Abstract
We have identified the gene for transcription termination factor Rho in Staphylococcus aureus. Deletion of rho in S. aureus reveals that it is not essential for viability or virulence. We also searched the available bacterial genomic sequences for homologs of Rho and found that it is broadly distributed and highly conserved. Exceptions include Streptococcus pneumoniae, Streptococcus pyogenes, Mycoplasma genitalium, Mycoplasma pneumoniae, Ureaplasma urealyticum, and Synechocystis sp. strain PCC6803, all of which appear not to possess a Rho homolog. Complementation studies indicate that S. aureus Rho possesses the same activity as Escherichia coli Rho and that the Rho inhibitor bicyclomycin is active against S. aureus Rho. Our results explain the lack of activity of bicyclomycin against many gram-positive bacteria and raise the possibility that the essentiality of rho may be the exception rather than the rule.
The rho gene codes for transcription termination factor Rho (19) and is essential for the viability of Escherichia coli (5). The function of Rho is to catalyze the release of RNA from a transcription complex after a Rho-dependent terminator sequence has been transcribed. Rho has an RNA-dependent NTPase activity, which is required for transcript release (10, 12), and an RNA-DNA helicase activity (1). Several Rho-dependent transcription terminators in Escherichia coli and coliphage λ are known (4, 20).
Rho is a hexamer of identical 47-kDa monomers (2, 9, 11, 17). Its ability to bind RNA is thought to be conferred by residues 22 to 116 of the 419-amino-acid protein (14). The ATP binding domain is located between residues 167 and 319 (7). No activity has been assigned to the essential C terminus of Rho, although it has been speculated as being involved in subunit interactions (1, 6, 8).
The antibiotic bicyclomycin inhibits Rho (23). With the exception of Micrococcus luteus (15, 16), gram-positive bacteria are resistant to bicyclomycin, and rho is not essential in the gram-positive bacterium Bacillus subtilis (13, 21). This study reports the sequence and characterization of the rho gene in the clinically important gram-positive pathogen Staphylococcus aureus.
MATERIALS AND METHODS
Bacteria and growth conditions.
Staphylococcus aureus was propagated in either tryptic soy broth (DIFCO) or Luria broth (LB). E. coli was grown in LB. For auxotrophy measurements with S. aureus, M9 minimal medium was supplemented with arginine, cysteine, glutamate, glycine, isoleucine, leucine, methionine, proline, and valine, all at 20 μg/ml. Nicotinate and thiamine were added at 0.2 μg/ml. All cultures were routinely incubated at 37°C. All strains used in this study are described in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| BLS107 | E. coli Δrho::Kanr | 22 |
| MG1655 | Wild-type E. coli strain | Laboratory collection |
| RN4220 | S. aureus laboratory strain | Laboratory collection |
| RSW100 | RN4220 Δrho::Tetr | This study |
| RSW101 | WCUH29 Δrho::Tetr | This study |
| WCUH29 | S. aureus clinical isolate | Laboratory collection |
| Plasmids | ||
| pAU2442 | 4.3-kb genomic clone from WCUH29 containing rho | This study |
| pPMrho | Temperature-sensitive replicon, 3.0-kb genomic clone of E. coli containing rho | 22 |
Identification of the S. aureus rho gene.
The rho gene from Staphylococcus aureus was identified from an S. aureus genomic sequence database by homology with E. coli rho. The plasmid pAU2442 used in this study is from a genomic library used in the sequencing project.
Deletion mutagenesis.
A rho deletion was introduced into S. aureus RN4220. This was achieved by electroporating S. aureus RN4220 to tetracycline resistance (Tcr) with pRW101, which has the rho gene with 500 bp of flanking sequence interrupted by a tetracyline resistance marker that replaces the rho gene from codon 1 to codon 313 (Fig. 1). The resulting Tcr colonies were then screened for erythromycin resistance (Emr), which is carried elsewhere on pRW101. Tcr Ems colonies indicated a double-crossover event occurred. The deletion of rho was confirmed by PCR analysis with primers flanking the Tcr marker. To construct a Δrho strain in a pathogenic background, the Δrho::tet mutation was moved into the S. aureus clinical isolate WCUH29 by bacteriophage Φ11 transduction. The resulting strain, RSW101, was used in virulence testing as described below.
