Abstract
The mitochondrial permeability transition pore (PTP) is a Ca2+-dependent megachannel that plays an important role in mitochondrial physiology and cell fate. Cyclophilin D (CyPD) is a well-characterized PTP regulator, and its binding to the PTP favors pore opening. It has previously been shown that p53 physically interacts with CyPD and opens the PTP during necrosis. Accumulating studies also suggest that the F-ATP synthase contributes to the regulation and formation of the PTP. F-ATP synthase IF1 (mitochondrial ATP synthase inhibitory factor 1) is a natural inhibitor of F-ATP synthase activity; however, whether IF1 participates in the modulation of PTP opening is basically unknown. Here, we demonstrate using calcium retention capacity assay that IF1 overexpression promotes mitochondrial permeability transition via opening of the PTP. Intriguingly, we show that IF1 can interact with the p53–CyPD complex and facilitate cell death. We also demonstrate that the presence of IF1 is necessary for the formation of p53–CyPD complex. Therefore, we suggest that IF1 regulates the PTP via interaction with the p53–CyPD complex, and that IF1 is necessary for the inducing effect of p53–CyPD complex on PTP opening.
Keywords: mitochondria, F-ATP synthase inhibitory factor 1, mitochondrial permeability transition, p53, cyclophilin D
Abbreviations: ANT, adenine nucleotide translocator; CRC, calcium retention capacity; CsA, cyclosporine A; CyPD, cyclophilin D; DMEM, Dulbecco’s modified Eagle’s medium; HA, hemagglutinin; HEK293T, human embryonic kidney 293T cell line; IF1, mitochondrial ATP synthase inhibitory factor 1; IP, immunoprecipitation; KCl, potassium chloride; OSCP, oligomycin sensitivity conferral protein; OXPHOS, oxidative phosphorylation; PDAC, pancreatic ductal adenocarcinoma; PEI, polyethylenimine; PPIF, peptidylprolyl isomerase F; PTP, the mitochondrial permeability transition pore; ROS, reactive oxygen species; sgRNA, single-guide RNA; tBHP, t-butyl hydroperoxide
Mitochondrial calcium overload and oxidative stress may trigger opening of the mitochondrial permeability transition pore (PTP), resulting in the rupture of outer mitochondrial membrane and the depletion of ATP, eventually apoptotic and necrotic cell death (1). Cyclophilin D (CyPD), encoded by PPIF (peptidylprolyl isomerase F) gene, is a most well-characterized regulator of PTP. Binding of CyPD to PTP sensitizes the pore to Ca2+, whereas its genetic ablation or detachment from PTP by cyclosporine A (CsA) suppresses the pore opening (2, 3, 4). CyPD modulates the activity of F-ATP synthase through its association to the lateral stalk of the enzyme (5, 6). CyPD interacts with oligomycin sensitivity conferral protein (OSCP) of F-ATP synthase and induces conformational changes transferred to FO sector, which could also favor Ca2+ binding to the catalytic core (7). The most recent molecular model of PTP formation is that the conformational change induced by Ca2+ binding to subunit β transmits to e-subunit and c-subunit ring via subunits b and g through OSCP (8, 9, 10, 11, 12).
The tumor suppressor p53, encoded by TP53 gene, is a well-known transcription factor that exerts diverse biological functions, such as DNA repair, cell cycle, apoptosis, and metabolism (13). Direct activation of Bax by p53 may induce mitochondrial membrane permeabilization, which participates in apoptotic program (14, 15). OSCP subunit has been identified as a new partner of mitochondrial matrix p53, which promotes assembly of F-ATP synthase and mitochondrial respiration (16). Oxidative stress induces PTP opening in a p53-dependent manner by its interaction with CyPD (17). A robust p53–CypD complex is induced by H2O2 treatment, and p53–CypD axis acts as an important contributor to ischemic stroke (17).
