Abstract
An assay was developed to determine the activity of peptide deformylase (PDF) inhibitors under conditions as close as possible to the physiological situation. The assay principle is the detection of N-terminal [35S]methionine labeling of a protein that contains no internal methionine. If PDF is active, the deformylation of the methionine renders the peptide a substrate for methionine aminopeptidase, resulting in the removal of the N-terminal methionine label. In the presence of a PDF inhibitor, the deformylation is blocked so that the N-formylated peptide is not processed and the label is detected. Using this assay, it is possible to determine the PDF activity under near-physiological conditions in a cell-free transcription-translation system as well as in intact bacterial cells.
All nascent polypeptides in eubacteria are synthesized with N-formyl-methionine at the N terminus, using the formyl-methionyl-tRNA for initiation (11). During elongation of the polypeptide chain, the formyl group is removed by the enzyme peptide deformylase (PDF; EC 3.5.1.27) (1, 5). The N-terminal methionine is removed by methionine aminopeptidase (MAP; EC 3.4.11.18) if the penultimate residue is small and uncharged, e.g., alanine, cysteine, glycine, proline, serine, threonine, and valine. N-blocked methionine polypeptides are not a substrate for the MAP, making deformylation a prerequisite for protein maturation (6, 13, 18). PDF and MAP are enzymes essential for growth in wild-type bacteria, as demonstrated by gene deletion in Escherichia coli (6, 12, 14). PDF mutants can only be constructed in an fmt (coding for formyltransferase; EC.2.1.2.9) mutant background (12). Formyltransferase formylates the methionyl-tRNA , and mutants for the corresponding gene show a strongly decreased growth rate (9).
, and mutants for the corresponding gene show a strongly decreased growth rate (9).
We have recently described the identification, optimization, and biological characterization of new PDF inhibitors (2). Since the antibacterial activities of these inhibitors were lower than expected from the inhibition at the enzyme level, we have performed several studies in order to better understand this discrepancy (3). As part of this effort, we developed new specific assays to determine the PDF activity in crude E. coli homogenates as well as in intact bacteria. The assay conditions reflect the physiological conditions in bacteria much more closely than an assay using the isolated enzyme, making them valuable tools for a meaningful assessment of the PDF-inhibitory activities of antibacterial compounds.
MATERIALS AND METHODS
Bacterial strains, plasmids, enzymes, and chemicals.
The E. coli strains used in this study were XL2-blue and BL21 (DE3) carrying pLysS (Stratagene, Basel, Switzerland). The strain was grown in Luria-Bertani medium (Difco Laboratories, Detroit, Mich.) with aeration at 37°C. The restriction enzymes were from New England Biolabs (Beverly, Mass.) or Amersham Pharmacia Biotech (Dübendorf, Switzerland) and were used in accordance with the specifications of the manufacturer. All other chemicals, including actinonin (Ro 06-1467), were from Sigma (St. Louis, Mo.). The synthesis of Ro 61-1811, Ro 66-0376, and Ro 66-6976 is described elsewhere (2) (Table 1).
TABLE 1.
Summary of chemical structure and potency of the tested PDF inhibitors in different assays
| Ro no. | Structure | IC50a (μM)
|
MIC (E. coli DC2)
|
||||
|---|---|---|---|---|---|---|---|
| Fe-PDF (E. coli) | Cell-free TC-TL
|
In vivo TC-TL (BL21) | |||||
| Gel | FlashPlate | μM | μg/ml | ||||
| 06-1467 | 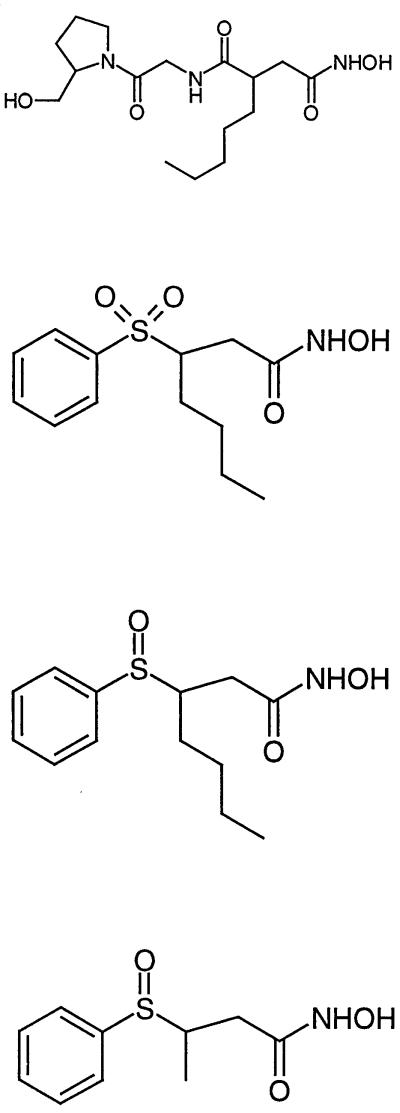 |
0.006 | 0.07 | 0.07 | 1 | 5.2 | 2 |
| 66-0376 | 0.04 | 0.3 | 0.35 | 0.7 | 28 | 4 | |
| 66-6976 | 0.1 | 0.35 | 0.5 | 0.3 | 1.9 | 0.5 | |
| 64-1811 | 20 | >100 | >100 | >500 | >263 | >64 | |
IC50 compound concentration required to inhibit 50% of specific activity in the assays. The results for the Fe-PDF enzyme assay and the MIC determination (calculated in micromolar) were taken from Apfel et al. (2). Inhibition of MAP was excluded (IC50 > 10 μM).
