Abstract
Subtyping Cryptosporidium parvum for outbreak investigations or epidemiological surveillance usually relies on DNA sequence analysis of a gene coding for a 60 KDa glycoprotein (gp60). Although gp60 can be useful for allelic discrimination and to help investigate sources and routes of transmission, the presence of common subtypes and recombination during the parasite's sexual life-cycle demand a multilocus-based method for more discriminatory genotyping. While whole genome sequencing would provide the ultimate approach, it is a time consuming and expensive option for faecal parasites such as Cryptosporidium that occur at low density and are difficult to propagate routinely. In this study, we selected and evaluated a panel of previously identified variable-number tandem-repeat (VNTR) markers, to establish a multilocus genotyping scheme based on fragment sizing, appropriate for inter-laboratory surveillance and outbreak investigations. Seven VNTR markers were validated in vitro and demonstrated typeability of 0.85 and discriminatory power of 0.99. The discriminatory power was much greater than the currently used gp60 sequencing (0.74), which identified 26 subtypes, compared to 100 different MLVA profiles within the same sample set. The assay was robust, with repeatable results and reproducibility across three laboratories demonstrating the scheme was suitable for inter-laboratory comparison of C. parvum subtypes. As the majority of genotypes (79%) were unique among epidemiologically unrelated samples, there was efficiency to infer linkage, and epidemiological concordance was observed in historical outbreaks. We propose that the multilocus variable number of tandem repeats analysis scheme is suitable to assist outbreak investigations.
Keywords: Cryptosporidium parvum, MLVA scheme, Multilocus, Outbreaks, Subtype, Tandem repeat, Validation
1. Introduction
The protozoan Cryptosporidium is one of the leading infectious causes globally of diarrheal disease (Kotloff et al., 2013; Khalil et al., 2018). Estimates of average prevalence in human infection have been reported at 4.3% in high-income countries and 10.4% in low-income countries (Dong et al., 2020). Disease can be particularly severe and prolonged in immunocompromised patients and in children under 5 years in developing countries.
Transmissible oocysts containing four infective sporozoites are shed in the faeces of infected hosts and are able to cause new infections without any maturation period. This enables transmission through direct contact with an infected person or animal, as well as through environmental routes such as water, food and fomites due to the robust oocyst wall that can protect the parasites in harsh conditions for long periods of time. These means of transmission facilitate varied risk factors for exposure and disease, and multiple outbreaks, caused by person-to-person or animal-to-person spread, and consumption of contaminated food or water have been reported (Cacciò and Chalmers, 2016; Smith et al., 2021).
There are over 40 species and many more genotypes of Cryptosporidium, some of which are host adapted, thus identifying the infecting species, genotype and subtype can give an indication of the source and route of transmission. It is therefore crucial to provide public health authorities and scientists with rapid and accurate microbiological information for risk assessment, to track sources of contamination, and enable implementation of appropriate control measures. Among the many Cryptosporidium species described to date, the potentially zoonotic species Cryptosporidium parvum poses a major challenge for surveillance and control due to the variety of sources of infection and routes of transmission (Cacciò and Chalmers, 2016).
The most widely used method for discriminating within Cryptosporidium spp. is DNA sequence analysis of the 60-kDa glycoprotein gene (gp60) (Strong et al., 2000). Analysis of the gp60 gene is based on counts of, and variation within, tandem poly-serine repeats and sequence variants downstream of this microsatellite region. However, this method can be limited by commonly found subtypes (Chalmers and Cacciò, 2016; Chalmers et al., 2019), and recombination between different lineages during the sexual stage of the life-cycle (Feng et al., 2002; Tanriverdi et al., 2007) contributing to heterogeneity inferred by the intragenic linkage disequilibrium of natural C. parvum populations (Mallon et al., 2003; Tanriverdi et al., 2008). This suggests that a multilocus approach may be more appropriate to characterise Cryptosporidium populations.
Numerous loci have been investigated in multilocus genotyping studies, however a standardised set of markers has yet to be universally adopted for Cryptosporidium (Robinson and Chalmers, 2012; Chalmers et al., 2018). Genetic loci containing a variable number of tandem repeats (VNTR) can enable rapid characterization of C. parvum isolates and infer linkage (Hotchkiss et al., 2015; Chalmers et al., 2017). However, one study, using nine markers, highlighted that some did not conform with preferred criteria for fragment sizing and may not be ideal for inter-laboratory surveillance and outbreak investigations (Chalmers et al., 2017). The selection of multilocus VNTR analysis (MLVA) loci should as far as possible be represented by a spread across chromosomal location, contain perfect homogenous repeats of ≥5 base pairs, not contain insertions or deletions in the repeat units and be flanked by 100% conserved sequences (Nadon et al., 2013). In an attempt to develop a harmonised approach to intra-species differentiation for Cryptosporidium the agreed processes and criteria were established during an expert workshop in 2016 (Chalmers and Cacciò, 2016; Chalmers et al., 2018). These processes cover the identification and selection of markers, evaluation and validation of the multilocus genotyping (MLG) scheme, and implementation and sustentation of the scheme (Chalmers et al., 2018).
In a previous study we identified 28 VNTR loci in C. parvum genomes that might be suitable for developing a robust MLVA scheme (Pérez-Cordón et al., 2016), and seven were selected using the published criteria (Chalmers and Cacciò, 2016; Chalmers et al., 2018). Initial analysis of a large panel of C. parvum samples using the QIAxcel Advanced capillary system (Qiagen, Germany) for fragment sizing showed that this platform provided insufficient resolution of our fragments and returned a high number of unresolved ambiguous fragment sizes (Pérez-Cordón et al., 2017). A similar finding was reported where this platform was used for Plasmodium vivax microsatellite genotyping (Manrique et al., 2015). By using an Applied Biosystems Genetic Analyzer (ThermoFisher Scientific, UK), more reliable fragment sizing was achieved. Here we describe the design, optimisation, and evaluation of PCR primers for VNTR markers and validation of a seven-marker MLVA scheme, including application in outbreak investigations.