F-ATP synthase IF1 (mitochondrial ATP synthase inhibitory factor 1), encoded by ATP5IF1 gene, is a natural inhibitor of F-ATP synthase activity (18). The oligomerization state of IF1 and its binding to F-ATP synthase depend on the matrix pH (19, 20). When the proton motive force is present, IF1 releases from F-ATP synthase and ATP synthesis recovers, indicating that IF1 contributes to preventing futile ATP hydrolysis (21). Phosphorylation of human IF1 by PKA abrogates its binding to F-ATP synthase (22). The active dimeric IF1 can bridge the F1–F1 part and promote dimerization of F-ATP synthase (23, 24). IF1 overexpression favors the formation of F-ATP synthase dimers and inhibits the enzyme activity, accompanied with increased density of mitochondrial cristae, thus, IF1 regulates mitochondrial biogenesis and morphology (25, 26). IF1 is upregulated in many human tumors and participates in oncogenesis, mediating the metabolic switch to a Warburg phenotype (27). IF1 overexpression in colon cancer results in generation of reactive oxygen species (ROS), which triggers a prosurvival and proliferative response (26). The relative expression level of IF1 varies between different tissues and cell types (25), which may contribute to heterogeneous phenotypes of tumors in metabolic phenotypes and chemoresistance (1, 28).
Despite the physiological roles of IF1 in regulating metabolic reprogramming and cell fate, the mechanism through which IF1 fulfils these functions is not fully understood. Whether IF1 plays a role in regulating PTP opening and its molecular mechanism awaits an answer. The present study has addressed the regulatory role of IF1 in PTP opening and cell death via its interaction with p53–CyPD complex.
Results
IF1 regulates mitochondrial membrane permeabilization
Many tumor tissues are not defective in mitochondrial oxidative phosphorylation (OXPHOS) but rely on oxidative respiration for their proliferation and metastasis (29, 30). Pancreatic carcinoma is a high OXPHOS subtype, and it is highly sensitive to an OXPHOS inhibitor phenformin that overcomes chemoresistance (31). In human pancreatic ductal adenocarcinoma (PDAC) cell line MIA PaCa-2, IF1 expression was notably upregulated (Fig. 1, A and B). The expression level of p53 in MIA PaCa-2 cells was significantly increased compared with other cancer cell lines like HCT116, HeLa, and HepG2 (Fig. 1, A and C). The expression level of CyPD in MIA PaCa-2 cells was slightly increased but not significant (Fig. 1, A and D). PDAC is a most lethal cancer. The regulatory mechanism and activity of PTP in PDAC is interesting to be known. Overexpression of IF1 caused activation of caspase 3 (Figs. 1, E and F and S1), which was diminished by the treatment of CsA (Fig. 1, G and H). These observations indicated that IF1 could play a role in modulation of mitochondrial membrane permeabilization.
Figure 1.
IF1 regulates mitochondrial membrane permeabilization.A and B, protein extracts of HEK293T, HCT116, CT26, HT29, HeLa, HepG2, Capan-1, PATU8988T, MIA PaCa-2, U937, and MEF cells were analyzed by Western blotting (WB). A, representative blots of five independent experiments. B, histograms represent the relative densitometry values of IF1 normalized to β-actin. Error bars indicated the mean ± SD of five independent experiments. ∗∗p < 0.01, ∗∗∗p < 0.001 versus MIA PaCa-2, one-way ANOVA with Bonferroni post hoc test. C, histograms represent the relative densitometry values of p53 normalized to β-actin. Error bars indicated the mean ± SD of at least five independent experiments. ∗p < 0.05 versus MIA PaCa-2, one-way ANOVA with Bonferroni post hoc test. D, histograms represent the relative densitometry values of CyPD normalized to β-actin. Error bars indicated the mean ± SD of at least four independent experiments. E and F, MIA PaCa-2 cells were transfected with pcDNA3.1+/C-HA empty vector (EV) or C terminus HA-tagged ATPIF1 plasmids (ATPIF1) extracted from two different Escherichia coli colonies for 24 h. Cell lysates were analyzed by WB. E, representative blots of three independent experiments. F, histograms represent the relative densitometry values of cleaved caspase 3 normalized to caspase 3. Error bars indicated the mean ± SD of three independent experiments. ∗p < 0.05, one-way ANOVA with Bonferroni post hoc test. G and H, MIA PaCa-2 cells were transfected with EV or ATPIF1 and treated with 2 μM CsA for 24 h. Cell lysates were analyzed by WB. G, representative blots of three independent experiments. H, histograms represent the relative densitometry values of cleaved caspase 3 normalized to caspase 3. Error bars indicated the mean ± SD of three independent experiments. ∗p < 0.05, ∗∗∗p < 0.001, one-way ANOVA with Bonferroni post hoc test. CsA, cyclosporine A; CyPD, cyclophilin D; HA, hemagglutinin; HEK293T, human embryonic kidney 293T cell line; IF1, mitochondrial ATP synthase inhibitory factor 1; MEF, mouse embryonic fibroblast.
