Abstract
Purpose
Form deprivation myopia (FDM) is an urgent public issue characterized by pathological changes, but the underlying mechanism remained unclear. The aim was to investigate bone morphogenetic proteins (BMPs) utilizing the pathogenesis of FDM.
Material and methods
Gene expression omnibus (GEO) database was used to analyze one mRNA profile (GSE89325) of FDM. Sixteen retina samples (8 FDM and 8 controls) were randomly divided into seven groups for differential gene expression analysis in R. software. The gene pathway and protein-protein interaction (PPI) analysis were performed by the DAVID and STRING databases. Cytoscape was used to draw the PPI network. The gene ontology (GO) enrichment and Kyoto encyclopedia of genes and Genomes (KEGG) analysis were determined to achieve gene annotation and visualization.
Results
A total of 18420 differentially expressed genes (DEGs) were identified associated with FDM. The only non-significant gene (BEND6) was separately analyzed between two groups. Thirteen hub genes were discovered, ACVR1, ACVR2A, ACVR2B, RGMB, BMPR2, BMPR1A, BMP2, BMPR1B, CHRD, PTH, PTH1R, PTHLH, and WNT9A. The expression alteration in FDM were mainly enriched in cytokine-cytokine, and neuroactive ligand receptor interaction pathways. BMP2 was the key gene in myopia progression.
Conclusions
Of clinical perspective, our findings reveal that expression of BMP2 as an underlying mechanism of FDM, providing an insight for therapeutic interventions.
Keywords: Bone morphogenetic protein 2, RNA sequence analysis, Retina, Form deviation myopia
Highlights
-
•
BMP2 is a novel biomarker of myopia.
-
•
Neuroactive ligand receptor interaction has some potential clinical applications.
-
•
Estimating gene changes could become one of methods for myopia prevention.
1. Introduction
The growth factor role of bone morphogenetic proteins (BMPs) has become an important area in numerous cellular functions, including chondrogenesis, cell proliferation, and extracellular matrix synthesis [1,2]. BMPs are crucially known for their salient sector in embryogenesis and development. Nevertheless, BMPs have been generally reported to promote early eye morphogenesis, collagen degeneration, and induce chondrogenic differentiation in human scleral fibroblasts [3]. Induction in chondrogenic differentiation has been regarded to be associated with some diseases, such as rheumatic arthritis and high myopia [[4], [5], [6]]. Apart from their efficiency in bone formation and repair, BMPs have other biological effects. For example, BMP2 is essential for retinal development. Both BMP2 and BMP4 are implicated in the formation of cardiac septa, and their deficiency might cause heart defects in mice. Moreover, BMP7 contributes to the deterioration of the kidney and the heart [[7], [8], [9]].
Myopia is a major cause of visual impairment among children. It has been related to ocular complications, such as macular degeneration, cataract, retinal detachment, and glaucoma. These comorbidities could increase the risk of irreversible visual loss [10,11]. The prevalence of myopia and high myopia is constantly rising globally and estimated to be 50% by the year 2050 [12,13]. Furthermore, early-onset myopic children have higher progression rates of high myopia [14,15]. However, the effect of BMP2 on the pathogenesis of myopia remains ambiguous. With the proliferation of low dose atropine [16], treatments for myopia have been largely evolved. Unfortunately, because low dose atropine induces various significant adverse events [17] and does not address the progression of high myopia, that is, there is an urgent need for new therapeutic methods. Therefore, a better understanding of the pathological mechanism for myopia progression will benefit the refreshment of clinical treatments.
Small mRNA suggests a role for post-transcriptional regulation that a light-dependent retina-to sclera signaling cascade and delineates possible pathobiological molecular drivers causing refractive errors [18]. Identification of differentially expressed mRNA and genes could be recognized through computational methods [19].
In this study, we identified the potential of BMPs as well as the difference in myopic retina between early and late timepoint recovery from FDM. Further analysis of the differentially expressed genes (DEGs) by functional annotation was performed to acknowledge the molecular basis and relevant pathways. Eventually, protein-protein interaction (PPI) network was analyzed to identify the key genes. We sought to explore the role of BMPs in FDM modulation to find a novel therapeutic target for myopia.