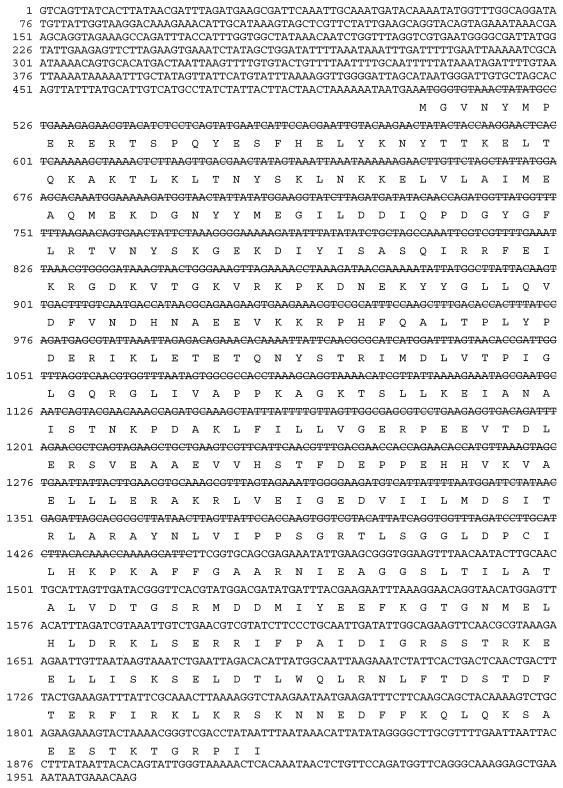
FIG. 1.
Sequence of S. aureus rho and flanking sequences used in this report. Shown is the DNA sequence of the region of the S. aureus chromosome that contains rho. The rho gene product is shown below its coding sequence. DNA sequence with a strikethrough line depicts the sequence deleted in the rho deletion-insertion strain described in this paper.
Complemention of E. coli rho.
E. coli rho was replaced with S. aureus rho by introducing the S. aureus rho-containing plasmid pAU2442 into the E. coli rho deletion strain BLS107(pPMrho). This strain has a deletion-insertion (rho::kan) mutation in the chromosomal copy of rho. A wild-type copy of rho is carried on the plasmid pPMrho, which has a temperature-sensitive replicon. This strain fails to grow at 42°C due to the loss of rho at this temperature (22). pAU2442 was introduced into BLS107(pPMrho) by electroporation and was plated at 30, 37, and 42°C. As a control, the parent strain was also plated at the different temperatures. While the parent strain failed to form isolated colonies at 37 and 42°C, the strain containing pAU2442 grew well and formed isolated colonies at all temperatures. The loss of pPMrho was confirmed by scoring for loss of chloramphenicol resistance, which is encoded by this plasmid. To determine if the S. aureus Rho protein is sensitive to bicyclomycin, strain BLS107(pAU2442) was tested for sensitivity by MIC determination. Liquid MICs were determined by a serial dilution method. Strain BLS107 was found to have an MIC of bicyclomycin of 125 μg/ml. This is twofold lower than that of the wild-type E. coli strain MG1655 (250 μg/ml). The MIC for the S. aureus strain RN4220 was found to be >2.5 mg/ml.
Virulence testing.
The rho deletion strain RSW101 and its wild-type parent, WCUH29, were tested for virulence with two mouse S. aureus infection models. The surgical wound model measures the ability to grow on sutures sewn in the skin of healthy mice, while the hematogenous pyelonephritis model measures the ability of the strains to cause kidney infections following parenteral infection with the bacteria.
Surgical wound infections.