Overexpression of IF1 sensitizes the PTP to Ca2+ and promotes cell death
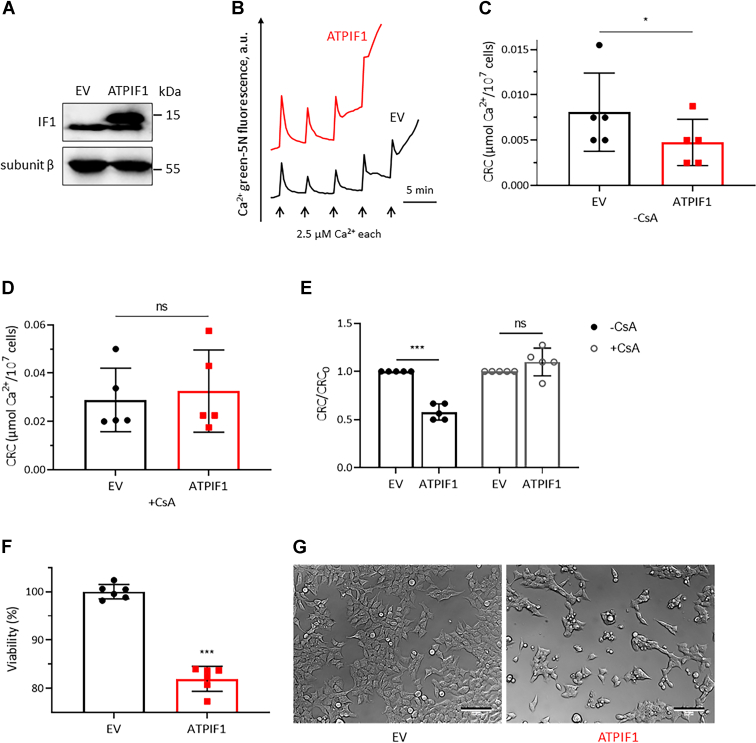
Calcium retention capacity (CRC) assay revealed that IF1 overexpression sensitized the pore to Ca2+ (Fig. 2, A–C), which was prevented by the addition of CsA (Fig. 2, D and E). Overexpression of IF1 favored cell death, which was evaluated by ATP assay (Figs. 2F and S2A) and by the observation of cell density and morphology under a microscope (Figs. 2G and S2B). IF1 overexpression inhibited cell growth and promoted cell death probably through induction of PTP opening.
Figure 2.
Overexpression of IF1 sensitizes the PTP to Ca2+and promotes cell death.A, MIA PaCa-2 cells were transfected with EV or ATPIF1 for 24 h. Cell lysates were analyzed by WB. The blots are representative of at least three independent experiments. B, representative CRC traces of digitonin-permeabilized MIA PaCa-2 cells transfected with EV (black) or ATPIF1 (red) for 24 h. C and D, CRC values of digitonin-permeabilized MIA PaCa-2 cells after transfection of EV (black column) or ATPIF1 (red column) in the absence of CsA (C) or in the presence of CsA (D). Error bars indicated the mean ± SD of five independent experiments. ∗p < 0.05, ns, Student’s t test. E, CRC/CRC0 ratio of digitonin-permeabilized MIA PaCa-2 cells transfected with ATPIF1 normalized to those transfected with EV. CRC values of permeabilized MIA PaCa-2 cells transfected with EV were set as one unit. Black columns and gray columns represent CRC/CRC0 ratio in the absence of CsA or in the presence of CsA, respectively. Error bars indicated the mean ± SD of five independent experiments. ∗∗∗p < 0.001, ns, two-way ANOVA with Bonferroni post hoc test. F, the effect of IF1 overexpression on cell viability in MIA PaCa-2 cells revealed by ATP assay. Error bars indicated the mean ± SD of six independent experiments. ∗∗∗p < 0.001 versus EV, Student’s t test. G, the images of MIA PaCa-2 cells transfected with EV or ATPIF1 for 24 h (the scale bar represents 100 μm). The figures are representative of at least three independent experiments. CRC, calcium retention capacity; CsA, cyclosporine A; EV, empty vector; IF1, mitochondrial ATP synthase inhibitory factor 1; ns, not significant; PTP, the mitochondrial permeability transition pore; WB, Western blotting.