2. Material and methods
2.1. Data source and processing
We downloaded RNA sequencing dataset GSE89325 [20] as well as the corresponding characteristics of myopic retina and controls from the Gene Expression Omnibus (GEO) website (http://www.ncbi.nlm.nih.gov/geo/) in October 2021. The profile of GSE89325 was based on the platform of GPL21476 Illumina HiSeq 1500 (Gallus gallus). The RNA sequencing data were sorted by controls and FDM male chicks (Leghorn/New Hampshire), which were left untreated, and treated at different timepoints (0 h, 6 h, 24 h).
The expression levels were initially standardized and filtered using Fast-QC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), multi-QC with matching reads beyond 80% [21], and Trim Galore (0.4.0), respectively. High quality sequence reads were aligned to chick genome (GalGal6) using Hisat2 [22] to obtain the counts for each sample.
2.2. Identification of DEGs
Our differential expression analysis based on RNA sequencing data was interpreted using the DEseq2 package (http://www.bioconductor.org/packages/release/bioc/html/DEseq2.html) in the R statistical environment (version 3.5.1) [23] to identify DEGs. The timepoint referred to occlusion (control vs myopia, 0 h vs control) and the 24 h recovery period (6 h vs 0 h, 24 h vs 6 h, and 24 h vs 0 h). Expression comparisons were conducted by moderated t-test method. The thresholds were |log 2fold change|>2, and false discovery rate (FDR) < 5% as the criteria for DEGs selection.
2.3. Gene annotation analysis
For those RNA target genes, Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) (http://www.genome.jp/kegg/pathway.html) analyses were performed on gene annotation. Pathway analysis was conducted through the Database for visualization, and integrated discovery (DAVID) (https://david.ncifcrf.gov/) [24,25]. Chick Ensemble Gene IDs were mapped to human gene symbol using BioMart (Ensemble release 101). While mapping detected no existing gene, they were connected to human gene orthologues (Homo sapiens GRCh38.p7).
For DEGs in myopic retina, PPI analysis was performed through the Search Tool for the Retrieval of Interacting Genes (STRING) database (http://www.string-db.org) and produced by Cytoscape software with a confidence score of >1.0 [26].
3. Results
3.1. Data quality assessment
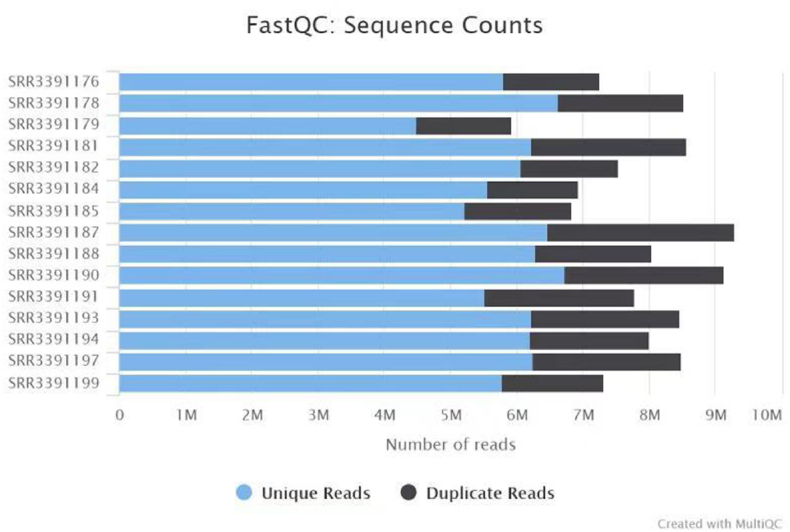
We searched 16 samples (8 myopia and 8 controls) from GEO database and then transferred into SRR documents using R software. Finally, a total of 18420 DEGs were identified between myopia and control groups. Through fast-QC and multi-QC assessment, we found 80% non-reproductive index sequence counts. The number of unique reads mostly centered on 6 reads, which demonstrated that RNA sequencing data were confidential. The plot was displayed in Fig. 1. We compared seven groups between controls and chicks recovery from FDM (0hrs, 6hrs, 24hrs).
Fig. 1.
Plot of quality control for 15 SRR documents using multi-QC. Duplicate reads were identified using trim-gallore. QC, quality control.
