, two-tailed unpaired t-test P = 0.68 for i, Mann-Whitney test P = 0.82 for j, Kruskal-Wallis test, ****P < 0.0001, Dunn’s multiple comparisons test: ****P < 0.0001 for hCS vs hStrS, ****P < 0.0001 for hCS vs hCS-hStrS, P = 0.80 for hStrS vs hCS-hStrS for k. (l) Representative traces of spontaneous EPSC (sEPSC) and (m) frequency of sEPSCs in hStrS and hCS-hStrS (n = 17 cells in hStrS, n =10 cells in hCS-hStrS from 3 hiPS cell lines; two-tailed Mann-Whitney test ***P = 0.0006. (n) Quantification of the number of dendritic spines (day 100–110: n = 9 neurons for hStrS, n = 27 neurons for hCS-hStrS from one hiPS cell line; two-tailed, Mann-Whitney test, P = 0.17).Data shown are mean ± s.e.m. Box plots show maximum, third quartile, median, first quartile, and minimum values.
Extended Data Fig. 10. Characterization of hiPS cells derived from subjects with 22q13.3 deletion syndrome.
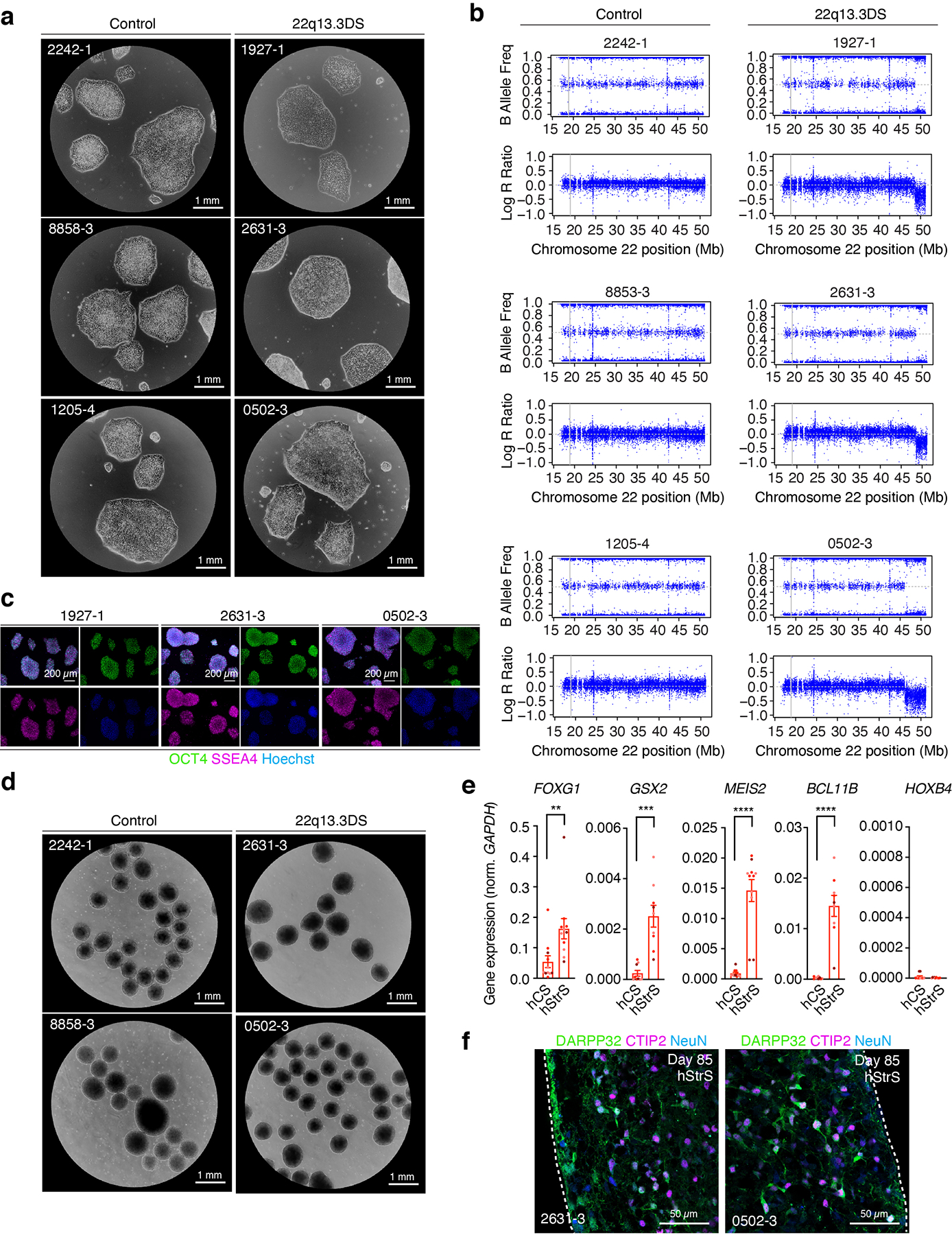
(a) Representative images showing morphology of hiPS cells from control and 22q13.3DS patients. Scale bars: 1 mm. Imaging was repeated in 2 independent differentiation experiments with similar results. (b) SNP array of hiPS cells showing the 22q13.3 deletion locus. Upper shows B Allele Frequency, and bottom Log R Ratio for chromosome 22q region. (c) Immunostaining for pluripotency stem cell markers OCT4 (green) and SSEA4 (magenta). Scale bars: 200 μm. (d) Representative images of 3D neural spheroids at day 5 of differentiation from control and 22q13.3DS hiPS cell lines. Scale bars: 1 mm. Imaging was repeated in spheroids from 3 independent differentiation experiments with similar results. (e) Gene expression (RT-qPCR) for the forebrain marker FOXG1, the LGE markers GSX2, MEIS2, BCL11B (CTIP2), and the spinal cord marker HOXB4 at day 22 of differentiation in hCS and hStrS