Construction of templates for TC-TL assays.
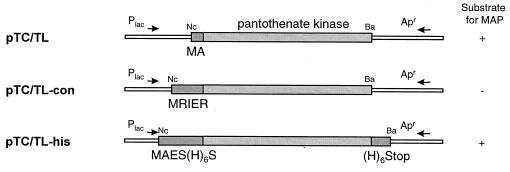
For further characterization of PDF-specific inhibitors, we developed a transcription-translation (TC-TL) assay to determine PDF activity in crude cell homogenates and in intact bacterial cells. Both assays are based on the N-terminal [35S]methionine labeling of a protein that contains no internal methionine. As PDF inhibitors prevent the deformylation of the [35S]methionine and the formyl-methionine-protein is not a substrate for MAP, the label is not removed from the protein. The proteins can be separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the label can be detected by autoradiography or radioactivity incorporated into His-tagged proteins can be detected on nickel chelate-coated FlashPlates (NEN Life Science Products, Boston, Mass.). For generating the templates for the TC-TL assays, we have identified several proteins from Haemophilus influenzae that contain only an N-terminal methionine. We have chosen pantothenate kinase as the indicator protein (35.6 kDa). We generated three different template vector constructs: pTC/TL (the standard template vector), pTC/TL-con (the control template vector; the translated protein is not a substrate for PDF, and the labeled methionine stays on the protein), and pTC/TL-his (the standard template vector with two additional six-His tags at the N and C termini) (Fig. 1). In all three cases, the full-length pantothenate kinase gene was amplified with the appropriate forward and reverse primers. The forward primer used introduced an NcoI site (5′-GCCATGG-3′) overlapping with the potential start codon, and the reverse primer carried a BamHI site downstream from the stop codon. The PCR products were digested with BamHI, and NcoI, separated on an agarose gel, eluted, and cloned into pDS56Δcat (a derivative of pDS56-RBS [19] carrying a deletion in the cat gene) digested with NcoI and BamHI, resulting in pTC/TL, pTC/TL-con, and pTC/TL-his, respectively (Fig. 1). The authenticity of each clone was verified by DNA sequencing.
FIG. 1.
Schematic representation of plasmids used in this study as templates in the cell-free or in vivo TC-TL assays. For details, see Materials and Methods. Abbreviations for restriction enzymes: Ba, BamHI; Nc, NcoI. +, yes; −, no.
TC-TL assay for determination of PDF activity in bacterial homogenates (cell-free TC-TL assay).