3.2. Total analyses (FDM versus controls)
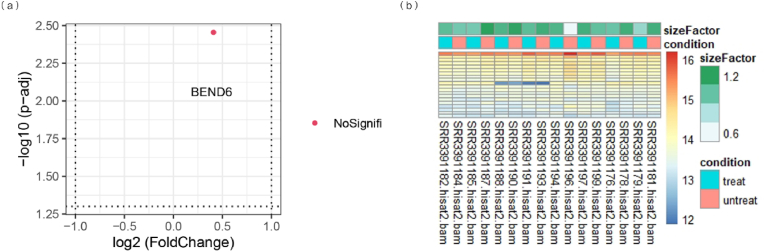
We compared control and FDM groups as shown in Fig. 2. The volcano plot indicated that only 1 gene (BEND 6) was detected to be differentially expressed (Fig. 2a). There was no statistically significant difference (p > 0.05). We found that 16 mRNAs expressions were obviously different from others. Among those mRNAs that 8 were upregulated with size Factor of 1.2 as shown in the heatmap (Fig. 2b).
Fig. 2.
Differentially expressed genes and mRNAs in myopic retina. The volcano plot (a) showed that a total of 1 non-significant gene was detected to be differentially expressed. The heat map (b) detected that 16 mRNAs expressed differentially between myopic retina and controls, and 15 of them were upregulated.
3.3. Comparisons between early and late FDM versus controls
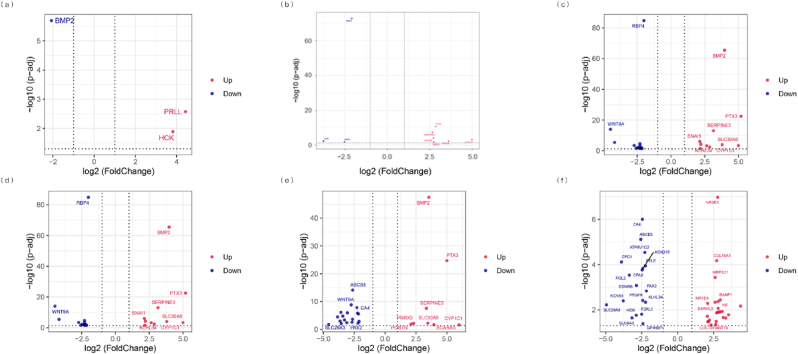
We compared the profiles of myopic and normal chicks, respectively. A total of 131 genes were detected as DEGs among seven groups. The volcano plots were displayed in Fig. 3. The volcano plot (a) showed that a total of 3 genes were detected to be differentially expressed between control and FDM 0 h; 2 were up-regulated and 1 was down-regulated. The volcano plot (b) displayed that 13 genes (up-regulated 8, down-regulated 5) differentially expressed between control and FDM 6 h. The volcano plot (c) indicated that 23 genes (up-regulated 7, down-regulated 15) differentially expressed between control and FDM 24 h. In early stage FDM, there were 23 DEGs about FDM 0 h vs FDM 6 h (d). Conversely, in late stage FDM, 26 and 42 DEGs were identified about FDM 0hrs vs FDM 24 h (e), and FDM 6hrs vs FDM 24 h, respectively (f). Altogether, only WNT9A was down-regulated in response to both FDM 0hrs vs FDM 6 h and FDM 0hrs vs FDM 24 h.
Fig. 3.
Differentially expressed genes in early and long term FDM retina. The volcano plot (a) showed that a total of 3 genes were detected to be differentially expressed between control and FDM 0 h; 2 were upregulated and 1 downregulated. The volcano plot (b) displayed that 13 genes expressed differentially between control and FDM 6 h, and 8 of them were upregulated. The volcano plot (c) displayed that 23 genes expressed differentially between control and FDM 24 h, and 7 of them were upregulated. In early FDM retina, 23 genes were differentially expressed in response to (FDM 0hrs vs FDM 6 h) (d), 26 genes in response to (FDM 0hrs vs FDM 24 h) (e), and 42 genes in response to (FDM 6hrs vs FDM 24 h) (f). Altogether, only WNT9A was downregulated in response to both FDM 0hrs vs FDM 6 h and FDM 0hrs vs FDM 24 h. FDM 0hrs: 0 h recovery from FD. FDM 6hrs: 6 h recovery from FD; FDM 24hrs: 24 h recovery from FD.
