Abstract
Background & Aims
The liver has a unique capacity to regenerate after injury in a highly orchestrated and regulated manner. Here, we report that O-GlcNAcylation, an intracellular post-translational modification regulated by 2 enzymes, O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), is a critical termination signal for liver regeneration following partial hepatectomy (PHX).
Methods
We studied liver regeneration after PHX on hepatocyte specific OGT and OGA knockout mice (OGT-KO and OGA-KO), which caused a significant decrease (OGT-KO) and increase (OGA-KO) in hepatic O-GlcNAcylation, respectively.
Results
OGA-KO mice had normal regeneration, but the OGT-KO mice exhibited substantial defects in termination of liver regeneration with increased liver injury, sustained cell proliferation resulting in significant hepatomegaly, hepatic dysplasia, and appearance of small nodules at 28 days after PHX. This was accompanied by a sustained increase in expression of cyclins along with significant induction in pro-inflammatory and pro-fibrotic gene expression in the OGT-KO livers. RNA-sequencing studies revealed inactivation of hepatocyte nuclear 4 alpha (HNF4α), the master regulator of hepatic differentiation and a known termination signal, in OGT-KO mice at 28 days after PHX, which was confirmed by both Western blot and immunohistochemistry analysis. Furthermore, a significant decrease in HNFα target genes was observed in OGT-KO mice, indicating a lack of hepatocyte differentiation following decreased hepatic O-GlcNAcylation. Immunoprecipitation experiments revealed HNF4α is O-GlcNAcylated in normal differentiated hepatocytes.
Conclusions
These studies show that O-GlcNAcylation plays a critical role in the termination of liver regeneration via regulation of HNF4α in hepatocytes.
Keywords: Cell Death, HNF4alpha, Proliferation, Partial Hepatectomy, Regeneration
Abbreviations used in this paper: ALT, alanine transaminase; H&E, hematoxylin and eosin; HNF4α, hepatocyte nuclear 4 alpha; IHC, immunohistochemistry; IP, immunoprecipitant; IPA, Ingenuity Pathway Analysis; KO, knockout; LR, liver regeneration; GlcNAc, N-acetylglucosamine; OGT, O-GlcNAc transferase; OGA, O-GlcNAcase; PHX, partial hepatectomy; p-Rb, phosphorylated Rb; PTM, post-translational modification; P1, promoter 1; P2, promoter 2; qPCR, quantitative polymerase chain reaction; RNA-Seq, RNA-sequencing; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling; WT, wild-type
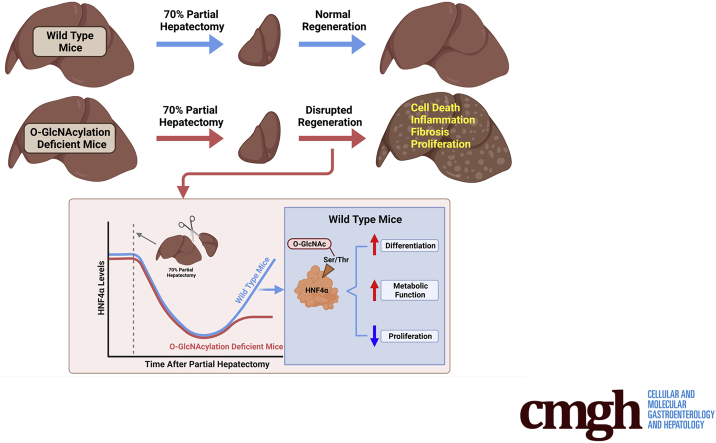
Graphical abstract
Summary.
O-GlcNAcylation is a protein modification that plays a critical role in various biological processes including cell proliferation, differentiation, and disease progression. These studies show that O-GlcNAcylation in hepatocytes is essential for proper liver regeneration. Without O-GlcNAcylation, hepatocytes keep on proliferating, eventually forming liver tumors.
The liver has a remarkable regenerative capacity after injury or surgical resection.1 Liver regeneration (LR) is tightly regulated by a plethora of redundant signals that regulate initiation and termination of cell proliferation as well as tissue remodeling. The most widely used model to study LR is partial hepatectomy (PHX) where approximately two-thirds of the liver is surgically removed.2 Liver cells, starting with hepatocytes and followed by other cells, enter the cell cycle and undergo a synchronized cell division to restore the lost liver mass. In rodents, this process takes between 3 and 5 days after which proliferation decreases, the newly divided cells undergo redifferentiation and tissue remodeling takes place. These latter events including inhibition of cell proliferation, redifferentiation, and tissue remodeling, are termed termination of liver regeneration. Whereas the initiation signals of LR are well-characterized, the mechanisms of termination of LR remain understudied.3 Proper termination of liver regeneration is essential because the loss of proper termination signals results in hepatomegaly, defects in redifferentiation of hepatocytes leading to either loss of liver function and liver failure or rapid preneoplastic changes in the liver leading to chronic liver disease.
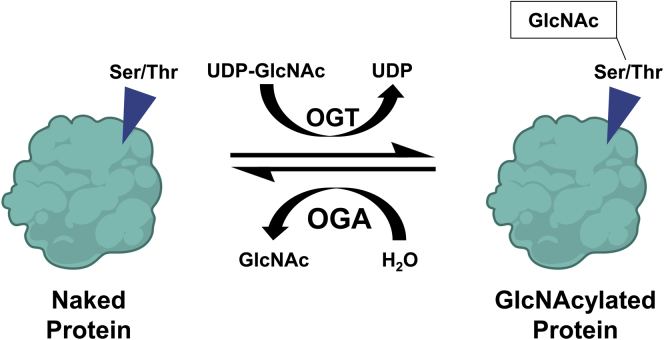
O-GlcNAcylation is an intracellular post-translational modification (PTM) that involves the addition of a single N-acetylglucosamine (GlcNAc) molecule on an exposed serine or threonine amino acid of a protein. The O-GlcNAcylation cycle is regulated by 2 enzymes, O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA). OGT catalyzes the transfer of GlcNAc from its carrier molecule UDP-GlcNAc to the protein. Whereas OGA catalyzes the hydrolysis and removal of the GlcNAc motif from the protein (Figure 1, A). In some cases, the residues that undergo O-GlcNAcylation on the target protein can also become phosphorylated providing the cell an additional mechanism of regulating downstream signaling.4 O-GlcNAcylation homeostasis is critical for healthy cells and aberrant O-GlcNAcylation has been linked to various diseases, including cancer, nonalcoholic fatty liver disease and alcoholic steatohepatitis.5, 6, 7 O-GlcNAcylation plays a major role in a myriad of cellular processes, including cell proliferation.8, 9, 10 Despite the role of O-GlcNAcylation in cell proliferation, little is known about its role in hepatocyte LR. In this study, we investigated the role of O-GlcNAcylation in the regulation of LR after PHX using control, OGA, and OGT hepatocyte-specific knockout mice (OGA-KO and OGT-KO respectively). Our studies revealed that decreasing O-GlcNAcylation leads to impaired termination of LR. Moreover, this is due to the loss of hepatocyte nuclear 4 alpha (HNF4α) function, a nuclear receptor critical for hepatocyte differentiation and function.
Figure 1.
The regulation of O-GlcNAcylation. A, Scheme of protein O-GlcNAcylation. OGT transfers a GlcNAc group, from the donor UDP-GlcNAc, onto an exposed serine or threonine of a protein. Conversely, OGA removes the O-GlcNAc modification reverting the protein back to a naked protein.
Results
Decrease O-GlcNAcylation Resulted in Hepatomegaly, Liver Injury, and Defective Termination of Liver Regeneration
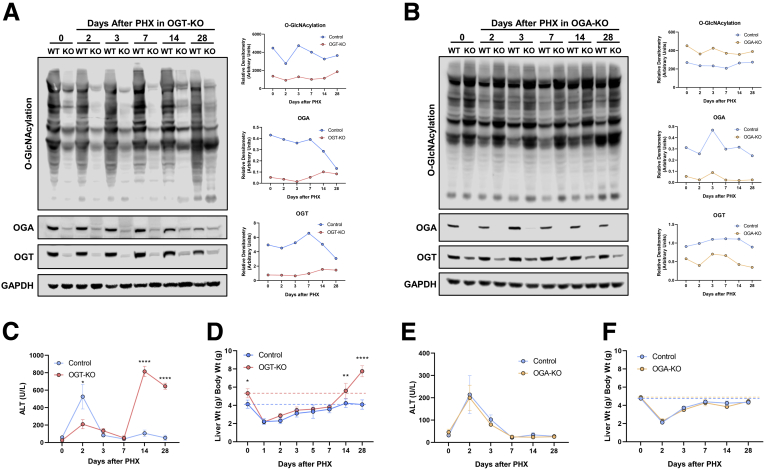
Hepatocyte-specific OGT-KO and OGA-KO mice were successfully generated by injecting OGTfl/Y and OGAfl/fl mice with AAV8-TBG-CRE, respectively (Figure 2, A-B). OGTfl/Y and OGAfl/fl mice treated with AAV8-TBG-GFP were used as controls (referred to as wild-type [WT]). As expected, deletion of OGT decreased global liver O-GlcNAcylation (Figure 2, A) and deletion of OGA enhanced O-GlcNAcylation levels after PHX (Figure 2, B). Further, deletion of either OGT or OGA decreased the reciprocal enzyme levels (Figure 2, A-B). A significant liver injury as demonstrated by serum alanine transaminase (ALT) levels was observed at 2 days after PHX in WT mice compared with OGT-KO mice. However, OGT-KO mice exhibited significantly higher liver injury at 14 and 28 days after PHX (Figure 2, C). Most importantly, the liver weight to body weight ratio indicated substantial hepatomegaly in the OGT-KO mice at 0, 14, and 28 days after PHX (Figure 2, D). At 28 days after PHX, the liver weight to body weight ratio was 80% higher in OGT-KO mice as compared with its 0-hour level indicating defective termination of regeneration. Interestingly, liver injury or liver weight to body weight ratios were not different between WT and OGA-KO mice, indicating that loss of OGA does not affect liver regeneration (Figure 2, E-F).
Figure 2.
Decreased hepatocyte O-GlcNAcylation lead to significant increase in liver weight to body weight ratios and liver injury. Western blot analysis of OGT-KO (A) and OGA-KO (B) mice of total liver O-GlcNAcylation levels, OGA, OGT, and GAPDH over a time course of 0 to 28 days after PHX with corresponding densitometry normalized to the loading control. Protein was pooled from 3 to 5 mice. Line graphs show serum ALT levels in OGT-KO mice (C) and OGA-KO mice (E) and liver weight to body weight ratio for OGT-KO (D) and OGA-KO (F) mice at various time points after PHX. Data are mean ± standard deviation. ∗P < .05; ∗∗P < .01; and ∗∗∗∗P < .0001.