derived from 22q13.3DS hiPS cells (n = 9 neural spheroids from 3 hiPS cell lines; two-tailed Mann-Whitney test **P < 0.002 for FOXG1, two-tailed unpaired t-test ***P = 0.0001 for GSX2, two-tailed Mann-Whitney test ****P < 0.0001 for MEIS2, two-tailed unpaired t-test ****P < 0.0001 for BCL11B (CTIP2), two-tailed Mann-Whitney test P = 0.99 for HOXB4. (f) Immunostaining for DARPP32 (green), CTIP2 (magenta) and NeuN (blue) in day 85 hStrS. Scale bar: 50 μm. Data shown are mean ± s.e.m. Immunostainings were repeated in spheroids from 2 independent differentiation experiments with similar results.
Supplementary Material
Supplementary Table 1 List of the top 100 genes in each of the clusters in hStrS single cell gene expression analyzed by the FindMarkers function of Seurat (non-parametric Wilcoxon rank sum test, two-sided)
Supplementary Table 2 List of the top 10 genes in each of the clusters in resolved hStrS single cell gene expression analyzed by the FindMarkers function of Seurat (non-parametric Wilcoxon rank sum test, two-sided)
Supplementary Video 3 Time lapse imaging on the hStrS side of cortico-striatal assembloids at 21 days after fusion (daf) (day 83 of differentiation) showing the growth cone of a hCS neuron labeled with eYFP (gray). Scale bar: 10 μm.
Supplementary Video 2 Time lapse imaging of axon projections (YFP+: green) from hCS into hStrS (mCherry: magenta) in cortico-striatal assembloids at 21 days after fusion (daf) (day 83 of differentiation). Scale bar: 100 μm.
Supplementary Video 1 Calcium imaging of hStrS neurons expressing the genetically encoded calcium indicator GCaMP6s at day 126 of differentiation. GCaMP6s was induced by iCre expression under a mini-promoter that included the regulatory region of the striatum enriched gene GPR88 (Ple94). Scale bar: 200 μm.