The mice used for these infections were 4- to 6-week-old male CD-1 mice. Approximately 15-cm sutures were soaked in overnight cultures of WCUH29 and RSW101 for 30 min at room temperature. The backs of the mice were shaved, the animals were anesthetized with isoflurane, and an ∼2-cm-long incision was made along the animals' backs. The infected sutures were secured at one end of the wound and tacked once midway along the wound on the underside of the skin. The suture was then secured at the other end of the wound, and the wound was closed with a single staple. After 5 days, the mice were sacrificed by CO2 overdose. The skin surrounding the wound was excised and then homogenized in 1 ml of phosphate-buffered saline (PBS) in a stomacher, and bacterial viable counts were enumerated. Both infections yielded over 106 bacteria after 5 days, with no significant difference observed between the strains. For the hematogenous pyelonephritis model, bacterial recovery rates (mean ± standard deviation; n = 5) were 5.35 ± 0.41 log10 CFU/ml for strain WCUH29 and 6.44 ± 0.81 log10 CFU/ml for strain WCUH29 Δrho. For the surgical wound model, the recovery rates were 6.58 ± 0.78 log10 CFU/ml for strain WCUH29 and 7.41 ± 0.57 log10 CFU/ml for strain WCUH29 Δrho. One hundred colonies isolated from the rho mutant infection were checked for tetracycline resistance, indicative of the presence of the mutation, and all were found to be tetracycline resistant.
Hematogenous pyelonephritis infections.
Six- to eight-week-old male CD-1 mice were used for hematogenous pyelonephritis infections. Cultures of WCUH29 and RSW101 cells were adjusted to an A600 of 0.6 or 0.3 per ml, and 200 μl was injected into each mouse via a tail vein. Mice were sacrificed on day 5 postinfection by CO2 overdose, and their kidneys were aseptically removed and then homogenized in 1 ml of PBS in a stomacher, and viable counts were enumerated (described above). The inoculum for both strains was 107 cells. The number of bacteria recovered from the kidneys infected with RSW101 after 5 days was not decreased. One hundred colonies from the RSW101 infection were checked for the tetracycline resistance marker, and 98% were found to be tetracycline resistant.
Nucleotide sequence accession number.
The nucleotide sequence of the S. aureus rho gene has been deposited in GenBank under accession no. AF333962.
RESULTS
The S. aureus Rho protein.
S. aureus Rho is predicted to code for a 443-amino-acid protein with a predicted molecular mass of 50 kDa. S. aureus Rho is 53% identical and 67% similar to E. coli Rho and 66% identical and 75% similar to B. subtilis Rho (Fig. 2). The predicted ATPase domain (residues 167 to 342 of E. coli Rho) is highly conserved among different bacterial species (18). The predicted ATPase domain of S. aureus Rho (residues 180 to 356) is 80% similar and 70% identical to E. coli Rho and 86% similar and 81% identical to B. subtilis Rho. Finally, the predicted RNA binding domain of S. aureus Rho (residues 40 to 132) has 63% similarity and 41% identity to E. coli Rho and 77% similarity and 60% identity to B. subtilis Rho.
FIG. 2.
Alignment of S. aureus Rho with B. subtilis and E. coli Rho. Sequences were aligned by using the MegAlign program of the DNAstar DNA analysis package. Identical residues are shaded.
Complementation.
The S. aureus rho gene can complement the lack of growth of an E. coli strain with rho deleted.. This was determined by curing a rho-deleted strain of a plasmid that carries E. coli rho in the presence of S. aureus rho carried on another plasmid. The sole Rho activity in the resulting strain, BLS107(pAU2442), is from S. aureus Rho. Strain BLS107(pAU2442) is sensitive to bicyclomycin, as indicated by the bicyclomycin MIC of 125 μg/ml, indicating that S. aureus Rho is inhibited by bicyclomycin. As further evidence of this, bicyclomycin was found to inhibit the ATPase activity of partially purified S. aureus Rho (data not shown).
The S. aureus rho gene is not essential for in vitro growth.