Mechanisms of Liver Injury on OGT-KO Mice After PHX
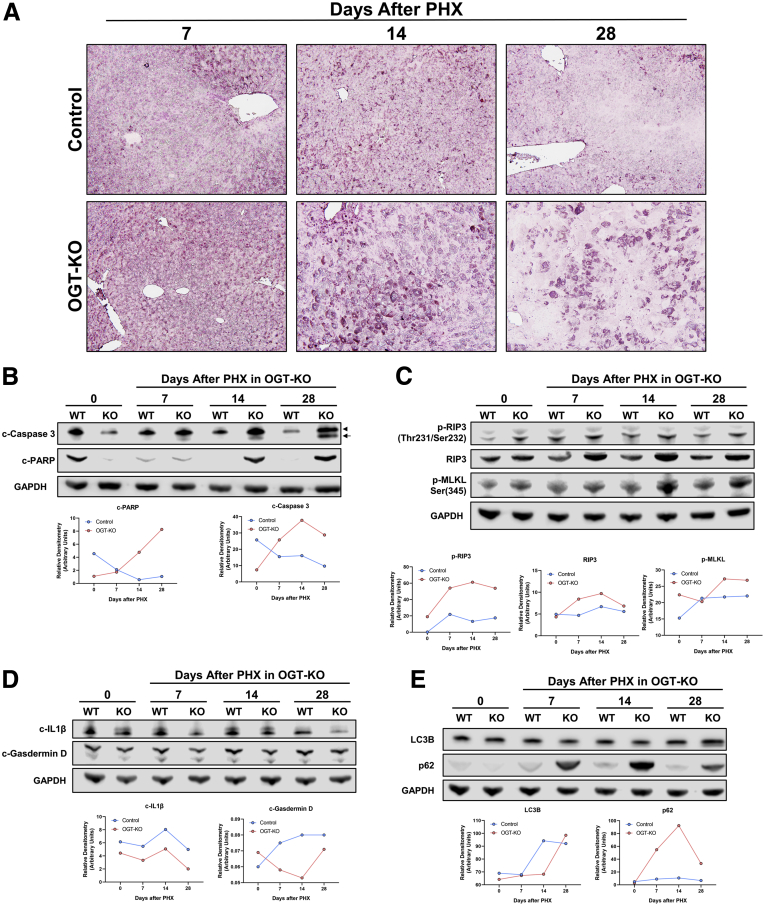
We performed terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining to determine the mechanisms of cell death in OGT-KO mice at 7-, 14-, and 28-day time points due to the extent of liver injury. At 14 and 28 days after PHX, the majority of the staining was localized in the cytosol with very limited nuclear staining indicating necrotic cell death and some apoptosis (Figure 3, A). There was no pattern in the TUNEL staining or ballooning hepatocytes in the hematoxylin and eosin (H&E), indicating no zonal distribution of these injured hepatocytes. Western blot analysis confirmed activation of both necroptosis and apoptosis at 14 and 28 days after PHX (Figure 3, B-C). Pyroptotic cell death mechanisms did not contribute to cell death determined by Western blot analysis (Figure 3, D). Interestingly, OGT-KO mice exhibited elevated p62, an autophagy marker (Figure 3, E). Taking these data together, OGT-KO mice exhibited liver injury due to necroptosis and apoptosis.
Figure 3.
OGT-KO mice exhibited necroptosis and apoptosis 14 and 28 days after PHX.A, TUNEL staining photomicrographs (200×) of WT and OGT-KO mice 7, 14, and 28 days after PHX to determine cell death. B, Western blot analysis of cleaved poly (ADP-ribose) polymerase (c-PARP), and cleaved caspase 3 (c-caspase 3), markers of apoptosis. Arrowhead indicates p-19 and arrow indicates p-17 c-caspase 3. C, Western blot analysis of necroptosis markers such as phosphorylated RIP3 (p-RIP3), RIP3, and phosphorylated MLKL (p-MLKL). D, Western blot analysis of pyroptosis markers such as cleaved IL1ß (c-IL1ß) and cleaved gasdermin d domain (c-Gasdermin D). E, Western blot analysis of autophagy markers such as SQSTM1 (p62) and LC3B. For each Western blot, GAPDH was used as the loading control, and samples were derived from pooled WT and OGT-KO mice (n = 3–5) per timepoint. Densitometry accompanied each Western blot with blue and red representing WT and OGT-KO mice, respectively.
Decreased O-GlcNAcylation Resulted in Sustained Cell Proliferation and Hepatic Dysplasia
We determined cell proliferation in the liver after PHX using PCNA immunohistochemistry (IHC). PCNA expression peaked at 2 days after PHX as expected in both WT and OGT-KO mice (Figure 4, A-B). After day 3, PCNA expression gradually declined in the WT, whereas it remained high in the OGT-KO mice till 28 days after PHX (Figure 4, A-B). Cell proliferation peaked in the WT and OGA-KO mice at 2 days after PHX before declining. Consistent with the liver weight to body weight data, we did not observe any difference in cell proliferation between WT and OGA-KO mice after PHX (Figure 5, A).
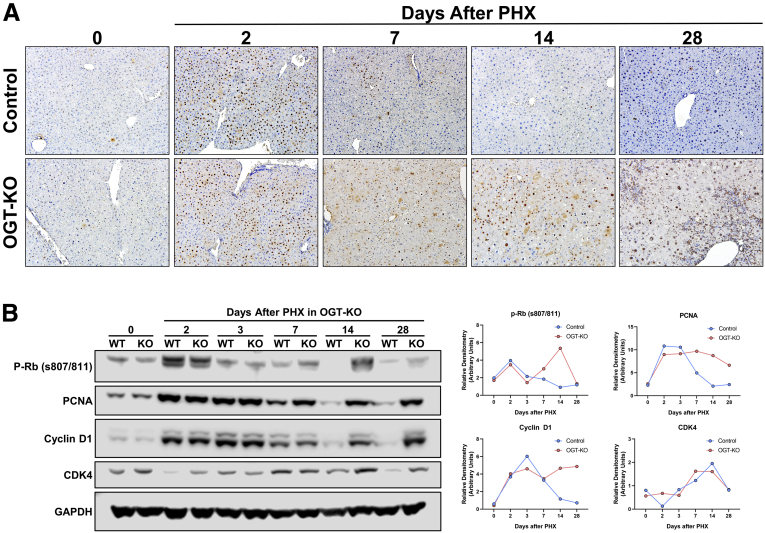
Figure 4.
Sustained cell proliferation in OGT-KO mice after PHX.A, Representative photomicrographs (200×) of PCNA-stained liver sections from OGT-KO and control mice throughout the PHX time course. B, Western blot analysis of the pro-mitogenic factors p-Rb, PCNA, Cyclin D1, and CDK4 (left panel) in pooled (n = 3–5) OGT-KO liver lysates at all time points post-PHX accompanied with the calculated densitometry normalized to GAPDH (right panel).
Figure 5.
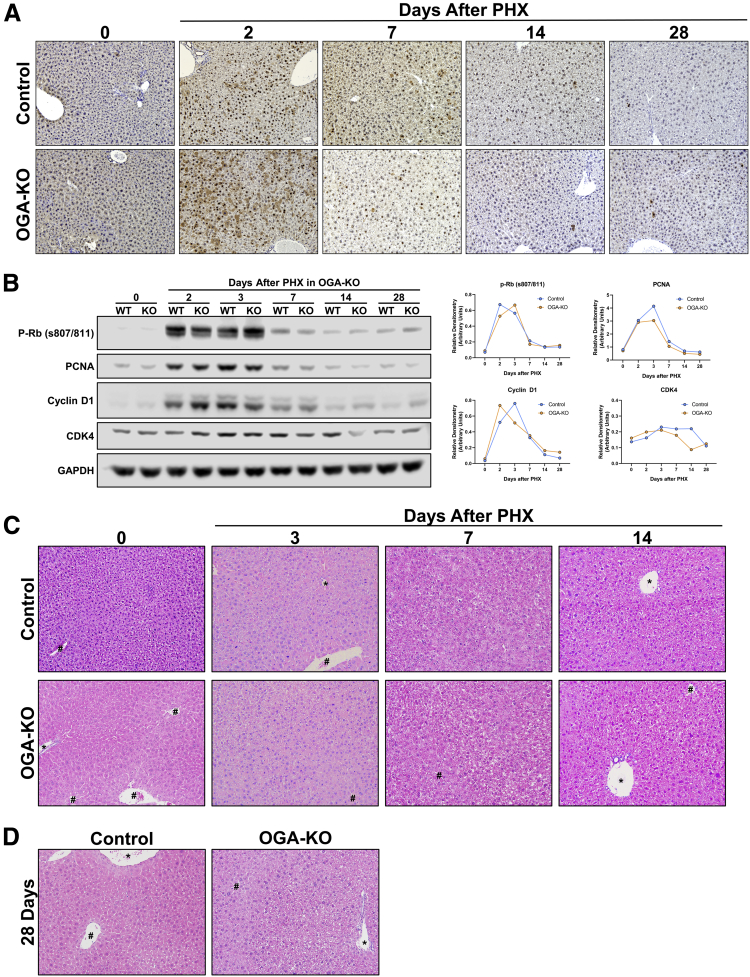
OGA-KO mice had no effect in proliferation and histology after PHX.A, Photomicrographs (200×) of PCNA staining for OGA-KO and WT mice after 2, 7, 14, and 28 days after PHX. B, Western blot analysis of promitogens (p-Rb, PCNA, Cyclin D1, and CDK4) in OGA-KO and WT mice after PHX accompanied with corresponding densitometry with blue and yellow representing WT and OGA-KO mice, respectively. Photomicrographs of H&E staining of OGA-KO mice at 0, 3, 7, 14 (C), and 28 (D) days after PHX. # marks central vein and ∗ marks portal triad.
Next, we investigated the expression of core cell cycle machinery that drives cell proliferation. Western blot analysis revealed that expression of cyclin D1, CDK4, and phosphorylated Rb (p-Rb) protein increased after PHX to similar levels in WT and OGT-KO livers at 2 days after PHX. A steady decline in expression of these proteins was observed in WT mice after the 2-day time point. However, OGT-KO mice exhibited sustained induction in cyclin D1, p-Rb, and CDK4 14 and 28 days after PHX. (Figure 6, B). In OGA-KO mice, expression of these proteins increased at 2 days post-PHX and decreased thereafter. There was no difference between the OGA-KO and WT mice, consistent with cell proliferation data (Figure 5, B).