Ablation of IF1 desensitizes the PTP to Ca2+ and prevents the cell toxicity of t-butyl hydroperoxide
IF1 is encoded by the ATPIF1 gene, and ΔATPIF1 mutant was generated to abolish the protein expression of IF1 (Fig. 3A). The role of IF1 in regulation of PTP opening was strengthened by the tests in ΔATPIF1 mutant. Ablation of IF1 strikingly desensitized the pore to Ca2+, resulting in a significant increase of CRC (Fig. 3, B–D). To further confirm the regulatory role of IF1 in PTP opening, the ΔPPIF mutant was generated to genetic disruption of CyPD expression, which is a well-characterized PTP regulator (Fig. 3E). t-Butyl hydroperoxide (tBHP) led to cell death in a mitochondrial permeability transition–dependent manner (32). About 20 micromolar of tBHP induced about 90% of MIA PaCa-2 cells dead after 24 h, and knockout of either IF1 or PPIF was able to prevent the cell toxicity of tBHP (Fig. 3F). These observations suggested that knockout of IF1 inhibited PTP opening and prevented the cell death induced by oxidative stress.
Figure 3.
Ablation of IF1 desensitizes the PTP to Ca2+and prevents the cell toxicity of tBHP.A, expression of IF1 in MIA PaCa-2 clonal cells after disruption of ATPIF1 gene (encoding IF1) using CRISPR/Cas9 technique. The blots are representative of at least three independent experiments. B, representative CRC traces of digitonin-permeabilized wild-type (WT, black) and ΔATPIF1 (red) MIA PaCa-2 cells. C, CRC values of digitonin-permeabilized WT (black column) and ΔATPIF1 (red column) MIA PaCa-2 cells. Error bars indicated the mean ± SD of seven independent experiments. ∗∗p < 0.01 versus WT, Student’s t test. D, CRC/CRC0 ratio of digitonin-permeabilized ΔATPIF1 MIA PaCa-2 cells normalized to WT. CRC values of digitonin-permeabilized WT MIA PaCa-2 cells were set as one unit. Error bars indicated the mean ± SD of seven independent experiments. ∗∗∗p < 0.001 versus WT, Student’s t test. E, expression of CyPD in MIA PaCa-2 clonal cells after disruption of PPIF gene using CRISPR/Cas9 technique. The blots are representative of at least three independent experiments. F, the effect of tBHP on cell viability in WT (black column), ΔATPIF1 (red column), and ΔPPIF (blue column) MIA PaCa-2 cells revealed by ATP assay after 24 h of treatment. Viability is presented as a percentage (%) of the control value without the treatment of tBHP. Error bars indicated the mean ± SD of at least two independent experiments. ∗p < 0.05 versus WT, ∗∗∗p < 0.001 versus WT, two-way ANOVA with Bonferroni post hoc test. CRC, calcium retention capacity; CyPD, cyclophilin D; IF1, mitochondrial ATP synthase inhibitory factor 1; PTP, the mitochondrial permeability transition pore; tBHP, t-butyl hydroperoxide.