2. Material and methods
2.1. Selection of VNTR markers
Seven markers were selected for inclusion in the C. parvum MLVA scheme, to be amplified in a four-plex and a three-plex PCR set-up (Table 1). Six markers were selected from 28 candidates identified previously, and shortlisted based on the number of VNTR alleles identified in silico compared to gp60 sequencing and their suitability for inclusion in a C. parvum MLVA scheme as described by Pérez-Cordón et al. (2016). The seventh marker was MM19 (Tanriverdi and Widmer, 2006) that was shown to be suitable and useful in a previous study (Chalmers et al., 2017).
Table 1.
VNTR loci and primers for MLVA analysis in Cryptosporidium parvum.
| PCR | Locus (alternative name) | Reference | Fragment sizing primer sequences and dyes used on forward primer | Sanger sequencing primer sequences |
|---|---|---|---|---|
| FOUR-PLEX | cgd1_470_1429 (GRH) | Herges et al., 2012 | F: PET-CTCAGGAAGAGGAAGATACGG | F: AGGAGGAAGTGGCGAATCAGG |
| R: GGAAGGTATGGCAGCAAAAG | R: GATAGGGTAGTTTTACCGGGGTTG | |||
| cgd4_2350_796 | Pérez-Cordón et al., 2016 | F: VIC-GGGTCAATCAGGCATGAGC | F: CAATGGATGCCAGACAAGCT | |
| R: TTGCATGTTTATCATATTATTTCCCAT | R: AAGGCTACGAGCAGATTAACG | |||
| cgd8_4440_NC_506 | Pérez-Cordón et al., 2016 | F: NED-CTCAATATTTTTTCCACACCTGAAC | F: CCATATTGAATGCAATGCCAAATC | |
| R: ACTGCCTGAGAAAGGAACCA | R: GCATGGATTAACGACCAGTTG | |||
| cgd8_4840_6355 (MM19) | Morrison et al., 2008 | F: FAM-GTTCCAGGAATATTTGATTCTGC | F: GCGGAGAAGGAGGATTTAATTC | |
| R: CTCCTACGCCAACTCCTA | R: TTACAACTCCAACTCCACCAC | |||
| THREE-PLEX | cgd5_10_310 (MSF) | Tanriverdi and Widmer, 2006 | F: VIC-AAGGTGAAGGAATCAAAGGC | F: TGACCCTTCTATTGAGCCAC |
| R: TTTGTCCTTCTTGCCCTCGG | R: ACTTCTTCCTCATCAGTAGC | |||
| cgd5_4490_2941 | Pérez-Cordón et al., 2016 | F: FAM- CAGTGAATAACTCTGAACGGAAC | F: ATCCAGTAATTTCTGACATTTCTGAG | |
| R: TTGATTTTGGGTTCGGTATTG | R: TAATTAACATTTTCAGGGTCTCTGG | |||
| cgd6_4290_9811 (MSC6–5) | Xiao and Ryan, 2008 | F: NED-CATTGGAACGTAAACAAAACCA | F: GACTTGGATTTGGACTTACACC | |
| R: CTAGCCGAATCTGGCGGTAT | R: TCCAAATGATGTAAATACTCCAGC |
2.2. Design, optimisation, and evaluation of primers
Primer 3 Plus software (Untergasser et al., 2012) was used to design primers under default parameters. For fragment sizing, it is desirable that primers are located as close as possible to the VNTR region, so each microsatellite together with 50 bp upstream and downstream was used as the input sequence to run the software. In some cases, the position of the primers was manually relocated a few bases downstream or upstream to prevent overlap with the repeat region or when melting temperatures differed by more than 3 °C between them. Sequence alignments from available genomes were used to make sure that primers are located in conserved regions and would amplify these loci from those C. parvum isolates. The selected primers were further analysed with OligoAnalyzer 3.1 software (Owczarzy et al., 2008) evaluating oligo Na++, Mg++ and dNTP concentrations as described previously (Chalmers et al., 2017), and were modified until predicted similar melting temperatures were achieved. In silico primer specificity was checked against the RefSeq Representative Genome Database using NCBI's Primer-BLAST tool (Ye et al., 2012).
In vitro optimization of primers was initially achieved using single round PCRs under the same conditions in 20 μL reactions with 1 μL of template DNA, 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate (dNTP), 200 nM of each primer, and 2.5 U of Taq DNA Polymerase (HotStar Taq, Qiagen, Germany). Amplification conditions consisted of an initial denaturation of 95 °C for 15 min, followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s, with a final extension of 72 °C for 5 min.
To investigate analytical sensitivity, C. parvum genomic DNA was extracted from salt-float purified oocysts or directly from stools, using Qiagen QIAamp extraction kits (blood and bodily fluid kit or stool kit, respectively), and stored at -20 °C until use as described previously (Robinson et al., 2020). Four dilutions of C. parvum genomic DNA, extracted from salt-float purified oocysts and quantified by qPCR to provide the equivalent to 25, 10, 5 and 1 oocysts/PCR were tested in triplicate with each primer pair. The primers were also tested on DNA extracted from a set of samples that were both semi-purified by salt flotation prior to extraction and directly extracted from the stools to ensure that the presence of excess non-Cryptosporidium DNA in the samples did not adversely affect the results.
The specificity of the primers was evaluated in vitro; first using DNA extracted directly from 22 known Cryptosporidium-negative stools and secondly from a panel of verified genomic DNA samples. This included DNA from 10 Cryptosporidium species commonly found in humans or ruminants (C. hominis, C. cuniculus, C. meleagridis, C. felis, C. canis, C. ubiquitum, C. xiaoi, C. andersoni, C. bovis, and C. ryanae), from other parasitic protozoa (Giardia duodenalis assemblages A and B, Toxoplasma gondii, Cyclospora cayetanensis, Neospora caninum, Sarcocystis tenella, Eimeria tenella, E. acervulina, E. maxima) and from those hosts we most commonly investigate in outbreaks; human, sheep and cattle.
The forward primers were labelled with fluorescent dyes PET, VIC, NED and 6-FAM (Life Technologies, UK) (Table 1). The seven primer sets were configured as two multiplex PCRs, one of four and one of three markers, according to amplification efficiency and similarity of annealing temperatures (Table 1). The multiplex PCR reactions were performed using the Type-it Microsatellite PCR kit (Qiagen, Germany) in final volumes of 25 μL, including 2 μL of template DNA. The PCR reactions were prepared on an epMotion 5075 machine (Eppendorf, Germany) in 96-well plates. PCR cycling conditions were those recommended by the manufacturer of the Type-it PCR kit (Qiagen, Germany); an initial Taq activation of 95 °C for 5 min followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 63 °C for 90 s and extension at 72 °C for 30 s with a final extension step at 60 °C for 30 min. Using 30 cycles reduced the number of spurious peaks seen with higher numbers of cycles.