Figure 6.
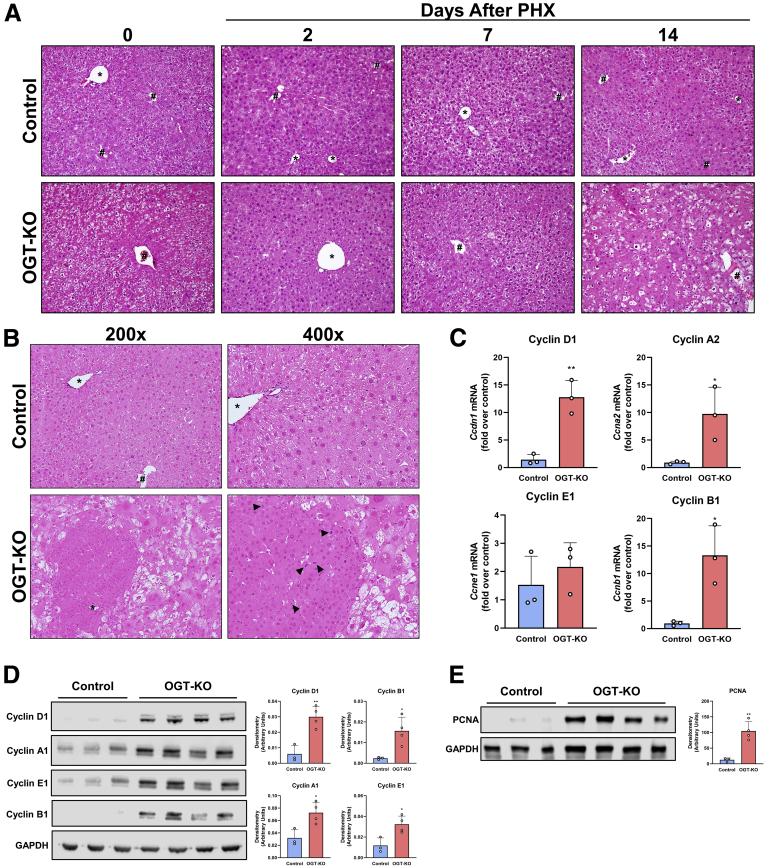
Hepatic dysplasia is exhibited in OGT-KO mice 28 days after PHX.A, H&E representative photomicrographs of liver sections from control and OGT-KO mice 0 hours and 2, 7, and 14 days after PHX at 200× magnification. B, H&E staining of 28 days after PHX at 200× and 400× magnification with arrow heads indicating mitotic figures. # represents central vein and ∗ represents portal triad. C, qPCR analysis of Cyclin D1, A2, E1, and B1 in OGT-KO and control livers from mice 28 days post-PHX, normalized to 18S and median of the control. Western blot analysis of liver lysate from 28 days after PHX OGT-KO mice of cyclin D1, A1, E1, B1 (D), and PCNA (E) with their corresponding densitometry. Bars represents the mean ± standard deviation. ∗P < .05 and ∗∗P < .01.
Hepatic Dysplasia, Inflammation, and Early Fibrosis Occurred in OGT-KO 28 Days After PHX
H&E staining was utilized to determine histopathological changes. As expected, OGA-KO mice had little to no effect in histological changes at all time points after PHX (Figure 5, C-D). In contrast, OGT-KO mice manifested significant histological changes at 14 and 28 days after PHX, characterized by ballooning hepatocytes (Figure 6, A-B). At 28 days after PHX, the OGT-KO liver showed presence of hepatic nodules containing mitotic figures, surrounded by extensive inflammatory and ductular cells (Figure 6, B). Quantitative polymerase chain reaction (qPCR) analysis revealed an induction of Cyclin D1, A2, and B1 gene expression and no change in Cyclin E2 in OGT-KO mice 28 days after PHX (Figure 6, C). Similarly, protein levels of Cyclin D1, A1, E1, and B1 were elevated 28 days after PHX (Figure 6, D). Western blot analysis revealed a significant increase in expression cell proliferation marker PCNA (Figure 6, E), corroborating. Additionally, PCNA IHC showed proliferation of both hepatocytes and the ductular cells (Figure 4, C).
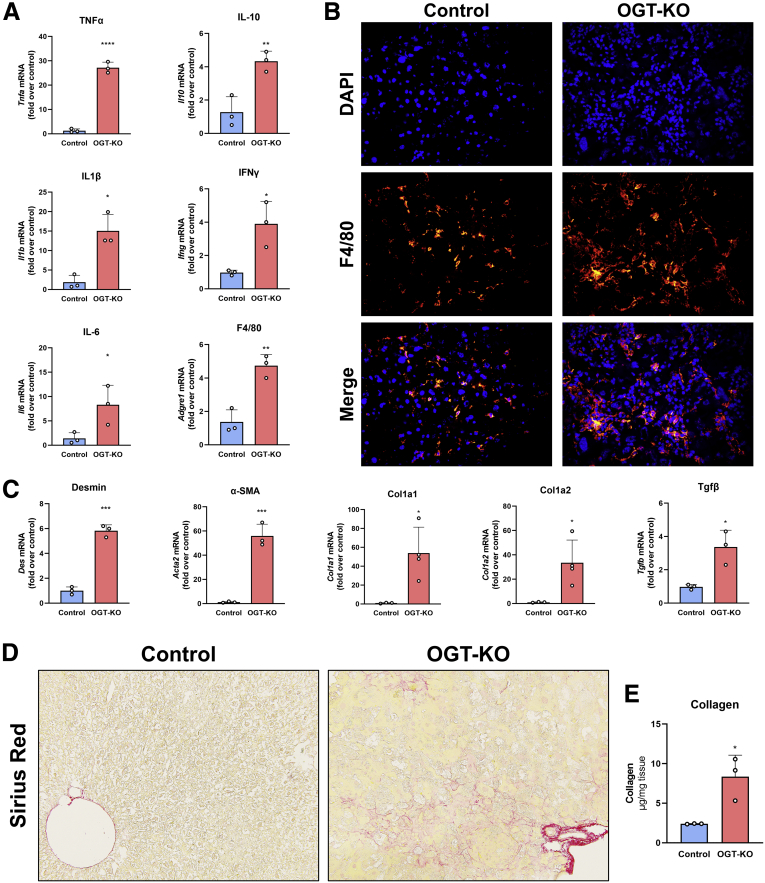
Next, we investigated the fibroinflammatory changes in the livers of WT and OGT-KO 28 days after PHX. qPCR analysis revealed a significant induction in the expression of pro-inflammatory genes (Tnfa, Il1b, Ifng, and Il6) and the anti-inflammatory gene Il10. OGT-KO mice showed induction in F4/80, a Kupffer cells marker (Figure 7, A), which was corroborated by immunofluorescence staining analysis showing increased F4/80+ cells in OGT-KO liver 28 days after PHX (Figure 7, B). qPCR analysis indicated a significant increase in the hepatic stellate cell marker desmin, its activation marker alpha-smooth muscle actin (Acta2) and Tgfb, a growth factor involved in activation of hepatic stellate cells (Figure 7, C). Consistent with these changes, OGT-KO livers also showed induction in fibrillar Col1a1 and Col1a2 expression (Figure 7, C). Picrosirius red staining showed an increased collagen deposition in OGT-KO livers compared with the control (Figure 7, D). Moreover, hydroxyproline assay showed a significant increase in collagen deposition in OGT-KO livers 28 days after PHX (Figure 7, E). These parameters of inflammation and fibrosis did not show induction at any time point in OGA-KO mice (Figure 8, B-C). These data indicate that OGT-KO 28 days after PHX have significant inflammation and fibrosis.
Figure 7.
Increased inflammation and fibrosis in OGT-KO mice after completion of LR.A, qPCR analysis of pro-inflammatory markers (Tnfa, Il6, Il1b, Ifng), anti-inflammatory marker (Il10) and Kupffer cell marker F4/80 (Adgre1). B, Representative immunofluorescence images (400×) of F4/80 staining, DAPI and merged images of liver sections in OGT-KO mice 28 days after PHX. C, qPCR analysis of markers of stellate cell activation Des, Acta2, Tgfb, Col1a1 and Col1a2 of livers from OGT-KO mice 28 days after PHX. D, Photomicrographs (200×) of picrosirius red stained liver sections showing significant fibrosis. E, Hydroxyproline assay of liver tissue from OGT-KO mice 28 days after PHX. qPCR was normalized to 18S and the median of the control group. Bars represents the mean ± standard deviation. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001.
Figure 8.
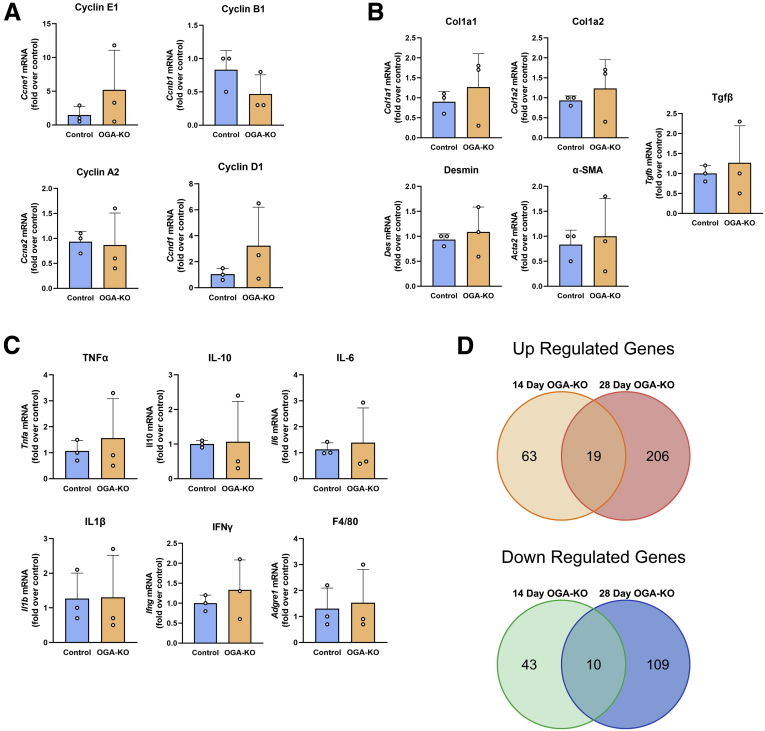
Minimal transcriptional changes occurred in OGA-KO mice at 14 and 28 days after PHX. qPCR analysis of gene expression of cell cyclins (Ccne1, Ccnb1, Ccna2 and Ccnd1) (A), fibrosis genes (Col1a2, Col1a1, Des, and Acta2) (B), and inflammatory genes (Tnfa, Il10, Il6, Il1b, Ifng, and Adgre1) (C) of livers from OGA-KO mice 28 days after PHX. qPCR was normalized to 18S and the median of the control group. Bars represent the mean ± SD. D, Venn diagrams of significantly changed genes using a ±2-fold change cutoff from RNA-seq from livers in OGA-KO mice, 14 and 28 days after PHX.