Ablation of CyPD blocks the inducing effect of IF1 overexpression on PTP
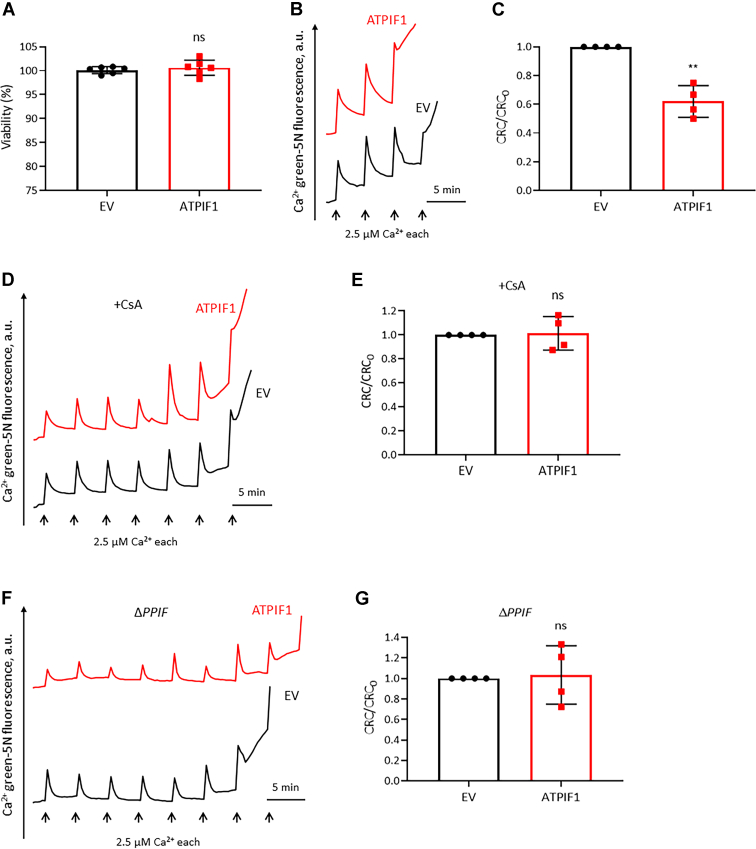
The role of IF1 in regulation of PTP opening was further investigated in the ΔPPIF mutant (Figs. 3, E and S2C). The inducing effect of cell death by IF1 overexpression was prevented in the ΔPPIF mutant (Figs. 4A and S2, C–E). Overexpression of IF1 sensitized the pore to Ca2+ in the WT cell lines (Figs. 4, B and C and S3, A and B), which was abolished by the presence of CsA (Figs. 4, D and E and S3, C and D) or the ablation of CyPD (Figs. 4, F and G and S3, E and F). These data suggested that IF1 facilitated Ca2+ release and cell death through induction of PTP opening.
Figure 4.
Ablation of CyPD blocks the inducing effect of IF1 overexpression on PTP.A, the effect of IF1 overexpression on cell viability in ΔPPIF MIA PaCa-2 cells revealed by ATP assay. Error bars indicated the mean ± SD of six independent experiments. ns, Student’s t test. B, representative CRC traces of digitonin-permeabilized WT MIA PaCa-2 cells transfected with EV (black) or ATPIF1 (red) for 24 h. C, CRC/CRC0 ratio of digitonin-permeabilized WT MIA PaCa-2 cells transfected with ATPIF1 normalized to those transfected with EV. CRC values of digitonin-permeabilized WT MIA PaCa-2 cells transfected with EV were set as one unit. Error bars indicated the mean ± SD of four independent experiments. ∗∗p < 0.01 versus EV, Student’s t test. D, representative CRC traces of digitonin-permeabilized WT MIA PaCa-2 cells transfected with EV (black) or ATPIF1 (red) for 24 h in the presence of CsA. E, CRC/CRC0 ratio of digitonin-permeabilized WT MIA PaCa-2 cells transfected with ATPIF1 normalized to those transfected with EV in the presence of CsA. CRC values of digitonin-permeabilized WT MIA PaCa-2 cells transfected with EV in the presence of CsA were set as one unit. Error bars indicated the mean ± SD of four independent experiments. ns, Student’s t test. F, representative CRC traces of digitonin-permeabilized ΔPPIF MIA PaCa-2 cells transfected with EV (black) or ATPIF1 (red) for 24 h. G, CRC/CRC0 ratio of digitonin-permeabilized ΔPPIF MIA PaCa-2 cells transfected with ATPIF1 normalized to those transfected with EV. CRC values of digitonin-permeabilized ΔPPIF MIA PaCa-2 cells transfected with EV were set as one unit. Error bars indicated the mean ± SD of four independent experiments. ns, Student’s t test. CRC, calcium retention capacity; CsA, cyclosporine A; CyPD, cyclophilin D; EV, empty vector; IF1, mitochondrial ATP synthase inhibitory factor 1; ns, not significant; PTP, the mitochondrial permeability transition pore.
