Abstract
Flavonoid metabolism in Camptotheca acuminate remained an untapped area for years. A tandem MS approach was used and focused on the mining and characterizing of flavonoids in mature C. acuminate. Fifteen new flavonoids and forty-three known flavonoids, including fifteen flavone analogs, sixteen flavonol analogs, seven flavanone analogs, six chalcone analogs, four xanthone analogs, ten flavane analogs were mined and identified based on their MS/MS fragments. Fifty-three of them were firstly characterized in C. acuminate. Eight biosynthetic precursors for these flavonoids were also identified. We constructed a specific metabolic map for flavonoids according to their relative contents in the flowers, fruits, stems, and leaves of C. acuminate. Furthermore, the most probable genes involved in chalcone biosynthesis, flavonoid hydroxylation, methylation, and glycosylation were further mined and fished in the gene reservoir of C. acuminate according to their conserved domains and co-expression analysis. These findings enable us to acquire a better understanding of versatile flavonoid metabolism in C. acuminate.
Keywords: Camptotheca acuminate, Flavonoids, Fragmentation pathway, Flavonoid metabolism
1. Introduction
Flavonoids, a type of well-known specialized metabolites, are widely distributed in nature. Numerous flavonoids serve as colored pigments and defensive agents for plants [1]. Usually, the C6–C3–C6 nucleus for flavonoids biogenetically originates from the condensation reaction between a 4-coumaroyl CoA unit (B-ring) and three malonyl CoA units (A-ring). In addition, a linkage ring (C-ring) usually forms through C3 unit cyclization. Based on the state of the central C3 unit, flavonoids could be further grouped into flavone, flavonol, flavanone, chalcone, xanthone, and flavane [1]. These specialized metabolites are widely distributed in fruits, cereals, vegetables, and medicinal plants. Flavonoids usually display anti-inflammatory [2], anticancer [3], antihypertension [4], anti-coronary heart disease bioactivities [5]. Although more than seven thousand flavonoids have been characterized up to 2017 [6], the number of flavonoids is still expanding due to the versatile flavonoid metabolism in plants.
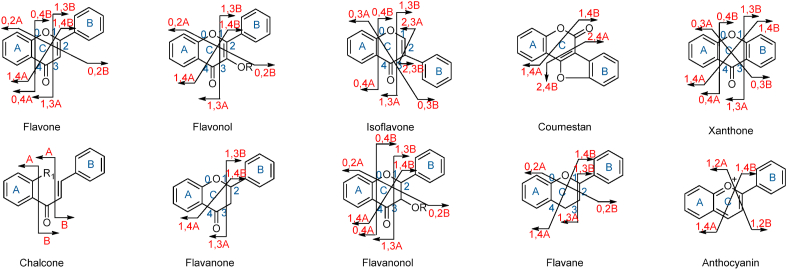
The structure elucidation for plant flavonoids was a fascinating work for natural product chemists. Nowadays, LC-MS/MS-based approach has been increasingly applied to characterize untapped secondary metabolites in plants, such as alkaloids [7]. Usually, the MS/MS spectra of flavonoids provide enough structural information due to their limited nucleus types and modified forms. Typical C-ring fragmentation patterns for flavonoids are summarized and illustrated in Fig. 1. Firstly, the cleavage of the central C3 unit through retro-Diels-Alder reaction in flavonoid produces several informative ions such as i,jA+ and i,jB+. These fragment ions have been used as the diagnostic ions for flavonoid characterization. The substituents on ring A could be determined according to the diagnostic ions of i,jA+, and the substituents on ring B could be confirmed according to the fragments of i,jB+. Furthermore, the detailed fragmentation spectra for flavone, flavonol, flavanone, chalcone, xanthone and flavane vary from each other due to their different skeleton types and substitution patterns. Secondly, the loss of methyl, hydroxyl, carbonyl, and glucosyl are frequently detected in the fragmentation spectra of flavonoids. These clues also provide practical information for the substituents on the three skeleton rings. With the help of these guiding clues, forty flavonoids were identified according to their key fragments using ESI-IT-MSn [8]. Seven flavonoids were characterized in Scutellaria baicalensis based on their typical cleavage patterns [9]. Thirty flavonoid aglycones were identified from Cornus officinalis using ESI-QTOF-MS/MS [10].
Fig. 1.
Typical skeleton cleavages for flavonoids.
Camptotheca acuminate, a native plant in China, is famous for its camptothecin-producing capability. Thirty-three alkaloids have been recently mined and characterized in our group through a UPLC-qTOF-HRMS based targeted metabolomics approach [7]. Hundreds of specialized metabolites have been detected in the flowers, fruits, stems, and leaves of this versatile plant. But a large amount of them remains uncharacterized. Numerous research efforts were poured on the isolation and characterization of alkaloids in C. acuminate due to their intriguing structure diversity and anticancer bioactivity. A metabolic map for indole alkaloids in C. acuminate has been constructed [7]. Interestingly, vigorous enzyme-mediated hydroxylation, methylation, and glycosylation modifications on the A, C or E ring of camptothecin analogs were observed in this plant. Therefore, we hypothesized that the indole alkaloid and flavonoid metabolic pathway in this plant might share common enzymatic catalysts, such as hydroxylase, glucosyltransferase, and O-methyltransferase. Some of these enzymes might be simultaneously responsible for the modification of camptothecin derivatives and flavonoids due to their substrate diversity. Therefore, identification and pathway elucidation of flavonoids in C. acuminate could also provide potential clues for uncovering the unknown steps in camptothecin biosynthesis. Only seven flavonoids were identified through HPLC-MS/MS analysis in C. acuminate up to now [11]. Flavonoid metabolism in C. acuminate remained an untapped area for years. Herein, purposefully mining and characterization of flavonoids and the genes probably involved in flavonoid metabolism were initiated with the help of our fundamental research. This study provides a comprehensive perspective on the versatile flavonoid metabolism in C. acuminate.
2. Materials and methods
2.1. Chemicals and reagents
Luteolin, quercetin, genistein, naringenin, isoliquiritigenin, mangiferin, and catechin standards were purchased from Weikeqi Biotech. Methanol, n-hexane, formic acid (analytical grade) were purchased from Chron Chemicals. Methanol (chromatographic grade) was purchased from Fisher.
2.2. Plant material collection
Fresh flower, fruit, stem, and leaf samples were collected from the mature C. acuminate trees in Sichuan Agricultural University (Yaan campus), sealed, and stored at −80 °C. The sampling interval was one week for flowers, two weeks for leaves and fruits, four weeks for stems.
2.3. Sample preparation and metabolite extraction
The frozen flower, fruit, stem, and leaf tissues were air-dried at 50 °C. Sample powder for each tissue was prepared through thoroughly grinding, and transferred into a conical flask, and soaked in n-hexane overnight. The n-hexane was removed through filtration, and the sample powder was dried, sealed, and stored. 500 μl of 80% methanol was added to re-suspend 0.1 g of sample powder and incubated at 30 °C for 10 min. The obtained extracts were centrifuged at 12,000 rpm for 5 min. The pipette was used to transfer the supernatants carefully. Then the obtained supernatants were filtrated by 0.22 μm filter in an Eppendorf tube through centrifugation. The final filtrate was used as the sample solution for subsequent LC-MS/MS analysis. An equal volume of the obtained filtrate from each sample was transferred into an Eppendorf tube and mixed by a vortex. The mixture was used as the QC sample, and the extraction solvent was designated as the blank sample.
2.4. UPLC-QTOF-MS/MS analyses
Metabolite separation was performed on a Vanquish UHPLC system (Thermo Fisher), coupled with an Orbitrap Q Exactive™ HF-X mass spectrometer (Thermo Fisher). The sample solution for the flowers, fruits, stems, and leaves was injected onto a Hyperil Gold C18 column (2.1 × 100 mm, 1.9 μm). A gradient program (phase A: 0.1% HCOOH; phase B: CH3OH; 0.0–1.5 min, 2%–2% B; 1.5–12.0 min, 2%–100% B; 12.0–14.0 min, 100%–100%; 14.0–14.1 min, 100%–2% B; 14.1–16.0 min, 2%–2%; flow rate: 0.2 mL/min) was used for metabolite separation. The mass spectrometer parameter was set as: positive mode, spray voltage 3.2 kV, sheath gas flow rate 35 arb, aux gas flow rate 10 arb, capillary temperature 320 °C, collision energy 20 eV–60 eV. To mine the fragmentation spectra of flavonoids and their biosynthetic precursors, the molecular ion peaks (200 Da–800 Da) were purposely extracted, and their structures were deduced according to their distinctive cleavage patterns.
2.5. Hierarchical clustering analysis
To construct the accumulation patterns for identified flavonoids in C. acuminate, hierarchical clustering analysis was performed via MetaboAnalyst 5.0 according to the semi-quantified data. The metabolic map for flavonoids in C. acuminate was constructed based on their biosynthetic origins and chemical structures.
2.6. Co-expression analysis and gene candidate screening
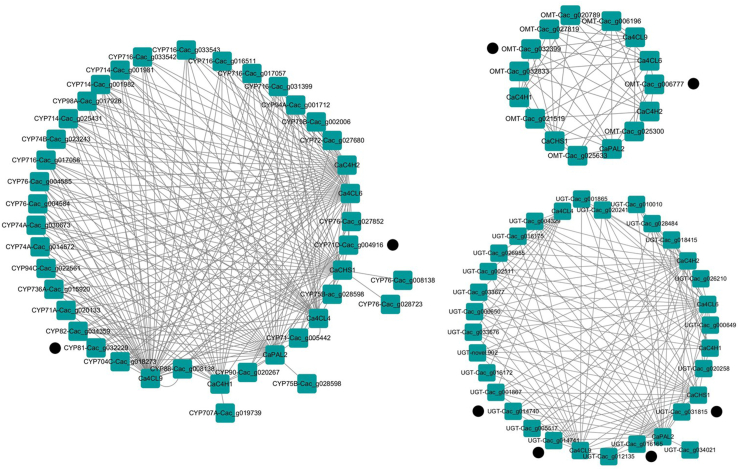
To screen the untapped CYP450, UDP-glucosyltransferase and O-methyltransferase probably involved in flavonoid metabolism, weighted correlation network analysis was performed with the R package WGCNA based on our transcriptomic data matrix for C. acuminate plantlets, which was deposited at NCBI (BioProject No. PRJNA704189). The expression data was prefiltered based on the built-in threshold, and the co-expression network was constructed using a soft threshold power of 8 and the mergeCutHeight parameter of 0.25. Cytoscape 3.8.2 was used to visualize the obtained network. The hierarchical clustering heat map for the bait genes, the newly mined CYP450, UDP-glucosyltransferase and O-methyltransferase, was generated based on their FPKM values in our transcriptomic data matrix using MeV 4.9.
