Abstract
Background & Aims
Pancreatic islet β-cells are factories for insulin production; however, ectopic expression of insulin also is well recognized. The gallbladder is a next-door neighbor to the developing pancreas. Here, we wanted to understand if gallbladders contain functional insulin-producing cells.
Methods
We compared developing and adult mouse as well as human gallbladder epithelial cells and islets using immunohistochemistry, flow cytometry, enzyme-linked immunosorbent assays, RNA sequencing, real-time polymerase chain reaction, chromatin immunoprecipitation, and functional studies.
Results
We show that the epithelial lining of developing, as well as adult, mouse and human gallbladders naturally contain interspersed cells that retain the capacity to actively transcribe, translate, package, and release insulin. We show that human gallbladders also contain functional insulin-secreting cells with the potential to naturally respond to glucose in vitro and in situ. Notably, in a non-obese diabetic (NOD) mouse model of type 1 diabetes, we observed that insulin-producing cells in the gallbladder are not targeted by autoimmune cells. Interestingly, in human gallbladders, insulin splice variants are absent, although insulin splice forms are observed in human islets.
Conclusions
In summary, our biochemical, transcriptomic, and functional data in mouse and human gallbladder epithelial cells collectively show the evolutionary and developmental similarities between gallbladder and the pancreas that allow gallbladder epithelial cells to continue insulin production in adult life. Understanding the mechanisms regulating insulin transcription and translation in gallbladder epithelial cells would help guide future studies in type 1 diabetes therapy.
Keywords: Islets, Insulin, Splice Variants, Gallbladder Development, Differentiation
Abbreviations used in this paper: bp, base pair; BSA, bovine serum albumin; ChIP, chromatin immunoprecipitation; CldU, 5-chloro-2-deoxyuridine; Ct, cycle threshold; E, embryonic day; ELISA, enzyme-linked immunosorbent assay; ER, endoplasmic reticulum; FACS, fluorescence-activated cell sorter; GFP/w, Green Flouroscent Protein/wild-type; GO, gene ontology; ICA, islet-like aggregate; IdU, 5-iodo-2-deoxyuridine; MIP, Mouse Insulin Promoter; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide; NOD/SCID, Non-obese Diabetic/severe combined immunodeficiency; PFA, paraformaldehyde; PBS, phosphate-buffered saline; PDX1, Pancreas and duodenal homeobox gene 1; qPCR, quantitative polymerase chain reaction; RNA-seq, RNA sequencing; SCB, sodium cacodylate buffer; SFM, serum-free medium; TLDA, TaqMan Low-Density Array; T1D, type 1 diabetes; WGA, weeks gestational age
Summary.
Insulin-producing cells are present in the epithelial lining of developing, as well as adult, mouse and human gallbladders. Although not physiologically relevant, these cells possess the capacity to transcribe and translate insulin. These cells were not destroyed in a mouse model of type 1 diabetes.
The regulation of mammalian gene expression is an important and complex issue in biology. Although pancreatic islet β-cells are the major insulin-producing cells in our body, we and others have shown ectopic expression of insulin in the gallbladder/biliary duct epithelium,1,2 thymus,3 and the brain.4,5 The gallbladder and pancreas arise from the foregut endoderm and share transcription factors during embryonic development.6 The occurrence of β-like cells in mouse biliary ducts was reported previously,2 and the inhibition of Hes1 in mouse gallbladder epithelial cells improved insulin production.7 A recent study also implicated their therapeutic potential by forced expression of pancreatic transcription factors to differentiate gallbladder-derived cells from individuals with type 1 diabetes (T1D) into insulin-producing cells.8
The development of the pancreas and its close neighbors (the liver and gallbladder) has been studied in mouse models. However, gallbladder development has not been reported as vastly as in the pancreas and the liver. Similarly, pancreas development in both rodents and human beings has not been compared with that of gallbladder. Here, using mouse models and human tissue/samples spanning embryonic to post-natal/adult life, we show a comparative molecular and functional analysis of insulin-transcribing cells in the gallbladder and pancreas. Although expression of genes and their splice variants across human tissues is fundamental to the understanding of tissue specificity and function, current tissue expression data sets, such as the Genotype-Tissue Expression (GTEx) project,9 lack gene expression data from the gallbladder. Our work adds new knowledge while showing the gallbladder to be an important and interesting naturally occurring source of insulin-producing cells. Considering the developmental similarity with the pancreas, natural occurrence of insulin-producing cells, and tissue availability, the gallbladder presents an alternative for cell-replacement therapy. Although the insulin secretion from these cells is physiologically not relevant, our data support the need for future work on understanding cellular and molecular mechanisms regulating insulin production in this functionally diverse tissue.
Results
Insulin Expression in Developing Mouse Gallbladder Cells
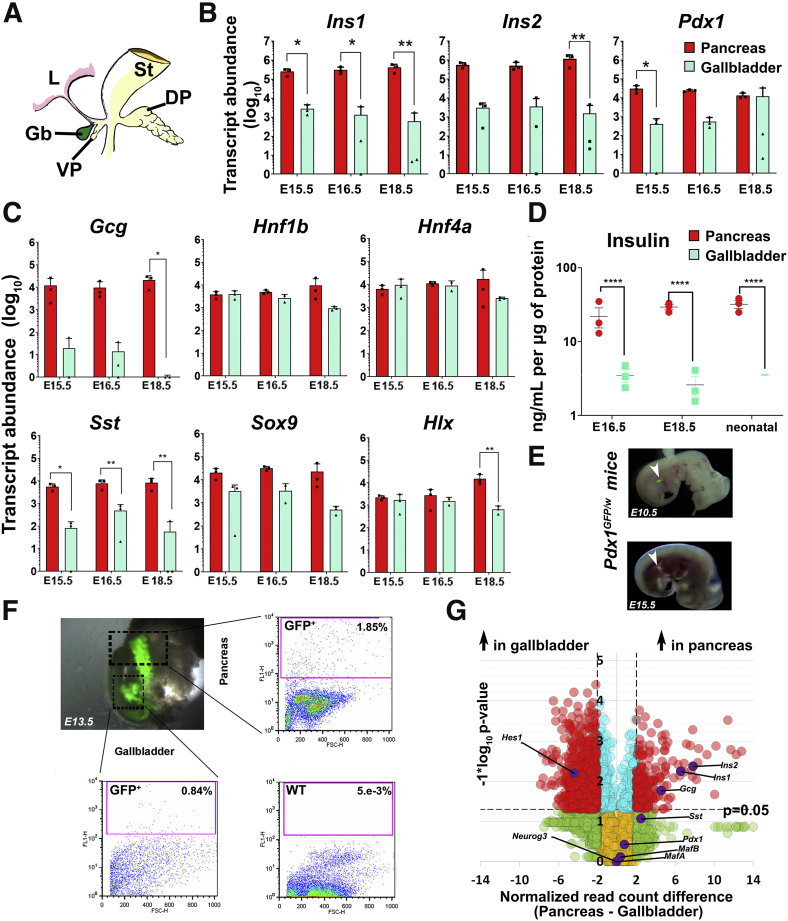
The gallbladder originates from the pancreatic bud that also gives rise to the ventral pancreas6 (Figure 1A) during embryonic development. We profiled key endocrine pancreatic gene transcripts in developing mouse pancreas and gallbladder on embryonic day (E)15.5, E16.5, and E18.5. The developing pancreas contained more copies of Ins1, Ins2 (Figure 1B), and Gcg, Sst (Figure 1C) gene transcripts compared with those in the gallbladder. Ins1, which is known to be more specific to mouse pancreas (but not the brain5), was significantly higher in the developing pancreas at all of the embryonic time points (Figure 1B). Ins2 expression levels were significantly different from those in the gallbladder closer to birth. Interestingly, the master regulatory pancreatic transcription factor Pdx1 was expressed in gallbladder cells at all times, and at levels similar to those in the pancreas closer to birth (E16.5 and E18.5) (Figure 1B). The expression of other gene transcripts (Hnf1b, Sox9, Hnf4a, and Hlx) did not show significant differences between developing pancreas and gallbladder (Figure 1C) during the time points assessed. Developing mouse gallbladders also contain immune-reactive insulin protein, albeit at significantly lower levels than those in the developing pancreas (Figure 1D). Using a Pdx1GFP/w reporter mouse10 (Figure 1E), we confirmed that cells within the prospective gallbladder and pancreatic buds show Pdx1 gene promoter activity (Figure 1F). As expected, wild-type gallbladder cells do not contain any GFP+ cells (Figure 1F, right bottom). We then performed bulk RNA sequencing (RNA-seq) on adult mouse pancreas and gallbladders to assess similarities and differences in pancreatic gene transcript levels across these functionally different adult tissues. Pancreatic hormone transcripts were significantly higher (≥2-fold difference; P ≤ .05) in the adult pancreas, whereas Hes1, a negative regulator of the pro-endocrine gene Neurog3 (also known as Ngn311) was expressed at significantly higher levels in gallbladder cells (Figure 1G). In line with the embryonic data (Figure 1B), the expression of Pdx1 was similar (Figure 1G) across adult gallbladders and the pancreas. Validation of RNA-seq data using real-time quantitative polymerase chain reaction (qPCR), confirmed similar Pdx1 expression in both adult tissues (Figure 2A). Relatively lower levels of Neurog3 transcripts in adult gallbladder presented with a significantly higher level of Hes1 in the qPCR-based validation (Figure 2A). Pancreatic islets and gallbladder epithelial cells of Pdx1GFP/w mice had insulin– as well as Pdx1–co-expressing cells (Figure 2B). Pancreatic β-cells had predominantly nuclear Pdx1 expression, whereas gallbladder epithelial cells from the same mice presented with cytoplasmic localization of Pdx1 (Figure 2B). The continued expression of Pdx1 in mouse gallbladder cells is intriguing. Pdx1 gene expression is controlled by 4 conserved sequence domains: area I (2761/-2457 base pairs [bp]), area II (-2153/-1923 bp), area III (-1879/-1600 bp), and area IV (-6529/-6047 bp).12 Chromatin immunoprecipitation (ChIP) analysis at the Pdx1 gene promoter region showed that these sites are active in the gallbladder (Figure 2C). Using flow cytometry and another mouse model (Mouse Insulin Promoter driving Green Flouroscent Protein; MIP-GFP),13 we confirmed mouse Ins1 promoter activity in adult mouse gallbladder cells. Here, we observed GFP-positive cells in both pancreas and gallbladder of MIP-GFP mice, with gallbladder tissue harboring approximately 9-fold less insulin-containing cells (Figure 2D). We then probed if the insulin produced by mouse gallbladder epithelial cells was packaged. Immune-electron microscopy of mouse gallbladder epithelial cells confirmed the presence (albeit a lower number) of electron-dense insulin secretory vesicles (Figure 2E) within gallbladder epithelium. Although the number and density of these granules was significantly lower, their existence in gallbladder is promising. Together, these studies show pancreatic endocrine gene and protein expression in embryonic and adult mouse gallbladder epithelium.
Figure 1.
Mouse gallbladder and pancreas development. (A) Mouse pancreatic gut schematic showing developing stomach (St), liver (L), dorsal pancreatic bud (DP), and the ventral pancreatic bud (VP) that gives rise to the gallbladder (Gb) and ventral pancreas. TaqMan-based real-time qPCR for rodent insulin genes (Ins1, Ins2), the master regulatory transcription factor (Pdx1) (B) as well as other genes (C) in developing mouse pancreas and gallbladder tissues harvested on E15.5, E16.5, and E18.5. Data in B and C represent means ± SD from 3 different litters of Friend Virus B NIH Jackson (FVB/NJ) mice, each with at least 6–7 embryos/litter. Transcript abundance was analyzed using 2-way analysis of variance with the Sidak adjustment for multiple comparisons. Each point in the scatter bar graph presents data from a single litter. (D) Insulin content in developing mouse pancreas and gallbladder tissues harvested at E16.5, E18.5, and neonatal day 1 pups. Data were obtained from pooled tissues from 3 different litters of FVB/NJ mice, each with at least 6–7 embryos/pups and presented after normalizing to total protein. Aligned dot plots present means ± SEM. Significance was calculated using 2-way analysis of variance with the Fisher's Least Significant Difference (LSD) test. (E and F) Pdx1GFP/w reporter mouse embryos were obtained from timed pregnancies (E10.5, E13.5, and E15.5) and GFP fluorescence (arrowheads) was used to identify pancreatic buds. Representative flow cytometry plots of pancreas and gallbladder tissues from Pdx1GFP/w (GFP+) and Pdx1w/w (wild-type) mice are presented with proportions of GFP+ cells as indicated. Acquisition and gating parameters were identical across WT and GFP+ tissues. Experiments were repeated at least 3 times with tissues pooled from 6 to 7 embryos. (G) Volcano plot for bulk RNA-seq data from the adult mouse gallbladder (n = 6) and pancreas (n = 6). Normalized read count difference between pancreas and gallbladder is plotted on the X-axis and statistical significance (−log10P value) is presented on the Y-axis. The dashed horizontal line represents P = .05, and the dashed vertical lines represent a 2-fold normalized DEseq value difference. Significantly altered (P ≤ .05 and ≥2-fold difference) transcripts are presented in red. Selected important pancreatic genes on the volcano plot are labeled and highlighted in purple. Throughout the figure, bars/dots for the pancreas are in red and for the gallbladder are in green. ∗P < .05, ∗∗P < .01
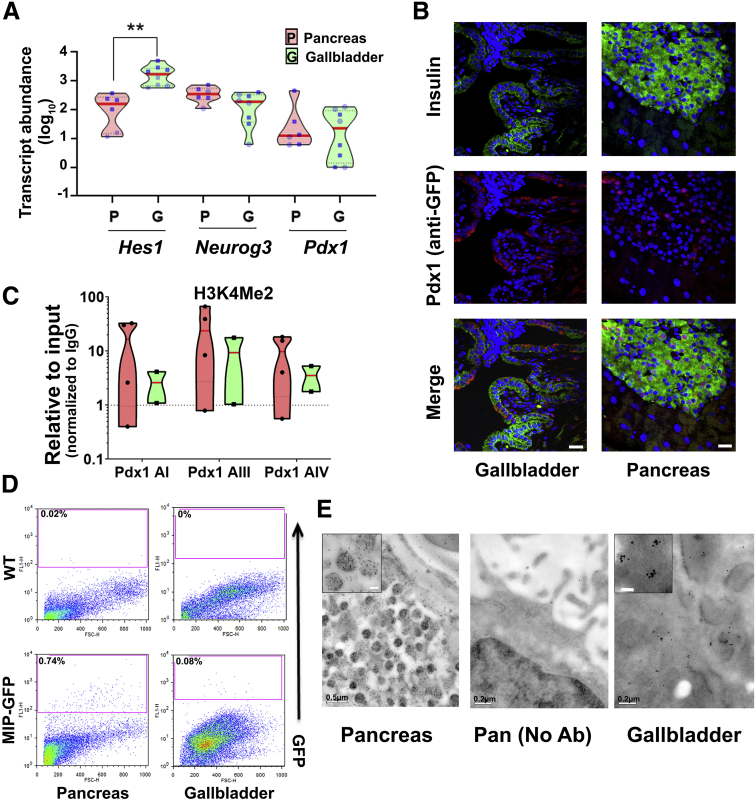
Figure 2.