IF1 interacts with p53–CyPD complex
The molecular mechanism through which IF1 regulates PTP was investigated. IF1 binds to a αDP–βDP catalytic interface of F-ATP synthase and also contacts subunit γ (33, 34, 35). Binding of Ca2+ to the catalytic core of F-ATP synthase causes a conformational change that transmits to the inner membrane subunits through the lateral stalk via OSCP, which triggers PTP formation (9, 36). OSCP subunit is one of the major interacting proteins with CyPD (7), and p53–CypD complex is required for PTP opening under oxidative stress (17). The possible interaction of IF1 with p53–CypD complex and its role in regulation of PTP were explored.
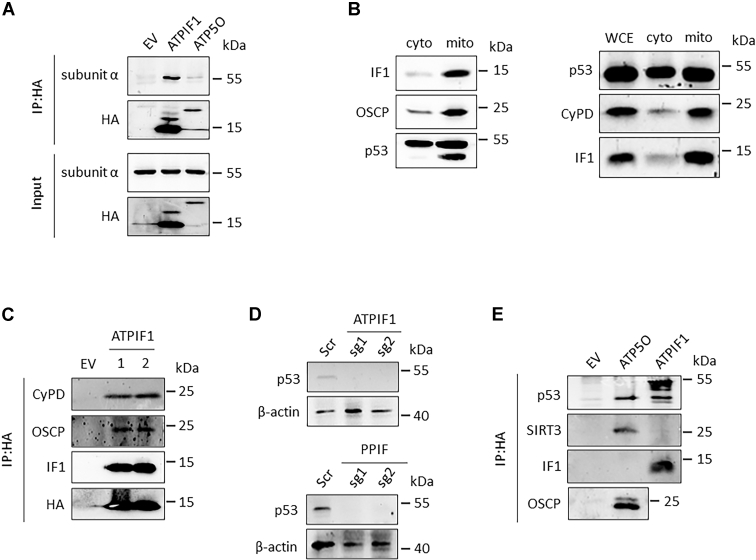
The mechanism through which IF1 regulates the permeability transition was investigated in human embryonic kidney 293T (HEK293T) cells. IF1 interacted with subunit α within mitochondria, but the interaction of exogenous OSCP with subunit α was not able to be detected (Fig. 5A), indicating that the conformational change induced by IF1 binding was transferred to PTP probably through an alternative pathway. IF1 and CyPD are two mitochondrial-specific proteins, and p53 is partly located in mitochondrial matrix under physiological condition (Fig. 5B). IF1 was able to interact with CyPD and OSCP (Fig. 5C). Disruption of either ATP1F1 or PPIF prevented the stable expression of p53 (Fig. 5D). The interaction of IF1 with p53 was able to be detected (Fig. 5E). The interaction between IF1 and p53–CyPD complex could not be enhanced by tBHP (Fig. S4), indicating that IF1 favored PTP opening through a mechanism different from that of tBHP. These results suggested that IF1 modulated PTP probably through p53–CypD axis.
Figure 5.
IF1 interacts with p53–CyPD complex.A, HEK293T cells were transfected with EV or plasmids carrying ATPIF1 (encoding IF1 protein) or ATP5O (encoding subunit OSCP) for 24 h. Isolated mitochondria were lysed for co-IP and WB. The blots are representative of three independent experiments. B, HEK293T cells were cultured for preparation of isolated mitochondria (mito) and cytosolic fraction (cyto), which were lysed for WB analysis. The blots are representative of three independent experiments. C, HEK293T cells were transfected with EV or plasmids carrying ATPIF1 extracted from two different Escherichia coli colonies for 24 h. Cell lysates were prepared for co-IP and analyzed by WB. The blots are representative of three independent experiments. D, HCT116 cells were transduced with lentiCRISPRv2 construct containing scrambled guide RNA (Scr) or two different single-guide RNAs targeting ATPIF1 or PPIF (sg1 or sg2) and selected by 2 μg/ml puromycin for about 7 days. Cell lysates were prepared to detect the expression level of p53 by WB analysis. The blots are representative of three independent experiments. E, HEK293T cells were transfected with EV or plasmids carrying ATP5O or ATPIF1 for 24 h. Cell lysates were prepared for co-IP and analyzed by WB. The blots are representative of at least three independent experiments. CyPD, cyclophilin D; EV, empty vector; HEK293T, human embryonic kidney 293T cell line; IF1, mitochondrial ATP synthase inhibitory factor 1; IP, immunoprecipitation; OSCP, oligomycin sensitivity conferral protein; WB, Western blotting.