3. Results and discussion
3.1. Mining and characterization of flavone, isoflavone, coumestan analogs
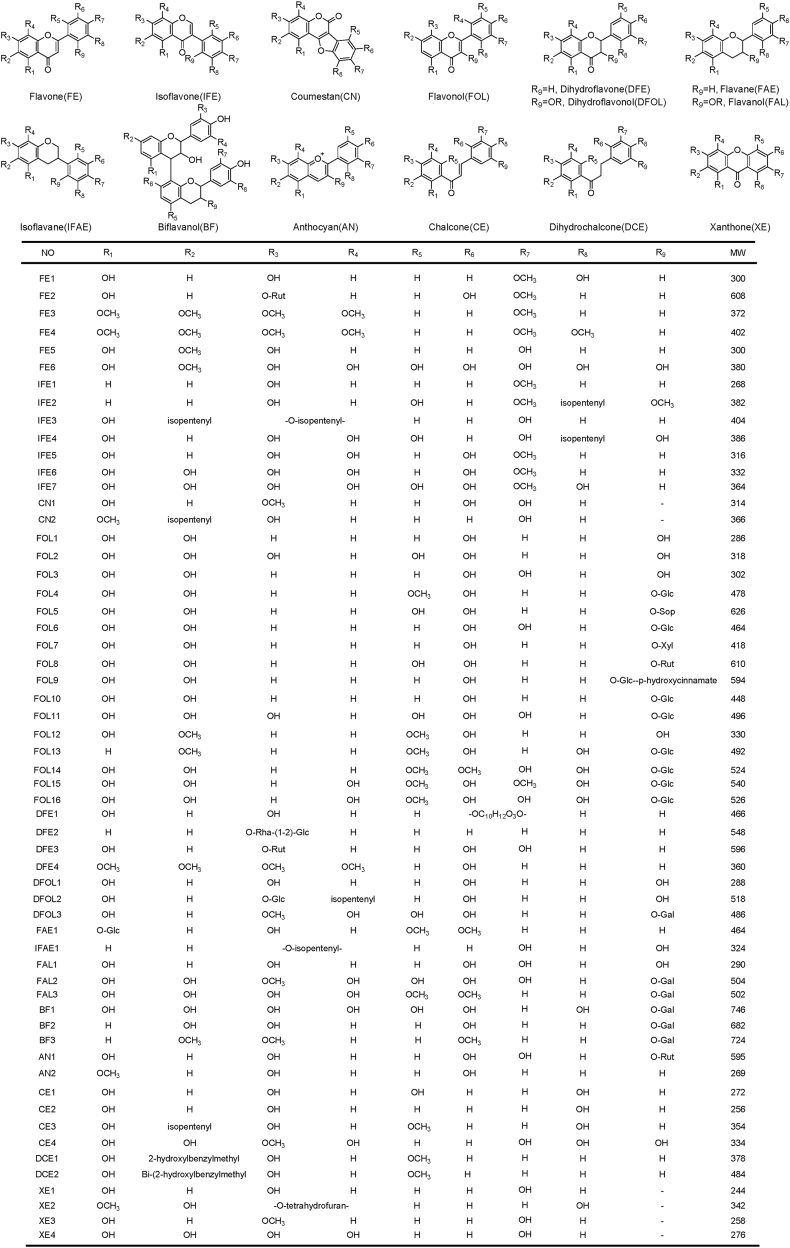
Six metabolites were identified and classified as flavone analogs (FE1-FE6) according to their informative diagnostic ions (Fig. 1) [[12], [13], [14], [15]]. Their fragmentation pathways are illustrated in Fig. S1-S6. The [M+H]+ ion formula of FE1 (301.0696, Tr 11.45 min) was annotated as C16H13O6. Five intermediate ions, including m/z 286.0490, 258.0527, 230.0583, 202.0606, and 184.0507, were produced through the commonly observed cleavage of 15 Da (-CH3), 28 Da (-CO), 28 Da (-CO), 28 Da (-CO), 18 Da (-H2O) in flavones. Furthermore, detecting two diagnostic ions, m/z 193.0140 (0, 4B+) and 153.0173 (1, 3A+), indicated two hydroxy located on ring A and a methoxy, along with a hydroxy, located on ring B. FE1 was identified as a flavone, diosmetin. The [M+H]+ ion formula of FE2 (609.2867, Tr 15.81 min) was annotated as C28H33O15. The cleavage of 15 Da (-CH3), 147 Da (-C6H11O4), and 28 Da (-CO) indicated the existence of methyl, rhamnose, and carbonyl. A same diagnostic fragment at m/z 137.0604 (0, 3A+-C12H20O9) was also observed, compared with that of FE1. And a rutinose was determined on ring A according to observing a diagnostic ion at m/z 433.1794 (1,4A+). Thus, FE2 was characterized as a derivative of diosmetin, diosmin. The [M+H]+ ion formula of FE3 (373.2209, Tr 12.39 min) was annotated as C20H21O7. The existence of five methoxys was verified according to the stepwise cleavage of 15 Da (-CH3), 31 Da (-OCH3), 15 Da (-CH3), 15 Da (-CH3), 15 Da (-CH3). The observation of a specific diagnostic ion at m/z 133.1015 (1, 3B+) indicated that only one of the five methoxys located on ring B and FE3 was a flavone analog. FE3 was finally characterized as tangeritin. The [M+H]+ ion formula of FE4 (403.1463, Tr 8.47 min), 30 Da higher than FE3, was annotated as C21H23O8 and defined as a methoxy derivative of FE3. It confirmed that four methoxys located on ring A according to detecting a diagnostic ion at m/z 241.1069 (1, 3A+) and two methoxys on ring B according to observing a diagnostic ion at m/z 167.0713 (0, 2B+). FE4 was characterized as nobiletin. The [M+H]+ ion formula of FE5 (301.0696, Tr 11.47 min) was annotated as C16H13O6. A series of intermediate ions at m/z 286.0490, 257.0461, 229.0504, 213.0543 formed through the cleavage of 15 Da (-CH3), or 44 Da (-CO2) and 28 Da (-CO), or 44 Da (-CO2). The substituent groups on ring B, and A were determined, respectively, according to two fragment ions at m/z 121.0285 (0, 2B+) and 167.0340 (0, 3A+). FE5 was characterized as a flavone analog, hispidulin. The [M+H]+ ion formula of FE6 (381.1384, Tr 11.17 min) was annotated as C16H13O11. Six intermediate ions at m/z 337.1436, 309.1490, 265.0916, 237.0925, 222.1108, 166.0509 formed through the stepwise loss of 44 Da (-CO2), 28 Da (-CO), 44 Da (-CO2), 28 Da (-CO), 15 Da (-CH3), 28 Da (-CO) and 28 Da (-CO). The existence of multiple hydroxy was confirmed according to the fragments mentioned above. Furthermore, three hydroxy and one methoxy were verified on ring A, and the other five hydroxy located on ring B according to the detection of two diagnostic fragments at m/z199.0836 (1, 3A+) and 211.1108 (1, 4B+). Finally, FE6 was characterized as a new flavone analog, 2′,5′,6’,8-tetrahydroxy-6-methoxy luteolin.
Seven metabolites were identified and classified as isoflavone analogs (IFE1-IFE7) according to their informative diagnostic ions (Fig. 1) [14,15]. Their fragmentation pathways are illustrated in Fig. S7-S13. The [M+H]+ ion formula of IFE1 (269.2265, Tr 10.44 min) was annotated as C16H13O4. The breakage of 28 Da (-CO), 28 Da (-CO), 42 Da (-C2H2O) were detected. Three fragments at m/z 241.1567, 213.1646, 171.1145 formed. IFE1 was classified as an isoflavone analog according to the diagnostic ion at m/z 149.1321 (0, 3B+). A methoxy on ring B was also confirmed based on this ion. It was finally characterized as formononetin according to the observation of four diagnostic fragments at m/z 149.1321 (0, 3B+), 133.1014 (1, 3B+), 121.1016 (0, 3A+), 93.0699 (0, 4A+), and the production of a series of intermediate ions. The [M+H]+ ion formula of IFE2 (383.3291, Tr 15.51 min) was annotated as C22H23O6. Three fragments at m/z 341.2543, 313.2514, 243.2106 formed through the breakage of 42 Da (-C2H2O), 28 Da (-CO), 70 Da (-C5H10). The existence of an isopentenyl on ring B was verified according to the observation of an ion at m/z 243.2106 produced through the cleavage of 70 Da (-C5H10) and two diagnostic fragments at m/z 191.1792 (0, 4B+- C5H8–2CH3) and 93.0700 (0, 4A+). Therefore, IFE2 was characterized as licoricone. The [M+H]+ ion formula of IFE3 (405.3188, Tr 12.24 min) was annotated as C25H25O5. The breakage of 28 Da (-CO), 28 Da (-CO), 70 Da (-C5H10), 14 Da (-CH2) were observed, and four intermediate fragments at m/z 377.3252, 349.2505, 279.2087, 265.1931 formed. IFE3 was annotated as an isoflavone analog. The existence of an isopentenyl, a hydroxy, and a cyclized isopentenyl on ring A was verified according to the observation of two diagnostic fragments at m/z 201.1632 (0, 3A+-C5H10) and 173.1316 (0, 4A+-C5H10). Another hydroxy on ring B was determined according to observing a diagnostic fragment at m/z 119.0853 (1, 3B+). IFE3 was finally characterized as osajin. The [M+H]+ ion formula of IFE4 (387.1444, Tr 10.88 min), 4 Da higher than IFE2, was annotated as C20H19O8. IFE4 was rapidly classified as an isoflavone analog according to detecting two diagnostic ions at m/z 235.0969 (0, 3B+) and 191.0711 (1, 3B+-CO). An isopentenyl and three hydroxy on ring B were also confirmed according to these diagnostic ions. Three hydroxy on ring A were further determined according to the observation of a diagnostic fragment at m/z 151.0390 (1, 3A+-H2O). Thus, IFE4 was determined as a new metabolite, 2′, 4′, 6′, 5, 7, 8-hexahydroxy-5′-isopentenyl isoflavone. The [M+H]+ ion formula of IFE5 (317.0655, Tr 9.73 min) was annotated as C16H13O7. The intermediate ions at m/z 285.0388, 257.0451, 229.0501, 201.0562 formed through the loss of 32 Da (-OCH4), 28 Da (-CO), 28 Da (-CO), 28 Da (-CO). Three hydroxy on ring A and one hydroxy, along with a methoxy, on ring B were determined according to two diagnostic fragments at m/z 153.0193 (0, 3A+) and 177.0544 (1,3B+). Thus, IFE5 was characterized as a new metabolite, 3′, 5, 7, 8-tetrahydroxy-4′-methoxy isoflavone. The [M+H]+ ion formula of IFE6 (333.1258, Tr 11.41 min) was annotated as C16H13O8. The intermediate ions at m/z 305.1272, 277.1316, 259.1238, 244.0970 formed through the breakage of 28 Da (-CO), 28 Da (-CO), 18 Da (-H2O), 15 Da (-•CH3). It indicated the existence of carbonyl, hydroxy, and methyl. The observation of a diagnostic ion at m/z 169.0760 (0,3A+) indicated the existence of four hydroxy groups on ring A. IFE6 was further identified as a new metabolite, 3′,5,6,7,8-pentahydroxy-4′-methoxy isoflavone. The [M+H]+ ion formula of IFE7 (365.1104, Tr 10.32 min), 32 Da higher than IFE6, was annotated as C16H13O10. Three hydroxy and one methoxy on ring B were confirmed according to two diagnostic fragments at m/z 197.0700 (0,3B+) and 181.0776 (1,3B+). It was characterized as a new derivative of IFE7, 2′,3′,5′,5,6,7,8-heptahydroxy-4′-methoxy isoflavone. The [M+H]+ ion formula of IFEP (289.1803, Tr 11.80 min) was annotated as C15H13O6. The breakage of 28 Da (-CO), 28 Da (-CO), 18 Da (-H2O), 18 Da (-H2O) were detected, and a series of intermediate ions at m/z 261.1011, 233.1078, 215.1430, 197.1321 formed. IFEP was classified as an isoflavone analog based on the diagnostic