RNA-Sequencing Revealed Decreased Activation of HNF4α in OGT-KO Mice 14 and 28 Days Post-PHX
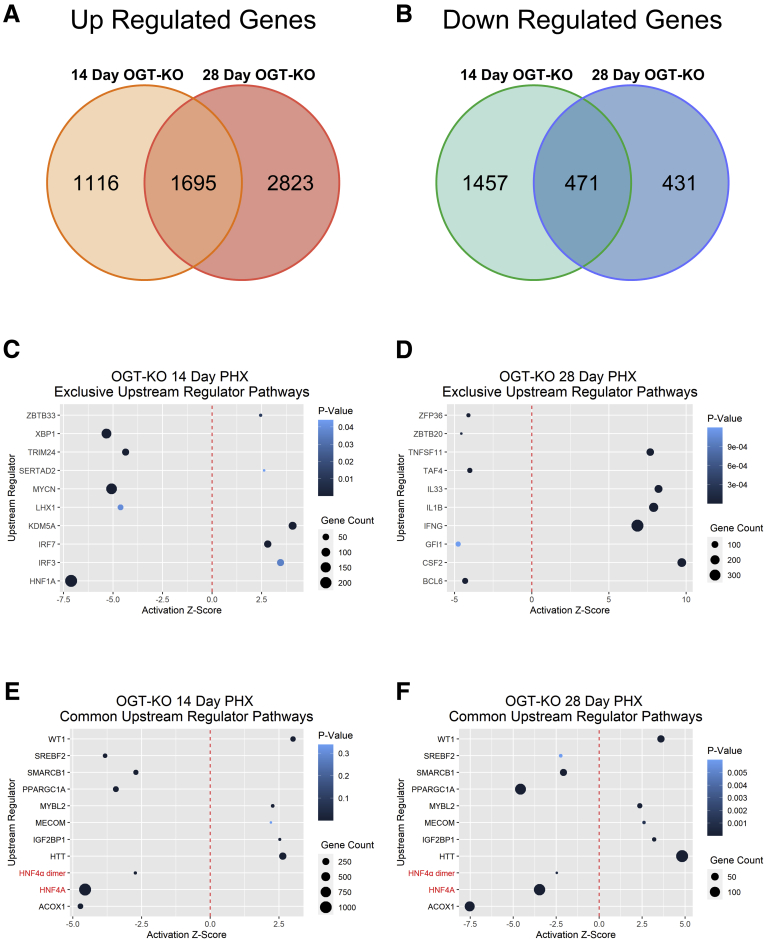
To gain insight into the mechanisms of the defective termination of LR, we performed RNA-sequencing (RNA-Seq) on 14 and 28 days after PHX in OGT-KO and WT mice. Using a 2-fold change expression cutoff, at 14 days after PHX, 2811 genes were upregulated, and 1928 were downregulated. A total of 4518 genes were upregulated, and 902 genes were downregulated at 28 days after PHX. A total of 1695 genes were commonly upregulated, and 471 genes were commonly downregulated between OGT-KO livers at 14 and 28 days after PHX (Figure 9, A-B; Table 1, Table 2). Conversely, RNA-Seq studies on WT and OGA-KO mice at 14 and 28 days after PHX revealed only 19 commonly upregulated and 10 commonly downregulated genes (Figure 9, D; Table 3, Table 4), which is consistent with no difference in liver regeneration between WT and OGA-KO mice.
Figure 9.
IPA of RNA-Seq data from 14 and 28 days after PHX revealed decrease activation of HNF4α. Venn diagrams showing upregulated (A) and downregulated (B) differentially expressed genes at 14 and 28 days after PHX using a 2-fold change cutoff. Dot plots of upstream regulators that are common between OGT-KO 14 days (C) and 28 days (D) post PHX. The red dotted line indicates Z-Score of 0, the size of the dot represents the number of altered genes within that pathway, and the color signifies the P-value.
Table 1.
Top Up- and Down-regulated Genes Shared Between OGT-KO 14 and 28 Days After PHX
| Gene symbol | OGT-KO 14 days | OGT-KO 28 days |
|---|---|---|
| Cacna1b | 48.94 | 4265.97 |
| Fmo3 | 2.01 | 2310.85 |
| Afp | 43.98 | 1709.08 |
| Krt20 | 122.85 | 708.81 |
| Phgdh | 12.63 | 593.99 |
| Hist1h3g | 9.33 | 528.96 |
| Liph | 2.03 | 471.22 |
| Zmynd12 | 31.80 | 438.22 |
| Fkbp1b | 2.90 | 429.97 |
| Gpc3 | 8.05 | 367.51 |
| Il1r2 | 3.37 | 347.48 |
| F13a1 | 6.32 | 318.95 |
| Lmtk3 | 2.50 | 297.98 |
| Sult2b1 | 2.50 | 289.73 |
| Plat | 7.81 | 280.15 |
| Mup16 | −180.25 | −1453.09 |
| Mup17 | −1272.05 | −790.11 |
| Mup15 | −1695.25 | −501.87 |
| Mup9 | −78.64 | −334.90 |
| Hsd3b5 | −81.71 | −316.31 |
| Mup19 | −403.60 | −265.02 |
| Irx1 | −7.26 | −257.25 |
| Thrsp | −3.21 | −243.32 |
| Car3 | −7.88 | −233.98 |
| Mup2 | −124.37 | −151.22 |
| Mup11 | −2402.40 | −129.03 |
| Moxd1 | −62.24 | −127.41 |
| Trhde | −7.66 | −109.71 |
| Spata22 | −2.17 | −94.18 |
| Lrit2 | −2.06 | −86.42 |
Table 2.
Top Up- and Down-regulated Genes in OGT-KO 14 and 28 Days After PHX
| OGT-KO 14 days |
OGT-KO 28 days |
||
|---|---|---|---|
| Gene | FC | Gene | FC |
| Slc7a11 | 318.93 | Cdh17 | 12,292.68 |
| Cbr3 | 289.26 | Cacna1b | 4265.97 |
| H19 | 198.39 | Nrk | 3729.75 |
| Mir675 | 198.39 | Sema3e | 2442.84 |
| Abcc12 | 187.61 | Fmo3 | 2310.85 |
| Krt20 | 122.85 | Cited1 | 2129.36 |
| Cyp2a4 | 121.49 | Afp | 1709.08 |
| Gpx2 | 110.23 | Ivl | 1560.15 |
| Pkp1 | 102.12 | Muc13 | 1246.67 |
| Ly6d | 88.14 | Psca | 1122.93 |
| Mup11 | −2402.40 | Mup16 | −1453.09 |
| Mup15 | −1695.25 | Mup17 | −790.11 |
| Mup17 | −1272.05 | Mup15 | −501.87 |
| Mup1 | −534.88 | Mup9 | −334.90 |
| Mup12 | −439.98 | Hsd3b5 | −316.31 |
| Mup19 | −403.60 | Mup22 | −285.38 |
| Mup7 | −201.78 | Lrrc19 | −280.55 |
| Mup16 | −180.25 | Mup19 | −265.02 |
| Gpr110 | −138.23 | Irx1 | −257.25 |
| Mup2 | −124.37 | Vmn2r20 | −257.25 |
Table 3.
Top Up- and Down-regulated Genes Shared Between OGA-KO 14 and 28 Days After PHX
| Gene symbol | OGA-KO 14 days | OGA-KO 28 days |
|---|---|---|
| Atp1a2 | 2.57 | 15.55 |
| Dbp | 2.38 | 8.81 |
| Rgs16 | 6.46 | 7.32 |
| Pkdrej | 2.31 | 4.87 |
| Fam110c | 3.35 | 4.37 |
| Gpc3 | 2.16 | 4.06 |
| Maff | 2.72 | 3.98 |
| Ttll8 | 2.46 | 3.42 |
| Hebp2 | 2.10 | 3.37 |
| Cxcr5 | 2.34 | 3.23 |
| Sema3e | 2.66 | 2.70 |
| Eif4e3 | 2.16 | 2.40 |
| Cyp2g1 | 2.91 | 2.34 |
| Itgax | 2.40 | 2.30 |
| Slc6a16 | 2.97 | 2.21 |
| Batf3 | 2.22 | 2.18 |
| Klf10 | 2.35 | 2.06 |
| Slc41a3 | 2.30 | 2.05 |
| Angptl7 | 2.25 | 2.03 |
| Oga | −5.98 | −5.26 |
| Fos | −3.69 | −3.81 |
| Tdrkh | −2.73 | −3.78 |
| Zfp773 | −2.87 | −3.44 |
| Gm12248 | −2.53 | −2.54 |
| Ica1l | −2.74 | −2.28 |
| Sgsm1 | −2.15 | −2.21 |
| Praf2 | −2.17 | −2.16 |
| Rpusd1 | −2.01 | −2.15 |
| Mypop | −2.13 | −2.06 |
Table 4.