Similarities and differences in mouse pancreatic and gallbladder cells. (A) Real-time qPCR data from postnatal mouse pancreas and gallbladder (N = 6–10 animals) for Hes1, Neurog3, and Pdx1 gene transcripts analyzed using Kruskal–Wallis with the Dunn multiple comparisons test. Each dot within the violins represents a different sample (circles represent the age group 1–7 days; squares represent the age group 3–8 weeks). The horizontal solid red line within each polygon represents the median, the horizontal black dotted line represents quartiles, and the polygons represent the density of individual data points and extend to minimum/maximum values. (B) Immunostaining of insulin (green) and Pdx1/GFP (red, using anti-GFP antibody) in the pancreatic islets and gallbladder epithelial cells of Pdx1GFP/w reporter mice. Nuclei (DNA) are shown in blue. Scale bars: 20 μm. (C) Three sites in the promoter region of mouse Pdx1 were assessed for the presence of H3K4 Me2 (active transcription mark) using the ChIP assay in adult C57BL/6J mice pancreas (n = 4) and gallbladders (n = 2 experiments with >6 mice gallbladders/experiment). Data are plotted relative to input and normalized to IgG. They are presented as individual points within violin plots presenting individual/pooled tissue. The dotted line represents the expression levels for isotype control (IgG) samples. The horizontal solid red line within each violin plot represents the median, and the polygons represent the density of data points and extend to minimum/maximum values. (D) Representative flow cytometry plots of pancreas and gallbladder tissues from adult (6- to 10-week-old) MIP-GFP reporter and wild-type (WT) mice. The percentage of GFP+ cells is indicated for each plot. Acquisition and gating parameters were identical across wild-type and GFP+ cells. Experiments were repeated at least 3 times with tissues pooled from 3 to 5 animals at each time. (E) Electron microscopy images from the same CD1 mouse pancreatic islet and gallbladder epithelial cells after immuno-gold labeling of insulin granules. Data were validated in 4 different preparations. Inset: Immuno-gold–labeled insulin secretory granules from higher-magnification scans. Scale bar: 0.1 μm. Lower-magnification panels show the secretory vesicles with a characteristic halo in pancreatic, but not as evident in gallbladder, cells. Middle: No-antibody pancreatic control. Scale bars as indicated in each image. Throughout the figure, violins for the pancreas are in red and for the gallbladder are in green. ∗∗P < .01.
Pancreatic Endocrine Gene Expression in Developing Human Gallbladder
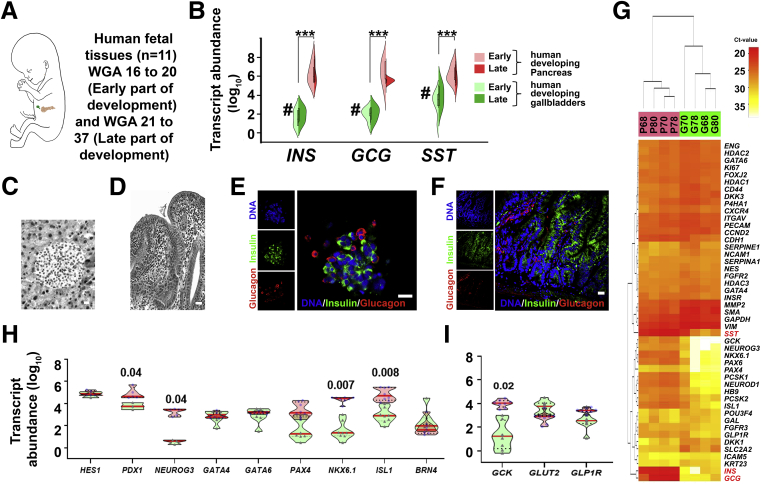
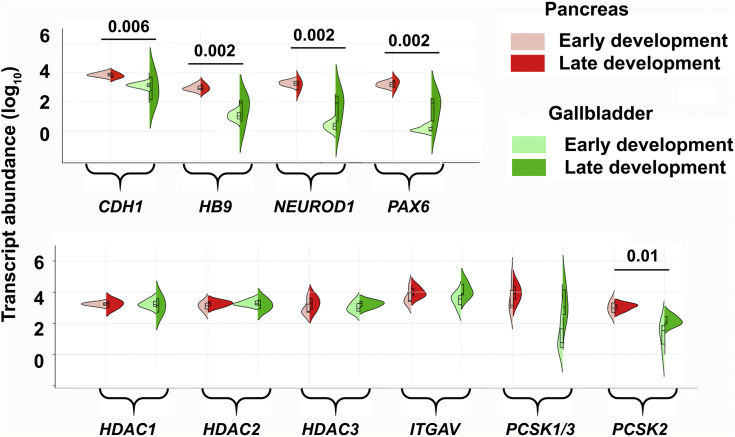
We extended our mouse studies to developing human pancreas and gallbladders accessible from 11 fetuses at early (16–20 weeks gestational age [WGA]; n = 5) or late stages (>20 WGA; n = 6) (Figure 3A) of pregnancy. In comparison with pancreatic tissue, all the islet hormones (INS, GCG, and SST) were expressed at significantly lower abundance (P < .001) (Figure 3B) in gallbladder tissues throughout the embryonic development. Differences between islet and gallbladder (pro)hormone transcript levels were significant across early (16–20 WGA; P < .01; n = 5) (Figure 3B), but not later, stages (>20 WGA; n = 6) (Figure 3B). Histologic (Figure 3C and D) and confocal microscopy analysis (Figure 3E and F) of >20 WGA human pancreas and gallbladders confirmed tissue-specific morphology and the presence of hormone-containing cells. Although insulin-producing cells are localized mostly to cell clusters within pancreatic islets, they are interspersed among columnar epithelial cells lining the gallbladder. We then compared the expression of pancreas-enriched transcription factors and receptors between matched pairs of gallbladder and pancreas samples from 4 human fetal donors (Figure 3G). Unsupervised bidirectional hierarchical clustering separated fetal pancreas and fetal gallbladders into 2 distinct groups (Figure 3G). We also validated the expression of selected pancreatic genes in remaining human pancreatic and gallbladder fetal tissues. Unlike mouse development, the significantly lower (P = .04) level of human NEUROG3 in developing gallbladder cells was not associated with any differences in HES1 expression (Figure 3H). Although levels of GCK gene transcripts were significantly higher in the developing pancreas (P = .02) (Figure 3I), GLP1R and the glucose transporter GLUT2 (also known as SLC2A2) were similar across these developing tissues (Figure 3I). Several other genes (CDH1, HB9, NEUROD1, PAX6, and PCSK2) were expressed at significantly higher levels in the developing human pancreas as compared with their levels in gallbladders (Figure 4). No significant differences were observed across the expression of zinc finger transcription factors (GATA4 and GATA6) (Figure 3H), islet integrin αV (ITGAV), and the histone deacetylases (HDAC1-3) (Figure 4).
Figure 3.
Human gallbladders show several markers of pancreatic endocrine lineage during embryonic development. (A) Schematic of developing human fetus. Pancreas and gallbladder tissues were available from a total of 11 different fetuses, which were classified as early (<20 WGA, n = 5) or late (>20 WGA, n = 6) development. (B) TaqMan-based real-time qPCR for key pancreatic endocrine hormones (INS, GCG, and SST) in developing human pancreas and gallbladder tissues are plotted based on their developmental stage. Split violin plots compared gene expression in early vs late gallbladders (shades of green) and pancreas (shades of red). The horizontal line within each bar of the split violins represents the median, bars extend to quartiles, and the polygons represent the density of data points and extend to minimum/maximum values. Data were analyzed using Kruskal–Wallis with the Dunn multiple comparisons tests. ∗∗∗P ≤ .001 denotes a significant difference between all pancreas and all gallbladder samples, #P ≤ .01 denotes a significant difference between the early pancreas and early gallbladder samples. (C and D) Representative H&E-stained images of human fetal pancreatic islet and gallbladder sections (in greyscale). (E and F) Immunostaining of insulin (green) and glucagon (red) in the human pancreatic islet and gallbladder epithelial cells from late development. Nuclei (DNA) are shown in blue. (C–F) Scale bar: 20 μm. (G) An unsupervised bidirectional hierarchical plot of 48 important pancreatic genes and transcription factors in the same human fetal pancreas and gallbladder tissues (n = 4; indicated by the sample number) using Euclidean distance metric and average linkage is presented. Heat map represents normalized qPCR Ct values (colored bar) of each gene (gene symbol listed on the right vertical axis) with low Ct values/high expression in orange–red color and higher Ct values/low expression in shades of yellow to white. (H and I) Real-time qPCR data from the human fetal pancreas (n = 5–11) and gallbladder (n = 3–11) for pancreatic genes and transcription factors, analyzed using 2-way analysis of variance with the Fisher's Least Significant Difference (LSD) test. The horizontal solid red line within each polygon represents the median, the horizontal black dotted line represents quartiles, and the polygons represent the density of data points extending to the minimum/maximum values.
Figure 4.
Similarities and differences in developing human pancreatic and gallbladder cells. Pancreas and gallbladder tissues from different (N = 11) human fetuses were classified as early (<20 WGA, n = 5) or late (>20 WGA, n = 6) development. TaqMan-based real-time qPCR for pancreatic/epithelial gene transcripts and histone deacetylation enzymes in developing human pancreas and gallbladder tissues are presented and analyzed using Kruskal–Wallis with the Dunn multiple comparisons test. Split violin plots compared the gene expression in early vs late gallbladders (shades of green) and pancreas (shades of red). The horizontal line within each bar of the split violins represents the median, bars extend to quartiles, and the polygons represent the density of data points and extend to minimum/maximum values. The P values represent a 2-tailed Welch test between all (early and late) gallbladder vs pancreas samples. No statistically significant differences were observed within the developmental groups (early vs late), for either of the pancreatic or gallbladder tissue samples for any of the gene transcripts shown.
Similarities in the Developing Human Pancreas and Gallbladders Are Retained in Adult Life
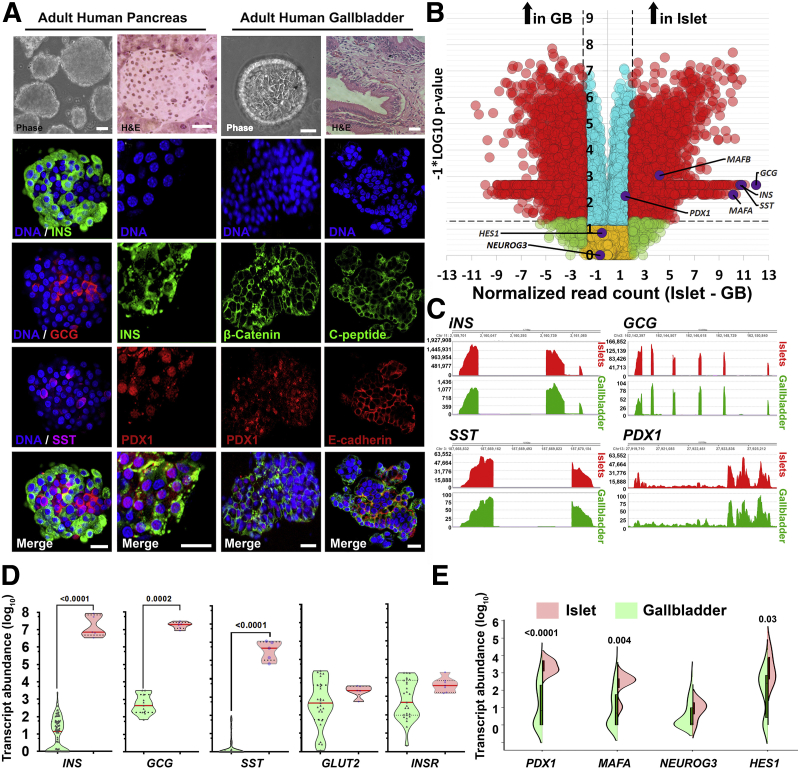
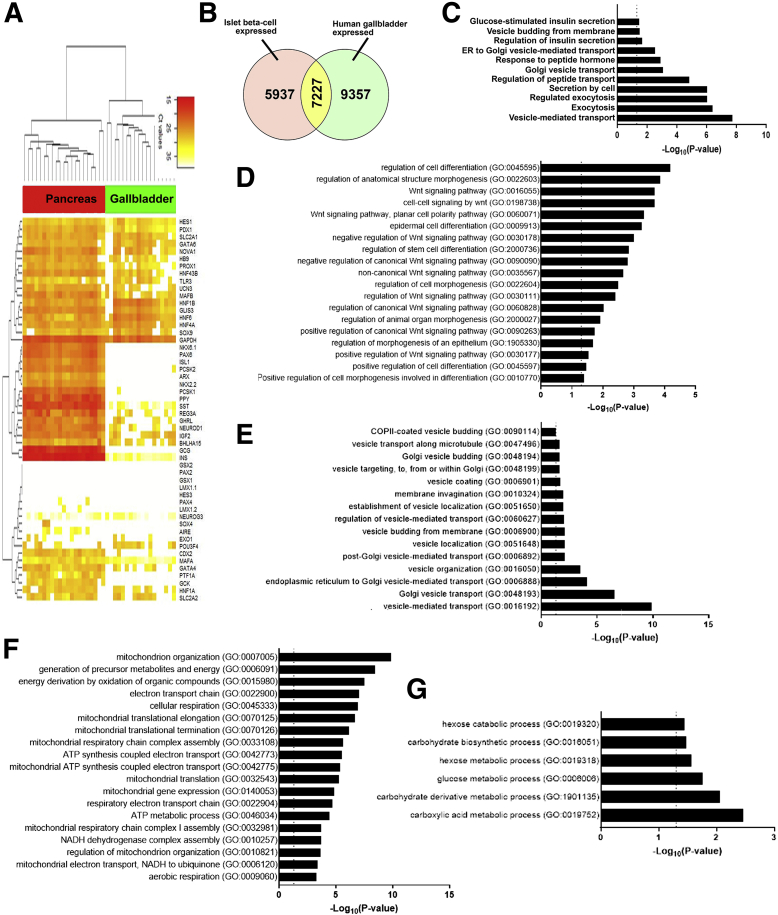
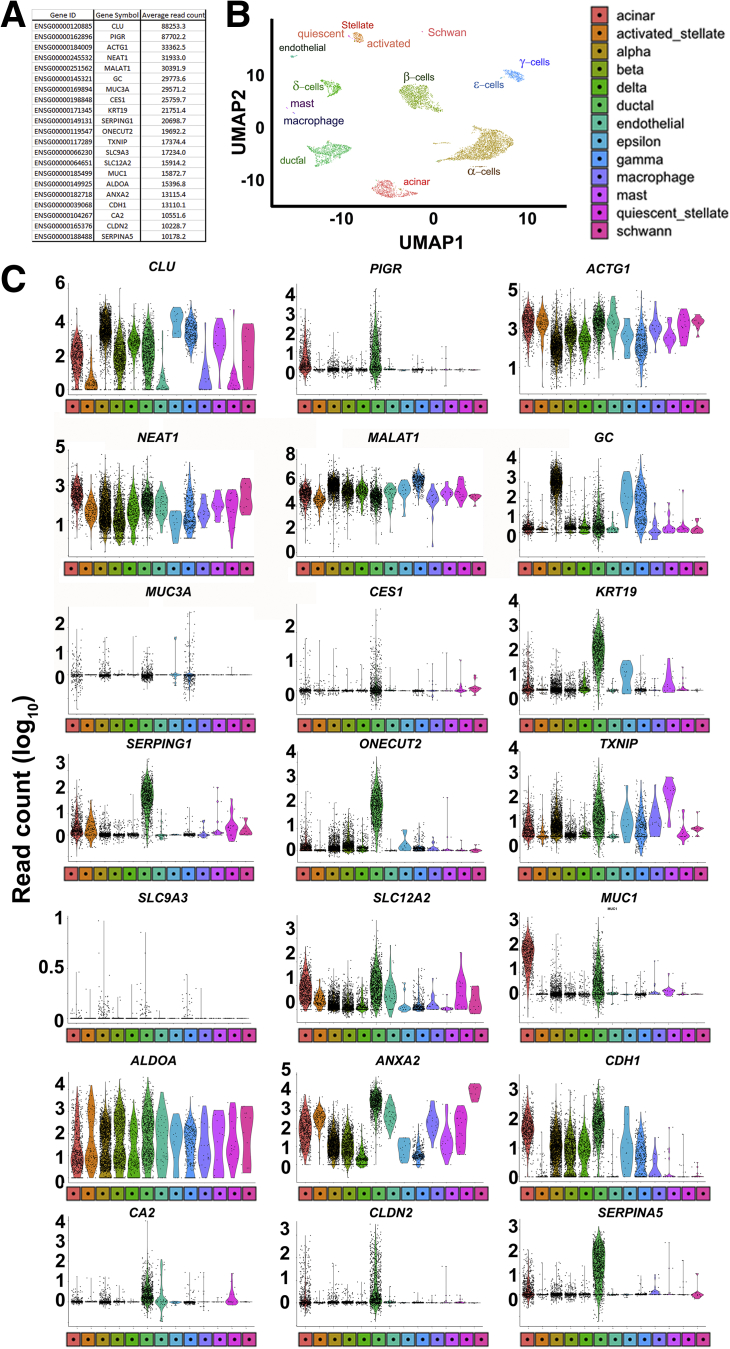
Bright-field and confocal microscopy of adult human pancreatic islets and gallbladder epithelial cells confirmed that the morphology and islet-specific protein production observed in fetal stages is retained in adult human gallbladder cells (Figure 5A). Adult human islets contain mature, hormone-producing cells that also present with Pancreas and Duodenal Homeobox gene 1 (PDX1)-immunopositivity, which is largely localized to the nuclei. Similar to isolated islets, adult human gallbladder epithelial cells isolated as epithelial sheets (see the Methods section) form hollow spheres of epithelial cells, which are immunopositive for C-peptide and PDX1 (Figure 5A), confirming the capacity of the gallbladder epithelial cells to process the prohormone into mature insulin and C-peptide. Similar to human pancreatic islets,14 gallbladder-derived epithelial cell clusters show immunopositivity for E-cadherin and β-catenin (Figure 5A). Bulk RNA-seq in freshly isolated adult human islets and gallbladder epithelial cells confirmed a large number of genes that are significantly different across these functionally diverse tissues (Figure 5B). RNA-seq coverage maps confirm that adult human gallbladder transcripts for INS, GCG, SST, and PDX1 mapped to the same genomic regions (Figure 5C) as in islets, although with a lot fewer copies in gallbladder cells. Validation in a different set of the freshly isolated adult human islet and gallbladder samples using TaqMan-based real-time qPCR (Figures 5D and E, and 6A) confirmed that islet hormones (INS, GCG, and SST) were at significantly higher (several hundred- to thousand-fold) levels in human islets, relative to those in gallbladder epithelium, while the expression of GLUT2 and INSR were comparable (Figure 5D). Pancreatic transcription factors (including PDX1 and MAFA), but not NEUROG3, were significantly more abundant in pancreatic islets (Figure 5E). We compared our human gallbladder RNA-seq data with a publicly available RNA expression data set of human laser capture microdissected β-cells from nondiabetic individuals (E-GEOD-20966).15 Although a large number of gene transcripts were significantly different, interestingly, several gene transcripts retained similar expression levels across these 2 functionally diverse tissues (Figure 6B, Supplementary Table 1). Gene ontology (GO) analysis of gallbladder transcripts filtered for β-cell expression indicated several GO terms that are relevant to insulin packaging and secretion (Figure 6C), cellular development/differentiation (Figure 6D), vesicle transport (Figure 6E), mitochondrial structure and function (Figure 6F), as well as carbohydrate metabolism including glucose metabolic processes (Figure 6G). Parallel with these analyses, we probed if gallbladder-enriched genes were expressed in human pancreatic islet cell subsets. We mapped the expression of 21 highly abundant human gallbladder-enriched gene transcripts identified through our RNA-seq data sets (Figure 7A) to publicly available pancreatic single-cell RNA-seq data sets (Figure 7B). Intriguingly, 17 of the 21 gene transcripts were present in acinar and/or ductal cells, while approximately 50% (10 of 21) of the gallbladder-enriched gene transcripts also were transcribed by pancreatic α- and/or β-cells (Figure 7C). In summary, our data show that the inherent capacity of human gallbladder epithelial cells for pro-endocrine gene transcript and protein expression (albeit at low levels), is retained in the post-natal stage and that the adult human α- and β-cells transcribe several of the gallbladder-enriched gene transcripts.