fragment at m/z 119.0852 (1, 3B+). One hydroxy on ring B was also determined based on this ion. Furthermore, the existence of a hydroxy on ring C was confirmed according to the observation of an ion at m/z 271.1670, formed through the cleavage of 18 Da in the initial cleavage step. It was identified as the critical bio-precursor for isoflavone biosynthesis according to its diagnostic ions and intermediate ions (Fig. S14). Furthermore, two metabolites were characterized as coumestan analogs (CN1–CN2) according to their diagnostic ions (Fig. 1) [16]. Their fragmentation pathways are illustrated in Fig. S15-S16. The [M+H]+ ion formula of CN1 (315.1107, Tr 11.10 min) was annotated as C16H11O7. The existence of one methoxy and carbonyl was verified according to the detection of a fragment at m/z 283.1098, formed through the cleavage of 32 Da (-OCH3), and another one at m/z 287.1166, produced through the cleavage of 28 Da (-CO). CN1 was classified into pterocarpan analog based on two specific diagnostic ions at m/z 107.0853 (1, 4A+-OCH4) and 181.0752 (1, 4B+). In addition, the existence of one methoxy and one hydroxy on ring A, two hydroxy on ring B were further confirmed based on detecting two diagnostic fragments at m/z 91.0540 (0, 4A+-OCH4) and 153.0178 (2, 4B+). Herein, CN1 was characterized as wedelolactone. The [M+H]+ ion formula of CN2 (367.2020, Tr 9.22 min) was annotated as C21H19O6. It was classified as a pterocarpan analog at first glance according to the observation of two fragments at m/z 221.1080 (2, 4A+-CH2) and 180.1026 (1, 4A+-•C2H3). One isopentenyl, one hydroxy, and one methoxy on ring A were also verified according to the observation of the fragments mentioned above. The specific cleavage of 18 Da (-H2O), 14 Da (-CH2), 28 Da (-CO), 14 Da (-CH2), 28 Da (-CO) were also detected in this metabolite. Thus, CN2 was characterized as glycyrol [16]. The structures of the identified flavone, isoflavone, and coumestan analogs (FE1-FE6, IFE1-IFE7, CN1–CN2) are illustrated in Fig. 2.
Fig. 2.
The chemical structures of the identified flavonoids in C. acuminate.
3.2. Mining and characterization of flavonol analogs
Sixteen metabolites were identified and classified as flavonol analogs (FOL1-FOL16) according to their informative diagnostic ions (Fig. 1) [12,17,18]. Their fragmentation pathways are illustrated in Fig. S17-S32. The [M+H]+ ion formula of FOL1 (287.0540, Tr 10.32 min) was annotated as C15H11O6. Three fragments at m/z 269.0463, 241.0509, 213.0544 formed through the cleavage of 18 Da (-H2O), 28 Da (-CO), 28 Da (-CO). FOL1 was rapidly classified as a flavonol analog according to the ion at m/z 165.0176 (0, 2A+). It confirmed two hydroxy located on ring A and one located on ring B based on observing two diagnostic fragments at m/z 153.0174 (1, 3A+) and 121.0287 (0, 2B+), respectively. FOL1 was characterized as kaempferol. The [M+H]+ ion formula of FOL2 (319.0508, Tr 2.49 min) was annotated as C15H11O8. The cleavage of 18 Da (-H2O), 28 Da (-CO), 28 Da (-CO) were observed, and three intermediate fragments at m/z 301.0429, 273.0465, 245.0164 formed. FOL2 was classified as a flavonol analog based on observing a diagnostic fragment at m/z 181.0038 (0,2A+). Three hydroxy on ring A was also confirmed according to this informative ion. FOL2 was further identified as gossypetin according to the cleavage of 28 Da (-CO) and 44 Da (-CO2). The [M+H]+ ion formula of FOL3 (303.0492, Tr 10.56 min), 16 Da higher than FOL1, was annotated as C15H11O7. The same diagnostic ion at m/z 165.0175 (0, 2A+) was also observed. Two hydroxy were verified on ring B and two on ring A according to observing a fragment at m/z 111.0074 (0,2B+-CO) and 153.0192 (1,3A+). Thus, FOL3 was determined as quercetin. The [M+H]+ ion formula of FOL4 (479.1510, Tr 9.74 min) was annotated as C22H23O12. The forming of two fragments at m/z 317.0656, 285.0385 through the breakage of 162 Da (-C6H10O5) and 32 Da (-OCH4) indicated the linkage of a glucosyl and methoxy on the skeleton ring. The methoxy was further verified on ring B, and the glucosyl was on ring C according to the diagnostic ions at m/z 355.1545 (1,4B+) and 153.0193 (0,2B+). Two hydroxy were determined on ring A according to detecting the fragment at m/z 137.0603 (0,3A+). Thus, FOL4 was determined as a glucosyl derivative of flavonol, isorhamnetin-3-O-glucoside. The [M+H]+ ion formula of FOL5 (627.2178, Tr 10.25 min) was annotated as C27H31O17. The forming of an ion at 303.0490 through the stepwise loss of two 162 Da (-C6H10O5) and the observation of a diagnostic fragment at m/z 165.0176 (0, 2A+-C12H20O10) indicated that FOL5 was a sophoroside derivative of the flavonol. The number of hydroxy on ring A and ring B was also determined according to the observation of two diagnostic fragments at m/z 153.0173 (1,3A+) and 137.0242 (0,2B+). Thus, FOL5 was determined as quercetin-3-O-sophoroside. The [M+H]+ ion formula of FOL6 (465.1641, Tr 9.39 min), 162 Da higher than FOL3, was annotated as C21H21O12. Three same diagnostic ions at m/z 165.0177 (0, 2A+-C6H10O5), 153.0174 (1,3A+), 137.0226 (0,3A+) were also detected. Therefore, FOL6 was rapidly characterized as a glucosyl derivative of FOL3, isoquercetin. The [M+H]+ ion formula of FOL7 (419.2183, Tr 9.88 min), which is 132 Da higher than FOL1, was annotated as C20H19O10. The fragments at m/z 287.0541, 241.0506, 213.0538 formed through the cleavage of 132 Da (-C5H8O4), 18 Da (-H2O) and 28 Da (-CO), 28 Da (-CO). Two same diagnostic ions at m/z 165.0176 (0, 2A+-C5H8O4) and 153.0174 (1,3A+), along with a diagnostic ion at m/z 121.0286 (0,2B+), were also detected, and the hydroxy on ring A and B were determined. Thus, FOL7 was characterized as kaempferol-3-O-arabinopyranoside. The [M+H]+ ion formula of FOL8 (611.2964, Tr 9.17 min) was annotated as C27H31O16. The existence of rutinoside in FOL8 was determined according to the cleavage of 146 Da (-C5H10O4) and 162 Da (-C6H10O5). Therefore, FOL8 was identified as rutin, according to detecting a fragment at m/z 303.0492 and three characteristic fragments at 165.0177 (0,2A+-C12H20O9), 153.0195 (1,3A+), 137.0241 (0,2B+). The [M+H]+ ion formula of FOL9 (595.2169, Tr 9.58 min) was annotated as C30H27O13. The forming of three fragment ions at m/z 449.1036, 287.0540, 259.0568 through the loss of 146 Da (-C9H6O2), 162 Da (-C6H10O5), 28 Da (-CO) indicated the existence of coumaroyl, glucosyl, and carbonyl. Two hydroxy on ring A and one on ring B were further confirmed according to observing the diagnostic fragments at m/z 165.0174 (0, 2A+-C15H16O9) and 121.0285 (0,2B+). Thus, FOL9 was characterized as tiliroside. The [M+H]+ ion formula of FOL10 (449.2143, Tr 9.67 min), 162 Da higher than FOL1, was annotated as C21H21O11. The cleavage of 162 Da (-C6H10O5) indicated the existence of a glucosyl. Same diagnostic fragments at m/z 165.0180 (0, 2A+-C6H10O5), 153.0174 (1,3A+) and 137.0225 (0,3A+) were detected. Thus, FOL10 was characterized as a glucosyl derivative of FOL1, astragalin. The [M+H]+ ion formula of FOL11 (497.1858, Tr 10.91 min) was annotated as C21H21O14. Three intermediate ions, including m/z 335.1399, 307.1399, 265.0967, were produced through the commonly observed loss of 162 Da (-C6H10O5), 28 Da (-CO), 42 Da (-C2H2O) in flavonols. FOL11 was classified as a flavonol analog according to the diagnostic fragments at m/z 169.0755 (1,3A+) and 195.0917 (1,4B+-C6H10O5). Three hydroxy on ring A and three on ring B was confirmed based on the two diagnostic ions. A glucosyl on ring C was verified according to detecting a diagnostic fragment at m/z 345.0787 (0,3B+) and an intermediate ion at m/z 335.1399 formed through the