The standard incubation mixture for the PDF cell-free TC-TL assay contained, in a final volume of 20 μl, 2 μl of amino acid mixture minus methionine, 8 μl of S30 premix without amino acids, 6 μl of S30 extract (all from Promega, Madison Wis.), 1 μg of plasmid template DNA (pTC/TL or pTC/TL-con), 2 μCi of l-[35S]methionine (1,000 Ci/mmol; Amersham), 0.5 μl of the test compound in dimethyl sulfoxide (DMSO) (final concentration, 1% DMSO), and nuclease-free water. After the mixture was incubated in tubes at 37°C for 1 h, the reaction was stopped by adding 100 μl of acetone and placing the tubes on ice for 15 min. The acetone-precipitated S30 sample was centrifuged at 12,000 × g for 5 min, the supernatant was removed, and the pellet was dried under vacuum for 15 min. The dry pellet was solubilized in 10 μl of water, and 10 μl of SDS sample buffer (0.2% bromphenol blue, 2% SDS, and 10% glycerol in 60 mM Tris-HCl, pH 6.8) was added. After being heated at 95°C for 5 min, the sample was loaded on an SDS-PAGE gel (10% acrylamide) using a discontinuous slab gel system as described by Laemmli (10). Electrophoresis was performed at room temperature using a Bio-Rad Protean II xi cell unit (20 by 20 cm). After electrophoresis and fixation, the gel was dried, and the labeled protein bands were visualized by fluorography (Amplify [Amersham]; X-OMAT autoradiography film [Kodak, Rochester, N.Y.]). In order to quantitate the protein produced, the corresponding bands were cut out, slices were transferred to a counting vial, scintillation fluid (Ultima Gold; Packard, Meriden, Conn.) was added, and counting was performed in a scintillation counter (Beta-counter; Packard).
In an alternative, higher-throughput assay, the in vitro-synthesized indicator proteins were measured via their binding to nickel chelate-coated FlashPlates. The standard incubation was done as described above with the following modifications: 2 μCi of l-[3H]methionine (85 Ci/mmol; Amersham) was used instead of l-[35S]methionine, the indicator protein had two six-His tags (N- and C-terminal pTC/TL-NC-his [Fig. 1A]), and 96-well plates were used instead of single tubes. After the 1-h incubation step, 130 μl of buffer (5 mM Na-phosphate, pH 7.0, and 300 mM NaCl) was added and the mixture was transferred quantitatively to a 96-well nickel chelate-coated FlashPlate. The plate was counted (TopCount · NXT; Packard) after an overnight incubation at room temperature.
TC-TL assay for determination of PDF activity in whole cells (in vivo TC-TL assay).
E. coli BL21 cells carrying pTC/TL or pTC/TL-con were grown at 37°C in M9 medium until they reached an optical density at 578 nm of 0.3 to 0.4. After induction of the indicator protein expression by the addition of 2 mM IPTG (isopropyl-β-d-thiogalactopyranoside and further incubation for 5 min, an aliquot (0.5 ml) was transferred to an Eppendorf tube and the test compounds in DMSO (1% final concentration) or DMSO alone (control) were added. After incubation for 30 min, the culture was pulse-labeled for 5 min with 2 μCi of [35S]methionine (1,000 Ci/mmol; Amersham) and chased with 50 μl of 1% methionine for 2 min. The sample was cooled on ice for 5 min and centrifuged at 8,000 × g for 5 min, the supernatant was removed, and the pellet was dissolved in 20 μl of SDS sample buffer and heated at 95°C for 5 min. The proteins were separated by electrophoresis on a 10% polyacrylamide gel. After electrophoresis and fixation, the gel was dried, and the labeled protein bands were visualized by fluorography.
General DNA techniques and transformation.
Chromosomal-DNA preparation was performed using the Qiagen (Hilden, Germany) genomic-DNA purification system. Preparation of plasmid DNA was performed using the Promega Wizard mini- or maxipurification system. Plasmids, PCR products, and chromosomal DNA were cleaved with the appropriate restriction enzymes, ligated, and transformed into E. coli XL2-blue cells (4, 16). Transformants were selected on Luria-Bertani agar plates containing ampicillin (100 μg/ml) for pDS56Δcat and its derivatives.
PCR and sequencing.
PCR was performed according to the manufacturer's recommendations using the Expand High-Fidelity DNA system (Roche Molecular Biochemicals, Rotkreuz, Switzerland) in a Perkin-Elmer (Foster City, Calif.) thermocycler. Sequencing was performed by the dideoxy chain termination method (17) with a modified DNA-sequencing kit (dye terminator cycle sequencing; PE Applied Biosystems, Foster City, Calif.). and an automated DNA sequencing system (ABI Prism 310 genetic analyzer; PE Applied Biosystems). Nucleotide and amino acid sequences were analyzed with the University of Wisconsin Genetics Computer Group sequence analysis package (8) and with the Lasergene program (DNASTAR, Madison, Wis.).