Figure 5.
Adult human gallbladder cells express low levels of pancreatic islet hormones. (A) Representative phase-contrast images, as well as H&E images, of adult human pancreatic islets and gallbladder epithelium, are shown in the top panel. Confocal images for insulin/C-peptide, glucagon, somatostatin, PDX1, β-catenin, or E-cadherin immunostaining are presented. Nuclei (DNA) are shown in blue. Scale bar: 20 μm. (B) Volcano plot for bulk RNA-seq data from the adult human gallbladder (n = 7) and islet samples (n = 6). Normalized read count difference between islet and gallbladder are plotted on the X-axis and statistical significance (−log10P value) is shown on the Y-axis. The dashed horizontal line represents P = .05, whereas the dashed vertical lines represent a 2-fold difference. Significantly altered (P ≤ .05 and ≥2-fold difference) transcripts are presented in red. Important pancreatic genes are labeled and highlighted in purple. (C) Coverage plots (obtained from bulk RNA-seq in panel B) for INS, GCG, SST, and PDX1 expression across their gene regions (shown at the top of each plot) in human islets and gallbladder. Y-axis presents read counts. (D and E) Real-time PCR data from human islets (n = 5–6) and gallbladder (n = 27–98) for pancreatic genes, analyzed using the Mann–Whitney test. (D) Each dot in the violin plot represents a different sample. The horizontal solid red line within each polygon represents the median, the horizontal black dotted line represents quartiles, and the polygons represent the density of data points, extending to minimum/maximum values. (E) The horizontal line within each bar of the split violins represents the median, bars represent quartiles, and the polygons represent the density of data points, extending to minimum/maximum values. The exact P value for the significant differences between pancreas and gallbladder are presented. GB, gallbladder.
Figure 6.
Gene expression pathways enriched in adult human gallbladder epithelial cells. (A) An unsupervised bidirectional hierarchical cluster of 51 genes known to be associated with or necessary for normal pancreas development/function, profiled in adult human pancreatic islets (red, n = 21) and human gallbladder epithelial cells (green, n = 18) was plotted using Euclidean distance metric and average linkage. The heat map representing normalized qPCR Ct values (color bar) for each gene (listed on right Y-axis) with low Ct values/high expression in orange–red color and higher Ct values/low expression in shades of yellow to white. Data consist of measurement on ViiA7 (INS, GCG, MAFA, NEUROG3, HES1, and PDX1) or using a custom TaqMan OpenArray platform. (B) Venn diagram showing the number of genes common between our bulk gallbladder RNA-seq (n = 6) and the publicly available data set of human islet β-cells (see the Methods section). (C-G) The most significant and relevant Biological Process GO categories enriched in gallbladder cells are presented. Data obtained from RNA-seq was used to identify the gallbladder-expressed genes and total gallbladder transcripts were filtered for β-cell–expressed transcripts obtained from E-GEOD-20966. The X-axis represents −log10 P value, the dotted vertical line represents the significant P value = .05, and relevant pathways are provided on the Y-axis. Pathways involved in (C) endocrine pancreatic β-cell function, (D) development and differentiation, (E) vesicle transport, (F) mitochondrial function, and (G) carbohydrate metabolism are presented. ATP, adenosine triphosphate; COPII, Coat protein complex II; NADH, Nicotinamide adenine dinucleotide phosphate.
Figure 7.
Human pancreatic single-cell RNA-seq shows the expression of gallbladder-enriched genes. (A) Gene ID, gene symbol, and average read count of 21 highly abundantly human gallbladder-enriched gene transcripts. (B) Uniform Manifold Approximation and Projection (UMAP) plot generated using Seurat (version 0.2.1) from human pancreatic single-cell RNA-seq (Panc8) data (see the Methods section for details). The single cells were clustered into 13 different pancreatic cell types, and each cluster is highlighted with a different color. (C) Violin plots showing the expression of each gene listed in panel A across all human pancreatic cell types presented in panel B. The Y-axis represents read counts (expression) of the gene transcript presented in each panel for the subset of human pancreatic cell type shown in panel B. The shape of the violin plot represents the density of individual data points and extends to minimum/maximum values. The color of the violin plot reflects the color and cell type from the UMAP plots in panel B.
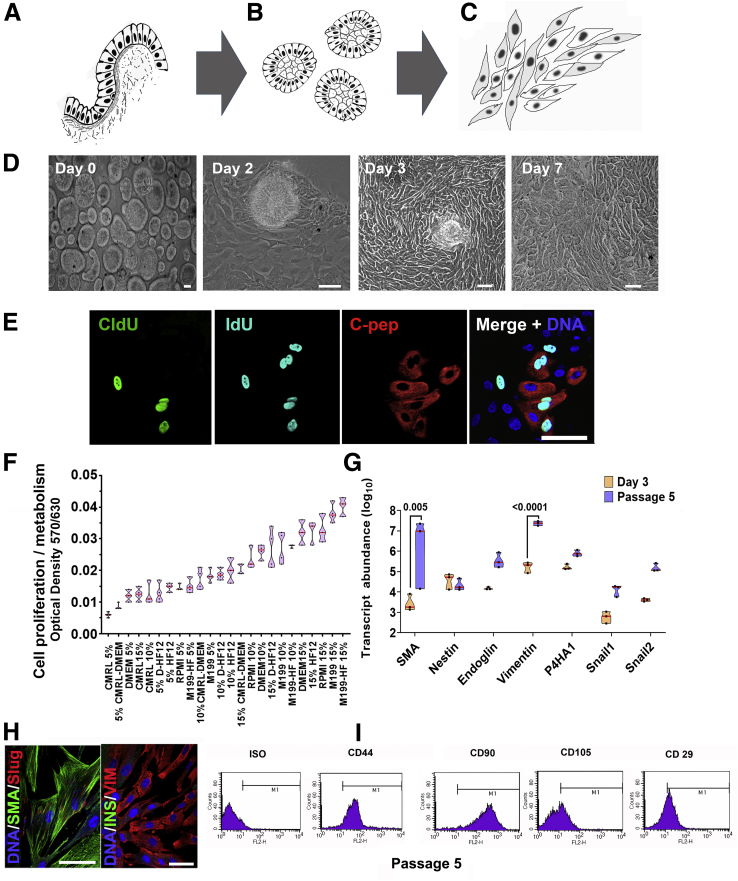
Adult Human Gallbladder Epithelial Cells Can Proliferate and Differentiate In Vitro
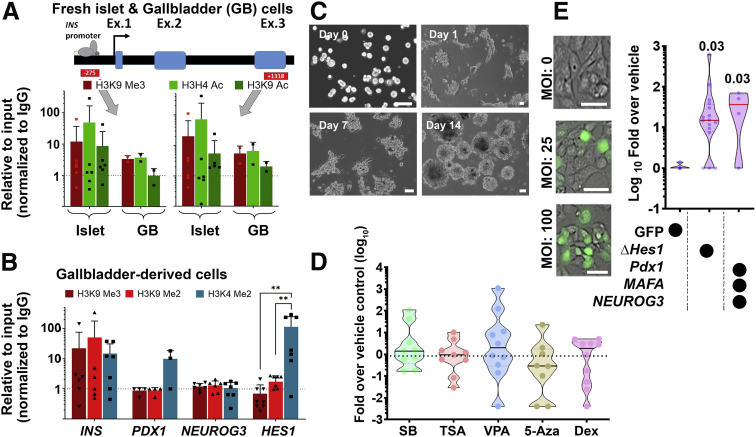
We established a unique protocol to isolate gallbladder epithelium without enzymatic digestion via gentle scraping of gallbladder epithelial cells using a sterile scalpel blade (Figure 8A). The epithelial sheets obtained by scraping, close on themselves to form hollow, epithelial clusters of cells (Figure 8B), which migrate and proliferate in vitro as mesenchymal-like cells (Figure 8C). Actual phase-contrast micrographs of this process are shown in Figure 8D. We exposed freshly isolated human gallbladder epithelial cells (as shown in Figure 8B or day 0 of 8D) to 2 consecutive pulses of different thymidine analogs; 5-chloro-2-deoxyuridine (CldU) and 5-iodo-2-deoxyuridine (IdU), added sequentially to cell culture media before their detection using specific antibodies. Using this unbiased, dual-thymidine, analogue-based, cell lineage tracing technique,14,16 we detected the presence of CldU+, IdU+, and C-peptide+ cells (Figure 8E) in gallbladder epithelial cells, propagating under optimal cell culture conditions (Figure 8F). This confirmed that insulin-producing cells in gallbladder epithelium contribute to the proliferating subsets of gallbladder-derived cells. Eventually, all proliferating populations of gallbladder epithelial cells acquire a mesenchymal-like phenotype and begin expressing mesenchymal gene transcripts (Figure 8G) and proteins such as smooth muscle actin and vimentin (Figure 8H). Interestingly, freshly isolated epithelial clusters had lower expression levels of these mesenchymal genes that significantly increased in abundance from passage 0 to passage 5 (Figure 8G). Flow cytometry–based analysis of passage 5 gallbladder-derived cells showed the presence of typical surface antigens expressed by mesenchymal stem cells such as for CD29, CD44, CD90, and CD105 (Figure 8I). We then assessed the chromatin landscape at the insulin gene in freshly isolated human islets and gallbladder clusters. At 2 different sites of insulin gene (-275 and +1318), human islets showed open chromatin conformation as confirmed by higher levels of H3H4Ac and H3K9Ac, histone modifications associated with active genes along with lower levels of H3K9 trimethylation, associated with silenced/inactive genes. In the freshly isolated gallbladder epithelial cells, these chromatin modifications were not significantly different from those observed in adult human islets (Figure 9A). Analysis of pancreatic gene promoters in passages 5–10 of gallbladder-derived mesenchymal cells showed a more inactive chromatin conformation (H3K9-Me3 and H3K9-Me2) at insulin and neurogenin 3 promoters, whereas an open/accessible chromatin conformation (significantly higher H3K4-Me2) was seen at the HES1 gene promoter (Figure 9B). Interestingly, the PDX1 gene promoter retained an active/open promoter conformation in these cells (Figure 9B). These data corroborate the lack of insulin expression in gallbladder-derived mesenchymal cells and support the potential for re-expression of insulin gene transcripts through chromatin conformational changes or via repressing HES1 expression.
Figure 8.
Adult human gallbladder-derived cells can be induced to differentiate in vitro. (A–C) Schematic of isolating and propagating adult human gallbladder epithelial cells in vitro. (A) The inner epithelial cell lining of the gallbladder is scraped off with a sterile scalpel. (B) The resulting epithelial sheet curls up into clusters, which then (C) adhere to culture plates and proliferate/propagate as mesenchymal cells. (D) Representative phase-contrast images of the process described through panels A–C; epithelial clusters from adult human gallbladders on day 0, attach (day 2), migrate out (day 3), and continue to grow/proliferate (day 7) in vitro. (E) Immunostaining of CldU (green), IdU (aqua), and C-peptide (red) in cultures of gallbladder epithelial cells after 7 days, involving 3-day exposure to CldU, 1 day of wash-out, and then 3 days of incubation in IdU-containing medium. Nuclei (DNA) are shown in blue as a representative from N = 10 preparations. (F) Optical density/absorbance after an MTT assay in isolated gallbladder epithelial cells in the presence of different growth-supporting conditions is shown on the Y-axis. Culture media and percentages of fetal bovine serum compositions are shown on the X-axis. Data were obtained from at least 3–4 different gallbladder preparations set up in a 96-well plate and each experiment is presented as a single data point. (G) TaqMan-based real-time qPCR for mesenchymal genes in freshly isolated gallbladder epithelial cells and in vitro propagated gallbladder-derived mesenchymal cells. Experiments were performed at least 3 times using different biological samples and correspond to individual data points within the violins. Transcript abundance was calculated as described in the Methods section and statistical significance between groups was assessed using 2-way analysis of variance with a P value presented after adjustment using the Sidak multiple comparisons test. (F and G) Polygons represent density distribution extending to minimum/maximum values with a horizontal red line at the median. (H) Proliferating human gallbladder-derived cells express mesenchymal proteins in vitro. Nuclei (DNA) are shown in blue. (I) Representative flow cytometry plots of gallbladder-derived mesenchymal cells at passage 5 for mesenchymal surface antigens. M1 gating indicates antigen-positive cells that are outside of the isotype control (ISO). The same acquisition and gating parameters were used while comparing isotype and antibody-stained cells. Experiments were repeated at least 3 times with different gallbladder donor cell preparations. Scale bars: 20 μm. C-pep, C-peptide; CMRL, Connaught Medical Research Laboratories; DMEM, Dulbecco’s modified Eagle medium; FL2-H, fluorescence channel 2 height; HF, Ham's F-12; SMA, smooth muscle actin.
Figure 9.