3.4. GO and KEGG analysis
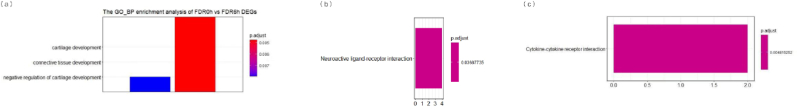
We conducted GO analysis of the DEGs in seven groups. Meanwhile, several comparisons were obtained among 0 h versus 6 h, 0 h versus 24 h, and 0 h versus control. A total of 44 biological processes (BP), 12 molecular functioning (MF), and 6 cellular components (CC) were found through the STRING database, and were mainly metabolism associated receptors, including signaling receptor activity, cell surface receptor signaling pathway, activin-activated receptor activity, and activin receptor complex. Moreover, the detected 3 KEGG pathways contributed to cytokine-cytokine receptor interaction, neuroactive ligand-receptor interaction, and TGF-beta signaling pathway (Fig. 4). The top 6 GO and KEGG pathways were shown in Table 1. Cell surface receptor signaling pathway was one of the top 6 BPs, and the remaining was BMP signaling pathway, positive regulation of developmental process, regulation of cell differentiation, positive regulation of angiogenesis, animal organ morphogenesis, axis specification, regulation of response to stimulus and regulation of animal organ morphogenesis.
Fig. 4.
Column plots showing all significant pathways from Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway comparisons during the induction of myopia and recovery from FDM. The significantly enriched pathways are visualized for (a) 0 h versus 6 h, (b) 0 h versus 24 h, and (c) 0 h versus Control. The size of each column is proportional to the number of core genes within the pathway.
Table 1.
The top 6 Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways of the differentially expressed mRNA targets between control and FDM 6 h.
| Category | GO term | Description | Count | Genes |
|---|---|---|---|---|
| BP | GO:0051239 | regulation of multicellular organismal process | 8 | WNT9A,ACVR2B,BMP2,BMPR1B,ACVR1,PTHLH, ACVR2A,RGMA |
| GO:0030154 | cell differentiation | 8 | WNT9A,ACVR2B,BMP2,BMPR1B,ACVR1,PTHLH, ACVR2A,RGMA | |
| GO:0007165 | signal transduction | 8 | WNT9A,ACVR2B,BMP2,BMPR1B,ACVR1,PTHLH, ACVR2A,RGMA | |
| GO:0048731 | system development | 8 | WNT9A,ACVR2B,BMP2,BMPR1B,ACVR1,PTHLH, ACVR2A,RGMA | |
| GO:2000026 | regulation of multicellular organismal development | 7 | WNT9A,BMP2,BMPR1B,ACVR1,PTHLH, ACVR2A,RGMA | |
| GO:0007166 | cell surface receptor signaling pathway | 7 | WNT9A,ACVR2B,BMP2,BMPR1B,ACVR1,ACVR2A,RGMA | |
| MF | GO:0005488 | binding | 7 | WNT9A,ACVR2B,BMP2,BMPR1B,ACVR1,PTHLH, ACVR2A |
| GO:0005515 | protein binding | 6 | WNT9A,BMP2,BMPR1B,ACVR1,PTHLH, ACVR2A | |
| GO:0038023 | signaling receptor activity | 5 | ACVR2B,BMPR1B,ACVR1,ACVR2A,RGMA | |
| GO:0004675 | transmembrane receptor protein serine/threonine kinase activity | 4 | ACVR2B,BMPR1B,ACVR1,ACVR2A | |
| GO:0017002 | activin-activated receptor activity | 3 | ACVR2B,ACVR1,ACVR2A | |
| GO:0048185 | activin binding | 3 | ACVR2B,ACVR1,ACVR2A | |
| CC | GO:0110165 | cellular anatomical entity | 8 | WNT9A,ACVR2B,BMP2,BMPR1B,ACVR1,PTHLH, ACVR2A,RGMA |
| GO:0005886 | plasma membrane | 5 | ACVR2B,BMPR1B,ACVR1,ACVR2A,RGMA | |