A Δrho strain, RSW100, was constructed by replacing nearly all of the rho gene with a tetracycline resistance cassette (Fig. 1). The resulting strain is viable and grows normally at 30, 37, 42, and 46°C. RSW100 also grows normally on rich medium (LB or tryptic soy broth) or minimal medium (supplemented M9 medium) and under anaerobic conditions (data not shown).
rho is not required for S. aureus virulence.
The rho deletion was transduced into the pathogenic clinical isolate WCUH29, yielding strain RSW101. Cultures of WCUH29 and RSW101 were used in the two in vivo models described in Materials and Methods. S. aureus strains mutated in a variety of different genes affecting cell metabolism or virulence have been examined in our laboratories. These strains have displayed reductions of up to 3 logs in the pyelonephritis model and 1 log in the surgical wound infection. In the experiments described here, the in vivo growth of RSW101 was not impaired compared to that of wild-type WCUH29 in either of the model systems. From these data, we conclude that, as with in vitro growth, rho is not essential for the virulence of the strain tested.
Review of Rho homologs present in public sequence database.
By using the S. aureus Rho sequence as a query, we searched the public databases and some proprietary sequence databases for homologs. Rho was found to be highly conserved; BLAST scores were 5 × 10−38 or lower for all full-length Rho sequences annotated as such. As indicated previously by Opperman and Richardson (18), Rho is broadly distributed among all the major phyla of the eubacteria (Table 2). A major exception to the universal presence of Rho among the eubacteria are the members of the low-GC, gram-positive bacteria Streptococus pyogenes, Streptococcus pneumoniae, Mycoplasma genitalium, M. pneumoniae, and Ureaplasma urealyticum, all of which do not possess a significant homolog. In the case of S. pneumoniae, we searched several proprietary databases in addition to publicly available databases and failed to identify a homolog. In addition to the mentioned bacteria, Synechocystis sp. strain PCC6803, a cyanobacterium, also apparently lacks a Rho homolog. As indicated previously (18), the ATP binding motif of Rho shares homology with the α and β subunits of F1 ATPases. This homology is conserved among the bacteria as well.
TABLE 2.
Bacterial phyla with members that possess a Rho homologa
| Aquificales |
| Aquifex aeolicus |
| Chlamydiales |
| Chlamydia muridarum |
| Chlamydia pneumoniae |
| Chlamydia trachomatis |
| Cytophaga-Flexibacter-Bacteroides |
| Porphyromonas gingivalis |
| Green sulfur |
| Chlorobium tepidum |
| Gram positive |
| Low G+C (not Streptococcus pyogenes, Mycoplasma pneumoniae, Mycoplasma genitalium, or Streptococcus pneumoniae) |
| Bacillus subtilis |
| Clostridium acetobutylicum |
| Listeria monocytogenes |
| High G+C (Actinobacteria) |
| Micrococcus luteus |
| Mycobacterium leprae |
| Mycobacterium tuberculosis |
| Streptomyces coelicolor |
| Streptomyces lividans |
| Proteobacteria |
| α Subdivision |
| Rhodobacter sphaeroides |
| Rickettsia prowazekii |
| β Subdivision |
| Neisseria animalis |
| Neisseria cinerea |
| Neisseria elongata |
| Neisseria gonorrhoeae |
| Neisseria lactamica |
| Neisseria meningitidis |
| Neisseria mucosa |
| Neisseria pharyngis |
| Neisseria polysaccharea |
| Neisseria subflava |
| ɛ Subdivision |
| Campylobacter jejuni |
| Helicobacter pylori |
| γ Subdivision |
| Actinobacillus actinomycetemcomitans |
| Actinobacillus pleuropneumoniae |
| Allochromatium vinosum |
| Buchnera aphidicola |
| Escherichia coli |
| Haemophilus influenzae |
| Pseudomonas aeruginosa |
| Pseudomonas fluorescens |
| Salmonella enterica serovar Typhimurium |
| Vibrio cholerae |
| Xylella fastidiosa |
| Yersinia pestis |
| Spirochaetales |
| Borrelia burgdorferi |
| Treponema pallidum |
| Thermotogales |
| Thermotoga maritima |
| Thermus/Deinococcus |
| Deinococcus radiodurans |
Groupings taken from http://www.ncbi.nih.gov/htbin-post/Taxonomy/wgetorg and determined by BLAST analysis with S. aureus Rho.