Characterizing adult human gallbladder epithelial cells in vitro. (A) One site (-275) in the insulin promoter region and another (+1318) in the exon 3 region were assessed for the presence of H3K9 Me3 (inactive mark) as well as H3H4 Ac and H3K9 Ac (active transcription marks) using ChIP assay in freshly isolated human islets (n = 6) and gallbladder epithelial cells (n = 2, pooled from 3 to 5). (B) Promoter regions of insulin, PDX1, NEUROG3, and HES1 were assessed for the presence of H3K9 Me3 and H3K9 Me2 (inactive marks) as well as H3K4 Me2 (active transcription mark) using ChIP assay in passages 5–10 of gallbladder-derived mesenchymal cells (n = 3–6 different biological preparations). Significance was calculated using 2-way analysis of variance with the Tukey multiple comparisons test. ∗∗P < .01. (A and B) Data are presented relative to input and normalized to IgG. The dotted line represents expression in isotype control (IgG) samples. Data in bar graphs present means ± SD and individual points present a different sample/pool. (C) Representative phase-contrast images of the differentiation process involving trypsinization (day 0), reaggregation (day 1), and cluster formation (days 7–14). (D) Real-time qPCR data for insulin genes in differentiated gallbladder cells using 5 different DNMT (DNA methyltransferase) and HDAC (Histone deacetylases) inhibitors/hypomethylation agents. Data are presented as fold-over vehicle control from 9 to 10 different biological samples/experiments. (E) Phase-contrast images of transduction efficiency using GFP-expressing adenoviral vectors. Real-time qPCR data for insulin genes in differentiated gallbladder cells using a vehicle (GFP alone) or a dominant-negative HES1 or a combination of PDX1, NEUROG3, and MAFA. Data are presented as fold-over vehicle control from 4 to 15 different biological samples and analyzed using the Kruskal–Wallis test. The exact P values after adjusting for multiple comparisons are presented. (D and E) Each dot in the violin plots represents a different sample. The horizontal solid line within each polygon represents the median, the horizontal black dotted line represents quartiles, and the polygons represent the density of individual data points and extend to minimum/maximum values. Scale bars: 20 μm. 5-Aza, 5-aza-2′-deoxycytidine; Dex, dexamethasone; GB, gallbladder; SB, sodium butyrate; TSA, trichostatin A; VPA, valproic acid.
We, therefore, assessed endocrine differentiation of gallbladder-derived mesenchymal cells using small molecules that are known to be DNA methyltransferase inhibitors or histone deacetylase inhibitors17, 18, 19 and/or via forced expression of a dominant-negative (Δ)HES1, or of the pro-endocrine transcription factors (PDX1, MAFA, and NEUROG3). Although differentiation to insulin-expressing islet-like clusters was observed over 14 days in the serum-free differentiation medium (Figure 9C), none of the small molecules (sodium butyrate, trichostatin A, valproic acid, 5-aza-2’-deoxycytidine, and dexamethasone) induced significantly higher insulin expression compared with vehicle controls (Figure 9D). On the other hand, we obtained a consistent and significant increase in INS transcript expression using both (ΔHES1 and pro-endocrine transcription factor) overexpression strategies (Figure 9E). Our data support the view that gallbladder/biliary duct cells can be differentiated to promote insulin expression.
Functional Analyses of Insulin-Producing Human Gallbladder Cells
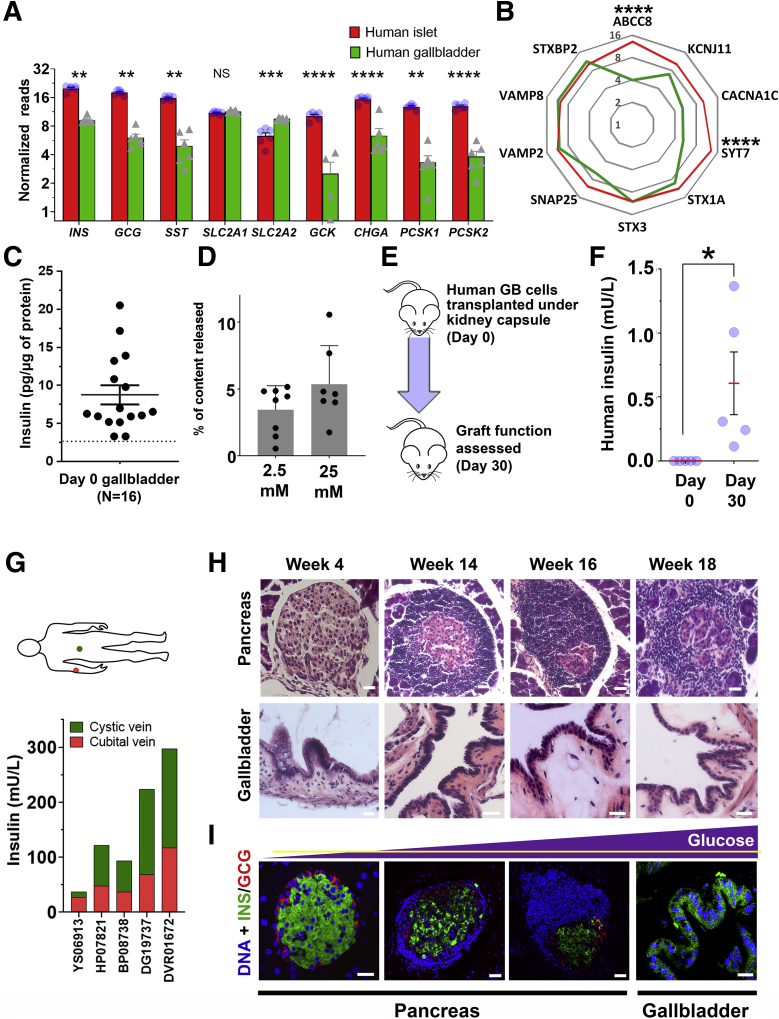
To extend our understanding of insulin secretion from gallbladder epithelial cells, we planned a series of experiments using freshly isolated adult human gallbladder epithelial cells. We compared genes known to be important in insulin sensing and exocytosis using bulk RNA-seq data of adult human islet and gallbladder epithelial cells. Although gene transcripts of islet hormones, chromogranin A, prohormone processing enzymes (PCSK1, PCSK2), and the glucose sensor (GCK) were significantly lower in gallbladder epithelial cells, the transcripts of the 2 glucose transporters were either similar (for SLC2A1) or significantly higher (for SLC2A2) in adult gallbladder epithelial cells (Figure 10A) than those in adult human islets. Except for pancreatic adenosine triphosphate–sensitive K+ channel ABCC8 (or SUR1) and the transmembrane protein synaptotagmin-7 (SYT7), gallbladder epithelial cells and human islets contained similar levels (Figure 10B) of gene transcripts for KCNJ11 (or KIR6.2), the L-type calcium channel CACNA1C (encoding CAV1.3 protein), the islet syntaxins and binding proteins (STX1A, STX3, and STXBP2), the synaptosomal-associated protein (SNAP25), and gene transcripts encoding the vesicle-associated membrane proteins (VAMP2 and VAMP8). These comparisons (Figures 5G and 10B) indicate that gallbladder epithelial cells express the set of genes necessary for insulin exocytosis. Freshly isolated gallbladder epithelial cells (Figure 10C) contain insulin protein and also release insulin/C-peptide in response to glucose (Figure 10D), indicating successful processing and secretion of insulin after glucose exposure in vitro. We then transplanted freshly isolated adult human gallbladder epithelial cells under the kidney capsule of immunocompromised Non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice (Figure 10E). A functional assessment performed on day 30 using intraperitoneal glucose injection resulted in detectable levels of human insulin in mouse circulation, which was not seen immediately after transplantation/day 0 (Figure 10F). To understand in situ insulin secretion from human gallbladder cells, we collected blood from the median cubital vein (peripheral) and cystic vein (gallbladder) at 30 minutes after intravenous glucose injection from individuals who were enrolled for cholecystectomy. Intriguingly, insulin levels in the cystic vein were higher than those in the cubital vein for 80% (n = 4 of 5) individuals assessed in this study (Figure 10G). Overall, we show that naturally occurring insulin-secreting cells within the gallbladder have relevant insulin secretory machinery and can respond to physiological changes in glucose concentrations. However the amount of insulin secreted from these cells (Figure 10C) is not physiologically relevant to maintain normal glucose concentrations, and is significantly lower than that reported in human islets.20
Figure 10.
Gallbladder epithelium contains functional insulin-producing cells. (A) Expression of key pancreatic hormones, chromogranin A, glucose transporters, and glucose-sensing, as well as (pro)insulin-processing enzyme gene transcripts in adult human islets (n = 6) and adult human gallbladder (n = 4–6) samples. Data were obtained from bulk RNA-seq and presented as normalized read counts. Bar graphs present means ± SEM and differences between groups were assessed using 2-tailed t tests. (B) Expression of secretory function-related genes in human islets (n = 6) and gallbladder (n = 7) is presented as normalized mean read counts in radar plot (red line, pancreas; green line, gallbladder). Data were analyzed using the Kruskal–Wallis test followed by adjustment for multiple comparisons using the Dunn test. (C) Insulin content in freshly isolated human gallbladder epithelial cells. Data were obtained from N = 16 different biological samples. Each dot in the scatter plot indicates a separate sample, with lines indicating means ± SEM. (D) C-peptide release in response to 2 different concentrations of glucose is presented as the percentage of total content. Data are from 7 to 8 different freshly isolated human gallbladder cells (individual points in scatter bar graph) and are presented as means ± SD. (E) Human gallbladder epithelial cells were isolated and transplanted in NOD/SCID mice and functional assessment was performed on day 0 and day 30. (F) Human insulin was measured in mouse serum. Data were obtained from N = 5 different gallbladder samples transplanted in mice. Each dot in the scatter plot represents a different sample and the red line and error bars represent means ± SEM. Significance was calculated using the Welch t test. (G) Circulating insulin concentrations in cystic (green) or median cubital (red) veins were measured in 5 individuals (labeled on the X-axis). Stacked bars represent the insulin concentration from cystic (gallbladder shown in green) and peripheral (cubital shown in red) veins. (H) Representative H&E images of pancreas and gallbladder sections obtained from NOD mice of different ages. (I) Immunostaining of insulin (green) and glucagon (red) in islet and gallbladder sections obtained from NOD mice at different ages with increasing concentration of circulating glucose (yellow horizontal line showing normoglycemic levels in the purple triangle representing increasing glucose concentrations). Nuclei (DNA) are shown in blue. Scale bars: 20 μm. ∗P ≤ .05, ∗∗P < .01, ∗∗∗P ≤ .001, and ∗∗∗∗P ≤ .0001. GB, gallbladder.
Insulin-Producing Gallbladder Epithelial Cells in Diabetes
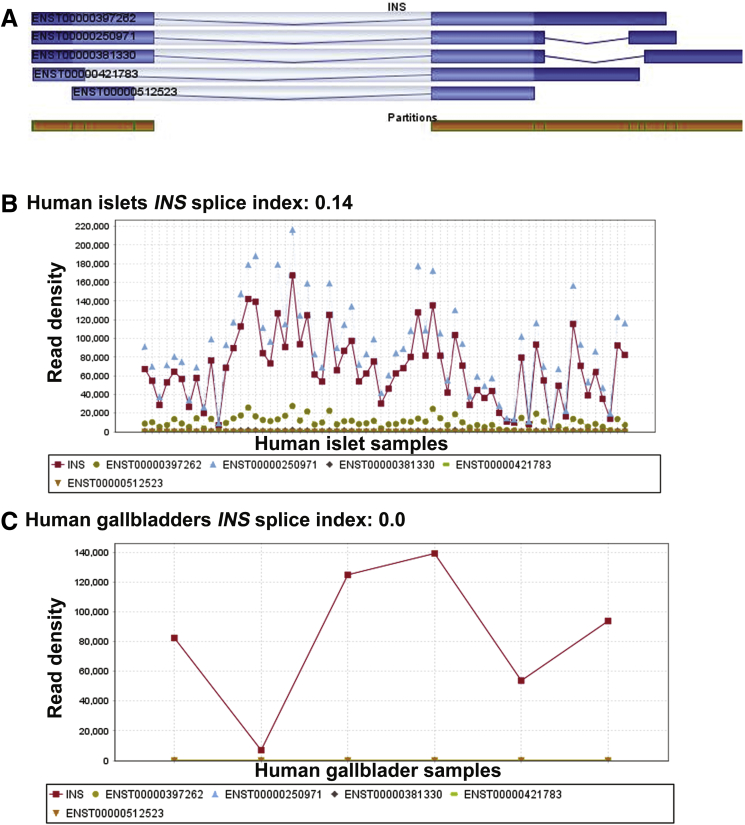
To understand if insulin-containing gallbladder epithelial cells also elicit an immune-mediated attack during diabetes progression, we examined immune infiltration in gallbladder and islets from female NOD mice. Because NOD females show significant infiltration at 12–14 weeks of age, we obtained gallbladder and pancreas from NOD mice at 4, 14, 16, and 18 weeks of age (Figure 10H). Predictably, the pancreas from these mice showed increasing infiltration of immune cells (Figure 10H) at 14–18 weeks of age, but no infiltration was observed in their gallbladders. Immunostaining for islet hormones confirmed the decreasing number (quantitative data not shown) of insulin-producing cells in the pancreas as plasma glucose increased (Figure 10I, yellow horizontal line for normoglycemic levels). Insulin-immunoreactive cells were present in gallbladder epithelial cells even when NOD mice showed high glucose levels at 18 weeks (Figure 10I), indicating that gallbladder epithelial cells can potentially escape immune recognition during type 1 diabetes progression in NOD mice. This interesting observation could be owing to a different autoimmune antigenic repertoire between islet β-cells and gallbladder insulin-producing cells. Analysis of splice variants in gallbladder (GSE152419, n = 7) and human islet (GSE152111, N = 66; and GSE134068, N = 18) RNA-seq data sets indicate a high differential splice index for human insulin variant (ENST00000250971) in islets (splicing index, 0.14, N = 66 islets or splicing index, 0.17, N = 18 islets) compared with no differential splicing observed in human gallbladder epithelial cells (ENST00000250971; splicing index, 0.0; n = 7) (Figure 11).
Figure 11.
Analysis of INS splice variants in human islet and gallbladder samples. Differential splicing was analyzed on the INS gene separately for human islet samples (GSE152111, N = 66) and gallbladder samples (GSE152419, n = 7). (A) The 5 INS transcript variants, along with partition coverage for INS gene (orange). The INS transcript splice variant profile is shown across (B) human islet samples and (C) gallbladder samples. (B and C) The 5 INS transcript splice variants, INS-201 (ENST00000250971, INS-202 (ENST00000381330), INS-203 (ENST00000397262), INS-204 (ENST00000421783), and INS-205 (ENST00000512523), along with the INS gene (red line, as per color legion), are shown. Data are presented in read density, which is the proportion of all reads associated with the gene computed for the quantification allocated to each transcript of the gene fractionally.
Discussion
The origin/proximity of the gallbladder endoderm to the developing pancreas is evolutionarily conserved across lower vertebrates.21,22 The toadfish has a single primary islet located in the dorsal region of the gallbladder,21 with smaller islets embedded in the mesenteric part of the gut. Zebrafish has one of its pancreatic lobes along the intestine–gallbladder–spleen tissue axis,22 while in Lake Van fish, pancreas is observed next to the gallbladder and bile duct.23 Ectopic pancreas in gallbladder,24 somatostatinomia in human extrahepatic biliary tract,25 and glucagon expression in liver and bile ducts26 are reported. All of these studies suggest evolutionary conserved common lineage of pancreas and gallbladder.
Insulin hormone immunoreactivity has been reported in body fluids other than blood.27 The presence of insulin in human gallbladder was reported in the 1960s, when insulin immunoassays were available to reliably measure insulin. These studies28,29 indicated significantly higher levels of insulin in the bile from the gallbladder, suggestive of a selective reabsorption or active production of insulin by gallbladder epithelium. Our quantitative studies (PCR and enzyme-linked immunosorbent assay [ELISA]) show that insulin is indeed produced in gallbladder epithelial cells. Immunostaining is a qualitative technique that provides a relative measure of immunopositive cells and the intensity of fluorescence depends on the acquisition settings. Our immunostaining data show that insulin protein is present in the gallbladder epithelium, while the quantitative data emphasize that it is significantly lower than in the islets. However, in a smaller number of individuals (N = 5) the presence of insulin in the cystic duct after glucose administration (Figure 10G) corroborates earlier observations28,29 of insulin immunoreactivity within the gallbladder.