| GO:0031224 | intrinsic component of membrane | 5 | ACVR2B,BMPR1B,ACVR1,ACVR2A,RGMA | |
| GO:0043235 | receptor complex | 4 | ACVR2B,BMPR1B,ACVR1,ACVR2A | |
| GO:0005887 | integral component of plasma membrane | 4 | ACVR2B,BMPR1B,ACVR1,ACVR2A | |
| GO:0048179 | activin receptor complex | 3 | ACVR2B,ACVR1,ACVR2A | |
| KEGG pathways | gga04350 | TGF-beta signaling pathway | 10 | BMPR1A,ACVR2B,BMPR2,BMP2,BMPR1B,ACVR1,RGMB, ACVR2A,CHRD, RGMA |
| gga04060 | Cytokine-cytokine receptor interaction | 7 | BMPR1A,ACVR2B,BMPR2,BMP2,BMPR1B,ACVR1,ACVR2A | |
| gga04080 | Neuroactive ligand-receptor interaction | 4 | PTGFR/F2RL1/PRLL/EDNRB |
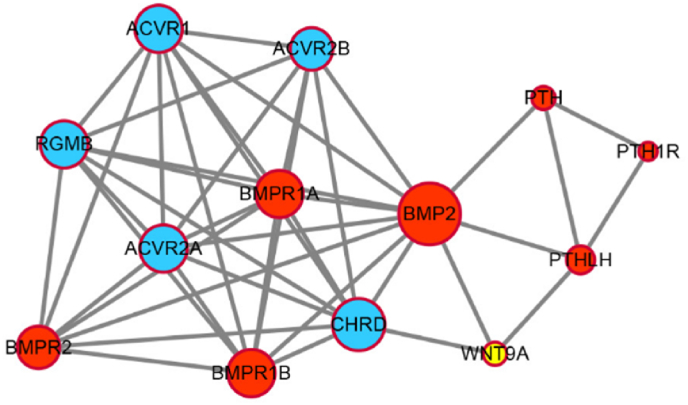
PPI analysis was performed on 13 DEGs as the following activin A receptor type 1 (ACVR1), activin A receptor type 2A (ACVR2A), activin A receptor type 2B (ACVR2B), repulsive guidance molecule BMP co-receptor b (RGMB), bone morphogenetic protein receptor type 2 (BMPR2), bone morphogenetic protein receptor type 1A (BMPR1A), bone morphogenetic protein 2 (BMP2), bone morphogenetic protein receptor type 1B (BMPR1B), chordin (CHRD), parathyroid hormone (PTH), parathyroid hormone 1 receptor (PTH1R), parathyroid hormone like hormone (PTHLH), and wnt family member 9A (WNT9A). To be specific, the size and color were influenced by the degree and the combined score dictating the edge size. Hence, low value corresponded to small size and light color. The result was shown in Fig. 5. BMP2 was identified as the key DEG between controls and recovery from FDM in our study.
Fig. 5.
Protein-protein interaction analysis of differentially expressed genes in response to control vs FDM 0hrs and bone morphogenetic protein 2 (BMP2) was identified as the key gene.
4. Discussion
In this study, we sought to identify DEGs in retina between early and late stage FDM and then further analyzed functional annotation and potential biomarkers as a reliable therapy for myopia progression. We investigated that BEND6 was the only DEG although it was not statistically significant. This finding was inconsistent with the previous study that identified BEND6 causing reciprocal effects on neural stem cell renewal and neurogenesis [27]. Thus, we inferred that BEND6 may inhibit neuron genesis and signal connection in cells though considering no significant statistical meaning.
BMPs have a profound impact in bone formation and repair. Recent studies have underestimated their vital role in normal organs proliferation and vascular homeostasis in chronic diseases. Elevated expression amounts of BMPs have been associated with the commodities of diabetes mellitus. Increasing evidence from recent study demonstrates the involvement of BMP signaling in retinal inflammation, hyperpermeability and pathological neovascularization in diabetic retinopathy [28]. Previous studies have found that cellular signal transduction pathway and ion ligand activate pathway were related to proliferative vitreoretinopathy, proliferative diabetic retinopathy and wet age-related macular degeneration in zebrafish and mouse retina [29,30]. However, general pathway mechanism corresponding to FDM was still far from investigated.