Top Up- and Down-regulated Genes in OGA-KO 14 and 28 Days After PHX
| OGA-KO 14 days |
OGA-KO 28 days |
||
|---|---|---|---|
| Gene | FC | Gene | FC |
| Mettl7a2 | 259.91 | Ctrb1 | 7430.51 |
| Rgs16 | 6.46 | Pnlip | 5975.84 |
| Pls1 | 4.20 | Cel | 3674.66 |
| Tcf24 | 4.08 | Cpa1 | 3066.49 |
| Cyp2c69 | 3.80 | 2210010C04Rik | 3000.75 |
| Ntrk2 | 3.76 | Cpb1 | 2515.86 |
| Fam110c | 3.35 | Prss2 | 2441.89 |
| Antxr1 | 3.26 | Cela3b | 2080.28 |
| Fam171b | 3.14 | Pnliprp1 | 2047.40 |
| Elmo1 | 2.99 | Clps | 1694.01 |
| Dmbt1 | −169.52 | Klhdc7b | −13.91 |
| Oga | −5.98 | Moxd1 | −9.48 |
| Nap1l3 | −4.36 | Oga | −5.26 |
| Cxcl1 | −4.22 | Npas2 | −5.07 |
| Cox7a1 | −3.77 | Pitx3 | −4.77 |
| Fos | −3.69 | Gm3776 | −4.67 |
| Mup9 | −3.38 | 1700010I14Rik | −4.17 |
| Aplp1 | −3.28 | Loxl4 | −3.84 |
| Kcnk10 | −2.91 | Fos | −3.81 |
| Zfp773 | −2.87 | Tdrkh | −3.78 |
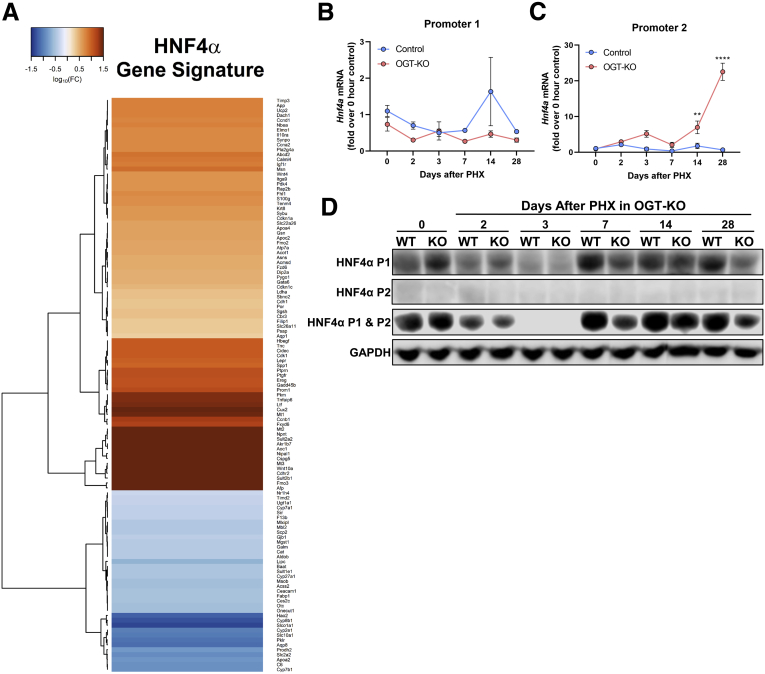
An upstream regulator analysis was performed using Ingenuity Pathway Analysis (IPA) to identify key transcription regulators involved in the transcriptomic changes observed at 14 and 28 days after PHX. Fourteen days after PHX had exclusive activation of the pathways involving interferon regulatory factor 3 and 7 and deactivation of the pathways driven by HNF1A (Figure 9, C). Activation of IFNG, IL1B, and TNF pathways were uniquely increased in 28 days after PHX, indicating an increase in inflammatory responses (Figure 9, D). Further, we identified transcription factors that were activated and inactivated at both 14 and 28 days after PHX in OGT-KO mice (Figure 9, E-F). Interestingly, OGT-KO mice showed significant deactivation in HNF4α function at both 14 and 28 days after PHX (Figure 9, E-F; Figure 10, A).
Figure 10.
HNF4α P1 and P2 isoform levels in OGT-KO mice.A, Heatmap of genes in the HNF4α pathway in OGT-KO mice 28 days after PHX. Color represents log10(fold change of gene) with Euclidean distance and ward.2 clustering. qPCR of Hnf4a in control and OGT-KO mice after PHX for P1 (B) and P2 (C) isoforms. qPCR was normalized to 18S and the median of the 0-hour control group. Point represents the mean ± standard deviation. ∗∗P < .01 and ∗∗∗∗P < .0001. D, Western blots of HNF4α P1, P2, and all isoforms from pooled control and OGT-KO liver lysates (n = 3–5) after PHX.
HNF4α Expression and Activity Declined in OGT-KO Livers After PHX
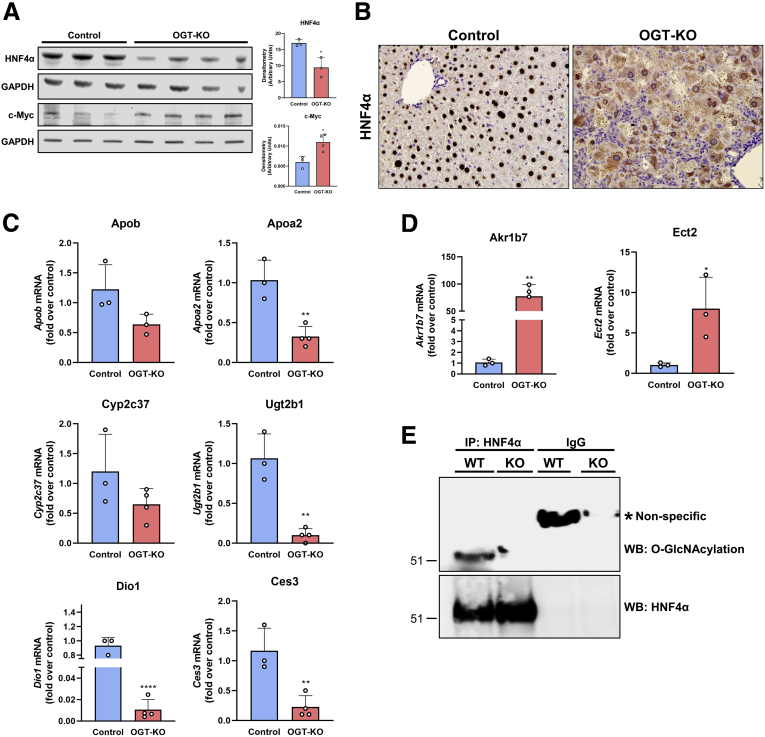
Hepatocyte HNF4α deficiency has been linked to spontaneous hepatocyte proliferation as well as defective termination of LR.11,12 Therefore, we investigated HNF4α expression and activity in OGT-KO mice 28 days after PHX. Western blot analysis of whole liver lysate indicated a significant decrease in HNF4α protein levels in OGT-KO mice 28 days after PHX (Figure 11, A). Previous studies have shown that HNF4α levels have reciprocal effects on the expression of the oncoprotein c-Myc.12,13 Western blot analysis showed induction in c-MYC expression in OGT-KO livers at 28 days after PHX (Figure 11, A). Interestingly, IHC analysis revealed marked cytoplasmic redistribution of HNF4α in OGT-KO mice (Figure 11, B). qPCR analysis of HNF4α target genes indicated the loss of HNF4α function with the decrease of positively regulated genes (Apob, Apoa2, Cyp2c37, Ugt2b1, Dio1, and Ces3) (Figure 11, C) and the increase of negatively regulated genes (Akr1b7 and Ect2) (Figure 11, D). HNF4α is governed by 2 promoters, promoter 1(P1) and promoter 2 (P2). P1 regulates HNF4α expression in adult hepatocytes, whereas P2 regulates HNF4α during early liver development.14 We utilized qPCR and Western blot analysis to determine the promoter activity in OGT-KO mice. We found that P1 isoforms had no changes in RNA level, whereas P2 had significant induction at 14 and 28 days after PHX in OGT-KO mice (Figure 10, B-C). However, no protein levels were detected for the P2 governed isoforms, whereas P1 had decreased amount in OGT-KO mice 7, 14, and 28 days after PHX (Figure 10, D). Finally, to determine if HNF4α undergoes O-GlcNAcylation, immunoprecipitant (IP) experiments were performed. Total liver lysates from WT mice were used for immunoprecipitation of HNF4α protein and then Western blotting was conducted to detect the fraction of O-GlcNAcylated HNF4α. The data indicate that in WT normal livers, HNF4α is heavily O-GlcNAcylated which is absent in OGT-KO mice (Figure 11, E).
Figure 11.
HNF4α expression and function is decreased in OGT-KO mice 28 days after PHX.A, Western blot analysis of HNF4α protein levels in livers of control and OGT-KO mice 28 days after PHX, with corresponding densitometry. B, Representative photomicrographs (400×) of HNF4α IHC of control and OGT-KO mice 28 days post PHX. qPCR analysis of HNF4α-positive target genes (C) and HNF4α-negative target genes (D) at 28 days after PHX. qPCR was normalized to 18S and median of the control group. Bars represents the mean ± standard deviation. ∗P < .05; ∗∗P < .01; and ∗∗∗∗P < .0001. E, IP pull-down of HNF4α from WT and OGT-KO mice. Western blot analysis was performed on HNF4α to show successful pulldown (upper blot) and O-GlcNAcylation (lower blot).
Methionine and Cysteine Metabolism is Altered in OGT-KO 14 and 28 Days After PHX
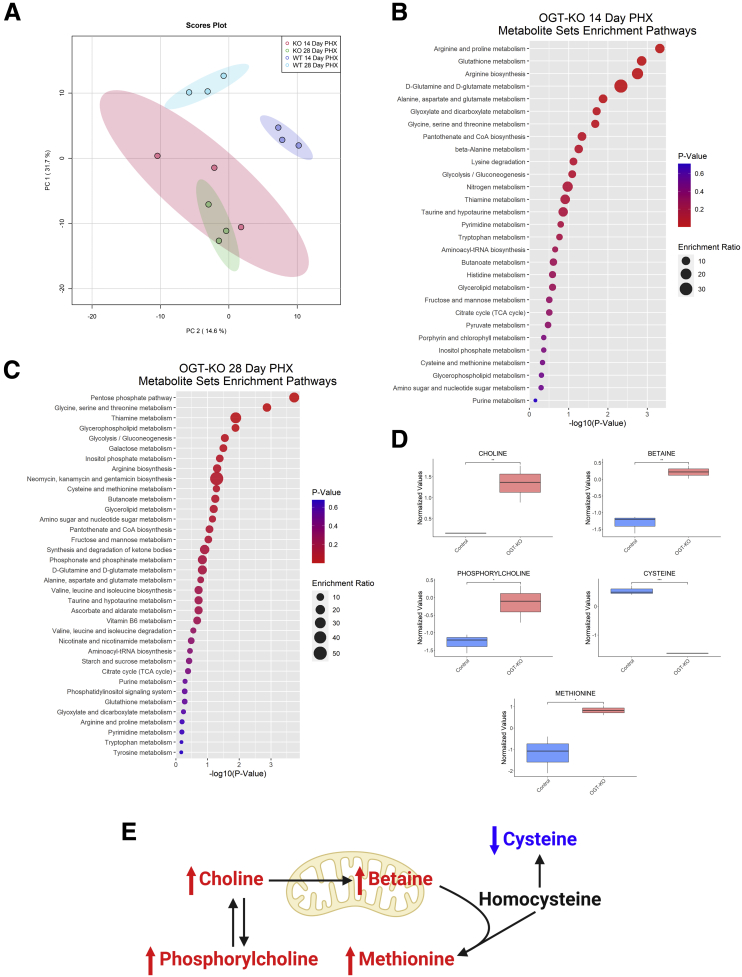
Lastly, we performed targeted metabolomics on WT and OGT-KO liver tissue lysates from 14 and 28 days after PHX. Principle component analysis revealed that both WT and OGT-KO clustering was driven by both PC1 (31.7% variance explained) and PC2 (14.6% variance explained) and with each time point similar to each other (Figure 12, A). To determine altered metabolic pathways, Metabolomic Set Enrichment Analysis was performed on both significantly altered metabolites from 14- and 28-day samples. A significant impact on amino acid metabolism and glutathione metabolism was observed at 14 days after PHX in OGT-KO mice (Figure 12, B). Likewise, OGT-KO mice at 28 days after PHX had significantly altered energy substrate metabolisms such as pentose phosphate and glucose metabolism (Figure 12, C). Specifically, altered methionine synthesis was observed in OGT-KO mice at 28 days after PHX (Figure 12, C-E). The methionine synthesis metabolites, phosphorylcholine, choline, betaine, and methionine were significantly increased in the OGT-KO 28 days after PHX, whereas cysteine was significantly decreased (Figure 12, D-E). Importantly, HNF4α regulates methionine and cysteine metabolism in hepatocytes.15
Figure 12.