DISCUSSION
In this report, we show that the S. aureus rho gene can complement an E. coli rho deletion strain. The ability of S. aureus rho to replace the essential E. coli rho gene raises the possibility that it performs the same functions in S. aureus and E. coli. rho is not essential in the low-GC, gram-positive bacterium Bacillus subtilis (13, 21), and several gram-positive species, including Staphylococcus aureus, are resistant to the Rho inhibitor bicyclomycin (15). Here we show that rho is not required for viability or virulence in S. aureus. The E. coli strain constructed here, which has had its own rho gene replaced with S. aureus rho, remains sensitive to bicyclomycin. This suggests that the resistance of gram-positive bacteria to bicyclomycin is a result of the nonessentiality of Rho. Supporting this conclusion and extending the lack of essentiality of rho to other genera, we found that the completed genomes of several members of the low-GC, gram-positive bacteria lack a Rho homolog. The cyanobacterium Synechocystis sp. strain PCC6803 was also found to lack a Rho homolog.
An open question is why rho should be essential in E. coli and other gram-negative bacteria, but not be essential in some gram-positive bacteria. It could be that Rho activity is redundant in the bacteria where it is not essential. This possibility cannot be readily discounted, but it is clear that the enzyme responsible for this redundant activity would have to have no sequence homology with Rho for this to be the case. A second possibility, that Rho is essential for organisms with a higher GC content, seems unlikely, given that Synechocystis sp. strain PCC6803 also lacks Rho, and yet its genome is 47% GC, close to that of E. coli. The lack of rho among several members of the low-GC, gram-positive bacteria and a member of the very distantly related cyanobacteria could indicate a trend that the essentiality of rho may be more the exception than the rule. Because of this, a more informative approach to the question is to ask why rho is essential in E. coli and other gram-negative bacteria. There are only a few Rho-dependent terminators in E. coli; the vast majority of E. coli operons contain factor-independent terminators. Furthermore, it is not obvious why inhibition of termination at Rho-dependent terminators would lead to a lethal event. It might be that there is some other function of Rho that better explains its essential nature in some bacteria.
REFERENCES
- 1.Bear D G, Andrews C L, Singer J D, Morgan W D, Grant R A, von Hippel P H, Platt T. Escherichia coli transcription termination factor Rho has a two-domain structure in its activated form. Proc Natl Acad Sci USA. 1985;82:1911–1915. doi: 10.1073/pnas.82.7.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bear D G, Hicks P S, Escudero K W, Andrews C L, McSwiggen J A, von Hippel P H. Interactions of Escherichia coli transcription termination factor Rho with RNA. J Mol Biol. 1988;199:623–635. doi: 10.1016/0022-2836(88)90306-3. [DOI] [PubMed] [Google Scholar]
- 3.Brennan C A, Dombroski A J, Platt T. Transcription termination factor Rho is an RNA-DNA helicase. Cell. 1987;48:945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- 4.Das A. Control of transcription termination by RNA binding proteins. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- 5.Das A, Court D, Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci USA. 1976;73:1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolan J W, Marshall N F, Richardson J P. Transcription termination factor Rho has three distinct structural domains. J Biol Chem. 1990;265:5747–5754. [PubMed] [Google Scholar]