cleavage of 162 Da. FOL11 was finally identified as a flavonol, 5′-hydroxy-gossypetin-3-O-glucoside. The [M+H]+ ion formula of FOL12 (331.1080, Tr 10.00 min) was annotated as C17H15O7. It confirmed that FOL12 was a flavonol analog according to detecting a diagnostic fragment at m/z 165.0699 (1, 3B+). The stepwise loss of two 15 Da (-•CH3) indicated the existence of two methoxys. One methoxy and hydroxy on ring A, and one methoxy and hydroxy on ring B were further verified according to the observation of two diagnostic ions at m/z 209.0542 (0,4B+) and 123.0799 (0,4A+). Thus, FOL12 was identified as 7-methyl isorhamnetin. The [M+H]+ ion formula of FOL13 (493.1665, Tr 10.11 min) was annotated as C23H25O12. Three intermediate fragments at m/z 331.1053, 313.0969, 285.1011 formed through the breakage of 162 Da (-C6H10O5), 18 Da (-H2O), 28 Da (-CO). FOL13 was classified as a flavonol analog according to the characteristic fragment at m/z 163.0395 (0, 2A+-C6H10O5). Besides, one methoxy on ring A was verified based on observing this ion. One methoxy and two hydroxy on ring B were further confirmed according to another ion at m/z 181.0747 (1,3B+-C6H10O5). Thus, FOL13 was characterized as a new flavonol, 3′,7-dimethyl-6′-hydroxy-fisetin-3-O-glucoside. The [M+H]+ ion formula of FOL14 (525.2978, Tr 11.53 min) was annotated as C23H25O14, 32 Da higher than FOL13. The fragments at m/z 363.1331, 335.1388, 320.1180 formed through the cleavage of 162 Da (-C6H10O5), 28 Da (-CO), 15 Da (-•CH3). It indicated the existence of glucosyl and methoxy. It was classified as a FOL13 derivative based on the diagnostic fragment at m/z 165.0540 (0,2A+-C6H10O5), 181.0774 (1,2A+-C6H10O5), and its cleavage pattern. Two hydroxy on ring A was also determined according to the diagnostic ion. FOL14 was characterized as a new flavonol, 3′,4′-dimethyl-5,5′, 6′-trihydroxy fisetin-3-O-glucoside. The [M+H]+ ion formula of FOL15 (541.1858, Tr 10.32 min) was annotated as C23H25O15, 16 Da higher than FOL14. The loss of 162 Da (-C6H10O5), 32 Da (-OCH4), 28 Da (-CO), 42 Da (-C2H2O), 28 Da (-CO) was also observed in its fragmentation spectra. The detection of the same ion at m/z 165.0542 (0,2A+-C6H10O5) indicated FOL15 was a hydroxy derivative of FOL14. Three hydroxy and two methoxys on ring B were confirmed according to detecting two diagnostic fragments at m/z 213.0647 (0,2B+) and 197.0700 (1,2B+). Thus, FOL15 was determined as a new flavonol, 3′-methyl-5′-methoxy-2′,5,6′-trihydroxy fisetin-3-O-glucoside. The [M+H]+ ion formula of FOL16 (527.2038, Tr 9.01 min) was annotated as C22H23O15, 14 Da lower than FOL15. Four fragments at m/z 365.1480, 333.1261, 305.1274, 277.0970 formed through the loss of 162 Da (-C6H10O5), 32 Da (-OCH4), 28 Da (-CO), 28 Da (-CO). It indicated the existence of glucosyl and methoxy. FOL16 was further annotated as a FOL15 derivative according to the observation of two characteristic fragments at m/z 165.0542 (0,2A+-C6H10O5) and 127.0388 (1,4A++2H). Two hydroxy on ring A and one glucosyl on ring C was also confirmed based on the detection of these diagnostic ions. FOL16 was characterized as a new flavonol, 3′-methyl-2′, 5, 5′, 6′-tetrahydroxy fisetin-3-O-glucoside. The structures of the identified flavonol analogs (FOL1-FOL16) are illustrated in Fig. 2.
3.3. Mining and identification of flavanone analogs
Four metabolites were characterized as flavanone analogs (DFE1-DFE4) according to their informative diagnostic ions (Fig. 1) [19]. Their fragmentation pathways are illustrated in Fig. S33-S36. The [M+H]+ ion formula of DFE1 (467.1845, Tr 7.48 min) was annotated as C25H23O9. A series of intermediate ions at m/z 287.1275, 203.0541, 143.0338 formed through the stepwise loss of 180 Da (-C10H12O3••), 84 Da (-C4H4O2), 60 Da (-CO2+O•). DFE1 was classified as flavanone analogue according to the specific diagnostic ions at m/z 153.0173 (1,3A+), 125.0231 (1, 4A+) and 163.0395 (1,4B+-180 Da). Two hydroxy on ring A was also confirmed based on these diagnostic ions. The large substituent on ring B was determined according to the cleavage of 180 Da (-C10H12O3••). Thus, DFE1 was characterized as silandrin. The [M+H]+ ion formula of DFE2 (549.3056, Tr 14.44 min) was annotated as C27H33O12. The cleavage of 18 Da (-H2O), 14 Da (-CH2), and 322 Da (-C12H18O10) from the mother ion indicated the existence of rhamnosyl (1–2) glucoside. This group was further verified on ring A based on two diagnostic fragments at m/z 271.0941 (1, 4A+-C6H10O4) and 133.1010 (1,4B+). Thus, DFE2 was characterized as flavanone-7-O-rhamnosyl (1–2) glucoside. The [M+H]+ ion formula of DFE3 (597.3744, Tr 14.88 min) was annotated as C27H33O15. A series of intermediate ions at m/z 579.4029, 565.4069, 537.2342 formed through the stepwise loss of 18 Da (-H2O), 14 Da (-CH2), 28 Da (-CO). A rutinosyl and a hydroxy were verified on ring A according to the observation of two fragments at m/z 461.2455 (1, 3A+) and 109.1013 (0, 4A+-C12H20O9). Finally, DFE3 was determined as eriocitrin. The [M+H]+ ion formula of DFE4 (361.1358, Tr 9.61 min) was annotated as C19H21O7. It confirmed that four methoxys located on ring A and one hydroxy on ring B according to the loss of 30 Da (−2•CH3), 30 Da (−2•CH3) and the detection of fragments at m/z 153.0562 (1, 4A+-4•CH3), 121.0655 (1, 3B+). DFE4 was identified as 4′-hydroxy-5,6,7,8-tetra methoxy flavanone.
Three metabolites were characterized as dihydroflavonol analogs (DFOL1-DFOL3) according to their informative diagnostic ions (Fig. 1) [20]. Their fragmentation pathways are illustrated in Figs. S37–S39. The [M+H]+ ion formula of DFOL1 (289.0696, Tr 7.95 min) was annotated as C15H13O6. The intermediate fragment at m/z 271.0616 was formed through the breakage of 18 Da (-H2O), then the loss of 28 Da (-CO), 28 Da (-CO), 28 Da (-CO) were detected, and a series of intermediate ions at m/z 243.0640, 215.0718, 187.0737 formed. The intermediate fragment at m/z 271.0616 could be determined as a flavone analog based on detecting an ion at m/z 163.0393 (0,4B+). Two hydroxy located on ring A, and one located on ring B according to the observation of two diagnostic ions at m/z 121.0286 (0, 2B+), 153.0192 (1, 3A+). But the fragment at m/z 271.0616 was produced through the cleavage of 18 Da (-H2O) from the mother ion. Thus, DFOL1 was finally characterized as 2,3-dihydrofisetin. The [M+H]+ ion formula of DFOL2 (519.1752, Tr 7.42 min) was annotated as C26H31O11. The cleavage of 18 Da (-H2O), 28 Da (-CO), 98 Da (–CO–C5H10), 162 Da (-C6H10O5) were detected, and four intermediate fragments at m/z 501.1748, 473.1940, 375.1469, 213.0545 formed. A fragment ion at m/z 151.0746 (1, 3A+-C6H10O5–C5H10) indicated a glucosyl and an isopentenyl located on ring A. It was classified as a flavanone analog according to the diagnostic ions at m/z 195.0652 (1, 4A+-C6H8O5), 181.0873 (0, 3B+), 105.0697 (0, 2B+). A hydroxy on ring B was also determined according to these fragment ions. Thus, DFOL2 was characterized as phellamurin. The [M+H]+ ion formula of DFOL3 (487.2125, Tr 10.09 min) was annotated as C23H19O12. The forming of ions at m/z 335.0926 and 317.0837 through the breakage of 152 Da (-C7H4O4) and 18 Da (-H2O) indicated the linkage of a gallate. It was classified as a flavanone analog according to the diagnostic ions at m/z 163.0395 (1,4B+-C7H6O5) and 135.0432 (1,3B+-C7H6O5). Two hydroxy on ring B and one gallate on ring C was also confirmed according to the diagnostic ions. DFOL3 was finally determined as a new metabolite, 5, 8-dihydroxy-7-methyl-2,3-dihydrofisetin-3-O-gallate ester. The structures of the identified flavanone analogs (DFE1-DFE4, DFOL1-DFOL3) are illustrated in Fig. 2.