Metabolomic analysis show significant change in OGT-KO mice at 28 days after PHX.A, Principal component analysis plot of control and OGT-KO mice for 14 and 28 days after PHX derived from targeted metabolomics. Ellipses represent clusters for each group. Metabolite Set Enrichment Analysis of OGT-KO mice 14 days (B) and 28 days (C) after PHX. Color represents P-value, and size corresponds to enrichment ration. D, Significantly altered metabolites in methionine and cysteine metabolism in OGT-KO mice 28 days after PHX. Bars represents the mean ± minimum and maximum normalized value. ∗P < .05; ∗∗P < .01; and ∗∗∗∗P < .0001. E, Metabolic pathway of methionine and cysteine with red and blue indicated increased or decreased metabolite in OGT-KO mice 28 days after PHX, respectively.
Discussion
LR after PHX, in mice, can be divided into 3 broad phases. The initiation of regeneration is where hepatocytes and other cells partially dedifferentiate and enter cell cycle, progression of regeneration is where cells go through mitosis and divide, and finally, the termination of regeneration is when cell proliferation declines, newly divided cells re-differentiate, and liver tissue is reorganized. The initiation and progression phases are relatively rapid and completed within 5 days after PHX. Termination of regeneration is more prolonged and can take up to a week. Most of the liver mass and function is restored by 2 weeks.16,17 After PHX, multiple signals have been well-characterized that initiate the cell cycle in LR, termed primary mitogens, such as the hepatocyte growth factor and epidermal growth factor.16,18, 19, 20 Auxiliary mitogens, consisting of tumor necrosis factor-α and interleukin-6, enhance the prolific effects of the primary mitogens during the initiation and progression of LR.16,21 However, much less is known about the termination of LR. Integrin-linked kinase, critical for hepatocyte interactions with extracellular matrix, has been identified as a termination signal.22,23 Recent studies from our laboratory have shown that HNF4α, the master regulator of hepatic differentiation,24,25 is critical for re-differentiation of hepatocytes after proliferation and plays a critical role in termination of liver regeneration.12, 13, 14 Previous studies have shown that defective termination of LR will result in significant hepatomegaly and death due to decline in liver function. We hypothesize that in cases where animals do not die due to acute liver failure, defective termination of liver regeneration will lead to hepatic dysplasia that can progress to liver cancer. Evidence presented here supports this hypothesis and indicates that hepatic O-GlcNAcylation is a major regulator of the termination of liver regeneration. After PHX, OGT-KO mice exhibited no differences during the initiation phase and progression of LR. However, during the termination of LR, we found sustained proliferation in hepatocytes indicating a termination defect (Figure 4, A). This was further exhibited by the significant increase in liver-weight to body-weight ratio 14 and 28 days after PHX (Figure 2, D). The foremost change was the development of hepatic nodules accompanied by significant inflammation and early fibrotic changes.
These data are consistent with previous observations that O-GlcNAcylation plays a critical role in cell cycle regulation.8,10,26,27 O-GlcNAcylation of proteins increases during the progression of G1 phase following a rapid decrease when cells enter S phase.9 This contributed to an immediate induction of OGA protein levels during S phase.9 Throughout G1 phase, Rb is O-GlcNAcylated and when the G1/S-checkpoint into S phase occurs, Rb is needed to be phosphorylated indicating cross-talk of PTMs during cell cycle progression.28 After S phase, during the G1 to M phase transition, increasing O-GlcNAcylation by OGA inhibition delays the advancement to M phase.10 During mitosis, global O-GlcNAcylation levels decrease.10 Either increasing or decreasing O-GlcNAc during mitosis will lead to aberrant spindle formation causing mitotic defects.8,29 We found that OGT deletion (reduction of O-GlcNAcylation) in hepatocytes for 7 days resulted in mild hepatomegaly due to hyperplasia as well as hypertrophy (Figure 2, B and D). Importantly, this indicates that OGT regulates hepatocyte proliferation without induction of proliferation by PHX.
We found ballooning hepatocytes accompanied with significant liver injury in OGT-KO mice at 14 and 28 days after PHX (Figure 2, C). Also, initial liver injury at 2 days post-PHX was higher in OGT-KO mice as compared with WT mice. These data indicate that loss of O-GlcNAcylation may make hepatocytes more vulnerable to cell death. These data are consistent with previous studies by Zhang et al, which demonstrated that OGT deletion in the liver using a CRE governed by the albumin promoter resulted in significant necroptosis and fibrosis.7 They further showed that RIP3K undergoes O-GlcNAcylation which reduces the stability of the protein.7 Our studies showed that delayed cell death in OGT-KO mice after PHX is both necroptosis as well as apoptosis (Figure 3, A-C). Interestingly, we observed an increase in p62, a marker of autophagy and Mallory-Denk bodies, in OGT-KO mice after PHX, which is commonly found in metabolic disorders and hepatocellular neoplasms (Figure 3, E).30 It is well-known that autophagy inhibits apoptosis, which suggests that the induction of p62 is probably due to Mallory-Denk body formation.31 To determine if autophagy is contributing to cell death, more studies will need to be done. However, the exact mechanisms of increased spontaneous cell death at 2 weeks after PHX in OGT-KO mice are not known. It is plausible that lack of hepatic redifferentiation, which is a critical component of termination of regeneration, may trigger the cell death at such a delayed time point.
This argument is supported by the RNA-Seq studies at the 14- and 28-day time points, which revealed significant decline in HNF4α target gene expression (Figure 10, A). HNF4α is extremely critical in maintaining hepatocyte function, and we have previously shown that deletion of HNF4α results in defective termination of liver regeneration.12,14 HNF4α protein levels and its function are regulated by PTMs. Most PTMs of HNF4α reduce function and stability. For instance, the C-terminal of HNF4α undergoes SUMOylation to promote degradation,32 acetylation of K458 attenuates the transcriptional activity, deactivating HNF4α,33 and phosphorylation occurs on multiple residues. Other studies have shown that protein kinase A-mediated S142 phosphorylation decrease HNF4α protein levels.34 Extracellular signal-regulated protein kinase 1/2 has been shown to phosphorylate multiple residues on HNF4α including S138/T139, S143, S147/S148, S151, T166/S167, and S313.35 Interestingly, the residue S142 was further found to be phosphorylated by extracellular signal-regulated protein kinase 1/2, indicating 2 kinases acting on the same residue.35 Here, we show that HNF4α is O-GlcNAcylated and without this, HNF4α function is lost. It is known that serine or threonine residue can be alternatively phosphorylated or O-GlcNAcylated.36 Our studies give rise to the possibility that normally HNF4α is O-GlcNAcylated, which prevents its degradation or cytoplasmic relocalization by inhibiting its phosphorylation. If this dynamic relationship occurs, this could explain why increasing O-GlcNAcylation in OGA-KO livers had no physiological phenotype. Further studies will need to be done to map the specific O-GlcNAcylated residue(s) on HNF4α. Nonetheless, our studies have uncovered a novel mechanism by which HNF4α stability and function are regulated. Further, our studies show that O-GlcNAc mediated regulation of HNF4α function is critical in termination of liver regeneration.
When OGA or OGT is deleted, a compensatory effect occurs, altering the expression of the reciprocal enzyme.37 We observed this phenomenon for the first time in hepatocyte-specific OGT-KO and OGA-KO mice (Figure 2, A-B). In OGT-KO mice, this compensatory effect cannot increase O-GlcNAcylation levels. However, in OGA-KO, the compensatory effect of decreasing OGT levels will reduce global O-GlcNAcylation closer to basal levels. This could potentially mitigate the adverse effects of exorbitant O-GlcNAcylation and explain why OGA-KO mice had normal LR. Future experiments will need to be done to artificially increase O-GlcNAcylation to prevent the compensatory effects in OGA-KO mice to determine the effects of augmented O-GlcNAcylation.
In summary, our studies are the first to examine and manipulate O-GlcNAcylation over a time course throughout the LR process after PHX. Our findings suggest that lack of O-GlcNAcylation causes defects in the termination of liver regeneration. Ablation of O-GlcNAcylation impedes the function and stability of HNF4α, which leads to decreased hepatocyte redifferentiation, significant necroinflammation, early fibrotic changes, and formation of dysplastic nodules. These results confirm the role of O-GlcNAcylation in termination of regeneration and preventing hepatic dysplasia and highlight O-GlcNAcylation as a therapeutic target in hepatic regenerative medicine.
Methods
Animal Care and Surgeries
All animal studies were approved by and performed in accordance with the Institutional Animal Care and Use Committee at the University of Kansas Medical Center. OGTfl/Y mice were developed by Dr Natasha Zachara at Johns Hopkins School of Medicine.38 OGAfl/fl mice were developed by Dr John Hanover at the National Institute of Diabetes and Digestive and Kidney Diseases.39 Two-month-old male OGTfl/Y and OGAfl/fl mice were injected intraperitoneally with AAV8-TBG-GFP or AAV8-TBG-CRE (Vector Biolabs) to generate control or hepatocyte specific OGT-KO or OGA-KO animal, as previously stated.40 PHX surgeries were performed on 8-week-old male C57BL/6J, OGTfl, OGT-KO, OGAfl/fl, and OGA-KO mice as previously described.22 Mice were euthanized at various time points between 0 to 28 days after PHX to collect blood and liver samples. Serum was isolated from clotted blood by centrifugation at 5000 rcf for 10 minutes at 4°C. A section of the liver was fixed in 10% neutral buffered formalin for 48 hours and then paraffin-embedded. A piece of liver was then cryopreserved in optimal cutting temperature compound. Liver injury was measured using serum ALT activity (Pointe Scientific ALT Assay by Fisher Scientific).