To confirm the number of VNTR repeats of new alleles and ensure accurate binning, these were Sanger sequenced (Source Bioscience, UK) using an alternative set of primers located ~50 bp distal to the alignment location of the fragment sizing primers (Table 1; Supplementary File 1) and performed as simplex PCR reactions. When sequence files were analysed, it was found to be easier to see the repeat units by translating into amino acids as described in Supplementary File 1. Representative sequences of different repeat lengths at each locus have been submitted to GenBank under accession numbers OM832408-OM832443 to enable future users to download and create a backbone alignment against which new alleles can be confirmed and the assay validated within their laboratory.
2.3. Validation panel of samples
The validation panel consisted of 259 C. parvum DNA samples confirmed by real-time PCR as described previously (Robinson et al., 2020) from humans and animals, representing sporadic cases with spatio-temporal variation, and cases and suspected sources in selected human outbreaks linked to animal premises (Fig. 1). Most of the sporadic cases were from a case control study in the north west of England and Wales in 2001–2002 (Hunter et al., 2004) and the animal cases were from the Animal and Plant Health Agency's diagnostic laboratories in the same area and timeframe. Additional sporadic human cases were from clinical microbiology laboratories local to the outbreaks around the time of each investigation. Samples from four outbreaks in England and Wales in 2016 and one in Wales in 2018 were submitted during the course of the investigation, thought at the time to be linked to the outbreak. All samples were previously characterised by sequence analysis of the gp60 gene as described elsewhere (Chalmers et al., 2019).
Fig. 1.
Samples included in the validation panel.
2.4. Verification of the MLVA typing scheme
Verification was undertaken to establish typeability, discriminatory power, epidemiologic concordance and robustness:
-
a)
Robustness (repeatability and reproducibility) was assessed using a subset of 24 samples (including controls) encompassing different gp60 genotypes from both humans and animals. A nuclease free water sample was used as a no template control and three C. parvum DNA samples extracted from stools were used as positive controls. The latter also provided known MLVA profiles enabling calibration. To assess repeatability, samples were tested and analysed at the Cryptosporidium Reference Unit (CRU) by two scientists using the same reagents and machines on different dates. Reproducibility was assessed by sending the same blind-coded DNA, a 96-well plate, and PCR reagents to the Moredun Research Institute (MRI, UK) and the Scottish Microbiology Reference Laboratories, Glasgow (SMiRL, UK) who tested, analysed and returned the results to the CRU.
-
b)
Typeability (T) (the number of isolates assigned a type within the total number of isolates) was assessed for each of the seven VNTR loci individually and for the seven-locus scheme as a whole using the full set of 259 samples.
-
c)
Discriminatory power (the probability that two unrelated strains will be placed into different types) was assessed using the Hunter Gaston Discriminatory Index (HGDI) (Hunter and Gaston, 1988) for 136 epidemiologically unrelated samples from within the panel that provided positive amplification for all seven markers. The maximum number of genotypes discriminated as a function of the number of loci used was investigated to identify the most discriminatory markers in combination using AMaCAID software (Caroli et al., 2011). A relative abundance curve (Whittaker plot) and minimum spanning tree were used to visualize the diversity of genotypes in these samples.
-
d)
Epidemiological concordance was assessed by testing 60 samples from five outbreaks including human cases and possible animal sources from the panel and minimum spanning trees were used to visualize the diversity of genotypes in these samples and local background cases.
2.5. Fragment analysis and subtyping
PCR products were sent for fragment sizing (DNA Sequencing and Services Unit, University of Dundee, UK) using an ABI 3500 Genetic Analyzer (Applied Biosystems, UK). The fluorescently-labelled amplicons enabled differentiation of the loci within the multiplex reaction mixes. Raw .fsa files were downloaded and analysed using Bionumerics software (Applied Maths, Belgium) (CRU and SMiRL) or Geneious software (Biomatters, New Zealand) (MRI) to obtain the number of repeats contained in the amplified fragment. MLGs were named by a profile formed of seven numbers each corresponding to the number of VNTR repeat motifs for each locus in the following chromosomal (cgd) order: cgd1_470_1429; cgd4_2350_796; cgd5_10_310; cgd5_4490_2941; cgd6_4290_9811; cgd8_4440_NC_506; cgd8_4840_6355. The number of repeats was used for allele naming rather than fragment size to enable consistency and comparison of results from alternative primer sets and sizing platforms.
Mixed alleles were defined as the presence of multiple peaks where the peak height ratio was >0.25 of the corresponding predominant allele, as described in MLVA assays used for Plasmodium (de Souza et al., 2015). Care was taken when determining whether multiple peaks were true, as bleed-through from highly fluorescent peaks in other channels was occasionally seen. Null alleles were defined where no peaks were detected.
MLVA profiles were expressed as shown by the following examples:
-
a)
a complete set of alleles, 4‐14‐5-7-27-28-17.
-
b)
with multiple alleles at loci cgd5_10_310 and cgd8_4440_NC_506, 4‐14-5/7‐7‐27-28/31‐17.
-
c)
with a null allele at cgd1_470_1429, Ø-14-5-7-27-28-17.
A detailed description of the assay and guidance on the interpretation of the fragment analysis and sequencing can be downloaded (Supplementary File 1).
2.6. Statistical analysis
HGDI was calculated using the equation described by Hunter and Gaston (1988). The maximum number of MLGs as a function of the number of VNTR loci included was calculated by AMaCAID run using Model 1 in R (Caroli et al., 2011). A Whittaker plot was used to visualize MLG richness distributions by calculating the number of samples with each MLG as a proportion of the total number of samples and plotting them from highest to lowest. Minimum spanning trees were created using the default settings in Bionumerics version 7.6.3 (Applied Maths, Belgium).
3. Results and discussion
The analytical sensitivity of the primers showed a mean of 81% replicates positive when DNA extracted from 25 C. parvum oocysts was tested per PCR (range 67–100% for each individual primer set) (Supplementary Table 1). This was considered acceptable as stool samples identified as Cryptosporidium-positive in diagnostic laboratories would be unlikely to contain such small numbers of oocysts and the assay was not developed for use as a detection method.