RESULTS
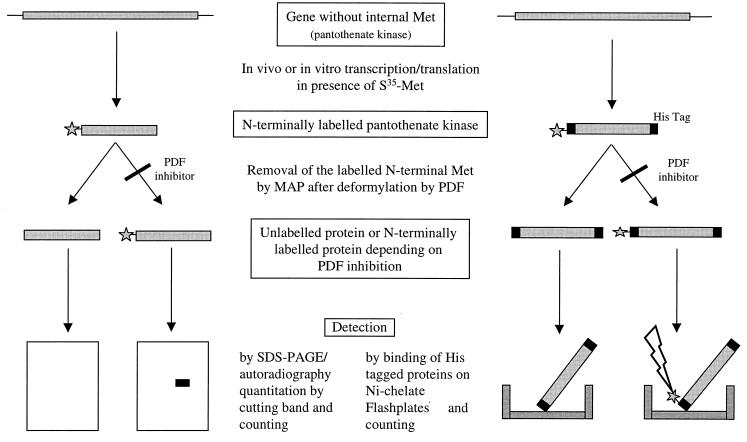
Using conventional enzyme assays for PDF activity to screen parts of our compound library, we have identified potent new inhibitors with activities in the nanomolar range (2). This high-throughput standard assay is based on the measurement of the hydrolysis of formyl-methionine-alanine-serine (f-MAS; Km, 4 to 10 mM) by recombinant purified E. coli PDF via detection of the formation of the new free amino group of MAS by reaction with fluorescamine (2). To further characterize the compounds, a PDF-specific TC-TL assay was developed that simulates the physiological conditions much better than the assay with the isolated enzyme. This assay system allowed us to determine PDF activity (i) in crude cell homogenates and (ii) in intact bacterial cells. Both assays are based on the N-terminal [35S]methionine labeling of a protein that contains no internal methionine. As PDF inhibitors prevent the deformylation of the [35S]methionine and the formyl-methionine-protein is not a substrate for MAP, the label is not removed from the protein, which can be separated by SDS-PAGE and detected by autoradiography. Alternatively, radioactivity incorporated into His-tagged proteins can be detected on nickel chelate-coated FlashPlates. An outline of the assay procedures is presented in Fig. 2. A prerequisite for using this assay as a specific test for inhibition of PDF is the absence of any inhibition of the enzyme MAP. Therefore, a MAP assay was performed with 4 mM Met-Ala-Ser as a substrate using recombinant E. coli enzyme in an adaptation of the method decribed by Chang et al. (7). No inhibition was observed at a compound concentration of 10 μM.
FIG. 2.
Schematic outline of the procedures for the detection of PDF inhibitor activity. The cell-free and whole-cell assays using detection of radioactivity from a polyacrylamide gel are shown on the left, and the higher-throughput assay using Ni-chelate-coated scintillant plates for detection is shown on the right. Incorporation of 35S- or 3H-labeled methionine, respectively, was used for labeling the N terminus.
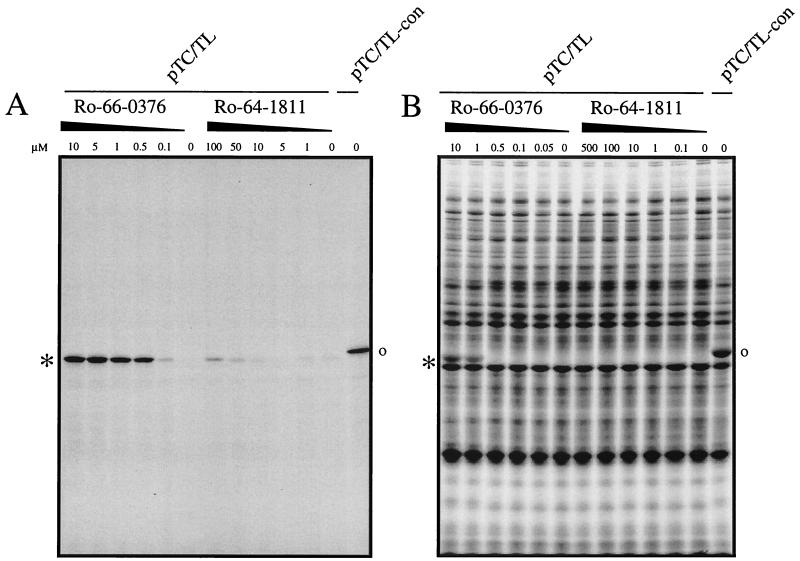
For generating the templates for the TC-TL assays, we have identified several proteins from H. influenzae that contain only one (the N-terminal) methionine. Pantothenate kinase (35.6 kDa) was chosen at random as the indicator, and plasmids for the expression of the corresponding gene were constructed, introducing N-terminal and/or C-terminal modifications in a lac repressor-based expression vector (pDS56Δcat). For generating pTC/TL (Fig. 1), the N-terminal sequence ATG was changed to ATG.GCA (encoding M-E). Previous N-terminal sequencing experiments with E. coli have shown that the methionine residues are effectively removed from proteins with M-E at the N terminus, even when the proteins are expressed at high levels (data not shown). As a positive control template, we have generated a second plasmid, pTC/TL-con, in which the ATG was replaced by a sequence encoding M-R-I-E-P, the N-terminal sequence from human kynureninase. In this construct, the penultimate residue is too big and charged for the N-terminal methionine to be removed by MAP. These two plasmids were used as templates. Figure 3 shows an example of fluorograms of polyacrylamide gels obtained by the whole-cell and the cell-free assays, using a strong and a weak inhibitor. With increasing amounts of the PDF inhibitor (Ro 66-0376), the removal of the N-terminal methionine is inhibited, and a single band or an additional protein band of the expected size becomes visible in the cell-free or the in vivo TC-TL assay, respectively (Fig. 3). For the weak inhibitor Ro 64-1811, even at high concentration (100 μM), no inhibition is detectable in the in vivo TC-TL assay, and only a slight inhibition is detectable in the cell-free assay (at 100 μM) (Fig. 3).