A significant body of evidence has shown that biliary-/gallbladder-derived stem/progenitor cells can produce insulin after recombinant PDX1,30 adenovirus-mediated transcription factor overexpression,7,31,32 or exposure to small molecules/growth factors.33,34 Several groups have reported their experience in propagating gallbladder epithelial cells from a range of species including mouse, rabbit, guinea pig, dog, bovine, and human beings.35, 36, 37, 38, 39 We used a nonenzymatic approach to isolate gallbladder epithelial cells. In line with previous reports, the overexpression of key pancreatic transcription factors (PDX1, MAFA, and NEUROG3)40 or HES1 inhibition7 in gallbladder-derived cells induced insulin expression.
Our studies in NOD mice, which naturally develop immune-mediated T1D, indicate that gallbladder epithelial cells do not show immune infiltration in T1D and that these cells may continue to produce small amounts of insulin. Considering recent reports of mechanisms contributing to autoimmunity in diabetes,41,42 a lower abundance of insulin, as well as the absence of alternate insulin splice forms (Figure 11) in the gallbladder, is encouraging. Genetic or environmental factors leading to endoplasmic reticulum (ER) stress are of critical importance in regulating the expression of alternative splice variants via alternative reading frames of insulin.43,44 Metabolic or inflammatory stress may lead to the generation of novel splice forms via recognition of a newly created splice site45 and the generation of defective ribosomal peptides.42,46 Such defective ribosomal peptides may act as neoantigens, to which central immune tolerance is absent. Another possibility is high levels of the taurine-conjugated bile acid tauroursodeoxycholic acid in the gallbladder, which is known to reduce ER stress and protect islet β-cells in T1D mouse models.47,48 Tauroursodeoxycholic acid is currently in clinical trials for T1D (NCT02218619), and further studies understanding the potential role of ER stress and defective ribosomal peptides in the gallbladder during T1D progression are merited. Interestingly, all cases of neonatal diabetes with undetectable levels of immunoreactive insulin also report gallbladder aplasia/hypoplasia,49 signifying the common developmental plans for these 2 functionally diverse organs. Future work also can focus on identifying immunoprotective mechanisms in the gallbladder.
The present study had several strengths. This study collectively showed the interspecies and age-related similarities across 2 functionally diverse, but developmentally related, organs: the gallbladder and the pancreas. We present a comparison between the gallbladder and pancreatic cells/samples using multiple techniques to show similarities in chromatin conformation (ChIP-PCR), gene transcription (bulk/single-cell RNA-seq, TaqMan qPCR, gene reporter analyses), insulin production (confocal microscopy, ELISA, immune-electron microscopy), and insulin release (animal models, human cells, and clinical in situ measurements). We also present direct analysis of insulin-producing cells in gallbladder and pancreas from NOD mice.
A limitation was that our studies do not explain the possible mechanisms underlying the biological variation observed in insulin transcript abundance across different samples. None of the gallbladder donors had type 1 or type 2 diabetes, although we do not have data on prediabetes or undiagnosed type 2 diabetes (hemoglobin A1c levels) in these individuals. Future studies would need to assess proinsulin and insulin ratios in the same gallbladder epithelial cell samples to understand the concentrations of processed and mature insulin and be statistically powered to understand if differences are related to ethnic, genetic, disease, or environmental components. There is a consistent presence of Pdx1 transcripts in the majority of gallbladder cells; while variable levels of insulin suggest that in some cells the endocrine differentiation has progressed selectively, while it is inhibited in other cells. Single-cell sequencing technologies for PDX+INS+ and PDX+INS- gallbladder cells would be informative to understand heterogeneity and to tease out regulatory pathways and molecules during gallbladder development. Although the differentiation of gallbladder-derived cells was not the principal aim of this study, differentiation strategies need improvement to enhance insulin production in gallbladder-derived progenitor cells followed by long-term functional validation in animal models of diabetes.
In summary, we show the inherent property of mouse and human gallbladder cells to transcribe, translate, package, and release insulin in response to glucose, although not at physiologically relevant levels. The capacity for insulin gene transcription in gallbladder epithelium appears to be driven by the developmental similarity, which programs pancreatic islet epigenetic and transcriptomic signatures in the gallbladder. The demonstration that this intrinsic insulin production escapes autoimmune damage during T1D progression renews interest in this functionally diverse organ.
Methods
Animals
Friend Virus B NIH Jackson (FVB/NJ) mice were used for embryonic gene expression and insulin content; CD1 mice were used for immune-gold labeling studies at the University of Bath; C57BL/6J mice were used for RNA-seq, qPCR, or ChIP studies; and Pdx1-GFP, MIP-GFP, and NOD mice were used for gene reporter and immunostaining studies. All animals were maintained at the experimental animal facilities in India, the United Kingdom, or Australia according to guidelines outlined by the respective institute’s animal care and use committee. Ethical approvals for the study were obtained from animal ethics committees at the National Centre for Cell Science in India, St. Vincent’s Hospital in Melbourne, and the University of Sydney. Breeding pairs were set, and pregnancy was confirmed by observing vaginal smears. Pregnant females and newborn mice were killed at predefined intervals and pancreatic buds or pancreas, as well as gallbladder tissue, was dissected carefully without cross-contamination using a stereomicroscope. Tissue samples at each of these time points were used for RNA isolation (in TRIzol, Thermo Fisher Scientific, Waltham, MA) and immunostaining (in 4% freshly prepared paraformaldehyde). Animal strain, numbers, age, and comparison groups for each experiment are provided in the respective Figure legends as per Animal Research: Reporting In Vivo Experiments (ARRIVE) guidelines.
Human Tissue Collection
Adult gallbladder, pancreas, and islet samples were obtained after human research ethics committee approvals from the Sydney Local Health District and St. Vincent’s Hospital in Melbourne. Fetal gallbladder and pancreas were obtained after informed consent from prospective parents who consented to fetal tissue for research after elective termination of pregnancy (<20 WGA) or after late abortions/miscarriage (>20 WGA) as per human research ethics committee approvals from the Shree Seva Medical Foundation (India) and the National Centre for Cell Science (India). Human cadaveric nondiabetic pancreas and islet samples were obtained as part of the research consented tissues through the Australian Islet Transplantation Program (at Westmead Hospital in Sydney and at St. Vincent’s Institute in Melbourne). Human gallbladders were obtained as surgical waste tissues (noncancerous) after cholecystectomies from the surgical teams (at Strathfield Private Hospital, Royal Prince Alfred Hospital [Sydney] and St. Vincent’s Hospital [Melbourne], and the National Centre for Cell Science [Pune, India]). Tissue samples were stored for RNA isolation (in TRIzol), immunostaining (in 4% freshly prepared paraformaldehyde), or processed for cell culture as detailed later. For in situ glucose stimulation study, we consented 5 individuals undergoing cholecystectomies for blood collection at the Asian Institute of Gastroenterology. Blood was collected before and after the glucose challenge. The study was performed according to the Declaration of Helsinki II. Study participants provided written informed consent before inclusion as per the ethical committee approval from the Asian Institute of Gastroenterology in India.
Gallbladder Cell Culture
Gallbladder tissues were collected in transport medium (M199 with 25 mmol/L HEPES and 2 mmol/L glutamine medium with 2× antibiotics) and processed in the laboratory. The gallbladder sample was washed thoroughly with the serum-free medium (M199 with 25 mmol/L HEPES and 2 mmol/L glutamine + Ham’s F12k) containing 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco, Carlsbad, CA) and fungizone (Gibco) until bile was removed. The inner surface of the gallbladder then was scraped off gently with a disposable aseptic scalpel blade to isolate the epithelial lining. These isolated cells were washed with the wash medium (described earlier), centrifuged at 1000g for 2 minutes, and the cell pellet was resuspended and plated in serum-containing medium (10% fetal bovine serum + M199 with 25 mmol/L HEPES and 2 mmol/L glutamine + Ham’s F12k) with penicillin and streptomycin (Gibco). This medium is referred to hereafter as a growth-promoting medium or serum-containing medium. Cells were maintained in an incubator at 37°C and with humidified 5% CO2 in the air and passaged 1:2 when confluent using trypsin (Gibco) + EDTA. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay was used to understand the media composition supporting the maximum viability and metabolic activity of isolated cells. Briefly, 50,000 cells were seeded in each well of 96-well plates and allowed to adhere overnight. Different media and serum concentrations were formulated and added to cells (24 conditions) in quadruplets. After 24 hours and 48 hours, MTT reagent (Sigma-Aldrich, St. Louis, MO) was added to each well, and after 2–3 hours conversion of MTT to formazan was measured using a spectrophotometer. The experiment was repeated with 3–4 different gallbladder preparations.
In Vitro Differentiation of Human Adult Gallbladder-Derived Cells
Human adult gallbladder-derived mesenchymal-like cells are trypsinized and differentiated after an optimized protocol in our laboratory.50, 51, 52 Briefly, these cells are plated on day 0 in serum-free medium (day 0 SFM) that consists of Dulbecco’s modified Eagle medium:F12 + 1% bovine serum albumin (BSA) +1× insulin-transferrin-selenium. Single cells on day 0 start to aggregate into islet-like clusters (ICAs). On day 1, the medium was changed with day 0 SFM to remove dead cells. On days 4 and 7, ICAs were exposed to a day 4 medium that contained 0.3 mmol/L taurine + day 0 SFM. On day 10, ICAs were exposed to day 10 SFM that contained nicotinamide (1 mmol/L) and exendin-4 (100 nmol/L), along with day 4 SFM. On day 14 these ICAs were harvested for RNA isolation. In some experiments, DNA methyltransferases (DNMT) and histone deacetylase (HDAC) inhibitors were added during the entire time of differentiation at the following final concentrations: 1 mmol/L sodium butyrate, 100 nmol/L trichostatin A, 1 mmol/L valproic acid, 2 μmol/L 5-aza-2’-deoxycytidine, and 1 μmol/L dexamethasone. Vehicle control (phosphate-buffered saline [PBS] or dimethyl sulfoxide) was used for comparison. Adenoviral vectors for PDX1, NEUROG3, and MAFA were used at 2 different multiplicities of infection in gallbladder-derived cells. Transductions were performed as described earlier.53 Transduced cells were differentiated following the protocol detailed earlier and GFP-alone adenovirus-transduced cells were used for gene expression comparisons.
RNA Isolation, Complementary DNA Synthesis, and Real-Time qPCR
RNA isolation from fresh human and mouse tissues at different stages (embryonic, adult) was performed using TRIzol (Invitrogen, Carlsbad, CA). RNA quality and quantity were measured on a ND-2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Complementary DNA synthesis was performed using a High-Capacity Complementary DNA Reverse-Transcription Kit (Thermo Fisher Scientific, Waltham, MA) as per the manufacturer’s recommendations. Real-time qPCR was performed with TaqMan primer and probes mix (Thermo Fisher Scientific, Foster City, CA) for genes listed in Supplementary Table 2 and TaqMan Fast Universal PCR Master Mix (Thermo Fisher Scientific, Foster City, CA) in a 5-μL reaction in 96-well optical clear plates. The cycle threshold (Ct) values of all the genes were normalized to the housekeeping gene 18s ribosomal RNA. Transcript abundance was calculated using normalized Ct values and the following formula” transcript abundance = 2(39-Ct value), where 39 is the limit of detection on the real-time PCR system.54 The TaqMan Low-Density Array (TLDA) cards were designed for selected pancreatic genes and housekeeping genes (Supplementary Table 3) and were obtained from Thermo Fisher Scientific (Waltham, MA). TLDA cards were used to assess gene expression in human fetal pancreas and gallbladder tissues using the manufacturer’s protocol standardized for TLDA cards on the 7900 HT system (Thermo Fisher Scientific, Waltham, MA). Normalized Ct values were used to plot hierarchical cluster heatmaps. A customized OpenArray Human Messenger RNA panel (Thermo Fisher Scientific, Waltham, MA) was designed to analyze the 45 selected pancreatic gene transcripts and housekeeping control genes in adult human islets and gallbladder (Supplementary Table 4). The customized panels were used following the manufacturer’s protocol standardized for gene expression using OpenArray on the QuantStudio 12K Flex Real-Time PCR platform (Thermo Fisher Scientific, Waltham, MA). Normalized Ct values were used to plot hierarchical cluster heatmaps.
Bulk RNA Sequencing
RNA-seq was performed on mouse tissues as detailed by Williams et al55 using the Ion Total RNA-Seq Kit v2 and the Ion OneTouch 200 Template Kit v2 DL (Thermo Fisher Scientific, Waltham, MA), and sequenced on the Ion Torrent PGM Instrument using the Ion PGM 316 Chip and the Ion PGM 200 Sequencing Kit v2 (Thermo Fisher Scientific, Waltham, MA). Adult human pancreatic islets and gallbladder epithelial cell samples (gallbladder data set GSE152419, n = 7; and human islet data set GSE152111, N = 66) were sequenced as 150 paired-end reads on the HiSeq4000 platform (Illumina, Singapore) as detailed elsewhere.20 Poly-T oligo-attached magnetic beads were used to purify the messenger RNA from total RNA for these samples.
Bulk RNA-Seq Analysis
The Strand next-generation sequencing version 2.5 software (Strand LS, Bengaluru, India) was used to analyze the RNA-seq data. For mouse samples, reads were aligned to the mouse mmu10 University of California Santa Cruz (UCSC) transcriptome and genome together with novel splice variants, using the Ensembl genes and transcript model. However, for human samples, reads were aligned to the human genome version 38 (hg38) transcriptome and genome together (with novel splice variants) using the Ensembl genes and transcript model. Raw reads were aligned with a minimum of 90% identity, a maximum of 5% gaps, and a minimum match length of 25 bps. The output of read-pairs with more than 5 valid matches was not reported. Quality trimming was applied to trim the 3’ end with an average quality less than 10 and on a poorly aligned portion at the 3’ end post alignment. After alignment, raw reads were filtered to remove reads with an average base quality below 20 and reads that have failed quality control. For mouse samples, an average of 2.57 million clean reads per sample were aligned to the reference transcriptome/genome (with novel splice variants; GSE152419). For human samples, an average of 29 million clean reads were generated through bulk RNA-seq of gallbladder epithelial cells (GSE152419, n = 7) and human islets (GSE152111, N = 66) and 78 million clean reads for another set of human islet samples (GSE134068, N = 18). DEseq was used to quantify and normalize the aligned reads, with the threshold normalized count set at 1.56 Baseline transformation was not applied to the preprocessing of the aligned read input data.
Pancreatic Single-Cell RNA Sequencing Analysis
Panc8 single-cell sequencing data (N = 14,890) were extracted from public data sets (GSE84133, GSE85241, E-MTAB-5061, GSE83139, and GSE81608). It was analyzed via R studio version 1.2.5033 (RStudio, Boston, MA) (built under R 3.6.1; R Foundation for Statistical Computing, Vienna, Austria) using SeuratData (version 0.2.1), Seurat, ggplot2, and cowplot packages. The original Seurat Panc8 package contains 8 different pancreas single-cell RNA-seq data sets from across 5 technologies (inDrop, CEL-Seq1, CEL-Seq2, Smart-Seq2, and Fluidigm C1). To improve the integrity of the data, low-read-count data sets obtained through inDrop technology were excluded. Analytical workflow included data preprocessing and feature selection, dimension reduction, and identification of anchor correspondences between data sets; filtering, scoring, and weighting of anchor correspondences; and data matrix correction or data transfer across experiments as described elsewhere.57,58
Differential Splicing Analysis
Differential splicing was analyzed using the Strand next-generation sequencing software. Differential splicing of the gene is based on changes in transcript proportion across the samples. The splice index of a gene refers to the variation associated with the proportion of transcript between each sample. The index is between 0 and 1, and a larger index indicates more differential expression/splicing.