No amplification was detected from either the panel of 22 known Cryptosporidium-negative DNA samples extracted directly from stool or the DNA from other non-Cryptosporidium organisms, suggesting the primers were specific for Cryptosporidium. Additionally, when fragments were sequenced to confirm new alleles, only Cryptosporidium sequences were identified. However, the organisms included in any specificity assessment will need to consider the endemicity of other pathogens in the local patient population. This scheme was designed and optimised specifically for subtyping C. parvum; however, the loci included may also be present and useful in other Cryptosporidium species. For example, all 7 markers were found in silico to be present in C. hominis, but with mismatches in the primer sequences. As may be expected due to the genetic similarity of the species, in vitro testing of three C. hominis and three C. cuniculus samples gave some limited amplification in several of the loci. C. hominis showed amplification in all but cgd1_470_1429 and MSF, with cgd4_2350_796 positive in two of the three samples. This concurs with previous work done investigating MSF, which required separate C. parvum and C. hominis species-specific primers (Tanriverdi and Widmer, 2006). C. cuniculus was negative in cgd1_470_1429, MSF, cgd4_2350_796 and cgd8_4840_6355 (MM19), positive in all samples at cgd5_4490_2941, and two of three samples at the remaining three loci (cgd4_2350_796, cgd6_4290_9811 and cgd8_4440_NC_506). All samples for each of the remaining eight Cryptosporidium species tested showed no amplification at any of the loci. Additional design and evaluation is required to determine whether there is potential for these loci to be included in a subtyping scheme for C. hominis or other Cryptosporidium species.
Comparison of the repeatability and reproducibility data between operators and laboratories showed a 100% agreement (Supplementary Table 2).
The overall typeability of the MLVA scheme for the full panel of 259 samples was T = 0.85 (Table 2; Supplementary Table 3); there was lack of amplification in 28 samples at 2–7 loci and 10 samples lacked amplification at one locus (Supplementary Table 4). The typeability of each individual marker ranged from 0.91 to 0.97, where locus cgd5_10_310 (MSF) showed the highest and both cgd1_470_1429 and cgd4_2350_796 the lowest (Table 2). This high typeability at each individual marker is desirable when considering a marker for use in a subtyping method (Struelens, 1996). The typeability of gp60 was 0.96, slightly below that of the marker with the greatest typeability.
Table 2.
Validation data for the markers and combined MLG scheme.
| VNTR locus | Typeability | Hunter-Gaston discrimination index |
|---|---|---|
| cgd1_470_1429 | 0.91 | 0.17 |
| cgd4_2350_796 | 0.91 | 0.62 |
| cgd5_10_310 | 0.97 | 0.17 |
| cgd5_4490_2941 | 0.93 | 0.42 |
| cgd6_4290 | 0.96 | 0.32 |
| cgd8_4440_NC_506 | 0.92 | 0.92 |
| cgd8_4840_6355 | 0.92 | 0.71 |
| Seven-locus MLG scheme | 0.85 | 0.99 |
The total combined HGDI considering all seven VNTR markers was 0.99, satisfying the requirements for a typing system (> 0.95) (van Belkum et al., 2007). This was higher than that provided for the same panel by gp60 sequencing alone (0.74). The highest HGDI was provided by locus cgd8_4440_NC_506 (0.92). The lowest HGDI was provided by cgd1_470_1429 (0.17) and cgd5_10_310 (0.17), with both loci showing a predominant allele in 91.2% of the unlinked samples; at cgd1_470_1429 this was four tandem repeats and at cgd5_10_310 this was five (Supplementary Table 3).
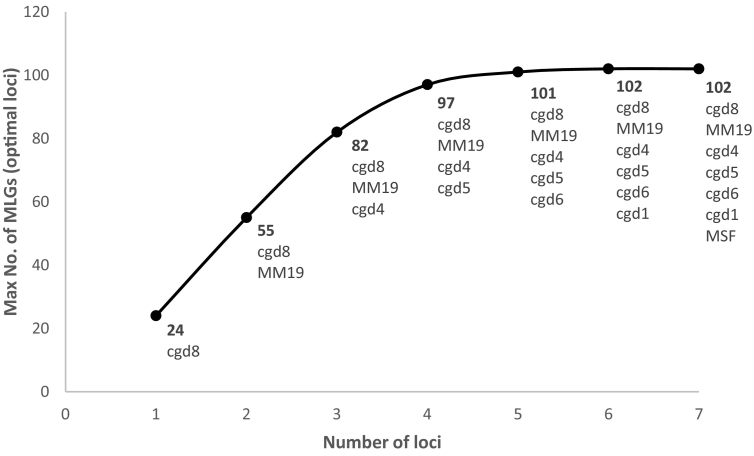
Increasing the number of markers provided an increasing number of MLGs but levelled off after the best four loci were combined, with only an additional five MLGs identified with the addition of the three least discriminatory loci (Fig. 2). The final combination of all seven markers returned 102 different MLGs from 136 epidemiologically unrelated samples (Fig. 2). The continued use of the least discriminatory markers will be reviewed following further testing with more samples as they may be found to be useful for certain types of C. parvum. For example, cgd5_10_310 was found previously to be highly conserved in gp60 subtype family IIa isolates, but more diversity was detected among subtype family IId (Chalmers et al., 2017). A reduction in the number of markers, if found not to compromise discriminatory power, could improve cost and time efficiency during epidemiological and especially outbreak investigations.
Fig. 2.
Maximum number of multilocus genotypes as a function of the number of VNTR loci included for 136 epidemiologically unrelated samples.
The Whittaker plot showed that the majority of MLGs (81/102, 79%) were unique among 136 epidemiologically unrelated samples, as shown by the long flat line on the X axis (Fig. 3) and individual nodes of the minimum spanning tree (Fig. 4). The higher relative abundance of a few genotypes may be due to some apparent unknown epidemiological relatedness between a few samples from the same areas and time period. This genetic clustering identified by MLVA could be useful in identifying unreported outbreaks and allow for improved public health strategies and management.
Fig. 3.
Relative abundance of multilocus genotypes among 136 epidemiologically unrelated samples.
Fig. 4.