3.4. Mining and identification of chalcone analogs
Four metabolites were identified and classified as chalcone analogs (CE1-CE4) according to the informative fragment ions [21]. Their fragmentation pathways are illustrated in Figs. S40–S43. Similar diagnostic ions with flavanone were sometimes observed in the fragmentation process of chalcone due to the C-ring cyclization rearrangement. The [M+H]+ ion formula of CE1 (273.0756, Tr 10.76 min) was annotated as C15H13O5. The breakage of 18 Da (-H2O), 28 Da (-CO), 28 Da (-CO) were observed, and three intermediate fragments at m/z 255.1754, 227.0682, 199.0755 formed. Two hydroxy located on ring A was confirmed according to the diagnostic fragment at m/z 153.0174 (1,3A+). The direct cleavage of 126 Da (-A ring) was also detected in the initial fragmentation step. Thus, CE1 was characterized as a chalcone, naringenin chalcone. The [M+H]+ ion formula of CE2 (257.1913, Tr 11.89 min) was annotated as C15H13O4. Four intermediate ions, including m/z 239.1809, 211.1475, 213.1639, 215.1430, were produced through the commonly observed cleavage of 18 (-H2O), 28 Da (-CO), or 44 Da (-CO2), or 42 Da (-C2H2O) in chalcones. Furthermore, two hydroxy was verified on ring A, and one hydroxy was confirmed on ring B according to observing two fragments at m/z 109.0644 and 147.1174. Thus, CE2 was rapidly identified as isoliquiritigenin. The [M+H]+ ion formula of CE3 (355.2335, Tr 13.05 min) was annotated as C21H23O5. The generating of an ion at m/z 287.1650 through the breakage of 68 Da(-C5H8) indicated the existence of an isopentenyl. A characteristic fragment at m/z 273.1870 was generated by breaking 14 Da (-CH2) and C-ring cyclization. The subsequent cleavage pathway of this ion was the same as flavanone. Two hydroxy on ring A and one on ring B were determined based on the diagnostic fragments at m/z 109.1010 (0,4A+) and 121.1014 (1,3B+). CE3 was identified as a chalcone, xanthohumol. The [M+H]+ ion formula of CE4 (335.1384, Tr 11.11 min) was annotated as C16H15O8. The stepwise cleavage of 15 Da (-CH3), 28 Da (-CO), 44 Da (-CO2) indicated the existence of methyl and multiple hydroxy. Furthermore, three hydroxy and one methoxy were verified on ring A according to the observation of two fragments at m/z 155.0595 and 183.0550. The forming of an ion at 198.0776 through the loss of 137 Da (-•C7H5O3) indicated three hydroxy were located on ring B. Thus, CE4 was finally characterized as a new chalcone analog, 2,3,3′,4′,5,5′-hexahydroxy-4-methoxy chalcone. Two metabolites were identified and classified as dihydrochalcones analogs (DCE1-DCE2) according to their fragmentation patterns. Their fragmentation pathways are illustrated in Figs. S44–S45. The [M+H]+ ion formula of DCE1 (379.2446, Tr 12.84 min) was annotated as C23H23O5. Four intermediate ions, including m/z 335.1394, 307.1457, 292.1170, 250.1100, were produced through the specific cleavage of 44 Da (-CO2), 28 Da (-CO), 15 Da (-CH3), 42 Da (-C2H2O) in chalcones, and it indicated the existence of hydroxy and methoxy. The production of a fragment at m/z 133.1015 meant no substituent was located on ring B. Thus, DCE1 was determined as uvaretin. The [M+H]+ ion formula of DCE2 (485.2557, Tr 13.88 min), 106 Da higher than DCE1, was annotated as C30H29O6. Four intermediate ions at m/z 467.3149, 439.3224, 397.2441, 333.2433 formed through the cleavage of 18 Da (-H2O), 28 Da (-CO), 42 Da (-C2H2O), 64 Da (-C5H4). The same ion at m/z 133.1016 was generated. Thus, DCE2 was identified as the hydroxy benzyl derivative of DCE1, angoluvarin. In addition, two biosynthetic precursors for chalcone, phenylalanine (CEP1) and 4-coumaric acid (CEP2) were also mined and identified according to their cleavage patterns (Figs. S46–S47) [22,23]. The structures of the identified chalcone analogs (CE1-CE4, DCE1-DCE2) are illustrated in Fig. 2.
3.5. Mining and identification of xanthone analogs
Four metabolites were identified and classified as xanthones analogs (XE1-XE4) according to their informative diagnostic ions (Fig. 1) [24]. Their fragmentation pathways are illustrated in Figs. S48–S51. The [M+H]+ ion formula of XE1 (245.1773, Tr 9.65 min) was annotated as C13H9O5. A series of intermediate ions at m/z 201.1133, 173.0950, 159.0788 formed through the cleavage of 44 Da (-CO2), 28 Da (-CO), or 42 Da (-C2H2O). Two hydroxy on ring A were verified according to the observation of a diagnostic fragment at m/z 125.0952 (1, 4A+). XE1 was identified as a xanthone analog, gentisein. The [M+H]+ ion formula of XE2 (343.1989, Tr 9.28 min) was annotated as C18H15O7. Four intermediate ions at m/z 325.1450, 310.1202, 200.1284, 144.0808 formed through the stepwise loss of 18 Da (-H2O), 15 Da (-•CH3), 110 Da (-C6H6O2), 56 Da (-2CO). A diagnostic ion at m/z 137.0604 (0, 4B+) indicated the existence of a hydroxy on ring B. The production of fragments at m/z 233.0962 and 200.1284 through the cleavage of 110 Da (-C6H6O2) from the mother ion and the ion at m/z 310.1202 indicated the existence of di-tetrahydrofuran moiety. The di-tetrahydrofuran moiety, along with a hydroxy and methoxy on ring A, was confirmed based on two fragments at m/z 207.0799 (0,4A+) and 251.1063 (1,3A+). Finally, XE2 was identified as 5-hydroxy sterigmatocystin based on its detailed cleavage tree. The [M+H]+ ion formula of XE3 (m/z 259.2046, Tr 14.60 min), 14 Da higher than XE1, was annotated as C14H11O5 and defined as a methoxy derivative of XE1. One methoxy and hydroxy was verified on ring A according to the observation of a diagnostic fragment at m/z 123.1166 (0,4A+). Furthermore, a hydroxy on ring B was also confirmed based on detecting a diagnostic ion at m/z 137.0970 (0,4B+). Thus, XE3 was identified as 3-methyl gentisein. The [M+H]+ ion formula of XE4 (277.2158, Tr 13.06 min), 32 Da higher than XE1, was annotated as C13H9O7. It was defined as a dihydroxy derivative of XE1 according to the observation of diagnostic ions at m/z 137.0603 (0,4B+), 109.0644 (0,3B+), 185.1319 (1,3A+), 157.1005 (1,4A+). XE4 was identified as a xanthone, 2,4-dihydroxy gentisein. In addition, three xanthone biosynthetic precursors XEP1-XEP3 were mined and identified according to their fragmentation spectra (Figs. S52–S54) [25]. The structures of the identified xanthone analogs (XE1-XE4) are illustrated in Fig. 2.
3.6. Mining and identification of flavane analogs
Ten metabolites were identified and classified as flavane analogs (FAE1, IFAE1, FAL1-FAL3, BF1-BF3, AN1-AN2) according to their informative diagnostic ions (Fig. 1) [26]. Their fragmentation pathways are illustrated in Figs. S55–S64. The [M+H]+ ion formula of FAE1 (465.1425, Tr 9.29 min) was annotated as C23H29O10. A series of intermediate ions at m/z 303.0492, 285.0388, 257.0447 formed through the cleavage of 162 Da (-C6H10O5), 18 Da (-H2O), 28 Da (-CO). A glucosyl and hydroxy on ring A and two methoxys on ring B were confirmed, and FAE1 was identified as a flavane, diffutin, according to the detection of four fragments at m/z 137.0226 (0, 2A+-C6H10O5), 153.0174 (1, 2A+-C6H10O5), 165.0177 (1, 3B+), 127.0393 (1, 4A+-C6H8O5). The [M+H]+ ion formula of IFAE1 (325.1954, Tr 14.41 min) was annotated as C20H21O4. The fragments at m/z 257.1865, 229.1917, 185.1337 formed through the breakage of 68 Da (-C5H8), 28 Da (-CO), 44 Da (-CO2). Two hydroxy on ring B was confirmed according to the diagnostic ion at m/z 151.1106 (1,4B+) and 137.0963 (1, 3B+). The substituent on ring A was determined based on the diagnostic ions at m/z 173.1312 (0,3A+), 189.1295 (1,3A+) and 203.1256 (2,3A+). IFAE1 was characterized as an isoflavane analog, glabridin, according to these fragment ions. The [M+H]+ ion formula of FAL1 (291.1971, Tr 11.32 min) was annotated as C15H15O6. The intermediate fragment at m/z 255.1748 generated through the cleavage of 36 Da (-2H2O), and two fragments at m/z 167.1061 (1,4B+), 135.0804 (0, 2A+-H2O) indicated that FAL1 was a flavanol derivative. Two hydroxy on ring A and two on ring B were determined based on two fragments at m/z 167.1061 (1,4B+) and 135.0804 (0,2A+-H2O). Thus, FAL1 was characterized as cianidanol. The [M+H]+ ion formula of FAL2 (505.3092, Tr 14.15 min) was annotated as C23H21O13. The existence of one gallate on ring C was verified based on the cleavage of 170 Da (-C7H6O5) or 152 Da (-C7H4O4) and the observation of an ion at m/z 277.2162 (1,3B+-CO2). One methoxy and three hydroxy on ring A were verified according to a characteristic fragment at m/z 171.1170 (1, 4A+). Three hydroxy on ring B were also determined according to detecting a diagnostic fragment at m/z 277.2162 (1,3B+-CO2). Thus, FAL2 was characterized as a new flavane, 6,8-dihydroxy-7-methyl epigallocatechin-3-O-gallate ester. The [M+H]+ ion formula of FAL3 (503.2908, Tr 13.94 min) was annotated as C24H23O12. The generating of a fragment at m/z 315.1897 through the breakage of 152 Da (-C7H4O4), 18 Da (-H2O), 18 Da (-H2O) indicated the existence of gallate and hydroxy. Two methoxys on ring B and a gallate on ring C were determined according to the detection of a diagnostic ion at 211.0961 (0,4B+-C7H4O4). FAL3 was identified as a new flavane, 6, 8-dihydroxy-3′,4′-dimethyl cianidanol-3-O-gallate ester. The [M+H]+ ion formula of BF1 (747.4274, Tr 14.91 min) was annotated as C37H31O17. The loss of 170 Da (-C7H6O5) or 152 Da (-C7H4O4) indicated the existence of a gallate moiety. A gallate on ring C and two hydroxy on ring B were further confirmed according to detecting a fragment at m/z 321.2093 (0,3B+). A fragment ion at m/z 125.0008 (1’, 4’A+) was detected. Thus, two hydroxy on ring A′ was also confirmed. BF1 was finally characterized as a biflavanol, epigallocatechin-(4–8)-epicatechin-3-O-gallate ester. The [M+H]+ ion formula of BF2 (683.3121, Tr 11.82 min) was annotated as C37H31O13. The existence of a gallate on ring C was determined according to the cleavage of 170 Da (-C7H6O5), 152 Da (-C7H4O4), and the diagnostic ion at m/z 303.1110 (1,4B+). One hydroxy on ring B was also verified based on this diagnostic ion. Interestingly, the generation of fragments at m/z 275.1163 and 411.1635 through a typical dimer break indicated that BF2 was a biflavanol analog. The hydroxy number on ring A and ring B was also determined. BF2 was finally identified as a new biflavanol, 3′,4′,7-trihydroxy flavanol-(4–8)-4′,7-dihydroxy flavanol-3-O-gallate ester. The [M+H]+ ion formula of BF3(725.4379, Tr 15.32 min), 42 Da higher than BF2, was annotated as C40H37O13. The loss of 152 Da (-C7H4O4), 18 Da (-H2O), 42 Da (-C2H2O) was observed in its MS spectra. Thus, the existence of a gallate and hydroxy was determined. BF3 was classified as a tri-methyl derivative of BF2 based on detecting a diagnostic fragment at m/z 451.1936 (1’,4’B+-C7H4O4). BF3 was characterized as a new biflavanol, 4′-hydroxy-3′,7-dimethoxyflavanol-(4–8)-4′-hydroxy-7-methoxyflavanol-3-O-gallate ester. The [M+H]+ ion formula of AN1 (595.2170, Tr 10.58 min) was annotated as C27H31O15+. A series of intermediate ions at m/z 287.0537, 241.0506, 213.0545 formed through the stepwise loss of 308 Da (-C12H20O9), 46 Da (-CH2O2), 28 Da (-CO). The rutinoside on ring C and two hydroxy on ring A were confirmed according to the fragments mentioned above and the detection of the fragment at m/z 165.0539 (1,2A+-C12H20O9). AN1 was further identified as an anthocyanin derivative, keracyanin, according to the detection of the diagnostic fragments and typical breakage of 18 Da (-H2O) and 28 Da (-CO). The [M+H]+ ion formula of AN2 (m/z 269.2265, Tr 14.53 min) was annotated as C16H13O4+. The breakage of 42 Da (-C2H2O), 28 Da (-CO), 42 Da (-C2H2O), were observed and a series of intermediate ions at m/z 227.1786, 199.1477, 157.1008 formed. A methoxy, along with a hydroxy on ring A and another on ring B, was verified, respectively, according to the characteristic fragments at m/z 105.0701 (0,4A+-H2O) and 147.1175 (0,4B+). AN2 was characterized as 7-hydroxy-2-(4-hydroxyphenyl)-5-methoxy chromenium. The [M+H]+ ion formula of FAEP1 (291.1921, Tr 10.94 min) was annotated as C15H15O6. Six intermediate fragments at m/z 263.1149, 235.1232, 217.1597, 189.1267, 147.0447 and 119.0492 formed through the loss of 28 Da (-CO), 28 Da (-CO), 18 Da (-H2O), 28 Da (-CO), 42 Da (-C2H2O), 26 Da (-C2H4). Two hydroxy on ring A and one on ring B were determined according to two diagnostic fragments at m/z 109.0644 (0,4A+) and 105.0701 (0,2B+-H2O). A loss of 18 Da (-H2O) was also detected in the first cleavage step. Thus, FAEP1 was determined as leucopelargonidin (Fig. S65). The [M+H]+ ion formula of FAEP2 (307.1457, Tr 11.18 min) was annotated as C15H15O7. An intermediate fragment at m/z 271.1679 was generated by breaking 18 Da (-H2O) and 18 Da (-H2O). Hereafter, two diagnostic ions for this fragment, 163.0398 (0,4B+) and 151.0751 (0,2A+) formed. It indicated the existence of two hydroxy on ring A and one on ring B. Thus, the ion at m/z 271.1679 was characterized as an anthocyanin derivative. Finally, FAEP2 was characterized as the biosynthetic precursor for anthocyanin, 2-hydroxy leucopelargonidin (Fig. S66). The structures of the identified flavane analogs (FAE1, IFAE1, FAL1-3, BF1-3, AN1-2) are illustrated in Fig. 2. The fragmentation spectra for six flavonoid standards are also provided in Figs. S67–S72 and compared with the identified flavonoids in C. acuminate to verify the accuracy of our results.