Staining Procedures and Imaging
Paraffin-embedded liver sections (5-μm thick) were used for H&E staining, IHC, immunofluorescence, and Picrosirius red staining as previously described.12,41 DNA strand breaks were determined by the TUNEL assay using the In Situ Cell Death Detection Kit (Roche Diagnostics). Optimal cutting temperature compound-embedded liver sections (5 μm thick) were used for immunofluorescence.
Protein Isolation, Western Blotting, and IP
Approximately 50 mg of liver tissue was homogenized in RIPA buffer (1% IgePAL, 40 mM GlcNAc, 0.5% Sodium Deoxycholate, 1% SDS, 1 × PBS, 1 × Halt Protease Inhibitor Cocktail [ThermoScientific], and 1 × Halt Phosphatase Inhibitor Cocktail [ThermoScientific]) to extract protein. Pierce BCA Protein Assay Kit (ThermoScientific) was used to measure protein concentration and analyzed on Biotek Synergy 2 microplate reader according to the manufacturer’s protocol. Western blot analysis was performed using individual or pooled samples of isolated protein, as previously described.12,42 Western blots were imaged using the Li-Cor Odyssey FC. Densitometry was done using Image Studio software. IP was performed as previously described.43 A total of 2 mg of protein from whole liver lysate and 2 μg of HNF4α antibody was used to precipitate the protein.
RNA Isolation and qPCR
Total RNA was isolated from livers of control, OGT-KO, and OGA-KO mice using TRizol Reagent according to the manufacturer’s protocol (ThermoFisher) and converted to cDNA as previously described.44 mRNA levels of various genes were measured using 100 ng of cDNA per reaction using PowerUp SYBR Green Master Mix according to the manufacturer’s protocol (ThermoFisher). A total of 384-well plate qPCR reactions were analyzed on the BioRad CFX384. CT values were normalized to 18S as previously discussed in depth.45
Hydroxyproline Assay
To quantitatively estimate collagen content in WT and OGT-KO mice liver, the hydroxyproline assay was used as previously described.46 Briefly, approximately 10 mg of liver was collected from WT and OGT-KO mice (n of 3). The livers were hydrolyzed by adding 100 μL of 12.1 N hydrochloric acid and 100 μL of water, then incubating for 3 hours at 120 °C. The samples were vortexed every 30 minutes during the incubation period to ensure complete tissue lysis. After incubation, the samples were centrifuged for 10 minutes at 10,000 × g. The liver supernatants were added to a 96 well plate by adding 5, 10, and 15 μL of sample per well, in duplicates, to which 100 μL of Chloramine T reagent (Millipore Sigma) was added and allowed to incubate for 25 minutes at room temperature. After incubation, 100 μL of Ehrlich’s Solution [1M 4- (dimethylamino)benzaldehyde in 1-propanol/60% perchloric acid (3:1, v/v)] (Millipore-Sigma) was added and incubated at 60 °C for 35 minutes in a non-humidified, laboratory oven. After the incubation, the absorbance of the samples was measured at 550 nm using a microplate reader (Synergy 2TM Multi-Mode microplate reader, Biotek, Winooski, VT). A standard curve was created using known concentrations from which hydroxyproline content in each sample was calculated. Collagen contains approximately 12.5% hydroxylated prolines47; therefore, collagen found per mg of ovarian tissue was extrapolated by dividing the hydroxyproline concentration by 12.5%.
RNA-Seq Analysis and IPA
RNA-Seq analysis was performed on pooled RNAs isolated from liver samples from OGTfl/Y, OGAfl/fl, OGT-KO, and OGA-KO mice at 14 and 28 days after PHX was analyzed by the Genomics Core Facility at KUMC using Illumina NovaSeq 6000 Sequencing System as previously stated.48 Datasets were deposited in the GEO database (GSE188882). For temporal analysis, up or down differentially expressed genes, using a 2-fold cutoff relative to the control, were uploaded into IPA. Predicted activation or inhibition of the upstream regulators based on changes in the downstream gene expression patterns was then compared between groups.
Metabolite Extraction
Liver (50 mg) was homogenized in precipitate solution (8:1:1 Acetonitrile: Methanol: Acetone), vortexed sample to ensure mixing, and the sample was kept on ice for 30 minutes to further precipitate proteins. Samples were then centrifuged at 15,000 rcf for 10 minutes to pellet proteins and lipids, supernatant was removed, and was dried in Speed Vacuum Centrifuge. Dried metabolites were reconstituted by adding 50μl μL H2O with 0.1% formic acid and vortexed, placed on ice for 15 minutes, and centrifuged again to remove any residual protein or lipid. Supernatants were transferred to labeled, glass liquid chromatography vials with glass insert and samples were then placed into Agilent HPLC 1100 series auto sampler.
Liquid Chromatography With Tandem Mass Spectrometry
An Agilent HPLC 1100 series was used with a Waters Acquity CSHTM Phenyl-Hexyl 1.7 μM 2.1 × 50-mm column. A Sciex API 4000 triple quadrupole mass spectrometer with an ESI source was used in positive mode with first scan event a full mass spectrometry scan at 55.0 to 1000 m/z. For positive polarity detection of 230 targeted metabolites with a gradient of 98% buffer A was set at 0.00 to 1.00 minute, buffer B increased to 15% at 4.00 minutes, then buffer B increased to 95% to 7.00 minutes, maintained to 8.00 minutes, then decreased to 2% buffer B at 8.50 minutes through 10.0 minutes. For negative polarity detection of metabolites, a gradient of 98% buffer A (H2O and 0.1% formic acid) and 2% buffer B (methanol) at 0 to 0.5 minutes, increased buffer B to 95% at 5 minutes, increased buffer B to 98% at 8.5 minutes, and then decreased buffer B to 2% to 9.0 to 10.0 minutes at a flow rate of 0.15 mL/min. In negative polarity mode for analysis, similar parameters were set at 55.0 to 1000 m/z.
Targeted Metabolomic Analysis
Metabolites were analyzed using the MetaboAnalyst 4.0 R package as previously stated.49 The unnormalized mass spectrometry peak area was used for the analysis. The metabolites were median-centered and log-transformed to produce a normal distribution. Metabolite enrichment analysis was performed on the significantly altered metabolites (criteria, P < .05).
Statistical Analysis
For all experiments not associated with RNA-seq or metabolomics, such as ALT measurements, results are expressed as mean ± standard error of the mean. GraphPad Prism 9 was used to graph and calculated statistics. The Student t test or analysis of variance with Tukey’s post-hoc was applied to all analyses, with a P-value < .05 being considered significant. Dot plots and heatmaps were produced in RStudio (R version 4.0.3; RStudio Team).
Acknowledgments
CRediT Authorship Contributions
Dakota Robarts, BS (Conceptualization: Supporting; Formal analysis: Lead; Methodology: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Steven R. McGreal, PhD (Data curation: Supporting; Methodology: Supporting)
David S. Umbaugh, BS (Methodology: Supporting)
Wendena S. Parkes, BS (Methodology: Supporting)
Manasi Kotulkar, MS (Methodology: Supporting)
Sarah Abernathy, BS (Methodology: Supporting)
Norman Lee, PhD (Methodology: Supporting)
Hartmut Jaeschke, PhD (Formal analysis: Supporting; Methodology: Supporting)
Sumedha Gunewardena, PhD (Data curation: Supporting; Formal analysis: Supporting)
Stephen A. Whelan, PhD (Formal analysis: Supporting; Methodology: Supporting)
John A. Hanover, PhD (Resources: Supporting)
Natasha E. Zachara, PhD (Resources: Supporting)
Chad Slawson, PhD (Conceptualization: Supporting; Formal analysis: Supporting)
Udayan Apte, PhD, DABT (Conceptualization: Lead; Formal analysis: Lead; Funding acquisition: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by National Institutes of Health (NIH)-Centers of Biomedical Research Excellence (P20 RR021940-03, P30 GM118247), National Institute of Environmental Health Sciences Toxicology Training Grant (T32 ES007079-34), NIH R01 DK0198414, and NIH R56 DK112768.