A minimum spanning tree showing the variability of full seven-locus MLVA profiles between a set of 136 epidemiologically unrelated samples. Differences between MLVA profiles: Thick solid lines = 1 locus, Thin solid line = 2 loci, Thick dashed line = 3 loci, Thin dashed line = 4 loci, No line > 4 loci. The size of the nodes depends on the number of isolates. Wedges in the nodes indicate the proportion of isolates from different sources with a particular MLVA profile. The distance between nodes in the tree do not reflect genetic distances between genotypes.
Different combinations of polymorphic markers have been used for subtyping Cryptosporidium in recent years (Robinson and Chalmers, 2012), and led to a better understanding of the population genetics of this parasite (Grinberg and Widmer, 2016). However, the lack of a standardised multilocus subtyping scheme led to reliance on gp60 sequencing during epidemiological investigations and while useful to some extent as we have found in our laboratory (Chalmers et al., 2019), the genetic advantages of a multilocus approach have been illustrated (Widmer and Lee, 2010). In this study, the discriminatory power of the multilocus scheme compared to gp60 alone has been shown, and in outbreaks discriminated those involved from background cases (Fig. 5).
Fig. 5.
Minimum spanning trees from the full seven-locus MLVA profiles identified from outbreak and local background cases. Differences between MLVA profiles: Thick solid lines = 1 locus, Thin solid line = 2 loci, Thick dashed line = 3 loci, Thin dashed line = 4 loci, No line > 4 loci. The sizes of the nodes depends on the number of isolates. Wedges in the nodes indicate the proportion of isolates from different sources with a particular MLVA profile. The distance between nodes in the tree do not reflect genetic distances between genotypes.
There was sufficient DNA for both MLVA and gp60 sequencing of 184/189 sporadic isolates of which 155 (84.2%) were typed at all 7 loci by MLVA compared with 174 (94.6%) typeable by gp60. The greater HGDI of MLVA compared to gp60 sequencing manifested as only 26 gp60 subtypes compared to 100 seven-locus MLGs by MLVA (excluding those with mixed alleles) (Supplementary Table 5). The two most common C. parvum gp60 subtypes within the UK, IIaA17G1R1 and IIaA15G2R1, were highly represented (Supplementary Table 5) and allowed us to look at variety within them. Of the 62 IIaA17G1R1 isolates in the panel, 42 had complete, unmixed MLVA profiles at all seven loci consisting of 39 different MLGs; 11 of the IIaA17G1R1 isolates had mixed allele profiles and 9 were negative at one or more loci (Supplementary Table 5). Of the 58 IIaA15G2R1 isolates, 51 had complete unmixed MLVA profiles consisting of 41 different MLGs; 2 of the IIaA15G2R1 isolates had mixed allele profiles and 5 were negative at one or more loci (Supplementary Table 5).
Of the 100 MLVA MLGs identified in sporadic cases, only 20 were found in more than a single sample and where more than one isolate shared an MLG it was not always within the same gp60 subtype (e.g. 4‐14‐5-7-27-28-16 was found in 4 isolates representing gp60 subtypes IIaA15G2R1, IIaA16G3R1 and IIaA13G2R1). This is to be expected, in the same way that any of the other single loci can vary between samples that are identical at all other loci, due to the recombination between isolates in a population, and adds strength to the argument for using multiple locus typing. This suggests that gp60 is still useful and could be used as an additional locus to further discriminate isolates in the multi-locus scheme but carries the additional cost and time of sequencing.
As expected, the variation in outbreaks was mainly much less than seen in sporadic cases. Therefore, clustering of MLGs in sporadic cases might indicate epidemiological linkage and warrant further investigation. Epidemiological concordance was observed, and in two of the outbreaks, A and E, all samples tested had the outbreak profile (Table 3). The samples tested from the other three outbreaks, B, C and D, displayed multiple subtypes (both at gp60 and MLVA), and two samples with mixed MLGs were also apparent (Table 3). Due to the nature of Cryptosporidium infections and outbreaks in farm settings, multiple infections might be expected in some outbreaks. An analysis of farm-related outbreaks has shown that a wide range of potential hosts may be present especially on open or petting farms (Smith et al., 2021). This variety may mean that different subtypes of C. parvum are able to recombine (Morris et al., 2019) resulting in a diversity of MLVA profiles and multiplicity of infection, as illustrated in the outbreaks C and D described below, and previously described by Grinberg and Widmer (2016). On livestock farms there seem to be a combination of unique farm genotypes indicating transmission within farms as well as intra-farm diversity (Ramo et al., 2016; Zhang et al., 2020). This has implications for sampling during investigations of outbreaks that needs to be sufficient to represent the diversity and “catching” the outbreak strains.
Table 3.
Multilocus genotyping and gp60 subtyping of Cryptosporidium parvum isolates linked to human outbreaks.
| Outbreak setting and year | gp60 subtype | MLVA profiles and number of isolates | |
|---|---|---|---|
| A. Open farm, England, 2016 | IIaA19G1R1 | 5-13-3-20-18-9-20 | x8 |
| B. Agricultural college farm, Wales, 2016 | IIaA15G2R1 | 4-13-5-7-27-32-15 (including 2 lambs) | x13 |
| 4-13-5-∅-∅-9-15 | x1 | ||
| ∅-∅-5-∅-27-∅-∅ (lamb) | x1 | ||
| ∅-∅-∅-∅-∅-∅-∅ | x2 | ||
| C. Open farm, England, 2016 | IIaA15G1R2 x 7 | 5-13-3-13-18-9-27 | x6 |
| 5-14-5-7-27-30-17 | x1 | ||
| IIaA17G1R1 x 6 | 4-14-5-7-27-30-17 | x3 | |
| 4-13-5-7-27-9-27 | x1 | ||
| 6-14-3-9-18-9-23 | x1 | ||
| 4/5-14-3/5-7-18/27-9/30-17 | x1 | ||
| D. Open farm at a theme park, England, 2016 | IIaA15G2R1 | 4-14-5-8-27-31-15 | x5 |
| 4-14-5-7-27-28-13 | x3 | ||
| 4-14-5-8-27-28-13 | x2 | ||
| 4-14-5-8-27-31-13 | x1 | ||
| 4-13-5-7-27-33-15 | x1 | ||
| ∅-14-∅-∅-∅-∅-∅ | x1 | ||
| ∅-∅-5-∅-27-9-∅ | x1 | ||
| ∅-∅-5-∅-27-∅-∅ | x1 | ||
| 4-14-3/5-8-18/27-31-15 | x1 | ||
| IIaA17G2R1 | 4-14-5-7-18-36-15 | x1 | |
| E. Residential activity farm, Wales, 2018 | IIdA24G1 | 6-13-3-12-18-14-4 | x5 |
Outbreak B presented a predominant MLVA profile (4‐13‐5-7-27-32-15) in 13 of 17 samples, including two lambs sampled to investigate the source of the outbreak. This improved the microbiological evidence, strengthening the hypothesis that the outbreak was linked to contact with lambs. One case sample differed at a single locus (cgd8_4440_NC_506), with 9 repeats seen instead of 32. A complete MLVA profile could not be obtained for four samples, including this one, due to the presence of negative amplification at one or more loci. Some of the negative amplification may be due to null alleles, particularly at cgd1_470_1429 where occasionally all other loci are strongly positive in certain samples (data not shown), and require further investigation to determine the reason for this lack of amplification.