Immunostaining
Tissue sections/cells were immunostained as described by Joglekar and Hardikar.59 For immunostaining, tissue sections or freshly isolated epithelial clusters, islets, or cultured gallbladder cells were fixed in 4% paraformaldehyde (PFA). Tissues were embedded in paraffin and 5- to 10-μm sections were placed on slides for further processing. H&E staining was performed on tissue sections using standard protocol after deparaffinization. For immunochemistry, permeabilization was performed in chilled methanol (50% v/v in water) or 0.5% Triton X-100 (Thermo Fisher Scientific, Waltham, MA) followed by blocking with 4% normal donkey serum (Sigma-Aldrich). Incubation with the primary antibody was at a dilution of 1:100 or 1:200 overnight at 4°C. Cells then were washed 5 times with 1× PBS containing Ca2+ and Mg2+ (Gibco) and incubated with secondary antibody at 37°C. Cells then were washed thoroughly and mounted in Vectashield mountant (Vector Laboratories, Peterborough, UK) containing Hoechst 33342 (Invitrogen). The primary antibodies used were rabbit polyclonal antibody to human C-peptide and guinea pig anti-insulin (both from Linco Research, Inc, St. Charles, MO), rabbit antisomatostatin (Dako, Santa Barbara, CA), mouse antiglucagon, mouse anti-GFP, mouse anti–smooth muscle actin, and rabbit anti-slug (all from Sigma-Aldrich), goat anti-Pdx1 (Abcam, Cambridge, UK), mouse anti–E-cadherin and mouse anti–β-catenin (BD Biosciences, Franklin Lakes, NJ), and mouse monoclonal antivimentin (Chemicon Int, Inc, Temecula, CA). Alexa-Fluor 488, 546, and 633 secondary antibodies (Invitrogen) were used at 1:200 dilution. Antibody concentrations and incubation times were identical across both tissue types (islets and gallbladder) and all samples. Images were scanned and assessed separately for islets and gallbladder cells using a Zeiss LSM 510 laser scanning confocal microscope (Zeiss, Oberkochen, Baden-Württemberg, Germany). Current, voltage applied to the photomultiplier tube, and laser intensities were identical across all samples within a tissue type and were adjusted so as to have below-saturation thresholds. Significantly lower laser power was needed to achieve below-saturation signal in the islet samples.
Immunoelectron Microscopy
CD1 mouse gallbladder and pancreas isolated from the same animal were fixed in 0.5% glutaraldehyde, 4% paraformaldehyde (Agar Scientifics, Stansted, Essex, England), and 2.5 mmol/L CaCl2 (Thermo Fisher Scientific, Waltham, MA) in 0.1 mol/L sodium cacodylate (Agar Scientifics, Stansted, Essex, England) buffer (SCB) at 4ºC overnight, rinsed the next day in a solution of 0.1 mol/L SCB and 2.5 mmol/L CaCl2 for 10 minutes, with 4 changes at room temperature. Samples were dehydrated in ethanol (70%, 90%, 100% dry X 2) for 15 minutes at room temperature, infiltrated with London reisin (LR) white (Agar Scientifics, Stansted, Essex, England) for 1 hour at room temperature, overnight at 4°C, and again at room temperature for 1 hour. Each tissue was transferred to a gelatin capsule (size 0; Agar Scientifics, Stansted, Essex, England) and the capsule was filled completely with LR white resin and allowed to polymerize at 50°C for 24 hours. Sections (100-nm) were cut and mounted on a formvar-carbon–coated nickel slot grid (Agar Scientifics, Stansted, Essex, England) and immunolabeled by floating the mounted grids on respective solutions. Sample (grids) were exposed to 0.05 mol/L glycine (Sigma) in PBS to block aldehyde sites for 15 minutes, rinsed briefly with PBS, blocked in blocking solution with 10% normal goat serum, 0.5% BSA in PBS for 15 minutes, rinsed briefly, and then incubated in guinea pig anti-insulin (Linco Research, Inc) at a dilution of 1:500 overnight at 4°C. Grids then were washed with PBS 3 times for 10 minutes each, and incubated in secondary antibodies (gold-tagged goat anti–guinea pig IgG; British Biolcell, Cardiff, UK) at 1:50 dilution for 2 hours at room temperature, covered, and washed in 0.1 mol/L SCB 3 times. The grids then were postfixed with 1% glutaraldehyde in 0.1 mol/L SCB for 15 minutes, rinsed twice with distilled water for 5 minutes each, and then dried. The grids then were stained in 2% uranyl acetate for 10 minutes, covered with lead citrate for approximately 4 minutes in a NaOH-containing chamber, washed with fresh distilled water, dried, and then imaged using transmission electron microscopy (Jeol 1200EX transmission electron microscopy, University of Bath, UK).
Flow Cytometry
Pdx1-GFP and MIP-GFP mice were killed at predetermined embryonic and adult stages. Pancreas and gallbladder tissues were stored on ice, finely chopped, followed by collagenase digestion (3 mg/mL, 10 minutes at 37°C) to generate a single-cell suspension. Cells were strained through a 70-μm cell strainer and then resuspended in fluorescence-activated cell sorting (FACS) buffer (Ca2+, Mg2+-free PBS +2% fetal calf serum) and acquired on a BD FACSCalibur (BD Biosciences,Franklin Lakes, NJ). Gallbladder-derived mesenchymal-like monolayer cells at passage 5 were harvested using trypsin. Cells were washed twice with FACS buffer, blocked with 5% BSA in 1× PBS (Ca2+ and Mg2+ free) for 30 minutes, incubated with fluorescently conjugated antibodies for 1 hour, washed twice with FACS buffer, fixed with 4% PFA, and then acquired using FACSCalibur. The antibodies used were phycoerythrin-labeled rat IgG2 (isotype control), anti-CD29, anti-CD44, anti-CD90, and anti-CD105 (all phycoerythrin-labeled; BD Biosciences, Franklin Lakes, NJ) at dilutions recommended by the manufacturer. The data were analyzed using CellQuest Pro software (BD Biosciences, Franklin Lakes, NJ). Isotype control or wild-type tissues were used to set up gating. Propidium iodide (PI)+ (dead) cells were excluded from analyses. All acquisition parameters were kept unchanged for the same type of tissue or cells.
Cell Lineage Tracing
To follow the propagation of the insulin-positive cells in vitro, we used 2 thymidine analogues, CldU and IdU (Sigma-Aldrich), as described previously.14 Freshly isolated gallbladder epithelial cells were seeded on Lab-Tek chamber slides (Nunc, Rochester, NY). CldU was added to freshly isolated cells at a concentration of 10 μmol/L and was incubated in a growth-promoting medium for 3 days, followed by 1 day of wash (without any analogue) and again a 3-day pulse of IdU at a concentration of 10 μmol/L. On day 7 the cells in culture were fixed in 4% PFA, washed once with PBS, and then 0.2% Triton X-100 was added for 5 minutes at room temperature. Antigen retrieval was performed by microwaving the slides in 0.01 mol/L sodium citrate, pH 6.0, for 6 minutes at high power until the citrate buffer started boiling, and then kept for 20 minutes in the same citrate buffer at low power in the microwave. After cooling down to room temperature, cells were washed once with 1× PBS containing Ca2+ and Mg2+ and then kept in 1.5 N HCL (diluted in 1× PBS containing Ca2+ and Mg2+) for the next 40 minutes at room temperature. This then was followed by blocking with 4% normal donkey serum at room temperature for 30 minutes. The rest of the steps involved an immunostaining procedure, as described earlier. Immunostaining was performed using antibodies for rat anti-CldU (Accurate Chemicals, Westbury, NY), mouse anti-IdU (BD Biosciences), and rabbit anti–C-peptide (Dako) at a concentration of 1:100. Alexa-Fluor 488, 546, and 633 secondary antibodies (Invitrogen) were used at 1:200 dilutions.
ChIP
Adult mouse gallbladder and pancreas, freshly isolated epithelial cells from the adult human gallbladder, freshly isolated human islets, and gallbladder-derived mesenchymal cells in culture at passage 5 were used in ChIP assay as described previously.59 Freshly isolated gallbladder epithelial cells (Figure 8B, and day 0 of 8D) from 3 to 5 donors had to be pooled in 2 experiments for ChIP to meet the requirement of high cell numbers. Briefly, cells were cross-linked, washed with buffers containing protease inhibitor cocktail (Sigma-Aldrich), and sonicated in lysis buffer to generate DNA fragments of 200–400 bps. Chromatins were immunoprecipitated using 2 μg specific dimethyl and trimethyl antibodies for H3K4 and H3K9, as well as for acetylation of H3K9 and H3 and H4 (Millipore, Billerica, MA). Precipitation cocktails included protein A/G plus beads (Pierce, Pittsburgh, PA), sonicated salmon sperm DNA (Amersham Biosciences, Pittsburgh, PA), and BSA (USB Corporations, Cleveland, OH). Rabbit and mouse IgG (Upstate, Millipore) were used as isotype controls. Chromatin was eluted using 2% sodium dodecyl sulfate, 0.1 mol/L NaHCO3, and 10 mmol/L dithiothreitol. Cross-links were reversed by incubating the eluted chromatin in 4 mol/L NaCl overnight at 65°C. This was followed by proteinase-K digestion and DNA extraction using phenol–chloroform–isoamyl alcohol. Input, immunoprecipitated, and isotype control DNA was resuspended in nuclease-free water and used for SYBR green or TaqMan qPCR with the primers listed in Supplementary Table 5 and with Fast SYBR green or TaqMan Fast master mix (Thermo Fisher Scientific, Waltham, MA) on the ViiA7 Real-Time PCR System platform (Thermo Fisher Scientific, Waltham, MA).
Glucose-Stimulated Insulin Secretion
Gallbladder epithelial clusters were obtained within 12–24 hours postsurgery after scraping of the epithelial layer (Figure 8B, and day 0 of 8D). They were kept in serum-free medium with 1% BSA for 3–4 hours before being handpicked under a phase-contrast microscope. These then were washed with Krebs–Ringer bicarbonate HEPES buffer containing 0.1% BSA (Sigma) and exposed in quadruplicate to basal (2.5 mmol/L glucose) or stimulated (25 mmol/L glucose) buffer, for 1 hour at 37°C. At the end of the incubation/exposure, cells were settled down or centrifuged at 300g for 1 minute to pellet. The supernatant was collected and assayed for insulin/C-peptide. The insulin content in the tissues/cells was measured by sonicating them in 200–500 μL acid ethanol, depending on the size of a tissue or cell pellet. The total protein concentration was measured using the Bradford assay (for mouse samples) or the Thermo Fisher Scientific (Waltham, MA) Qubit protein assay (for human samples). Insulin or C-peptide concentrations were measured by an ELISA kit (Mercodia, Winston Salem, NC). Circulating human insulin was measured using the same kits (Mercodia) after plasma separation. Plasma was separated by centrifugation at 1000g for 10 minutes and stored at -80°C before performing insulin ELISAs. Blood glucose was measured using the Accu-Chek glucometer (Roche Diagnostics, Basel, Switzerland).
Transplantation of Human Gallbladder Epithelial Cells
Transplantation of freshly isolated gallbladder epithelial cells was performed on 8- to 12-week-old male NOD/SCID to assess their function in response to glucose stimulation as described previously.51,60 During this procedure, animals were placed on their back after anesthesia with isoflurane. A total of 500–700 freshly isolated gallbladder epithelial clusters were added into approximately 20 μL animal blood (obtained from the tail) to form a blood clot, which then was transplanted under the kidney capsule of NOD/SCID mice without losing any of the cell clusters. Animals were opened by making a left lateral incision to expose the left kidney. The kidney gently was pulled out and a superficial cut was made in the kidney capsule. The blood clot containing gallbladder cells was placed below the kidney capsule. The capsule then was massaged gently to close the cut. The kidney then was placed back in its original position. The incision was closed using 3–4 absorbable sutures (Davis-Geck, Manati, PR) and an autoclip wound clipper (BD Biosciences). Topical ointment (Soframycin; Sanofi-aventis, NSW, Australia) was applied over the sutured wounds after surgery and animals were administered analgesics (0.05 mg/kg buprenorphine every 12 hours for 3 days). On the 30th day after surgery, blood was collected at 30 minutes after a 2 g/kg body weight glucose load.
Pathway Analysis
To analyze enrichment for β-cell pathways, lists of gallbladder-expressed genes (GSE152419) were compared with β-cell–expressed genes (from E-GEOD-20966) using GO analysis on Pantherdb.org.61 Pre-analytic workflows included cleaning up entries not mapping to protein-coding gene symbols. Gene lists of 16,584 gallbladder transcripts and 13,164 β-cell transcripts were compared for overlap in transcripts using Venn diagrams (https://bioinfogp.cnb.csic.es/tools/venny/index.html), identifying 7227 genes present in both the gallbladder and β-cell expression data set. GO analysis was performed using the list of 7227 gallbladder transcripts also present in β-cells and compared with the list of all human genes as reference. Statistical over-representation was calculated using the Fisher exact test, using Bonferroni correction for multiple testing.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 8.4.1 (GraphPad Software, San Diego, CA) or R software (version 3.6.2; R Foundation for Statistical Computing, Vienna, Austria), SPSS Statistics 27 (Chicago, IL), or Microsoft Excel (version 2016; Microsoft, Redmond, WA). R software was used to perform unsupervised hierarchical clustering maps using heatmap.2 function in gplots. Add-on and other R packages XLconnect (with ActivePerl software) and RColorBrewer were used for data set import and visualization along with R package gplots. GraphPad Prism was used to perform all remaining analyses using appropriate statistical tests and corrected for multiple comparisons if required. Details of each statistical test, the number of replicates, and the number of animals/biological preparations are provided in the respective Figure legends. Split violin and spider/radar plots were created using the BioVinci data visualization package (BioTuring Inc., San Diego, CA). The Kolmogorov–Smirnov test was used to check for data normality in SPSS. The F-test was performed to check for variance in Excel. For non-normally distributed data, a 2-tailed Mann–Whitney test was used to calculate the P value with no ties computed as performed in R. For normally distributed data with equal or unequal variance, a 2-tailed Student or Welch t test was used to calculate the P value in Excel, respectively.
All authors had access to the study data and reviewed and approved the final manuscript.
Data and Materials Availability
The Biospecimen Reporting for Improved Study Quality guidelines62 was followed. Single-cell sequencing (Panc8) data sets (N = 14,890) were extracted from already available data sets (GSE84133, GSE85241, E-MTAB-5061, GSE81076 and GSE86469). Our data on bulk RNA-seq for the human gallbladder data set (GSE152419, n = 7) and human islet data sets (GSE152111, N = 66 and GSE134068, N = 18), have been uploaded to the Gene Expression Omnibus database. Data for mouse gallbladder and pancreas bulk RNA-seq also are available through GSE152419.
Acknowledgments
The authors acknowledge and are thankful for the support from surgical teams, consenting donors or families of the donors, as well as infrastructure support obtained through the Rebecca L Cooper Foundation (A.A.H.) and provided by the National Centre for Cell Science, India and the Faculty of Medicine and Health, University of Sydney. All authors acknowledge the assistance from Dr Andrew M. Holland and Dr Suzanne J. Micallef in the provision of the Pdx1GFP/w reporter mice, Professor Manami Hara for the MIP-GFP mice, and the support from Professor Jonathan Slack and Professor Harry Heimberg for the viral vectors used in the study. The authors also acknowledge support from Dr Ramesh Dumbre, Dr Pabitra Sahoo, Ms Fahmida Khan, Ms Sophie Breedveld, Ms Mariah Taleb, RPA Pathology Services, Bosch Facilities (Bosch Mass Spectometry Facility and Live Cell Analysis Facility) at the University of Sydney. A.G.E. and E.G.S were supported by Australian National and Health and Medical Research Council grants (GNT1079004, GNT1117596, GNT1129861, GNT1138717, GNT1123277), and by the Stafford Fox Medical Research Foundation. Additional infrastructure funding to the Murdoch Children's Research Institute was provided by the Australian Government National Health and Medical Research Council Independent Research Institute Infrastructure Support Scheme and the Victorian Government's Operational Infrastructure Support Program.