3.7. Metabolic pathway mapping of flavonoids and untapped biosynthetic gene discovery
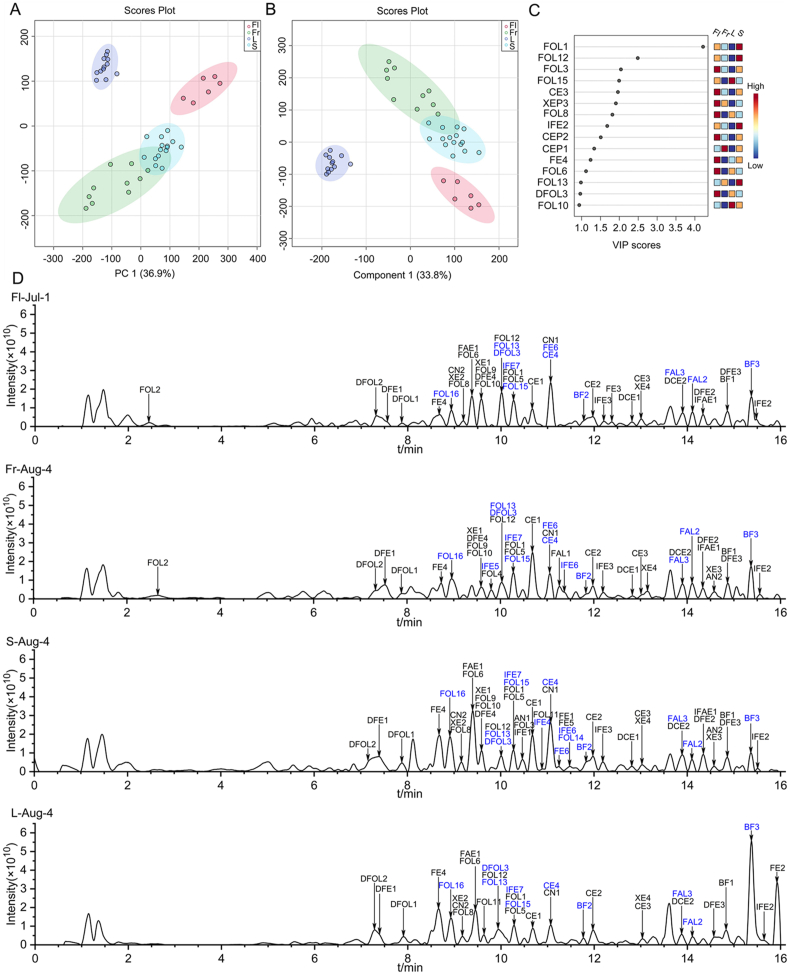
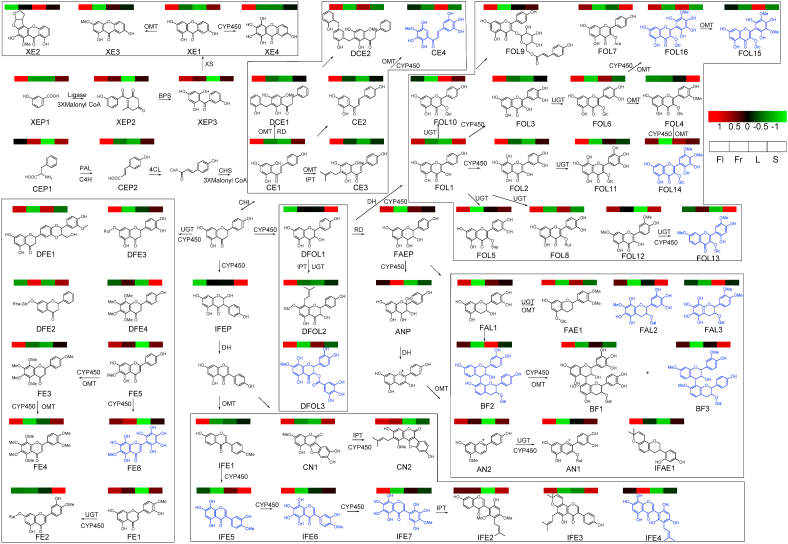
To screen the discrimination flavonoids in C. acuminate, the ion abundance of the fifty-eight identified flavonoid metabolites in the flower, fruit, stem, and leaf samples was extracted from the raw dataset matrix and submitted to MetaboAnalyst 5.0 for multivariate analysis and enrichment analysis. PCA analysis in Fig. 3A indicated that fruit groups were not separated from the stem group. Better separation between the four groups were obtained based on the PLS-DA analysis and illustrated in Fig. 3B. Five discrimination flavonoids, including CE3, FOL1, FOL3, FOL12, FOL15 (VIP>2), were obtained (Fig. 3C). Interestingly, four of them were classified into flavonol analogs. Furthermore, CE3 and FOL3 could be designated as the marker metabolites for flowers, FOL15 for leaves, FOL1 and FOL12 for stems in C. acuminate based on their enrichment patterns. To unveil the flavonoid metabolism in C. acuminate, a metabolic map for all the identified flavonoids was constructed and depicted in Fig. 4 based on their biogenetic originations, chemical structures, and enrichment pattern in different tissues. Undoubtedly, this ancient tree species could be designated as a versatile flavonoid manufacturer. Numerous untapped CYP450, glucosyltransferase, and O-methyltransferase probably involved in flavonoid metabolism could be mined in the gene reservoir of C. acuminate, based on our metabolic pathway for flavonoids.
Fig. 3.
Multivariate statistical analysis (A) PCA analysis, (B) PLS-DA score plots of flavonoids in the flowers, fruits, stems, and leaves of C. acuminate, (C) The obtained discriminating flavonoids (VIP score >2). (D) The TICs for the flowers, fruits, stems, and leaves of C. acuminate (New flavonoids are marked in blue).
Fig. 4.
The detailed version of metabolic pathway for flavonoids in C. acuminate. New flavonoids are depicted in blue. PAL: phenylalanine ammonia-lyase, C4H: cinnamate 4-hydroxylase, 4CL: 4-Coumarate coenzyme A ligase, CHS: chalcone synthase, CHI: chalcone isomerase, BPS: benzophenone synthase, XS: xanthone synthase, CYP450: cytochrome P450, OMT: O-methyltransferase, UGT: UDP-glucosyltransferase, PT: prenyltransferase, RD: reductase, DH: dehydrase
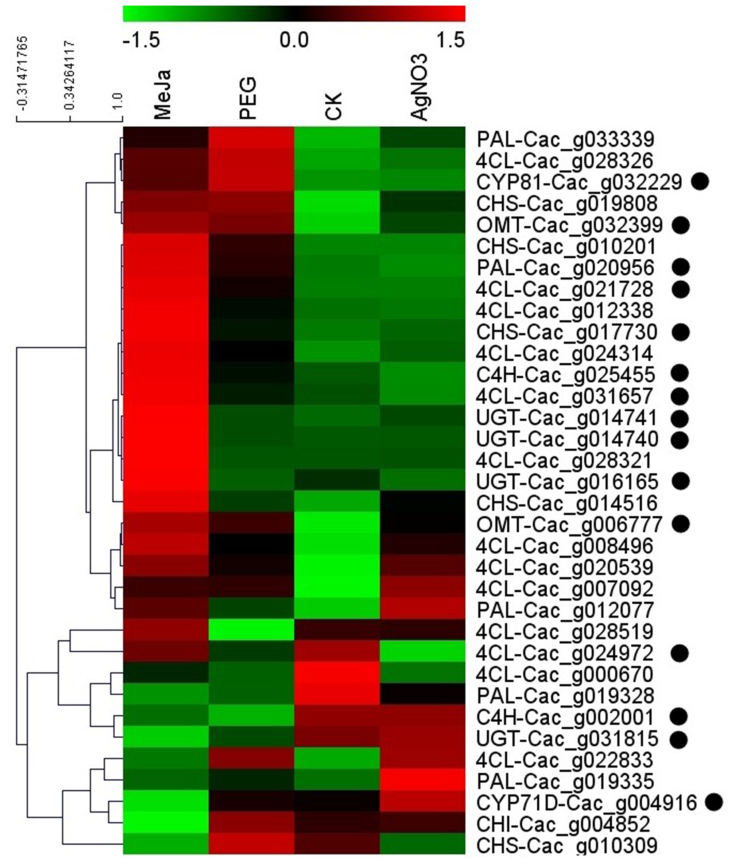
Firstly, the genes involved in the biosynthesis of chalcone, the common precursor for flavonoids, were purposefully mined according to their conserved domains. Five phenylalanine ammonia-lyase, two cinnamate 4-hydroxylase, five chalcone synthase, one chalcone isomerase, thirteen 4-coumarate coenzyme A ligase, were obtained from the generated genomic dataset. The identities and coverages were calculated through sequence alignment with the characterized ones from other plants and listed in Table 1. The distribution pattern for these bait genes was generated according to our co-expression analysis results. Although the twenty-six bait genes mentioned above were broadcast in eleven different clusters, seven of them, including CaPAL-Cac_g020956, CaC4H-Cac_g002001, CaC4H-Cac_g025455, Ca4CL-Cac_g024972, Ca4CL-Cac_g021728, Ca4CL-Cac_g031657, and CaCHS-Cac_g017730, which were responsible for the upstream biosynthetic pathway for flavonoids, were concentratedly distributed in cluster turquoise. And the unique CaCHI-Cac_g004852, along with CaCHS-Cac_g010309, appeared in cluster tan. The mining of the CYP450, OMT, and UGT candidate genes probably involved in the biosynthetic modification of flavonoids was initiated and focused on the turquoise cluster.