FIG. 3.
Autoradiography of SDS-PAGE gels from cell-free and in vivo TC-TL assays. The cell-free (A) or the intact-cell in vivo TC-TL standard (B) assay was performed using pTC/TL as a template in the presence of increasing amounts of potential PDF inhibitors (as indicated). As a positive control, the reaction using pTC/TL-con as a template is also shown. ∗, expected protein for pTC/TL; o, expected protein for pTC/TL-con. A representative experiment is shown.
In an alternative assay allowing an increased throughput, we measured the in vitro-synthesized His-tagged indicator protein by its binding to a nickel chelate-coated FlashPlate. As a template for that assay, a plasmid was constructed so that a six-His tag was added to the N-terminus and C-terminus of the indicator protein (pTC/TL-his) (Fig. 1). The conditions for the binding of the indicator protein to the nickel chelate-coated FlashPlate were optimized as described in Materials and Methods. The 50% inhibitory concentrations detected by this method were very similar to those obtained by the gel method, the signal-to-noise ratio being slightly higher with FlashPlate Attempts to use this nickel chelate-coated FlashPlate system for bacterial lysates were not successful.
Table 1 summarizes the comparison of the results of the isolated enzyme assay, those of cell-free TC-TL experiments with detection on gels and on FlashPlate, and those of in vivo TC-TL.
DISCUSSION
We have optimized new PDF inhibitors that were very active against the purified enzyme but displayed only weak antibiotic activity (2). In order to establish the extent to which these inhibitors do indeed inhibit the enzyme in bacteria, we designed TC-TL assays that allowed the measurement of PDF inhibition in bacterial-cell homogenates as well as in intact bacteria. We observed a general tendency toward less potent inhibition in the homogenate system (cell-free TC-TL) by a factor of 4 to 12 (Table 1). This is not surprising, since the assay using the isolated enzyme is run under somewhat artificial conditions compared to the assays described here. The use of a nonphysiological tripeptide substrate (f-MAS; 5 mM) in the enzyme assay is only one example (2). There was no significant difference between the values obtained from the cell-free TC-TL assays using the gel and the FlashPlate. Comparing the cell-free TC-TL data with data from intact bacteria in vivo TC-TL), the activity remains nearly identical for two inhibitors (Ro 66-6976 and Ro 66-0376), while it drops for Ro 06-1467 (actinonin). Differences in stability or penetration may account for this phenomenon. The results of the in vivo TC-TL assay and the MIC determination differ by a factor of 5 to 40. This could indicate that the enzyme activity has to be inhibited to a great extent (much more than 50%) to obtain a significant effect on the growth rate. Theoretically, we cannot completely exclude the possibility that the inhibitors might also interfere with additional, unknown targets to result in growth inhibition, but our results (presented in the accompanying article) indicate that this is not the case. The assay we describe here could be of general use for the characterization of inhibitors of the enzymes PDF and MAP.