CRediT Authorship Contributions
Mugdha V. Joglekar (Conceptualization: Supporting; Data curation: Equal; Formal analysis: Equal; Investigation: Lead; Methodology: Equal; Software: Equal; Supervision: Equal; Writing – original draft: Lead; Writing – review & editing: Equal)
Subhshri Sahu (Data curation: Equal; Formal analysis: Equal; Methodology: Equal; Software: Supporting; Writing – review & editing: Equal)
Wilson KM Wong (Data curation: Equal; Formal analysis: Equal; Methodology: Supporting; Software: Equal; Validation: Equal; Writing – review & editing: Equal)
Sarang N. Satoor (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting)
Charlotte X. Dong (Software: Supporting; Validation: Supporting)
Ryan J Farr (Data curation: Supporting; Methodology: Supporting)
Michael D. Williams (Data curation: Supporting)
Prapti Pandya (Data curation: Supporting)
Gaurang Jhala (Data curation: Supporting; Resources: Supporting)
Sundy N.Y. Yang (Data curation: Supporting)
Yi Vee Chew (Data curation: Supporting)
Nicola Hetherington (Data curation: Supporting)
Dhan Thiruchevlam (Resources: Supporting)
Sasikala Mitnala (Data curation: Supporting; Resources: Supporting)
Guduru V Rao (Resources: Supporting)
Duvvuru Nageshwar Reddy (Resources: Supporting)
Thomas Loudovaris (Resources: Supporting)
Wayne J. Hawthorne (Resources: Supporting)
Andrew G. Elefanty (Resources: Supporting)
Vinay M. Joglekar (Resources: Supporting)
Edouard G. Stanley (Resources: Supporting)
David Martin (Resources: Supporting)
Helen E. Thomas (Resources: Supporting)
David Tosh (Conceptualization: Supporting; Funding acquisition: Supporting; Supervision: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Louise T. Dalgaard (Software: Equal; Validation: Equal; Writing – review & editing: Supporting)
Anandwardhan A. Hardikar (Conceptualization: Lead; Data curation: Supporting; Funding acquisition: Lead; Investigation: Supporting; Project administration: Lead; Supervision: Lead; Visualization: Supporting; Writing – original draft: Equal; Writing – review & editing: Equal)
Data Availability Statement
Sequencing data are available through GEO data sets cited in text.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported mainly by a 2008–2009 British Council (UK–India Educational Research Initiative) exchange program (A.A.H. and D.T.), a 2012–2015 National Health and Medical Research Council project grant (A.A.H.), and the Danish Diabetes Academy visiting professorship (A.A.H. and L.T.D.). Also funded by the Australian Research Council (2012–2016) and the Juvenile Diabetes Research Foundation (JDRF) Australian Type 1 Diabetes Clinical Research Network (grant 4-CDA2016-228-MB; a special initiative of the Australian Research Council) (A.A.H.), the JDRF USA postdoctoral fellowship (2012–2014), advanced post-doctoral fellowship (2015–2018), and the JDRF career transition award (2019–2021) (M.V.J.). The University of Sydney, Australian postgraduate award (University Post-graduate Award or Australian Post-graduate Award), and JDRF Australia PhD top-up awards (W.K.M.W. and R.J.F.), and Council of Scientific and Industrial Research fellowships (Government of India) (S.S.) are acknowledged. W.K.M.W. currently is funded through a grant from the Leona M. and Harry B. Helmsley Charitable Trust (grant 2018PG-T1D009) in collaboration with the JDRF Australian Type 1 Diabetes Clinical Research Network grant 3-SRA-2019-694-M-B (A.A.H.). Also supported by the Summer Scholarship from the National Health and Medical Research Council Clinical Trials Centre, University of Sydney (P.P.).
Supplementary Material
Supplementary Table 2.
Gene Assay IDs Used for qPCR
| Target species | Assay ID | Gene symbol | Gene name | Dye |
|---|---|---|---|---|
| Human | Hs02741908_m1 | INS | Insulin | FAM-MGB |
| Human | Hs00174967_m1 | GCG | Glucagon | FAM-MGB |
| Human | Hs00174949_m1 | SST | Somatostatin | FAM-MGB |
| Human | Hs01651425_s1 | MAFA | V-Maf (v- musculoaponeurotic fibrosarcoma) avian musculoaponeurotic fibrosarcoma oncogene homolog A | FAM-MGB |
| Human | Hs00360700_g1 | NGN3 | Neurogenin 3 | FAM-MGB |
| Human | Hs00172878_m1 | HES1 | Hairy and enhancer of split 1 (hairy and enhancer of split family basic helix-loop-helix (BHLH) transcription factor 1) | FAM-MGB |
| Human | Hs00236830_m1 | PDX1 | Pancreatic and duodenal homeobox 1 | FAM-MGB |
| Human | Hs03003631_g1 | 18S | Eukaryotic 18S ribosomal RNA | VIC-MGB |
| Mouse | Mm01259683_g1 | Ins1 | Insulin-1 | FAM-MGB |
| Mouse | Mm00731595_gH | Ins2 | Insulin-2 | FAM-MGB |
| Mouse | Mm00801712_m1 | Gcg | Glucagon | FAM-MGB |
| Mouse | Mm00436671_m1 | Sst | Somatostatin | FAM-MGB |
| Mouse | Mm00437606_s1 | Neurog3 | Neurogenin 3 | FAM-MGB |
| Mouse | Mm01342805_m1 | Hes1 | Hairy and enhancer of split 1 | FAM-MGB |
| Mouse | Mm00435565_m1 | Pdx1 | Pancreatic and duodenal homeobox 1 | FAM-MGB |
| Mouse | Mm00447452_m1 | Hnf1b | Hepatocyte nuclear factor 1-β | FAM-MGB |
| Mouse | Mm00448840_m1 | Sox9 | Sex-determining gene on the Y chromosome (SRY)-box transcription factor 9 | FAM-MGB |
| Mouse | Mm00433964_m1 | Hnf4a | Hepatocyte nuclear factor 4-α | FAM-MGB |
| Mouse | Mm00468656_m1 | Hlx | H2.0-like homeobox | FAM-MGB |
| Mouse | Mm03302249_g1 | Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | FAM-MGB |
NOTE. List of TaqMan primer/probe assays selected for real-time qPCR on the ViiA7 platform.
rRNA, ribosomal RNA.
FAM-MGB, Fluorescein amidites-minor groove binder;
Supplementary Table 3.
List of TaqMan Primer/Probe Gene Expression Assays for Real-Time qPCR on the TLDA Platform
| Assay ID | Gene symbol | Gene name |
|---|---|---|
| Hs00169631_m1 | INSR | Insulin receptor |
| Hs00218236_m1 | FOXJ2 | Forkhead box J2 |
| Hs00174949_m1 | SST | Somatostatin |
| Hs00607978_s1 | CXCR4 | C-X-C (Cysteine-X-cysteine) motif chemokine receptor 4 |
| Hs00426835_g1 | ACTA2 | Actin α 2, smooth muscle |
| Hs00183740_m1 | DKK1 | Dickkopf WNT signaling pathway inhibitor 1 |
| Hs00175619_m1 | PCSK1 | Proprotein convertase subtilisin/kexin type 1 |
| Hs00707120_s1 | NES | Nestin |
| Hs00355773_m1 | INS | Insulin |
| Hs00174967_m1 | GCG | Glucagon |
| Hs00240792_m1 | FGFR2 | Fibroblast growth factor receptor 2 |
| Hs00274931_s1 | GAL | Galanin and GMAP (galanin message associated peptide) prepropeptide |
| Hs00174139_m1 | CD44 | CD44 molecule (Indian blood group) |
| Hs00171403_m1 | GATA4 | GATA binding protein 4 |
| Hs00264887_s1 | POU3F4 | POU class 3 homeobox 4 |
| Hs00240871_m1 | PAX6 | Paired box 6 |
| Hs00185584_m1 | VIM | Vimentin |
| Hs00165775_m1 | GLUT2 (SLC2A2) | Solute carrier family 2 member 2 |
| Hs00153380_m1 | CCND2 | Cyclin D2 |
| Hs00232018_m1 | GATA6 | GATA binding protein 6 |
| Hs99999901_s1 | 18S | Eukaryotic 18S ribosomal RNA |
| Hs00606262_g1 | HDAC1 | Histone deacetylase 1 |
| Hs00173014_m1 | PAX4 | Paired box 4 |
| Hs00360700_g1 | NEUROG3 | Neurogenin 3 |
| Hs00232355_m1 | NKX6-1 | NK6 homeobox 1 |
| Hs00232128_m1 | HLXB9 | Motor neuron and pancreas homeobox 1 |
| Hs00192380_m1 | SERPINI1 | Serpin family I member 1 |
| Hs00170285_m1 | ICAM5 | Intercellular adhesion molecule 5 |
| Hs00168575_m1 | P4HA1 | Prolyl 4-hydroxylase subunit α 1 |
| Hs00164438_m1 | ENG | Endoglin |
| Hs00159922_m1 | PCSK2 | Proprotein convertase subtilisin/kexin type 2 |
| Hs00179829_m1 | FGFR3 | Fibroblast growth factor receptor 3 |
| Hs00210096_m1 | KRT23 | Keratin 23 |
| Hs00247426_m1 | DKK3 | Dickkopf WNT (Wingless-related integration site) signaling pathway inhibitor 3 |
| Hs00187320_m1 | HDAC3 | Histone deacetylase 3 |
| Hs00170423_m1 | CDH1 | Cadherin 1 |
| Hs00606991_m1 | MKI67 | Marker of proliferation Ki-67 |
| Hs00167155_m1 | SERPINE1 | Serpin family E member 1 |
| Hs00159598_m1 | NEUROD1 | Neuronal differentiation 1 |
| Hs00231032_m1 | HDAC2 | Histone deacetylase 2 |
| Hs00169777_m1 | PECAM1 | Platelet and endothelial cell adhesion molecule 1 |
| Hs99999905_m1 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| Hs00234422_m1 | MMP2 | Matrix metallopeptidase 2 |
| Hs00157705_m1 | GLP1R | Glucagon-like peptide 1 receptor |
| Hs00169851_m1 | NCAM1 | Neural cell adhesion molecule 1 |
| Hs00158126_m1 | ISL1 | ISL LIM homeobox 1 |
| Hs00277220_m1 | GCK | Glucokinase |
| Hs00233790_m1 | ITGAV | Integrin subunit α V |
NOTE. All assays use FAM-MGB (Fluorescein amidites-minor groove binder) dye at 20× stock concentration. 18S is the housekeeping gene.
Supplementary Table 4.
List of TaqMan Primer/Probe Gene Expression Assays for Real-Time qPCR on the OpenArray Platform
| Assay ID | Gene symbol | Gene name |
|---|---|---|
| Hs00173014_m1 | PAX4 | Paired box 4 |
| Hs00271378_s1 | MAFB | V-Maf (v- musculoaponeurotic fibrosarcoma) avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| Hs00359592_m1 | NOVA1 | Neuro-oncologic ventral antigen 1 |
| Hs00240858_m1 | PAX2 | Paired box 2 |
| Hs03003631_g1 | 18S | Eukaryotic 18S ribosomal RNA |
| Hs00240871_m1 | PAX6 | Paired box 6 |
| Hs00268388_s1 | SOX4 | SRY (sex-determining gene on the Y chromosome)-box transcription factor 4 |
| Hs01001343_g1 | SOX9 | SRY (sex-determining gene on the Y chromosome)-box transcription factor 9 |
| Hs00603586_g1 | PTF1A | Pancreas associated transcription factor 1a |
| Hs00413554_m1 | HNF6 (ONECUT1) | Hepatocyte nuclear factor 6 (1 cut homeobox 1) |
| Hs01001602_m1 | HNF1β (TCF2) | Hepatocyte nuclear factor 1-β (transcription factor 2) |
| Hs00167041_m1 | HNF1α (TCF1) | Hepatocyte nuclear factor 1-α (transcription factor 1) |
| Hs00230853_m1 | HNF4α | Hepatocyte nuclear factor 4-α |
| Hs00541450_m1 | GLIS3 | GLIS family zinc finger 3 |
| Hs00232355_m1 | NKX6.1 | NK6 transcription factor related, locus 1 |
| Hs00159616_m1 | NKX2.2 | NK2 transcription factor related, locus 2 |
| Hs00896294_m1 | PROX1 | Prospero-related homeobox 1 |
| Hs01367669_g1 | HES3 | Hairy and enhancer of split 3 (Hes family BHLH (basic helix-loop-helix) transcription factor 3) |
| Hs01922995_s1 | NEUROD1 | Neuronal differentiation 1 |
| Hs00892681_m1 | GLUT1 (SLC2A1) | Solute carrier family 2 (facilitated glucose transporter), member 1 |
| Hs01096908_m1 | GLUT2 (SLC2A2) | Solute carrier family 2 (facilitated glucose transporter), member 2 |
| Hs01564555_m1 | GCK | Glucokinase |
| Hs00846499_s1 | UCN3 | Urocortin 3 |
| Hs00158126_m1 | ISL1 | ISL LIM homeobox 1 |
| Hs00171403_m1 | GATA4 | GATA binding protein 4 |
| Hs00232018_m1 | GATA6 | GATA binding protein 6 |
| Hs00292465_m1 | ARX | Aristaless related homeobox |
| Hs01005963_m1 | IGF2 | Insulin-like growth factor 2 |
| Hs00264887_s1 | POU3F4 | POU class 3 homeobox 4 |
| Hs00703572_s1 | BHLHA15 (MIST1) | Basic helix-loop-helix family member A15 |
| Hs00907365_m1 | HB9 (MNX1) | Homeobox HB9 (motor neuron and pancreas homeobox 1) |
| Hs00158750_m1 | LMX1.2 (LMX1B) | LIM homeobox transcription factor 1β |
| Hs00892663_m1 | LMX1.1 (LMX1A) | LIM homeobox transcription factor 1α |
| Hs01078080_m1 | CDX2 | Caudal type homeobox 2 |
| Hs00793699_g1 | GSX1 | Genomic Screened homeobox 1 |
| Hs00370195_m1 | GSX2 | Genomic Screened homeobox 2 |
| Hs01116195_m1 | EXO1 | Exonuclease 1 |
| Hs00170171_m1 | REG3A | Regenerating family member 3α |
| Hs01551078_m1 | TLR3 | Toll-like receptor 3 |
| Hs00358111_g1 | PPY | Pancreatic polypeptide |
| Hs00230829_m1 | AIRE | Autoimmune regulator |
| Hs01074053_m1 | GHRL | Ghrelin and obestatin prepropeptide |
| Hs01026107_m1 | PCSK1 | Proprotein convertase subtilisin/kexin type 1 |
| Hs00159922_m1 | PCSK2 | Proprotein convertase subtilisin/kexin type 2 |
| Hs00232764_m1 | HNF3β (FOXA2) | Hepatocyte nuclear factor 3β (forkhead box A2) |
NOTE. All assays use FAM-MGB (Fluorescein amidites-minor groove binder) dye at 20× stock concentration. 18S is the housekeeping gene.
Supplementary Table 5.
List of Primer Sets Used for ChIP DNA qPCR
| Species | Gene locus | Forward primer | Reverse primer | Probe |
|---|---|---|---|---|
| Mouse | Pdx1 A-I | CCAGTATCAGGGAGGACTATCA | TACCCAGCCATTAGGCAAGA | |
| Mouse | Pdx1 A-III | ACCGTGTCACCAAGTCAACCC | AGAGCCACCTGTGCCCGTCAA | |
| Mouse | Pdx1 A-IV | CTCTTCCTGATTCCCTGAAGTC | ACTAAGAGTGCTCTGGGCTCTG | |
| Human | INS pro | GTGGAAAGTGGTTTAGGTGAGGGT | ACCTGCTTGATGGCCTCTTCTGAT | |
| Human | PDX1 pro | CACACAACGAATGCCAGAGTTTCG | ACTGATCTCAGAGGGAACCCACA | |
| Human | NEUROG3 pro | AAGAGAGGCAGTGAAACACCAGGA | AACTCTCGGTTCCTCAAAGAGCCT | |
| Human | HES1 pro | TCCTCCTCCCATTGGCTGAAAGT | TTGGTGATCAGTAGCGCTGTTCCA | |
| Human | INS -275 | TGTGAGCAGGGACAGGTCTG | TCCTCAGGACCAGCGGG | 6FAM-CCACCGGGCCCCTGGTTAAGACTCTA |
| Human | INS +1318 | CAGCTGGAGAACTACTGCAACTAGA | GCTGGTTCAAGGGCTTTATTCC | 6FAM-CCGCCTCCTGCACCGAGAGAGA |
NOTE. Sequences start from 5' to 3'. Gene loci for which a probe sequence is not provided were performed using SYBR green chemistry. TaqMan qPCR was used for human INS-275 and INS+1318.
Pro, promoter.