References
- 1.Michalopoulos G.K. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michalopoulos G.K. Hepatostat: Liver regeneration and normal liver tissue maintenance. Hepatology. 2017;65:1384–1392. doi: 10.1002/hep.28988. [DOI] [PubMed] [Google Scholar]
- 3.Kang L.I., Mars W.M., Michalopoulos G.K. Signals and cells involved in regulating liver regeneration. Cells. 2012;1:1261–1292. doi: 10.3390/cells1041261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hart G.W., Slawson C., Ramirez-Correa G., Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu W., Zhang X., Wu J.L., Fu L., Liu K., Liu D., Chen G.G., Lai P.B., Wong N., Yu J. O-GlcNAc transferase promotes fatty liver-associated liver cancer through inducing palmitic acid and activating endoplasmic reticulum stress. J Hepatol. 2017;67:310–320. doi: 10.1016/j.jhep.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 6.Jozwiak P., Forma E., Brys M., Krzeslak A. O-GlcNAcylation and metabolic reprograming in cancer. Front Endocrinol (Lausanne) 2014;5:145. doi: 10.3389/fendo.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B., Li M.D., Yin R., Liu Y., Yang Y., Mitchell-Richards K.A., Nam J.H., Li R., Wang L., Iwakiri Y., Chung D., Robert M.E., Ehrlich B.E., Bennett A.M., Yu J., Nathanson M.H., Yang X. O-GlcNAc transferase suppresses necroptosis and liver fibrosis. JCI Insight. 2019;4 doi: 10.1172/jci.insight.127709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanza C., Tan E.P., Zhang Z., Machacek M., Brinker A.E., Azuma M., Slawson C. Reduced O-GlcNAcase expression promotes mitotic errors and spindle defects. Cell Cycle. 2016;15:1363–1375. doi: 10.1080/15384101.2016.1167297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drougat L., Olivier-Van Stichelen S., Mortuaire M., Foulquier F., Lacoste A.S., Michalski J.C., Lefebvre T., Vercoutter-Edouart A.S. Characterization of O-GlcNAc cycling and proteomic identification of differentially O-GlcNAcylated proteins during G1/S transition. Biochim Biophys Acta. 2012;1820:1839–1848. doi: 10.1016/j.bbagen.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 10.Slawson C., Zachara N.E., Vosseller K., Cheung W.D., Lane M.D., Hart G.W. Perturbations in O-linked beta-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J Biol Chem. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- 11.Walesky C., Gunewardena S., Terwilliger E.F., Edwards G., Borude P., Apte U. Hepatocyte-specific deletion of hepatocyte nuclear factor-4alpha in adult mice results in increased hepatocyte proliferation. Am J Physiol Gastrointest Liver Physiol. 2013;304:G26–G37. doi: 10.1152/ajpgi.00064.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huck I., Gunewardena S., Espanol-Suner R., Willenbring H., Apte U. Hepatocyte nuclear factor 4 alpha activation is essential for termination of liver regeneration in mice. Hepatology. 2019;70:666–681. doi: 10.1002/hep.30405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang-Verslues W.W., Sladek F.M. Nuclear receptor hepatocyte nuclear factor 4alpha1 competes with oncoprotein c-Myc for control of the p21/WAF1 promoter. Mol Endocrinol. 2008;22:78–90. doi: 10.1210/me.2007-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walesky C., Apte U. Role of hepatocyte nuclear factor 4alpha (HNF4alpha) in cell proliferation and cancer. Gene Expr. 2015;16:101–108. doi: 10.3727/105221615X14181438356292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Q., Li Y., Gao X., Kang K., Williams J.G., Tong L., Liu J., Ji M., Deterding L.J., Tong X., Locasale J.W., Li L., Shats I., Li X. HNF4alpha regulates sulfur amino acid metabolism and confers sensitivity to methionine restriction in liver cancer. Nat Commun. 2020;11:3978. doi: 10.1038/s41467-020-17818-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalopoulos G.K., Bhushan B. Liver regeneration: biological and pathological mechanisms and implications. Nat Rev Gastroenterol Hepatol. 2021;18:40–55. doi: 10.1038/s41575-020-0342-4. [DOI] [PubMed] [Google Scholar]
- 17.Michalopoulos G.K., DeFrances M.C. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 18.Stolz D.B., Mars W.M., Petersen B.E., Kim T.H., Michalopoulos G.K. Growth factor signal transduction immediately after two-thirds partial hepatectomy in the rat. Cancer Res. 1999;59:3954–3960. [PubMed] [Google Scholar]
- 19.Block G.D., Locker J., Bowen W.C., Petersen B.E., Katyal S., Strom S.C., Riley T., Howard T.A., Michalopoulos G.K. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. J Cell Biol. 1996;132:1133–1149. doi: 10.1083/jcb.132.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bucher N.L., Patel U., Cohen S. Hormonal factors concerned with liver regeneration. Ciba Found Symp. 1977:95–107. doi: 10.1002/9780470720363.ch5. [DOI] [PubMed] [Google Scholar]
- 21.Webber E.M., Bruix J., Pierce R.H., Fausto N. Tumor necrosis factor primes hepatocytes for DNA replication in the rat. Hepatology. 1998;28:1226–1234. doi: 10.1002/hep.510280509. [DOI] [PubMed] [Google Scholar]
- 22.Apte U., Gkretsi V., Bowen W.C., Mars W.M., Luo J.H., Donthamsetty S., Orr A., Monga S.P., Wu C., Michalopoulos G.K. Enhanced liver regeneration following changes induced by hepatocyte-specific genetic ablation of integrin-linked kinase. Hepatology. 2009;50:844–851. doi: 10.1002/hep.23059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gkretsi V., Apte U., Mars W.M., Bowen W.C., Luo J.H., Yang Y., Yu Y.P., Orr A., St-Arnaud R., Dedhar S., Kaestner K.H., Wu C., Michalopoulos G.K. Liver-specific ablation of integrin-linked kinase in mice results in abnormal histology, enhanced cell proliferation, and hepatomegaly. Hepatology. 2008;48:1932–1941. doi: 10.1002/hep.22537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morimoto A., Kannari M., Tsuchida Y., Sasaki S., Saito C., Matsuta T., Maeda T., Akiyama M., Nakamura T., Sakaguchi M., Nameki N., Gonzalez F.J., Inoue Y. An HNF4alpha-microRNA-194/192 signaling axis maintains hepatic cell function. J Biol Chem. 2017;292:10574–10585. doi: 10.1074/jbc.M117.785592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunewardena S., Huck I., Walesky C., et al. Progressive loss of hepatocyte nuclear factor 4 alpha activity in chronic liver diseases in humans. Hepatology. 2022 doi: 10.1002/hep.32326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan E.P., Duncan F.E., Slawson C. The sweet side of the cell cycle. Biochem Soc Trans. 2017;45:313–322. doi: 10.1042/BST20160145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akan I., Love D.C., Harwood K.R., Bond M.R., Hanover J.A. Drosophila O-GlcNAcase deletion globally perturbs chromatin O-GlcNAcylation. J Biol Chem. 2016;291:9906–9919. doi: 10.1074/jbc.M115.704783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wells L., Slawson C., Hart G.W. The E2F-1 associated retinoblastoma-susceptibility gene product is modified by O-GlcNAc. Amino Acids. 2011;40:877–883. doi: 10.1007/s00726-010-0709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan E.P., Caro S., Potnis A., Lanza C., Slawson C. O-linked N-acetylglucosamine cycling regulates mitotic spindle organization. J Biol Chem. 2013;288:27085–27099. doi: 10.1074/jbc.M113.470187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strnad P., Zatloukal K., Stumptner C., Kulaksiz H., Denk H. Mallory-Denk-bodies: lessons from keratin-containing hepatic inclusion bodies. Biochim Biophys Acta. 2008;1782:764–774. doi: 10.1016/j.bbadis.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Thorburn A. Apoptosis and autophagy: regulatory connections between two supposedly different processes. Apoptosis. 2008;13:1–9. doi: 10.1007/s10495-007-0154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou W., Hannoun Z., Jaffray E., Medine C.N., Black J.R., Greenhough S., Zhu L., Ross J.A., Forbes S., Wilmut I., Iredale J.P., Hay R.T., Hay D.C. SUMOylation of HNF4alpha regulates protein stability and hepatocyte function. J Cell Sci. 2012;125:3630–3635. doi: 10.1242/jcs.102889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokoyama A., Katsura S., Ito R., Hashiba W., Sekine H., Fujiki R., Kato S. Multiple post-translational modifications in hepatocyte nuclear factor 4alpha. Biochem Biophys Res Commun. 2011;410:749–753. doi: 10.1016/j.bbrc.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 34.Viollet B., Kahn A., Raymondjean M. Protein kinase A-dependent phosphorylation modulates DNA-binding activity of hepatocyte nuclear factor 4. Mol Cell Biol. 1997;17:4208–4219. doi: 10.1128/mcb.17.8.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veto B., Bojcsuk D., Bacquet C., Kiss J., Sipeki S., Martin L., Buday L., Balint B.L., Aranyi T. The transcriptional activity of hepatocyte nuclear factor 4 alpha is inhibited via phosphorylation by ERK1/2. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z., Gucek M., Hart G.W. Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc Natl Acad Sci U S A. 2008;105:13793–13798. doi: 10.1073/pnas.0806216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kazemi Z., Chang H., Haserodt S., McKen C., Zachara N.E. O-linked beta-N-acetylglucosamine (O-GlcNAc) regulates stress-induced heat shock protein expression in a GSK-3beta-dependent manner. J Biol Chem. 2010;285:39096–39107. doi: 10.1074/jbc.M110.131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shafi R., Iyer S.P., Ellies L.G., O'Donnell N., Marek K.W., Chui D., Hart G.W., Marth J.D. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keembiyehetty C., Love D.C., Harwood K.R., Gavrilova O., Comly M.E., Hanover J.A. Conditional knock-out reveals a requirement for O-linked N-Acetylglucosaminase (O-GlcNAcase) in metabolic homeostasis. J Biol Chem. 2015;290:7097–7113. doi: 10.1074/jbc.M114.617779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGreal S.R., Bhushan B., Walesky C., McGill M.R., Lebofsky M., Kandel S.E., Winefield R.D., Jaeschke H., Zachara N.E., Zhang Z., Tan E.P., Slawson C., Apte U. Modulation of O-GlcNAc levels in the liver impacts acetaminophen-induced liver injury by affecting protein adduct formation and glutathione synthesis. Toxicol Sci. 2018;162:599–610. doi: 10.1093/toxsci/kfy002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang L., Fang P., Septer S., Apte U., Pritchard M.T. Inhibition of mast cell degranulation with cromolyn sodium exhibits organ-specific effects in polycystic kidney (PCK) rats. Int J Toxicol. 2018;37:308–326. doi: 10.1177/1091581818777754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhushan B., Poudel S., Manley M.W., Jr., Roy N., Apte U. Inhibition of glycogen synthase kinase 3 accelerated liver regeneration after acetaminophen-induced hepatotoxicity in mice. Am J Pathol. 2017;187:543–552. doi: 10.1016/j.ajpath.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z., Costa F.C., Tan E.P., Bushue N., DiTacchio L., Costello C.E., McComb M.E., Whelan S.A., Peterson K.R., Slawson C. O-Linked N-Acetylglucosamine (O-GlcNAc) transferase and O-GlcNAcase interact with Mi2beta protein at the agamma-globin promoter. J Biol Chem. 2016;291:15628–15640. doi: 10.1074/jbc.M116.721928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borude P., Edwards G., Walesky C., Li F., Ma X., Kong B., Guo G.L., Apte U. Hepatocyte-specific deletion of farnesoid X receptor delays but does not inhibit liver regeneration after partial hepatectomy in mice. Hepatology. 2012;56:2344–2352. doi: 10.1002/hep.25918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Briley S.M., Jasti S., McCracken J.M., Hornick J.E., Fegley B., Pritchard M.T., Duncan F.E. Reproductive age-associated fibrosis in the stroma of the mammalian ovary. Reproduction. 2016;152:245–260. doi: 10.1530/REP-16-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards C.A., O’Brien W.D., Jr. Modified assay for determination of hydroxyproline in a tissue hydrolyzate. Clin Chim Acta. 1980;104:161–167. doi: 10.1016/0009-8981(80)90192-8. [DOI] [PubMed] [Google Scholar]
- 48.Walesky C., Edwards G., Borude P., Gunewardena S., O'Neil M., Yoo B., Apte U. Hepatocyte nuclear factor 4 alpha deletion promotes diethylnitrosamine-induced hepatocellular carcinoma in rodents. Hepatology. 2013;57:2480–2490. doi: 10.1002/hep.26251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chong J., Wishart D.S., Xia J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinformatics. 2019;68:e86. doi: 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]