ACKNOWLEDGEMENTS
We thank members of the Pasca laboratory at Stanford University for scientific inputs and the Stanford Wu Tsai Neurosciences Virus Core for production of AAVs. This work was supported by the US National Institute of Health (NIH) BRAINS Award (MH107800) (to S.P.P.), the MQ Fellow Award (to S.P.P.), the NYSCF Robertson Stem Cell Investigator Award (to S.P.P.), the Stanford Human Brain Organogenesis Program in the Wu Tsai Neurosciences Institute (to S.P.P.), the Kwan Research Fund (to S.P.P.), the Coates Foundation (to S.P.P.), the Senkut Foundation (to S.P.P.), the Uytengsu Research fund (to S.P.P.), the Chan Zuckerberg Initiative Ben Barres Investigator Award (to S.P.P.), Stanford Medicine Dean’s Fellowship (to Y.M., F.B), Stanford Maternal & Child Health Research Institute (MCHRI) Postdoctoral Fellowship (to Y.M., F.B., and O.R.), and the American Epilepsy Society Postdoctoral Research Fellowship (to F.B.).
Footnotes
COMPETING INTERESTS STATEMENT
Stanford University has filed a provisional patent application covering the protocol and methods for the generation of striatal spheroids and cortico-striatal assembloids. M.H.P. is a consultant for and has equity interest in CRIPR Tx. During the duration of this study, K.I. was an employee of Daiichi-Sankyo Co., Ltd, although the company had no input on the design, execution, interpretation, or publication of the data.
Code availability
The codes used for calcium imaging and electrophysiology analyses in this study are available on request from the corresponding author.
Data availability
Gene expression data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE149931. Human Brain Transcriptome (https://hbatlas.org/) was used to explore transcriptome data of the developing and adult human brain27.The data in this study are available on request from the corresponding author.
REFERENCES
- 1.Alexander GE, DeLong MR & Strick PL Parallel organization of functionally segregated circuits linking basal ganglia and cortex. Annu Rev Neurosci 9, 357–381 (1986). [DOI] [PubMed] [Google Scholar]
- 2.Shepherd GM Corticostriatal connectivity and its role in disease. Nat Rev Neurosci 14, 278–291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milad MR & Rauch SL Obsessive-compulsive disorder: beyond segregated cortico-striatal pathways. Trends Cogn Sci 16, 43–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peca J et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 472, 437–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welch JM et al. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature 448, 894–900 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amin ND & Pasca SP Building Models of Brain Disorders with Three-Dimensional Organoids. Neuron 100, 389–405 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Sasai Y Cytosystems dynamics in self-organization of tissue architecture. Nature 493, 318–326 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Pasca SP The rise of three-dimensional human brain cultures. Nature 553, 437–445 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Eiraku M et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Mariani J et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci U S A 109, 12770–12775 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lancaster MA et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasca AM et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nature methods 12, 671–678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian X et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 165, 1238–1254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marton RM et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat Neurosci 22, 484–491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bershteyn M et al. Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell 20, 435–449 e434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klaus J et al. Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat Med 25, 561–568 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Blair JD, Hockemeyer D & Bateup HS Genetically engineered human cortical spheroid models of tuberous sclerosis. Nat Med 24, 1568–1578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasca AM et al. Human 3D cellular model of hypoxic brain injury of prematurity. Nat Med 25, 784–791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birey F et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L et al. Human embryonic stem cell-derived GABA neurons correct locomotion deficits in quinolinic acid-lesioned mice. Cell Stem Cell 10, 455–464 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delli Carri A et al. Developmentally coordinated extrinsic signals drive human pluripotent stem cell differentiation toward authentic DARPP-32+ medium-sized spiny neurons. Development 140, 301–312 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Arber C et al. Activin A directs striatal projection neuron differentiation of human pluripotent stem cells. Development 142, 1375–1386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watson C, Paxinos G & Puelles L The mouse nervous system, Edn. 1st. (Elsevier Academic Press, Amsterdam; Boston; 2012). [Google Scholar]