Outbreak C was a mixed subtype outbreak by both gp60 and MLVA. Six of the seven IIaA15G1R2 cases showed the same MLVA profile (5‐13‐3-13-18-9-27), with the remaining sample differing at 6 of the 7 loci (Table 3). However, these differences matched the alleles in the predominant MLVA profile (4‐14‐5-7-27-30-17) from the other gp60 subtype, IIaA17G1R1. This illustrates that recombination may have occurred between the different subtypes within the population of parasites. Of the remaining three IIaA17G1R1 samples, one had three alleles also found in the main IIaA15G2R1 MLVA profile and four that were present in the IIaA17G1R1 profile; one sample showed mixed alleles at four loci, with the predominant alleles found in both of gp60 subtypes present; and the third sample differed from both the MLVA profiles at three loci, looking quite different with rarer alleles present at two loci. The finding of two gp60 subtypes as well as the several MLVA profiles may be explained by the exposure (e.g. petting, bottle and dry feeding) to a variety of animals at the farm (including sheep, lambs, cows, calves, rabbits, pigs, guinea pigs and meerkats) together with the long period of time (> 6 weeks) over which the outbreak occurred. It is possible that if a greater proportion of outbreak case samples had been available for testing (only 13 of the 33 case samples were available), more consistency might have emerged in the genotyping.
Outbreak D showed the highest genotypic variability. All but one of the 17 samples were gp60 subtype IIaA15G2R1 with the remaining case IIaA17G2R1and its own MLVA profile (although it shared 5 of the alleles with different IIaA15G2R1 samples from the outbreak). There were 5 different complete MLVA profiles among the IIaA15G2R1 cases. There were two main alleles at each of three loci, present in different combinations. Three samples had null alleles at multiple loci and one sample was mixed at two loci. This outbreak was linked to a petting farm within a theme park which, similarly to outbreak C, involved a high number of visitors and exposure to a variety of animals.
In this study, we showed that the seven-locus MLVA scheme for C. parvum is a useful method for subtyping and characterization of outbreak cases with a higher discriminatory capability than the commonly used subtyping method based on the gp60 sequence analysis. Previously, Mattsson et al. (2008) showed the valuable utility of a MLG approach for the investigation of outbreaks of cryptosporidiosis using a three-locus method (MS1, TP14 and GP15) to identify two different sources of exposure in a swimming-pool outbreak. In our study, even where multiple MLVA profiles were identified there was commonality of alleles, although some outbreaks were more complex than others. On the other hand, it is possible that some cases from outbreaks A and C were misclassified using historical information and background cases may have had links to the farm (outbreak A) (for example, as unidentified secondary cases) or the source of their infection was elsewhere (outbreak C). When the assay is used during an on-going outbreak, this information would be pursued further in real-time as part of the investigation.
Despite the benefits of using several loci for subtyping and inferring linkage, no standard set of loci has been adopted for use in epidemiological surveillance (Robinson and Chalmers, 2012; Chalmers et al., 2018). However, following the discussions at the 2016 expert workshop (Chalmers and Cacciò, 2016; Chalmers et al., 2018), the development and validation of the seven-locus scheme described in this study aims to achieve a more standardised approach for the interspecies investigation of C. parvum. The high level of discrimination among sporadic samples, and relatively conserved alleles within outbreak settings, enables not only a greater understanding of the outbreak subtypes and dynamics, but also allows for additional cases within the local community to be identified and followed up. This was apparent in Outbreaks A and C, where the outbreak MLVA profiles were also seen within the background samples from the local community (Fig. 5) but not in any of the other samples tested during this study. If identical MVLA profiles in background samples were identified in real-time during an outbreak further epidemiological investigation into these cases would be warranted.
Although whole genome sequencing would provide the highest discriminatory capabilities, its integration in the routine workflow of clinical laboratories poses a series of practical issues for its implementation particularly when analysing Cryptosporidium (Morris et al., 2019). Given the high discriminatory power of this MLVA scheme, relative low cost (the seven locus MLVA scheme has a similar cost to sequencing a single gene, e.g. gp60) and fast turnaround time, it appears to be a useful tool to supplement epidemiological investigations.
4. Conclusions
The MLVA scheme provided a much greater discrimination than gp60 sequencing. In general, the majority of sporadic cases had unique MLGs indicating that the MLVA scheme could be a useful means of identifying potentially linked cases and ascertaining transmission pathways. The epidemiological concordance showed some variation among the isolates thought to be from the same outbreak in some instances, but there may have been misclassification of cases associated with the outbreak, multiplicity of infection and/or genetic recombination. However, there was generally a predominant outbreak subtype and where mixed subtypes were identified the MLGs occasionally demonstrated mixed profiles including the outbreak strain(s). The high discriminatory nature of the MLVA also clearly identified cases that may not be part of the outbreaks and would have merited further follow-up. Besides its high discriminatory power, this C. parvum MLVA subtyping scheme is cost-effective and rapid, being able to provide results from DNA within a working day, which is crucial for outbreak investigations.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Prof. Giovanni Widmer and Dr. Willie Weir for their helpful advice on data analysis and Dr. Harriet Risby for her assistance with the manuscript formatting and additional laboratory testing. This work was part funded by the European Union Seventh Framework Programme ([FP7/2007-2013] [FP7/2007-2011] under Grant agreement no: 311846).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fawpar.2022.e00151.