Numerous mechanisms were detected to be associated with FDM through functional annotation of the DEGs. Among them, some have been reported to be related to myopia, including immunity [31,32], inflammation [[32], [33], [34]], and BMP2 upregulation [35,36]. We further analyzed the efficiency of BMP2 dysregulation on myopic retina by comparing early and late recovery from FDM. Because BMP2 dysregulation plays a pathogenic role in promoting complex growth-modulatory signal pathways. In early myopia, scleral fibroblast is a risk factor of disease progression, thus, our study reporting the influence of BMP2 on retina is vital to understand the scleral fibroblast function in myopia. In addition, we proposed that BMP2 was up-regulated in FDM 0hrs, and 4 DEGs were detected to skew myopic retina into a disease-promoting phenotype through scleral fibroblasts metabolism. BMP2 has been revealed to be up-regulated in FDM [36] and breast cancers [37]. Besides, it has been reported that BMP2 was involved with the adhesion of monocytes to fibronectin and endothelial cells [38], suggesting that BMP2 may contribute to the inflammatory system. However, the role of BMP2 in FDM and myopia pathogenesis has never been reported. Among the 14 mRNA target genes, BMP2, interacting with remaining six genes, was the most significant up-regulated gene. Meanwhile, our study observed that BMP2 disrupted the endothelial barrier function, induced endothelial-leukocyte adhesion and up-regulated retinal vascular endothelial growth factor levels, inflammatory markers and cytokines such as ICAM-1, IL-6 and IL-8 [28]. BMP2 has been found to be enriched in Smad pathway and up-regulated in myopia by Wang et al. [39]. They inferred that BMP2 may boost myopia by elevating the proliferation and differentiation of Human sclera fibroblast (HSF), as well as to benefit extracellular matrix synthesis potentially through classical pathway, which coincides with our results.
To the best of our knowledge, this is the first study to explore the DEGs by RNA sequencing analysis aiming to obtain deeply understanding of the pathophysiological mechanism of myopia. Besides, our study is of significance for other pathway-related diseases in adolescence such as refractive error and anisometropia. It should be noted that the present study has limitations. First, the relatively small sample size of this study may limit our ability. Therefore, we established many comparisons in multiple aspects to avoid possible bias. Secondly, our study was initially performed based on animal retina not human subjects. The main reason was little clinical data for human could be acquired in the present database. Last but not least, there is a lack of relevant experiments to identify the accuracy of bioinformatic results. To avoid potential risks, we implemented supreme criteria while selecting samples by setting |log 2fold change|>2, and FDR <5% in the GEO database. Further studies should be conducted on BMP2 to prove valuable functions in myopia prevention, as modulation of HSF may be a novel strategy for indulging myopia progression.
5. Conclusion
Taken together, the findings of this study suggest that cellular signal transduction plays a significant role in both myopic retina and sclera fibroblast about occurrence and progression of myopia. From the perspective of myopia prevention, it is an urgent method to inhibit BMP2 expression as well as proliferation of sclera fibroblast, which would induce risks of high myopia in myopic subjects, especially young children. To explore a supplementary bioinformatical signal pathway in BMP2 provides an original treatment of myopia progression that is based on molecular mechanism of myopia in clinical practice. In addition, BMP2 potentially represents a novel marker for myopia and may modulate extracellular matrix homeostasis in FDM.
Funding statement
This research did not receive a specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare no conflicts of interest regarding the publication of this article.
Contributor Information
Chun-Wen Chen, Email: wy56525169@163.com.
Jing-Yan Yao, Email: yjysdfy@163.com.
Data availability
The data used and analyzed for the present study are available from the corresponding author.