ACKNOWLEDGMENTS
We acknowledge the expert technical assistance of Christian Lacoste, Olivier Partouche, and Bernard Rutten.
REFERENCES
- 1.Adams J M. On the release of the formyl group from nascent protein. J Mol Biol. 1968;33:571–589. doi: 10.1016/0022-2836(68)90307-0. [DOI] [PubMed] [Google Scholar]
- 2.Apfel C M, Banner D W, Bur D, Dietz M, Hirata T, Hubschwerlen C, Locher H, Page M G P, Pirson W, Rossé G, Specklin J-L. Hydroxamic acid derivatives as potent peptide deformylase inhibitors and antibacterial agents. J Med Chem. 2000;43:2324–2331. doi: 10.1021/jm000018k. [DOI] [PubMed] [Google Scholar]
- 3.Apfel C M, Locher H, Evers S, Takács B, Hubschwerlen C, Pirson W, Page M G P, Keck W. Peptide deformylase as an antibacterial drug target: target validation and resistance development. Antimicrob Agents Chemother. 2001;45:1058–1064. doi: 10.1128/AAC.45.4.1058-1064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1997. [Google Scholar]
- 5.Ball L A, Kaesberg P. Cleavage of the N-terminal formylmethionine residue from a bacteriophage coat protein in vitro. J Mol Biol. 1973;79:531–537. doi: 10.1016/0022-2836(73)90404-x. [DOI] [PubMed] [Google Scholar]
- 6.Chang S-Y P, McGary E C, Chang S. Methionine aminopetidase gene of Escherichia coli is essential for cell growth. J Bacteriol. 1989;171:4071–4072. doi: 10.1128/jb.171.7.4071-4072.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang Y H, Teichert U, Smith J. Purification and characterization of a methionine aminopeptidase from Saccharomyces cerevisiae. J Biol Chem. 1990;265:19892–19897. [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
9.Guillon J-M, Mechulam Y, Schmitter J-M, Blanquet S, Fayat G. Disruption of the gene for Met-tRNA
 formyltransferase severely impairs growth of Escherichia coli. J Bacteriol. 1992;174:4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
formyltransferase severely impairs growth of Escherichia coli. J Bacteriol. 1992;174:4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar] - 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Marcker K, Sanger F. N-Formyl-methionyl-tRNA. J Mol Biol. 1963;8:835–840. doi: 10.1016/s0022-2836(64)80164-9. [DOI] [PubMed] [Google Scholar]
- 12.Mazel D, Pochet S, Marliere P. Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO J. 1994;13:914–923. doi: 10.1002/j.1460-2075.1994.tb06335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meinnel T, Mechulam Y, Blanquet S. Methionine as translation start signal: a review of the enzymes of the pathway in Escherichia coli. Biochimie. 1993;75:1061–1075. doi: 10.1016/0300-9084(93)90005-d. [DOI] [PubMed] [Google Scholar]
- 14.Meinnel T, Blanquet S. Characterization of the Thermus thermophilus locus encoding peptide deformylase and methionyl-tRNA(fMet) formyltransferase. J Bacteriol. 1994;176:7387–7390. doi: 10.1128/jb.176.23.7387-7390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newton D T, Creuzenet C, Mangroo D. Formylation is not essential for initiation of protein synthesis in all eubacteria. J Biol Chem. 1999;274:22143–22146. doi: 10.1074/jbc.274.32.22143. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook R, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 17.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;71:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solbiati J, Chapman-Smith A, Miller J L, Miller C G, Cronan J E., Jr Processing of the N termini of nascent polypeptide chains requires deformylation prior to methionine removal. J Mol Biol. 1999;290:607–614. doi: 10.1006/jmbi.1999.2913. [DOI] [PubMed] [Google Scholar]
- 19.Stüber D, Matile H, Garotta G. System for high-level production in E. coli and rapid purification of recombinant proteins: application to epitope mapping, preparation of antibodies, and structure-function analysis. In: Lefkovits J, Pernis B, editors. Immunologic methods. Vol. 4. Orlando, Fla: Academic Press; 1990. pp. 121–152. [Google Scholar]