Appendix A. Supplementary data
Supplementary File 1: Assay Transfer Document
Supplementary Tables 1, 2, 4 and 5
Supplementary Table 3: Details and results from the panel of 259 samples used in the validation of the MLVA scheme.
References
- Cacciò S.M., Chalmers R.M. Human cryptosporidiosis in Europe. Clin Microbiol Infect. 2016;22(6):471–480. doi: 10.1016/j.cmi.2016.04.021. [DOI] [PubMed] [Google Scholar]
- Caroli S., Santoni S., Ronfort J. AMaCAID: a useful tool for accurate marker choice for accession identification and discrimination. Mol Ecol Resour. 2011;11(4):733–738. doi: 10.1111/j.1755-0998.2011.02993.x. [DOI] [PubMed] [Google Scholar]
- Chalmers R.M., Cacciò S. Towards a consensus on genotyping schemes for surveillance and outbreak investigations of Cryptosporidium, Berlin, June 2016. Euro Surveill. 2016;21(37):30338. doi: 10.2807/1560-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers R.M., Robinson G., Hotchkiss E., Alexander C., May S., Gilray J., Connelly L., Hadfield S.J. Suitability of loci for multiple-locus variable-number of tandem-repeats analysis of Cryptosporidium parvum for inter-laboratory surveillance and outbreak investigations. Parasitology. 2017;144(1):37–47. doi: 10.1017/S0031182015001766. [DOI] [PubMed] [Google Scholar]
- Chalmers R.M., Pérez-Cordón G., Cacció S.M., Klotz C., Robertson L.J., Participants of the Cryptosporidium Genotyping Workshop (EURO-FBP) Cryptosporidium genotyping in Europe: the current status and processes for a harmonised multi-locus genotyping scheme. Exp Parasitol. 2018;191:25–30. doi: 10.1016/j.exppara.2018.06.004. [DOI] [PubMed] [Google Scholar]
- Chalmers R.M., Robinson G., Elwin K., Elson R. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasit Vectors. 2019;12(1):95. doi: 10.1186/s13071-019-3354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza A.M., de Araújo F.C.F., Fontes C.J.F., Carvalho L.H., de Brito C.F.A., de Souza T.N. Multiple-clone infections of Plasmodium vivax: definition of a panel of markers for molecular epidemiology. Malar J. 2015;14:330. doi: 10.1186/s12936-015-0846-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Yang Y., Wang Y., Yang D., Yang Y., Shi Y., Li C., Li L., Chen Y., Jiang Q., Zhou Y. Prevalence of Cryptosporidium infection in the global population: a systematic review and meta-analysis. Acta Parasit. 2020;65:882–889. doi: 10.2478/s11686-020-00230-1. [DOI] [PubMed] [Google Scholar]
- Feng X., Rich S.M., Tzipori S., Widmer G. Experimental evidence for genetic recombination in the opportunistic pathogen Cryptosporidium parvum. Mol Biochem Parasitol. 2002;119(1):55–62. doi: 10.1016/s0166-6851(01)00393-0. [DOI] [PubMed] [Google Scholar]
- Grinberg A., Widmer G. Cryptosporidium within-host genetic diversity: systematic bibliographical search and narrative overview. Int J Parasitol. 2016;46(8):465–471. doi: 10.1016/j.ijpara.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Herges G.R., Widmer G., Clark M.E., Khan E., Giddings C.W., Brewer M., McEvoy J.M. Evidence that Cryptosporidium parvum populations are panmictic and unstructured in the upper Midwest of the United States. Appl Environ Microbiol. 2012;78(22):8096–8101. doi: 10.1128/AEM.02105-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss E.J., Gilray J.A., Brennan M.L., Christley R.M., Morrison L.J., Jonsson N.N., Innes E.A., Katzer F. Development of a framework for genotyping bovine-derived Cryptosporidium parvum, using a multilocus fragment-typing tool. Parasit Vectors. 2015;8(1):500. doi: 10.1186/s13071-015-1107-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P.R., Gaston M.A. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26(11):2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P.R., Hughes S., Woodhouse S., Syed Q., Verlander N.Q., Chalmers R.M., Morgan K., Nichols G., Beeching N., Osborn K. Sporadic cryptosporidiosis case-control study with genotyping. Emerg Infect Dis. 2004;10(7):1241–1249. doi: 10.3201/eid1007.030582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil I.A., Troeger C., Rao P.C., Blacker B.F., Brown A., Brewer T.G., Colombara D.V., De Hostos E.L., Engmann C., Guerrant R.L., Haque R., Houpt E.R., Kang G., Korpe P.S., Kotloff K.L., Lima A.A.M., Petri W.A., Jr., Platts-Mills J.A., Shoultz D.A., Forouzanfar M.H., Hay S.I., Reiner R.C., Jr., Mokdad A.H. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob Health. 2018;6(7):e758–e768. doi: 10.1016/S2214-109X(18)30283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff K.L., Nataro J.P., Blackwelder W.C., Nasrin D., Farag T.H., Panchalingam S., Wu Y., Sow S.O., Sur D., Breiman R.F., Faruque A.S., Zaidi A.K., Saha D., Alonso P.L., Tamboura B., Sanogo D., Onwuchekwa U., Manna B., Ramamurthy T., Kanungo S., Ochieng J.B., Omore R., Oundo J.O., Hossain A., Das S.K., Ahmed S., Qureshi S., Quadri F., Adegbola R.A., Antonio M., Hossain M.J., Akinsola A., Mandomando I., Nhampossa T., Acácio S., Biswas K., O’Reilly C.E., Mintz E.D., Berkeley L.Y., Muhsen K., Sommerfelt H., Robins-Browne R.M., Levine M.M. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the global enteric multicenter study, GEMS): a prospective, case-control study. Lancet. 2013;382(9888):209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Mallon M., MacLeod A., Wastling J., Smith H., Reilly B., Tait A. Population structures and the role of genetic exchange in the zoonotic pathogen Cryptosporidium parvum. J Mol Evol. 2003;56(4):407–417. doi: 10.1007/s00239-002-2412-3. Erratum in: J. Mol. Evol. 56 (6), 778. [DOI] [PubMed] [Google Scholar]