Table 1.
The most probable genes involved in flavonoid metabolism in C. acuminate.
| Description | C. acuminate gene number | Top-hit gene GenBank number |
Identity (%) | Coverage (%) | Distribution |
|---|---|---|---|---|---|
| Phenylalanine ammonia-lyase | Cac_g033339 | ASU87402.1 | 90.86 | 99 | purple |
| Cac_g020956 | ASG81459.1 | 88.86 | 99 | turquoise | |
| Cac_g019328 | ACC63889.1 | 86.12 | 99 | brown | |
| Cac_g019335 | AJR20994.1 | 85.83 | 99 | pink | |
| Cac_g012077 |
CAA3029115.1 |
84.67 |
97 |
lightcyan |
|
| Cinnamate 4-hydroxylase | Cac_g002001 | ANR76395.1 | 99.60 | 99 | turquoise |
| Cac_g025455 |
AAT39513.1 |
99.01 |
99 |
turquoise |
|
| 4-Coumarate coenzyme A ligase | Cac_g008496 | PSS08186.1 | 85.90 | 99 | blue |
| Cac_g022833 | QGI57467.1 | 84.74 | 99 | red | |
| Cac_g007092 | QGN03528.1 | 84.69 | 99 | blue | |
| Cac_g024972 | KAG5243652.1 | 84.68 | 99 | turquoise | |
| Cac_g012338 | CAA2996027.1 | 76.98 | 81 | yellow | |
| Cac_g021728 | PON90428.1 | 79.74 | 99 | turquoise | |
| Cac_g028326 | PSS05058.1 | 79.63 | 93 | darkgreen | |
| Cac_g028321 | PSR98869.1 | 75.00 | 88 | yellow | |
| Cac_g031657 | PSR89508.1 | 74.42 | 92 | turquoise | |
| Cac_g024314 | AGE10595.1 | 71.79 | 99 | salmon | |
| Cac_g000670 | PON94724.1 | 70.50 | 53 | brown | |
| Cac_g028519 | QGI57467.1 | 69.88 | 99 | red | |
| Cac_g020539 |
KAG5236492.1 |
62.59 |
98 |
blue |
|
| Chalcone synthase | Cac_g017730 | ARO92270.1 | 100.00 | 99 | turquoise |
| Cac_g010309 | ADD74168.1 | 93.78 | 98 | tan | |
| Cac_g010201 | AQZ26783.1 | 85.01 | 95 | yellow | |
| Cac_g014516 | KAB1207574.1 | 84.65 | 99 | yellow | |
| Cac_g019808 |
KAB1207574.1 |
81.38 |
94 |
yellow |
|
| Chalcone isomerase |
Cac_g004852 |
AUT30534.1 |
84.95 |
99 |
tan |
| CYP450 | Cac_g004916 | AGB55868.1 | 57.14 | 99 | turquoise |
| Cac_g032229 |
AB001379.1 |
41.04 |
98 |
turquoise |
|
| O-methyltransferase | Cac_g006777 | AGQ21571.1 | 66.67 | 94 | turquoise |
| Cac_g032399 |
AAA86982.1 |
52.19 |
98 |
turquoise |
|
| UDP-glucosyltransferase | Cac_g014740 | AAS94329.1 | 56.21 | 98 | turquoise |
| Cac_g014741 | AAS94329.1 | 57.23 | 98 | turquoise | |
| Cac_g016165 | JQ070807.1 | 54.72 | 98 | turquoise | |
| Cac_g031815 | BAR88078.1 | 56.33 | 96 | turquoise |
Secondly, thirty-five CYP450 candidate genes, nine OMT candidate genes, twenty-five UGT candidate genes, which were co-expressed with CaPAL-Cac_g020956, CaC4H-Cac_g002001, CaC4H-Cac_g025455, Ca4CL-Cac_g024972, Ca4CL-Cac_g021728, Ca4CL-Cac_g031657, and CaCHS-Cac_g017730, were extracted from the co-expression matrix of the turquoise cluster. The co-expressed network was constructed with Cytoscape 3.8.2 and depicted in Fig. 5. Interestingly, only nineteen of the twenty-five UGT candidates, eight of the nine OMT candidates, twenty-four of the thirty-five CYP450 candidates were synchronously correlated with the four kinds of genes involved in flavonoid biosynthesis. Thus, the full amino acid sequence of these candidates was further extracted from the genomic and transcriptomic dataset and used to perform sequence alignment analysis with the characterized ones. Encouragingly, two of the obtained OMT candidates, Cac_g006777 and Cac_g032399, shared 66.67% and 52.19% identities with flavone-8-O-methyltransferase in Ocimum basilicum (AGQ21571.1) [27] and flavonoid 3′-O-methyltransferase in Chrysosplenium americanum (AAA86982.1) [28], respectively. Two of the obtained CYP450 candidates, Cac_g004916 and Cac_g032229, were classified as CYP71D and CYP81 family, respectively, according to their conserved domains and catalytic residues. They shared 57.14% and 41.04% identities with the flavonoid 6-hydroxylase in Glycine max (AGB55868.1) [29] and isoflavone 2′-hydroxylase in Glycyrrhiza echinate (AB001379.1) [30], respectively. Four UGT candidates, Cac_g031815, Cac_g016165, Cac_g014741, and Cac_g014740, were screened from the obtained UGT candidates according to their conserved domains and catalytic residues. They shared 56.33%, 54.72%, 57.23 and 56.21% identities with the flavonol 3-O-glucosyltransferase (BAR88078.1) in Glycine max [31], flavonol 5-O-glucosyltransferase (AF171902.1) in Paeonia lactiflora [32], flavonoid 7-O-glucosyltransferases (AAS94329.1) in Beta vulgaris [33], respectively. The identities and coverages for these newly fished CYP450s, OMTs, UGTs, are listed in Table 1. Sequence alignment analysis with the characterized ones is provided in Figs. S73–S79 . The expression levels for the newly fished CYP450s, OMTs, UGTs, along with the bait genes, are visualized in Fig. 6based on our transcriptomic data for C. acuminate plantlets after different elicitor treatments. Encouragingly, most of our targeted genes cluster together in the heat map. Five of the seven bait genes distributed in cluster turquoise, and six of the eight newly fished genes, were significantly upregulated after MeJa treatment. The expression level of CYP71-Cac_g004916 was significantly upregulated after AgNO3 treatment. Minor upregulation was observed for the targeted genes distributed in cluster turquoise after PEG and AgNO3 treatment. These results indicated that treatment with MeJa could trigger flavonoid metabolism in C. acuminate plantlets. Functional characterization of these newly fished gene candidates involved in flavonoid metabolism in C. acuminate could be initiated with the help of these guiding results.
Fig. 5.
Co-expression analysis for the bait genes and the probable CYP450, OMT, UGT candidate genes involved in flavonoid biosynthesis.
Fig. 6.
The hierarchical clustering heat map for the bait genes and the newly fished CYP450, OMT, UGT candidate genes involved in flavonoid biosynthesis. The genes distributed in cluster turquoise are marked with black circle.
4. Conclusions
Except for the fascinatingly alkaloid-producing capability, the Chinese native plant, C. acuminate, was herein proved to be a versatile flavonoids producer through a UPLC-Orbitrap MS-based targeted metabolomics. Fifteen new flavonoids, along with forty-three known flavonoids and eight biosynthetic precursors, were carefully mined and identified from mature C. acuminate. Furthermore, fifty-three of them were firstly characterized in C. acuminate. Five discriminating flavonoid metabolites, CE3, FOL1, FOL3, FOL10, FOL8, were also obtained based on the multivariate analysis. The full metabolic map for flavonoids in C. acuminate was constructed based on their biogenetic origination and tissue enrichment patterns. The most probable genes involved in chalcone biosynthesis, flavonoid hydroxylation, methylation, and glycosylation were further mined and fished in the gene reservoir of C. acuminate. These findings provide detailed biosynthetic insights into the versatile flavonoid metabolism in C. acuminate.
CRediT authorship contribution statement
Xiang Pu: Conceptualization, Project administration, Funding acquisition, Data curation, Formal analysis, Analysis, Writing – review & editing. Jia Li: Methodology, Data curation, Analysis, Formal analysis, Software, Visualization. Ziang Guo: Methodology, Data curation, Analysis, Formal analysis, Software, Visualization. Minji Wang: Methodology, Data curation, Formal analysis, Analysis, Software, Visualization. Ming Lei: Methodology, Data curation, Formal analysis, Analysis, Software, Visualization. Shengnan Yang: Methodology, Formal analysis. Jun Yang: Methodology, Formal analysis, Software. Hanguang Wang: Validation, Formal analysis. Li Zhang: Supervision, Validation. Qianming Huang: Investigation, Project administration, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors wish to acknowledge the financial support provided by the Department of Science and Technology of Sichuan Province, PR China (Project No. 2021ZYD0059), the National Natural Science Foundation of China (Project No. 21708028) and the National College Students Innovation and Entrepreneurship Training Program, PR China (Project No. 201910626009).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2022.03.007.