References
- 1.Li H.H., Huo L.J., Gao Z.Y., Zhao F., Zeng J.W. Regulation of scleral fibroblast differentiation by bone morphogenetic protein-2. Int. J. Ophthalmol. 2014;7(1):152–156. doi: 10.3980/j.issn.2222-3959.2014.01.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H.H., Sun Y.L., Cui D.M., Wu J., Zeng J.W. Effect of dopamine on bone morphogenesis protein-2 expression in human retinal pigment epithelium. Int. J. Ophthalmol. 2017;10(9):1370–1373. doi: 10.18240/ijo.2017.09.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seko Y., Azuma N., Takahashi Y., Makino H., Morito T., Muneta T., et al. Human sclera maintains common characteristics with cartilage throughout evolution. PLoS One. 2008;3(11) doi: 10.1371/journal.pone.0003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L., Frost M.R., Siegwart J.T., Jr., Norton T.T. Altered gene expression in tree shrew retina and retinal pigment epithelium produced by short periods of minus-lens wear. Exp. Eye Res. 2018;168:77–88. doi: 10.1016/j.exer.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H., Cui D., Zhao F., Huo L., Hu J., Zeng J. BMP-2 is involved in scleral remodeling in myopia development. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0125219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H.P., Lin Y.J., Lin W.Y., Wan L., Sheu J.J., Lin H.J., et al. A novel genetic variant of BMP2K contributes to high myopia. J. Clin. Lab. Anal. 2009;23(6):362–367. doi: 10.1002/jcla.20344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeRubertis F.R., Craven P.A., Melhem M.F., Salah E.M. Attenuation of renal injury in db/db mice overexpressing superoxide dismutase: evidence for reduced superoxide-nitric oxide interaction. Diabetes. 2004;53(3):762–768. doi: 10.2337/diabetes.53.3.762. [DOI] [PubMed] [Google Scholar]
- 8.Katagiri T., Watabe T. Bone morphogenetic proteins. Cold Spring Harbor Perspect. Biol. 2016;8(6) doi: 10.1101/cshperspect.a021899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang R.N., Green J., Wang Z., Deng Y., Qiao M., Peabody M., et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 2014;1(1):87–105. doi: 10.1016/j.gendis.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarez-Peregrina C., Sánchez-Tena M., Martinez-Perez C., Santiago-Dorrego C., Yvert T., Andreu-Vazquez C., et al. The influence of genetics in myopia control: a pilot study. J. Clin. Med. 2021;10(4):808. doi: 10.3390/jcm10040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bremond-Gignac D. [Myopia in children] Med. Sci. (Paris) 2020;36(8–9):763–768. doi: 10.1051/medsci/2020131. [DOI] [PubMed] [Google Scholar]
- 12.Flitcroft D.I., He M., Jonas J.B., Jong M., Naidoo K., Ohno-Matsui K., et al. IMI - defining and classifying myopia: a proposed set of standards for clinical and epidemiologic studies. Invest. Ophthalmol. Vis. Sci. 2019;60(3):M20–M30. doi: 10.1167/iovs.18-25957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holden B.A., Fricke T.R., Wilson D.A., Jong M., Naidoo K.S., Sankaridurg P., et al. Global prevalence of myopia and high myopia and temporal trends from 2000 through 2050. Ophthalmology. 2016;123:1036–1042. doi: 10.1016/j.ophtha.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Chua S.Y., Sabanayagam C., Cheung Y.B., Chia A., Valenzuela R.K., Tan D., et al. Age of onset of myopia predicts risk of high myopia in later childhood in myopic Singapore children. Ophthalmic Physiol. Opt. : J. Br. Coll. Ophthalmic Opticians (Optometrists) 2016;36:388–394. doi: 10.1111/opo.12305. [DOI] [PubMed] [Google Scholar]
- 15.Tideman J.W.L., Polling J.R., Vingerling J.R., Jaddoe V.W.V., Williams C., Guggenheim J.A., et al. Axial length growth and the risk of developing myopia in European children. Acta Ophthalmol. 2018;96:301–309. doi: 10.1111/aos.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li F.F., Yam J.C. Low-concentration atropine eye drops for myopia progression. Asia Pac J Ophthalmol (Phila). 2019;8(5):360–365. doi: 10.1097/APO.0000000000000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong Q., Janowski M., Luo M., Wei H., Chen B., Yang G., et al. Efficacy and adverse effects of atropine in childhood myopia: a meta-analysis. JAMA Ophthalmol. 2017;135:624–630. doi: 10.1001/jamaophthalmol.2017.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tedja M.S., Wojciechowski R., Hysi P.G., Eriksson N., Furlotte N.A., Verhoeven V.J.M., et al. Genome-wide association meta-analysis highlights light-induced signaling as a driver for refractive error. Nat. Genet. 