- 24.Onorati M et al. Molecular and functional definition of the developing human striatum. Nat Neurosci 17, 1804–1815 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Phelan K & McDermid HE The 22q13.3 Deletion Syndrome (Phelan-McDermid Syndrome). Mol Syndromol 2, 186–201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yun K, Potter S & Rubenstein JL Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development 128, 193–205 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Kang HJ et al. Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fleck JS, He Z, Boyle MJ, Camp JG & Treutlein B Resolving brain organoid heterogeneity by mapping single cell genomic data to a spatial reference. bioRxiv, 2020.2001.2006.896282 (2020). [Google Scholar]
- 29.Waclaw RR, Ehrman LA, Pierani A & Campbell K Developmental origin of the neuronal subtypes that comprise the amygdalar fear circuit in the mouse. J Neurosci 30, 6944–6953 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu JS, Vogt D, Sandberg M & Rubenstein JL Cortical interneuron development: a tale of time and space. Development 144, 3867–3878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silberberg SN et al. Subpallial Enhancer Transgenic Lines: a Data and Tool Resource to Study Transcriptional Regulation of GABAergic Cell Fate. Neuron 92, 59–74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steiner H & Tseng K-Y Handbook of basal ganglia structure and function, Edn. Second edition. (Elsevier/Academic Press, Amsterdam; Boston; 2017). [Google Scholar]
- 33.Dimidschstein J et al. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat Neurosci 19, 1743–1749 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen TW et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Leeuw CN et al. rAAV-compatible MiniPromoters for restricted expression in the brain and eye. Mol Brain 9, 52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blaesse P, Airaksinen MS, Rivera C & Kaila K Cation-chloride cotransporters and neuronal function. Neuron 61, 820–838 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Peixoto RT, Wang W, Croney DM, Kozorovitskiy Y & Sabatini BL Early hyperactivity and precocious maturation of corticostriatal circuits in Shank3B(−/−) mice. Nat Neurosci 19, 716–724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wickersham IR, Finke S, Conzelmann KK & Callaway EM Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods 4, 47–49 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Etessami R et al. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J Gen Virol 81, 2147–2153 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Wilson CJ Morphology and synaptic connections of crossed corticostriatal neurons in the rat. J Comp Neurol 263, 567–580 (1987). [DOI] [PubMed] [Google Scholar]
- 41.Sohur US, Padmanabhan HK, Kotchetkov IS, Menezes JR & Macklis JD Anatomic and molecular development of corticostriatal projection neurons in mice. Cereb Cortex 24, 293–303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo L, Callaway EM & Svoboda K Genetic Dissection of Neural Circuits: A Decade of Progress. Neuron 98, 865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klapoetke NC et al. Independent optical excitation of distinct neural populations. Nature methods 11, 338–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belleau ML & Warren RA Postnatal development of electrophysiological properties of nucleus accumbens neurons. J Neurophysiol 84, 2204–2216 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Peixoto RT et al. Abnormal Striatal Development Underlies the Early Onset of Behavioral Deficits in Shank3B(−/−) Mice. Cell Rep 29, 2016–2027 e2014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Y et al. Atypical behaviour and connectivity in SHANK3-mutant macaques. Nature 570, 326–331 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Misceo D et al. A translocation between Xq21.33 and 22q13.33 causes an intragenic SHANK3 deletion in a woman with Phelan-McDermid syndrome and hypergonadotropic hypogonadism. Am J Med Genet A 155A, 403–408 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Shcheglovitov A et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature 503, 267–271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plenz D & Aertsen A Neural dynamics in cortex-striatum co-cultures--I. anatomy and electrophysiology of neuronal cell types. Neuroscience 70, 861–891 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Bloem B, Huda R, Sur M & Graybiel AM Two-photon imaging in mice shows striosomes and matrix have overlapping but differential reinforcement-related responses. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi M et al. Effects of NR2A and NR2B-containing N-methyl-D-aspartate receptors on neuronal-firing properties. Neuroreport 22, 762–766 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Lieberman OJ et al. Dopamine Triggers the Maturation of Striatal Spiny Projection Neuron Excitability during a Critical Period. Neuron 99, 540–554 e544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graybiel AM & Ragsdale CW Jr. Histochemically distinct compartments in the striatum of human, monkeys, and cat demonstrated by acetylthiocholinesterase staining. Proc Natl Acad Sci U S A 75, 5723–5726 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cederquist GY et al. Specification of positional identity in forebrain organoids. Nat Biotechnol 37, 436–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yi F et al. Autism-associated SHANK3 haploinsufficiency causes Ih channelopathy in human neurons. Science 352, aaf2669 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pecho-Vrieseling E et al. Transneuronal propagation of mutant huntingtin contributes to non-cell autonomous pathology in neurons. Nat Neurosci 17, 1064–1072 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Yoon SJ et al. Reliability of human cortical organoid generation. Nature methods 16, 75–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ikeda K et al. Efficient scarless genome editing in human pluripotent stem cells. Nat Methods 15, 1045–1047 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Cradick TJ, Qiu P, Lee CM, Fine EJ & Bao G COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol Ther Nucleic Acids 3, e214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sloan SA, Andersen J, Pasca AM, Birey F & Pasca SP Generation and assembly of human brain region-specific three-dimensional cultures. Nat Protoc 13, 2062–2085 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stuart T et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wertz A et al. Single-cell-initiated monosynaptic tracing reveals layer-specific cortical network modules. Science 349, 70–74 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Susaki EA et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Tainaka K et al. Chemical Landscape for Tissue Clearing Based on Hydrophilic Reagents. Cell Rep 24, 2196–2210 e2199 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Barry PH JPCalc, a software package for calculating liquid junction potential corrections in patch-clamp, intracellular, epithelial and bilayer measurements and for correcting junction potential measurements. J Neurosci Methods 51, 107–116 (1994). [DOI] [PubMed] [Google Scholar]
- 67.Paz JT et al. Closed-loop optogenetic control of thalamus as a tool for interrupting seizures after cortical injury. Nat Neurosci 16, 64–70 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sorokin JM et al. Bidirectional Control of Generalized Epilepsy Networks via Rapid Real-Time Switching of Firing Mode. Neuron 93, 194–210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Makinson CD et al. Regulation of Thalamic and Cortical Network Synchrony by Scn8a. Neuron 93, 1165–1179 e1166 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 List of the top 100 genes in each of the clusters in hStrS single cell gene expression analyzed by the FindMarkers function of Seurat (non-parametric Wilcoxon rank sum test, two-sided)
Supplementary Table 2 List of the top 10 genes in each of the clusters in resolved hStrS single cell gene expression analyzed by the FindMarkers function of Seurat (non-parametric Wilcoxon rank sum test, two-sided)
Supplementary Video 3 Time lapse imaging on the hStrS side of cortico-striatal assembloids at 21 days after fusion (daf) (day 83 of differentiation) showing the growth cone of a hCS neuron labeled with eYFP (gray). Scale bar: 10 μm.
Supplementary Video 2 Time lapse imaging of axon projections (YFP+: green) from hCS into hStrS (mCherry: magenta) in cortico-striatal assembloids at 21 days after fusion (daf) (day 83 of differentiation). Scale bar: 100 μm.
Supplementary Video 1 Calcium imaging of hStrS neurons expressing the genetically encoded calcium indicator GCaMP6s at day 126 of differentiation. GCaMP6s was induced by iCre expression under a mini-promoter that included the regulatory region of the striatum enriched gene GPR88 (Ple94). Scale bar: 200 μm.
Data Availability Statement
Gene expression data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE149931. Human Brain Transcriptome (https://hbatlas.org/) was used to explore transcriptome data of the developing and adult human brain27.The data in this study are available on request from the corresponding author.