Contributor Information
Xiang Pu, Email: puxiang@sicau.edu.cn.
Qianming Huang, Email: hqming@sicau.edu.cn.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Cuyckens F., Claeys M. Mass spectrometry in the structural analysis of flavonoids. J Mass Spectrom. 2004;39:1–15. doi: 10.1002/jms.585. [DOI] [PubMed] [Google Scholar]
- 2.Spagnuolo C., Moccia S., Russo G.L. Anti-inflammatory effects of flavonoids in neurodegenerative disorders. Eur J Med Chem. 2018;153:105–115. doi: 10.1016/j.ejmech.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Imran M., Rauf A., Abu-Izneid T., Nadeem M., Shariati M.A., Khan I.A., et al. Luteolin, a flavonoid, as an anticancer agent: a review. Biomed Pharmacother. 2019;112:108612. doi: 10.1016/j.biopha.2019.108612. [DOI] [PubMed] [Google Scholar]
- 4.Maaliki D., Shaito A.A., Pintus G., EI-Yazbi A., Eid A.H. Flavonoids in hypertension: a brief review of the underlying mechanisms. Curr Opin Pharmacol. 2019;45:57–65. doi: 10.1016/j.coph.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Hertog M.G., Feskens E.J., Hollman P.C., Katan M.B., Kromhout D. Dietary antioxidant flavonoids and risk of coronary heart disease: the Zutphen Elderly Study. Lancet. 1993;342:1007. doi: 10.1016/0140-6736(93)92876-u. [DOI] [PubMed] [Google Scholar]
- 6.Akimoto N., Ara T., Nakajima D., Suda K., Ikeda C., Takahashi S., et al. Flavonoidsearch: a system for comprehensive flavonoid annotation by mass spectrometry. Sci Rep. 2017;7:1243. doi: 10.1038/s41598-017-01390-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pu X., Zhang C.R., Gao H.C., Huang L., Zhu L., Rao Y., et al. Biosynthesis-inspired mining and identification of untapped alkaloids in Camptotheca acuminate for enzyme discovery using ultra-high performance liquid chromatography coupled with quadrupole-time of flight-mass spectrometry. J Chromatogr A. 2020;1620:461036. doi: 10.1016/j.chroma.2020.461036. [DOI] [PubMed] [Google Scholar]
- 8.Yang W.Z., Ye M., Qiao X., Wang Q., Bo T., Guo D.A. Collision-induced dissociation of 40 flavonoid aglycones and differentiation of the common flavonoid subtypes using electrospray ionization ion-trap tandem mass spectrometry and quadrupole time-of-flight mass spectrometry. Eur J Mass Spectrom. 2012;18:493–503. doi: 10.1255/ejms.1206. [DOI] [PubMed] [Google Scholar]
- 9.Wu W., Yan C.Y., Li L., Liu Z.Q., Liu S.Y. Studies on the flavones using liquid chromatography-electrospray ionization tandem mass spectrometry. J Chromatogr A. 2004;1047:213–220. doi: 10.1016/j.chroma.2004.06.128. [DOI] [PubMed] [Google Scholar]
- 10.Qin Y., Gao B.Y., Shi H.M., Cao J., Yin C.L., Lu W.Y., et al. Characterization of flavonol mono-, di-, tri- and tetra-O-glycosides by ultra-performance liquid chromatography-electrospray ionization-quadrupole time-of-flight mass spectrometry and its application for identification of flavonol glycosides in Viola tianschanica. J Pharm Biomed Anal. 2017;142:113–124. doi: 10.1016/j.jpba.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Montoro P., Maldini M., Piacente S., Macchia M., Pizza C. Metabolite fingerprinting of Camptotheca acuminata and the HPLC-ESI-MS/MS analysis of camptothecin and related alkaloids. J Pharm Biomed Anal. 2010;51:405–415. doi: 10.1016/j.jpba.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 12.Ma Y.L., Li Q.M., Heuvel H.V.D., Claeys M. Characterization of flavone and flavonol aglycones by collision-induced dissociation tandem mass spectrometry. Rapid Commun Mass Spectrom. 1997;11:1357–1364. doi: 10.1002/(SICI)1097-0231(19991015)13:19<1932::AID-RCM735>3.0.CO;2-W. 1364. [DOI] [PubMed] [Google Scholar]
- 13.Fabre N., Rustan I., Hoffmann E., Quetin-Leclercq J. Determination of flavone, flavonol, and flavanone aglycones by negative ion liquid chromatography electrospray ion trap mass spectrometry. J Am Soc Mass Spectrom. 2001;12:707–715. doi: 10.1016/S1044-0305(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 14.Madeira P.J.A., Borges C.M., Florêncio M.H. Electrospray ionization Fourier transform ion cyclotron resonance mass spectrometric and semi‐empirical calculations study of five isoflavone aglycones. Rapid Commun Mass Spectrom. 2010;24:3432–3440. doi: 10.1002/rcm.4791. [DOI] [PubMed] [Google Scholar]
- 15.Kang J.G., Hick L.A., Price W.E. A fragmentation study of isoflavones in negative electrospray ionization by MSn ion trap mass spectrometry and triple quadrupole mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:857–868. doi: 10.1002/rcm.2897. [DOI] [PubMed] [Google Scholar]
- 16.Kim Y., Shrestha R., Kim S., Kim J.A., Lee J., Jeong T.C., et al. In vitro characterization of glycyrol metabolites in human liver microsomes using HR-resolution MS spectrometer coupled with tandem mass spectrometry. Xenobiotica. 2020;50:380–388. doi: 10.1080/00498254.2019.1636418. [DOI] [PubMed] [Google Scholar]
- 17.Parejo I., Jáuregui O., Viladomat F., Bastida J., Codina C. Characterization of acylated flavonoid-O-glycosides and methoxylated flavonoids from Tagetes maxima by liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2010;18:2801–2810. doi: 10.1002/rcm.1697. [DOI] [PubMed] [Google Scholar]
- 18.March R.E., Miao X.S., Metcalfe C.D. A fragmentation study of a flavone triglycoside, kaempferol-3-O-robinoside-7-O-rhamnoside. Rapid Commun Mass Spectrom. 2004;18:931–934. doi: 10.1002/rcm.1428. [DOI] [PubMed] [Google Scholar]
- 19.Xu T.T., Yang M., Li Y.F., Chen X., Wang Q.R., Deng W.P., et al. An integrated exact mass spectrometric strategy for comprehensive and rapid characterization of phenolic compounds in licorice. Rapid Commun Mass Spectrom. 2013;27:2297–2309. doi: 10.1002/rcm.6696. [DOI] [PubMed] [Google Scholar]
- 20.Khan N.A., Wu H.F. Analysis of silymarin extracted from a commercial dosage by combining liquid-liquid extraction with negative electrospray tandem mass spectrometry. Rapid Commun Mass Spectrom. 2004;18:2960–2962. doi: 10.1002/rcm.1700. [DOI] [PubMed] [Google Scholar]
- 21.Tai Y.P., Pei S.F., Wan J.P., Cao X.J., Pan Y.J. Fragmentation study of protonated chalcones by atmospheric pressure chemical ionization and tandem mass spectrometry. Rapid Commun Mass Spectrom. 2006;20:994–1000. doi: 10.1002/rcm.2404. [DOI] [PubMed] [Google Scholar]
- 22.MassBank of North America 2021. https://mona.fiehnlab.ucdavis.edu/spectra/display/PR100032
- 23.MassBank of North America 2021. https://mona.fiehnlab.ucdavis.edu/spectra/display/PR100261
- 24.Du X.G., Wang W., Zhang Q.Y., Cheng J., Avula B., Khan I.A., et al. Identification of xanthones from Swertia punicea using high-performance liquid chromatography coupled with electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2012;26:2913–2923. doi: 10.1002/rcm.6419. [DOI] [PubMed] [Google Scholar]
- 25.Masters K.S., Bräse S. Xanthones from fungi, lichens and bacteria: the natural products and their synthesis. Chem Rev. 2012;112:3717–3776. doi: 10.1021/cr100446h. [DOI] [PubMed] [Google Scholar]
- 26.Lee R.J., Lee V.S.Y., Tzen J.T.C., Lee M.R. Study of the release of gallic acid from (-)-epigallocatechin gallate in old oolong tea by mass spectrometry. Rapid Commun Mass Spectrom. 2010;24:851–858. doi: 10.1002/rcm.4442. [DOI] [PubMed] [Google Scholar]
- 27.Berim A., Gang D.R. Characterization of two candidate flavone 8-O-methyltransferases suggests the existence of two potential routes to nevadensin in sweet basil. Phytochemistry. 2013;92:33–41. doi: 10.1016/j.phytochem.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Gauthier A., Gulick P.J., Ibrahim R.K. Characterization of two cDNA clones which encode O-methyltransferases for the methylation of both flavonoid and phenylpropanoid compounds. Arch Biochem Biophys. 1998;351:243–249. doi: 10.1006/abbi.1997.0554. [DOI] [PubMed] [Google Scholar]
- 29.Artigot M.P., Baes M., Dayde J., Berger M. Expression of flavonoid 6-hydroxylase candidate genes in normal and mutant soybean genotypes for glycitein content. Mol Biol Rep. 2013;40:4361–4369. doi: 10.1007/s11033-013-2526-2. [DOI] [PubMed] [Google Scholar]
- 30.Akashi T., Aoki T., Takahashi T., Kameya N., Nakamura I., Ayabe S. Cloning of cytochrome P450 cDNAs from cultured Glycyrrhiza echinata L. cells and their transcriptional activation by elicitor-treatment. Plant Sci. 1997;126:39–47. doi: 10.1016/S0168-9452(97)00091-5. [DOI] [Google Scholar]
- 31.Di S., Yan F., Rodas F.R., Rodriguez T.O., Murai Y., Iwashina T., et al. Linkage mapping, molecular cloning and functional analysis of soybean gene Fg3 encoding flavonol 3-O-glucoside/galactoside (1-2) glucosyltransferase. BMC Plant Biol. 2015;15:126. doi: 10.1186/s12870-015-0504-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao D., Hao Z., Tao J. Effects of shade on plant growth and flower quality in the herbaceous peony (Paeonia lactiflora Pall.) Plant Physiol Biochem. 2012;61:187–196. doi: 10.1016/j.plaphy.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Isayenkova J., Wray V., Nimtz M., Strack D., Vogt T. Cloning and functional characterisation of two regioselective flavonoid glucosyltransferases from Beta vulgaris. Phytochemistry. 2006;67:1598–1612. doi: 10.1016/j.phytochem.2006.06.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.