2018;50(6):834–848. doi: 10.1038/s41588-018-0127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verhoeven V.J., Hysi P.G., Wojciechowski R., Fan Q., Guggenheim J.A., Höhn R., et al. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat. Genet. 2013;45(3):314–318. doi: 10.1038/ng.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vocale L.G., Crewther S., Riddell N., Hall N.E., Murphy M., Crewther D. RNA-seq and GSEA identifies suppression of ligand-gated chloride efflux channels as the major gene pathway contributing to form deprivation myopia. Sci. Rep. 2021;11(1):5280. doi: 10.1038/s41598-021-84338-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai Q., Andreu-Agullo C., Insolera R., Wong L.C., Shi S.H., Lai E.C. BEND6 is a nuclear antagonist of Notch signaling during self-renewal of neural stem cells. Development. 2013;140(9):1892–1902. doi: 10.1242/dev.087502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elmasry K., Habib S., Moustafa M., Al-Shabrawey M. Bone morphogenetic proteins and diabetic retinopathy. Biomolecules. 2021;11(4):593. doi: 10.3390/biom11040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conedera F.M., Quintela Pousa A.M., Presby D.M., Mercader N., Enzmann V., Tschopp M. Diverse signaling by TGFβ isoforms in response to focal injury is associated with either retinal regeneration or reactive gliosis. Cell. Mol. Neurobiol. 2021;41(1):43–62. doi: 10.1007/s10571-020-00830-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan J., Shen W., Lee S.R., Mathai A.E., Zhang R., Xu G., et al. Targeting the Notch and TGF-β signaling pathways to prevent retinal fibrosis in vitro and in vivo. Theranostics. 2020;10(18):7956–7973. doi: 10.7150/thno.45192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleiszig S.M.J., Kroken A.R., Nieto V., Grosser M.R., Wan S.J., Metruccio M.M.E., et al. Contact lens-related corneal infection: intrinsic resistance and its compromise. Prog. Retin. Eye Res. 2020;76 doi: 10.1016/j.preteyeres.2019.100804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei C.C., Kung Y.J., Chen C.S., Chang C.Y., Lin C.J., Tien P.T., et al. Allergic conjunctivitis-induced retinal inflammation promotes myopia progression. EBioMedicine. 2018;28:274–286. doi: 10.1016/j.ebiom.2018.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue M., Ke Y., Ren X., Zhou L., Liu J., Zhang X., et al. Proteomic analysis of aqueous humor in patients with pathologic myopia. J. Proteonomics. 2021;234 doi: 10.1016/j.jprot.2020.104088. [DOI] [PubMed] [Google Scholar]
- 34.Lin H.J., Wei C.C., Chang C.Y., Chen T.H., Hsu Y.A., Hsieh Y.C., et al. Role of chronic inflammation in myopia progression: clinical evidence and experimental validation. EBioMedicine. 2016;10:269–281. doi: 10.1016/j.ebiom.2016.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Azmoun S., Hang A., Zeng J., Eng E., Wildsoet C.F. Retinal defocus and form-deprivation induced regional differential gene expression of bone morphogenetic proteins in chick retinal pigment epithelium. J. Comp. Neurol. 2020;528(17):2864–2873. doi: 10.1002/cne.24957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Liu Y., Hang A., Phan E., Wildsoet C.F. Differential gene expression of BMP2 and BMP receptors in chick retina & choroid induced by imposed optical defocus. Vis. Neurosci. 2016;33:E015. doi: 10.1017/S0952523816000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scimeca M., Giocondo R., Montanaro M., Granaglia A., Bonfiglio R., Tancredi V., et al. BMP-2 variants in breast epithelial to mesenchymal transition and microcalcifications origin. Cells. 2020;9(6):1381. doi: 10.3390/cells9061381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pardali E., Makowski L.M., Leffers M., Borgscheiper A., Waltenberger J. BMP-2 induces human mononuclear cell chemotaxis and adhesion and modulates monocyte-to-macrophage differentiation. J. Cell Mol. Med. 2018;22(11):5429–5438. doi: 10.1111/jcmm.13814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q., Xue M.L., Zhao G.Q., Liu M.G., Ma Y.N., Ma Y. Form-deprivation myopia induces decreased expression of bone morphogenetic protein-2, 5 in Guinea pig sclera. Int. J. Ophthalmol. 2015;8(1):39–45. doi: 10.3980/j.issn.2222-3959.2015.01.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used and analyzed for the present study are available from the corresponding author.