References
- 1.Sahu S., Joglekar M.V., Dumbre R., Phadnis S.M., Tosh D., Hardikar A.A. Islet-like cell clusters occur naturally in human gall bladder and are retained in diabetic conditions. J Cell Mol Med. 2009;13:999–1000. doi: 10.1111/j.1582-4934.2008.00572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dutton J.R., Chillingworth N.L., Eberhard D., Brannon C.R., Hornsey M.A., Tosh D., Slack J.M. Beta cells occur naturally in extrahepatic bile ducts of mice. J Cell Sci. 2007;120:239–245. doi: 10.1242/jcs.03330. [DOI] [PubMed] [Google Scholar]
- 3.Oven I., Brdickova N., Kohoutek J., Vaupotic T., Narat M., Peterlin B.M. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol. 2007;27:8815–8823. doi: 10.1128/MCB.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heller R.S., Tsugu H., Nabeshima K., Madsen O.D. Intracranial ectopic pancreatic tissue. Islets. 2010;2:65–71. doi: 10.4161/isl.2.2.10580. [DOI] [PubMed] [Google Scholar]
- 5.Mehran A.E., Templeman N.M., Brigidi G.S., Lim G.E., Chu K.Y., Hu X., Botezelli J.D., Asadi A., Hoffman B.G., Kieffer T.J., Bamji S.X., Clee S.M., Johnson J.D. Hyperinsulinemia drives diet-induced obesity independently of brain insulin production. Cell Metab. 2012;16:723–737. doi: 10.1016/j.cmet.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Sahu S., Tosh D., Hardikar A.A. New sources of beta-cells for treating diabetes. J Endocrinol. 2009;202:13–16. doi: 10.1677/JOE-09-0097. [DOI] [PubMed] [Google Scholar]
- 7.Coad R.A., Dutton J.R., Tosh D., Slack J.M. Inhibition of Hes1 activity in gall bladder epithelial cells promotes insulin expression and glucose responsiveness. Biochem Cell Biol. 2009;87:975–987. doi: 10.1139/o09-063. [DOI] [PubMed] [Google Scholar]
- 8.Galivo F., Benedetti E., Wang Y., Pelz C., Schug J., Kaestner K.H., Grompe M. Reprogramming human gallbladder cells into insulin-producing beta-like cells. PLoS One. 2017;12 doi: 10.1371/journal.pone.0181812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holland A.M., Micallef S.J., Li X., Elefanty A.G., Stanley E.G. A mouse carrying the green fluorescent protein gene targeted to the Pdx1 locus facilitates the study of pancreas development and function. Genesis. 2006;44:304–307. doi: 10.1002/dvg.20214. [DOI] [PubMed] [Google Scholar]
- 11.Lee J.C., Smith S.B., Watada H., Lin J., Scheel D., Wang J., Mirmira R.G., German M.S. Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes. 2001;50:928–936. doi: 10.2337/diabetes.50.5.928. [DOI] [PubMed] [Google Scholar]
- 12.Gerrish K., Van Velkinburgh J.C., Stein R. Conserved transcriptional regulatory domains of the pdx-1 gene. Mol Endocrinol. 2004;18:533–548. doi: 10.1210/me.2003-0371. [DOI] [PubMed] [Google Scholar]
- 13.Hara M., Wang X., Kawamura T., Bindokas V.P., Dizon R.F., Alcoser S.Y., Magnuson M.A., Bell G.I. Transgenic mice with green fluorescent protein-labeled pancreatic beta-cells. Am J Physiol Endocrinol Metab. 2003;284:E177–E183. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- 14.Joglekar M.V., Joglekar V.M., Joglekar S.V., Hardikar A.A. Human fetal pancreatic insulin-producing cells proliferate in vitro. J Endocrinol. 2009;201:27–36. doi: 10.1677/JOE-08-0497. [DOI] [PubMed] [Google Scholar]
- 15.Marselli L., Thorne J., Dahiya S., Sgroi D.C., Sharma A., Bonner-Weir S., Marchetti P., Weir G.C. Gene expression profiles of beta-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teta M., Rankin M.M., Long S.Y., Stein G.M., Kushner J.A. Growth and regeneration of adult beta cells does not involve specialized progenitors. Dev Cell. 2007;12:817–826. doi: 10.1016/j.devcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Gnyszka A., Jastrzebski Z., Flis S. DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res. 2013;33:2989–2996. [PubMed] [Google Scholar]
- 18.Eckschlager T., Plch J., Stiborova M., Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int J Mol Sci. 2017;18:1414. doi: 10.3390/ijms18071414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myasoedova V.A., Sukhorukov V., Grechko A.V., Zhang D., Romanenko E., Orekhov V., Orekhov A.N. Inhibitors of DNA methylation and histone deacetylation as epigenetically active drugs for anticancer therapy. Curr Pharm Des. 2019;25:635–641. doi: 10.2174/1381612825666190405144026. [DOI] [PubMed] [Google Scholar]
- 20.Wong W.K., Jiang G., Sorensen A.E., Chew Y.V., Lee-Maynard C., Liuwantara D., Williams L., O'Connell P.J., Dalgaard L.T., Ma R.C., Hawthorne W.J., Joglekar M.V., Hardikar A.A. The long noncoding RNA MALAT1 predicts human pancreatic islet isolation quality. JCI Insight. 2019;5 doi: 10.1172/jci.insight.129299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palazon-Fernandez J.L., Peiro Suso M., Mancera J.M., Sarasquete C. Immunohistochemical study of the principal pancreatic islet of the toadfish, Halobatrachus didactylus (Pisces: Batrachoididae) Acta Histochem. 2011;113:256–261. doi: 10.1016/j.acthis.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Chen S., Li C., Yuan G., Xie F. Anatomical and histological observation on the pancreas in adult zebrafish. Pancreas. 2007;34:120–125. doi: 10.1097/01.mpa.0000246661.23128.8c. [DOI] [PubMed] [Google Scholar]
- 23.Kaptaner B. Immunohistochemical distribution of insulin-, glucagon- and somatostatin-containing cells in the pancreas of Lake Van fish (Alburnus tarichi Guldenstadt, 1814) (Cyprinidae) Eur J Histochem. 2019;63:2999. doi: 10.4081/ejh.2019.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami M., Tsutsumi Y. Aberrant pancreatic tissue accompanied by heterotopic gastric mucosa in the gall-bladder. Pathol Int. 1999;49:580–582. doi: 10.1046/j.1440-1827.1999.00905.x. [DOI] [PubMed] [Google Scholar]
- 25.Goodman Z.D., Albores-Saavedra J., Lundblad D.M. Somatostatinoma of the cystic duct. Cancer. 1984;53:498–502. doi: 10.1002/1097-0142(19840201)53:3<498::aid-cncr2820530321>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 26.Barbezat G.O. Glucagon and the pancreas, liver and bile ducts. S Afr J Surg. 1974;12:213–217. [PubMed] [Google Scholar]
- 27.Henderson J.R. Insulin in body fluids other than blood. Physiol Rev. 1974;54:1–22. doi: 10.1152/physrev.1974.54.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Quijada C., Goni P.M. Liver and insulin: presence of insulin in bile. Metabolism. 1967;16:514–521. doi: 10.1016/0026-0495(67)90080-7. [DOI] [PubMed] [Google Scholar]
- 29.Daniel P.M., Henderson J.R. Insulin in bile and other body fluids. Lancet. 1967;1:1256–1257. doi: 10.1016/s0140-6736(67)92718-3. [DOI] [PubMed] [Google Scholar]
- 30.Cardinale V., Puca R., Carpino G., Scafetta G., Renzi A., De Canio M., Sicilia F., Nevi L., Casa D., Panetta R., Berloco P.B., Reid L.M., Federici G., Gaudio E., Maroder M., Alvaro D. Adult human biliary tree stem cells differentiate to beta-pancreatic islet cells by treatment with a recombinant human Pdx1 peptide. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hickey R.D., Galivo F., Schug J., Brehm M.A., Haft A., Wang Y., Benedetti E., Gu G., Magnuson M.A., Shultz L.D., Lagasse E., Greiner D.L., Kaestner K.H., Grompe M. Generation of islet-like cells from mouse gall bladder by direct ex vivo reprogramming. Stem Cell Res. 2013;11:503–515. doi: 10.1016/j.scr.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagaya M., Katsuta H., Kaneto H., Bonner-Weir S., Weir G.C. Adult mouse intrahepatic biliary epithelial cells induced in vitro to become insulin-producing cells. J Endocrinol. 2009;201:37–47. doi: 10.1677/JOE-08-0482. [DOI] [PubMed] [Google Scholar]
- 33.Eberhard D., Tosh D., Slack J.M. Origin of pancreatic endocrine cells from biliary duct epithelium. Cell Mol Life Sci. 2008;65:3467–3480. doi: 10.1007/s00018-008-8427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen F., Li T., Sun Y., Liu Q., Yang T., Chen J., Zhu H., Shi Y., Hu Y.P., Wang M.J. Generation of insulin-secreting cells from mouse gallbladder stem cells by small molecules in vitro. Stem Cell Res Ther. 2019;10:289. doi: 10.1186/s13287-019-1407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auth M.K., Keitzer R.A., Scholz M., Blaheta R.A., Hottenrott E.C., Herrmann G., Encke A., Markus B.H. Establishment and immunological characterization of cultured human gallbladder epithelial cells. Hepatology. 1993;18:546–555. [PubMed] [Google Scholar]
- 36.Gunter-Smith P.J., Abdulkadir O., Hammonds-Odie L., Scanlon M., Terrell R. A primary culture of guinea pig gallbladder epithelial cells that is responsive to secretagogues. Am J Physiol Gastrointest Liver Physiol. 2000;279:G866–G874. doi: 10.1152/ajpgi.2000.279.5.G866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawamura Y., Yoshida K., Nakanuma Y. Primary culture of rabbit gallbladder epithelial cells in collagen gel matrix. Lab Invest. 1989;61:350–356. [PubMed] [Google Scholar]
- 38.Oda D., Lee S.P., Hayashi A. Long-term culture and partial characterization of dog gallbladder epithelial cells. Lab Invest. 1991;64:682–692. [PubMed] [Google Scholar]
- 39.Plevris J.N., Walker S.W., Harrison D.J., Dhariwal A., Hayes P.C., Bouchier I.A. Primary culture of bovine gall bladder epithelial cells. Gut. 1993;34:1612–1615. doi: 10.1136/gut.34.11.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D.A. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purcell A.W., Sechi S., DiLorenzo T.P. The evolving landscape of autoantigen discovery and characterization in type 1 diabetes. Diabetes. 2019;68:879–886. doi: 10.2337/dbi18-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kracht M.J., van Lummel M., Nikolic T., Joosten A.M., Laban S., van der Slik A.R., van Veelen P.A., Carlotti F., de Koning E.J., Hoeben R.C., Zaldumbide A., Roep B.O. Autoimmunity against a defective ribosomal insulin gene product in type 1 diabetes. Nat Med. 2017;23:501–507. doi: 10.1038/nm.4289. [DOI] [PubMed] [Google Scholar]
- 43.Oslowski C.M., Urano F. The binary switch that controls the life and death decisions of ER stressed beta cells. Curr Opin Cell Biol. 2011;23:207–215. doi: 10.1016/j.ceb.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eizirik D.L., Cnop M. ER stress in pancreatic beta cells: the thin red line between adaptation and failure. Sci Signal. 2010;3:pe7. doi: 10.1126/scisignal.3110pe7. [DOI] [PubMed] [Google Scholar]
- 45.Carmody D., Park S.Y., Ye H., Perrone M.E., Alkorta-Aranburu G., Highland H.M., Hanis C.L., Philipson L.H., Bell G.I., Greeley S.A. Continued lessons from the INS gene: an intronic mutation causing diabetes through a novel mechanism. J Med Genet. 2015;52:612–616. doi: 10.1136/jmedgenet-2015-103220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomaidou S., Zaldumbide A., Roep B.O. Islet stress, degradation and autoimmunity. Diabetes Obes Metab. 2018;20(Suppl 2):88–94. doi: 10.1111/dom.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brozzi F., Nardelli T.R., Lopes M., Millard I., Barthson J., Igoillo-Esteve M., Grieco F.A., Villate O., Oliveira J.M., Casimir M., Bugliani M., Engin F., Hotamisligil G.S., Marchetti P., Eizirik D.L. Cytokines induce endoplasmic reticulum stress in human, rat and mouse beta cells via different mechanisms. Diabetologia. 2015;58:2307–2316. doi: 10.1007/s00125-015-3669-6. [DOI] [PubMed] [Google Scholar]
- 48.Engin F., Yermalovich A., Nguyen T., Hummasti S., Fu W., Eizirik D.L., Mathis D., Hotamisligil G.S. Restoration of the unfolded protein response in pancreatic beta cells protects mice against type 1 diabetes. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3006534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitchell J., Punthakee Z., Lo B., Bernard C., Chong K., Newman C., Cartier L., Desilets V., Cutz E., Hansen I.L., Riley P., Polychronakos C. Neonatal diabetes, with hypoplastic pancreas, intestinal atresia and gall bladder hypoplasia: search for the aetiology of a new autosomal recessive syndrome. Diabetologia. 2004;47:2160–2167. doi: 10.1007/s00125-004-1576-3. [DOI] [PubMed] [Google Scholar]
- 50.Gershengorn M.C., Hardikar A.A., Wei C., Geras-Raaka E., Marcus-Samuels B., Raaka B.M. Epithelial-to-mesenchymal transition generates proliferative human islet precursor cells. Science. 2004;306:2261–2264. doi: 10.1126/science.1101968. [DOI] [PubMed] [Google Scholar]
- 51.Parekh V.S., Joglekar M.V., Hardikar A.A. Differentiation of human umbilical cord blood-derived mononuclear cells to endocrine pancreatic lineage. Differentiation. 2009;78:232–240. doi: 10.1016/j.diff.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 52.Phadnis S.M., Joglekar M.V., Dalvi M.P., Muthyala S., Nair P.D., Ghaskadbi S.M., Bhonde R.R., Hardikar A.A. Human bone marrow-derived mesenchymal cells differentiate and mature into endocrine pancreatic lineage in vivo. Cytotherapy. 2011;13:279–293. doi: 10.3109/14653249.2010.523108. [DOI] [PubMed] [Google Scholar]
- 53.Thowfeequ S., Li W.C., Slack J.M., Tosh D. Reprogramming of liver to pancreas. Methods Mol Biol. 2009;482:407–418. doi: 10.1007/978-1-59745-060-7_25. [DOI] [PubMed] [Google Scholar]
- 54.Hardikar A.A., Farr R.J., Joglekar M.V. Circulating microRNAs: understanding the limits for quantitative measurement by real-time PCR. J Am Heart Assoc. 2014;3 doi: 10.1161/JAHA.113.000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams M.D., Joglekar M.V., Satoor S.N., Wong W., Keramidaris E., Rixon A., O'Connell P., Hawthorne W.J., Mitchell G.M., Hardikar A.A. Epigenetic and transcriptome profiling identifies a population of visceral adipose-derived progenitor cells with the potential to differentiate into an endocrine pancreatic lineage. Cell Transplant. 2019;28:89–104. doi: 10.1177/0963689718808472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902 e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joglekar M.V., Hardikar A.A. Isolation, expansion, and characterization of human islet-derived progenitor cells. Methods Mol Biol. 2012;879:351–366. doi: 10.1007/978-1-61779-815-3_21. [DOI] [PubMed] [Google Scholar]
- 60.Joglekar M.V., Patil R.R., Satoor S.N., Wong W.K.M., Karandikar M.S., Hardikar A.A. Promoting pro-endocrine differentiation and graft maturation following surgical resection of the mouse pancreas. Methods Mol Biol. 2021;2224:87–98. doi: 10.1007/978-1-0716-1008-4_6. [DOI] [PubMed] [Google Scholar]
- 61.Mi H., Muruganujan A., Huang X., Ebert D., Mills C., Guo X., Thomas P.D. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0) Nat Protoc. 2019;14:703–721. doi: 10.1038/s41596-019-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moore H.M., Kelly A.B., Jewell S.D., McShane L.M., Clark D.P., Greenspan R., Hayes D.F., Hainaut P., Kim P., Mansfield E.A., Potapova O., Riegman P., Rubinstein Y., Seijo E., Somiari S., Watson P., Weier H.U., Zhu C., Vaught J. Biospecimen reporting for improved study quality (BRISQ) Cancer Cytopathol. 2011;119:92–101. doi: 10.1002/cncy.20147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available through GEO data sets cited in text.