- Manrique P., Hoshi M., Fasabi M., Nolasco O., Yori P., Calderón M., Gilman R.H., Kosek M.N., Vinetz J.M., Gamboa D. Assessment of an automated capillary system for Plasmodium vivax microsatellite genotyping. Malar J. 2015;14:326. doi: 10.1186/s12936-015-0842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J.G., Insulander M., Lebbad M., Björkman C., Svenungsson B. Molecular typing of Cryptosporidium parvum associated with a diarrhoea outbreak identifies two sources of exposure. Epidemiol Infect. 2008;136:1147–1152. doi: 10.1017/S0950268807009673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A., Robinson G., Swain M.T., Chalmers R.M. Direct sequencing of Cryptosporidium in stool samples for public health. Front Public Health. 2019;7:360. doi: 10.3389/fpubh.2019.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison L.J., Mallon M.E., Smith H.V., MacLeod A., Xiao L., Tait A. The population structure of the Cryptosporidium parvum population in Scotland: a complex picture. Infect Genet Evol. 2008;8:121–129. doi: 10.1016/j.meegid.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadon, C.A., Trees, E., Ng, L.K., Møller Nielsen, E., Reimer, A., Maxwell, N., Kubota, K.A., Gerner-Smidt, P., 2013. MLVA Harmonization Working Group. Development and application of MLVA methods as a tool for inter-laboratory surveillance. Euro Surveill18 (35), 20565. doi: 10.2807/1560-7917.es2013.18.35.20565. [DOI] [PMC free article] [PubMed]
- Owczarzy R., Tataurov A.V., Wu Y., Manthey J.A., McQuisten K.A., Almabrazi H.G., Pedersen K.F., Lin Y., Garretson J., McEntaggart N.O., Sailor C.A., Dawson R.B., Peek A.S. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36(Suppl. 2):W163–W169. doi: 10.1093/nar/gkn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cordón G., Robinson G., Nader J., Chalmers R.M. Discovery of new variable number tandem repeat loci in multiple Cryptosporidium parvum genomes for the surveillance and investigation of outbreaks of cryptosporidiosis. Exp Parasitol. 2016;169:119–128. doi: 10.1016/j.exppara.2016.08.003. [DOI] [PubMed] [Google Scholar]
- Pérez-Cordón G., Robinson G., Troell K., Morris A., Chalmers R.M. Conference Proceedings of VI International Giardia & Cryptosporidium Conference; 2017 April 26–28; Havana, Cuba. 2017. Evaluation of newly discovered variable number repeats (VNTR) in Cryptosporidium parvum. [Google Scholar]
- Ramo A., Monteagudo L.V., Del Cacho E., Sánchez-Acedo C., Quílez J. Intra-species genetic diversity and clonal structure of Cryptosporidium parvum in sheep farms in a confined geographical area in northeastern Spain. PLoS One. 2016;11(5) doi: 10.1371/journal.pone.0155336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G., Chalmers R.M. Assessment of polymorphic genetic markers for multi-locus typing of Cryptosporidium parvum and Cryptosporidium hominis. Exp Parasitol. 2012;132(2):200–215. doi: 10.1016/j.exppara.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Robinson G., Elwin K., Chalmers R.M. Cryptosporidium diagnostic assays: molecular detection. Methods Mol Biol. 2020;2052:11–22. doi: 10.1007/978-1-4939-9748-0_2. [DOI] [PubMed] [Google Scholar]
- Smith R.P., Newton K., Rimdap E., Wight A., Robinson G., Chalmers R.M. Review of investigations of premises housing animals that were linked to human outbreaks of cryptosporidiosis in England and Wales between 2009 and 2019. Vet Rec. 2021;189 doi: 10.1002/vetr.246. [DOI] [PubMed] [Google Scholar]
- Strong W.B., Gut J., Nelson R.G. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect Immun. 2000;68(7):4117–4134. doi: 10.1128/IAI.68.7.4117-4134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struelens M.J. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin Microbiol Infect. 1996;2(1):2–11. doi: 10.1111/j.1469-0691.1996.tb00193.x. [DOI] [PubMed] [Google Scholar]
- Tanriverdi S., Widmer G. Differential evolution of repetitive sequences in Cryptosporidium parvum and Cryptosporidium hominis. Infect Genet Evol. 2006;6(2):113–122. doi: 10.1016/j.meegid.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Tanriverdi S., Blain J.C., Deng B., Ferdig M.T., Widmer G. Genetic crosses in the apicomplexan parasite Cryptosporidium parvum define recombination parameters. Mol Microbiol. 2007;63(5):1432–1439. doi: 10.1111/j.1365-2958.2007.05594.x. [DOI] [PubMed] [Google Scholar]
- Tanriverdi S., Grinberg A., Chalmers R.M., Hunter P.R., Petrovic Z., Akiyoshi D.E., London E., Zhang L., Tzipori S., Turnwine J.K., Widmer G. Inferences about the global population structures of Cryptosporidium parvum and Cryptosporidium hominis. Appl Environ Microbiol. 2008;74(23):7227–7234. doi: 10.1128/AEM.01576-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40(15) doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A., Tassios P.T., Dijkshoorn L., Haeggman S., Cookson B., Fry N.K., Fussing V., Green J., Feil E., Gerner-Smidt P., Brisse S., Struelens M., European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM) Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect. 2007;13(Suppl. 3):1–46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- Widmer G., Lee Y. Comparison of single- and multilocus genetic diversity in the protozoan parasites Cryptosporidium parvum and C. hominis. Appl Environ Microbiol. 2010;76(19):6639–6644. doi: 10.1128/AEM.01268-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Ryan U.M. In: Cryptosporidium and Cryptosporidiosis. Fayer R., Xiao L., editors. CRC Press; London: 2008. Molecular epidemiology; pp. 119–171. [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Hu S., Zhao W., Guo Y., Li N., Zheng Z., Zhang L., Kváč M., Xiao L., Feng Y. Population structure and geographical segregation of Cryptosporidium parvum IId subtypes in cattle in China. Parasit Vectors. 2020;13(1):425. doi: 10.1186/s13071-020-04303-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1: Assay Transfer Document
Supplementary Tables 1, 2, 4 and 5
Supplementary Table 3: Details and results from the panel of 259 samples used in the validation of the MLVA scheme.