- 7.Dombroski A J, LaDine J R, Cross R L, Platt T. The ATP binding site on rho protein. Affinity labelling of Lys181 by pyridoxal 5′-diphospho-5′-adenosine. J Biol Chem. 1988;263:18810–18815. [PubMed] [Google Scholar]
- 8.Dombroski A J, Platt T. Structure of ρ factor: an RNA-binding domain and a separate region with strong similarity to ATP-binding domains. Proc Natl Acad Sci USA. 1988;85:2538–2542. doi: 10.1073/pnas.85.8.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finger L R, Richardson J P. Stabilization of the hexameric form of Escherichia coli protein rho under ATP hydrolysis conditions. J Mol Biol. 1982;156:203–219. doi: 10.1016/0022-2836(82)90467-3. [DOI] [PubMed] [Google Scholar]
- 10.Galluppi G R, Lowery C, Richardson J P. Nucleoside triphosphate requirement for termination of RNA synthesis by rho factor. In: Losick R, Chamberlin M, editors. RNA polymerase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1976. pp. 657–665. [Google Scholar]
- 11.Gogol E P, Seifred S E, von Hippel P H. Structure and assembly of the Escherichia coli transcription termination factor Rho and its interactions with RNA. I. Cryoelectron microscopic studies. J Mol Biol. 1991;221:1127–1138. doi: 10.1016/0022-2836(91)90923-t. [DOI] [PubMed] [Google Scholar]
- 12.Howard B H, de Crombrugghe B. ATPase activity required for termination of transcription by the Escherichia coli protein factor ρ. J Biol Chem. 1976;251:2520–2524. [PubMed] [Google Scholar]
- 13.Ingham C J, Dennis J, Furneaux P A. Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol Microbiol. 1999;31:651–653. doi: 10.1046/j.1365-2958.1999.01205.x. [DOI] [PubMed] [Google Scholar]
- 14.Modrak D, Richardson J P. The RNA-binding domain of transcription termination factor Rho: isolation, characterization and determination of sequence limits. Biochemistry. 1994;33:8292–8299. doi: 10.1021/bi00193a016. [DOI] [PubMed] [Google Scholar]
- 15.Nishida M, Mine Y, Matsubara T. Bicyclomycin, a new antibiotic. III. In vitro and in vivo antimicrobial activity. J Antibiot. 1972;25:582–593. [PubMed] [Google Scholar]
- 16.Nowatzke W L, Keller E, Koch G, Richardson J P. Transcription termination factor Rho is essential for Micrococcus luteus. J Bacteriol. 1997;179:5238–5240. doi: 10.1128/jb.179.16.5238-5240.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oda T, Takanami M. Observations on the structure of the termination factor Rho and its attachment to DNA. J Mol Biol. 1972;71:799–802. doi: 10.1016/s0022-2836(72)80041-x. [DOI] [PubMed] [Google Scholar]
- 18.Opperman T, Richardson J P. Phylogenetic analysis of sequences from diverse bacteria with homology to the Escherichia coli rho gene. J Bacteriol. 1994;176:5033–5043. doi: 10.1128/jb.176.16.5033-5043.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinkham J L, Platt T. The nucleotide sequence of the rho gene of E. coli K-12. Nucleic Acids Res. 1983;11:3531–3545. doi: 10.1093/nar/11.11.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Platt T, Richardson J P. Escherichia coli Rho factor: protein and enzyme of transcription termination. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 365–388. [Google Scholar]
- 21.Quirk P G, Dunkley E A, Jr, Lee P, Krulwich T A. Identification of a putative Bacillus subtilis rho gene. J Bacteriol. 1993;175:647–654. doi: 10.1128/jb.175.3.647-654.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Washburn R S, Stitt B L. In vitro characterization of transcription termination factor Rho from Escherichia coli rho(nusD) mutants. J Mol Biol. 1996;260:332–346. doi: 10.1006/jmbi.1996.0404. [DOI] [PubMed] [Google Scholar]
- 23.Zwiefka A, Kohn H, Widger W R. Transcription termination factor rho: The site of bicyclomycin inhibition in Escherichia coli. Biochemistry. 1993;32:3564–3570. doi: 10.1021/bi00065a007. [DOI] [PubMed] [Google Scholar]