IF1 combined with p53–CyPD complex facilitates cell death
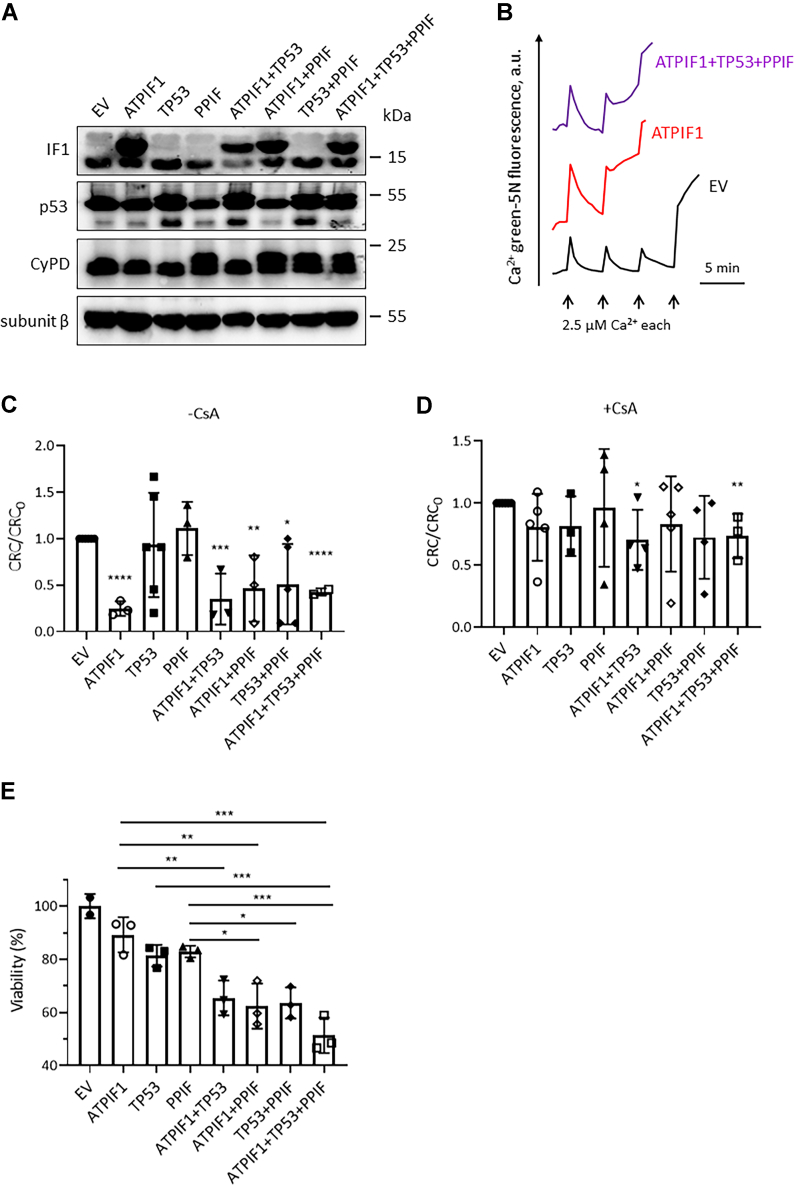
Overexpression of IF1 sensitized PTP to Ca2+ in MIA PaCa-2 (Fig. 2, A–E) and HEK293T cells (Fig. 6, A–C). IF1 was able to interact with p53–CyPD complex; therefore, p53 and CyPD were also overexpressed to investigate their effects on mitochondrial CRC and cell death. Either overexpression of p53 or CyPD alone did not influence the CRC unless their combination was used, suggesting that p53 or CyPD was not sufficient for the pore opening (Fig. 6C). IF1 alone or its combination with p53 and CyPD favors the pore opening (Fig. 6C). Unexpectedly, IF1 and p53–CyPD complex did not exhibit synergistic effect on the CRC (Fig. 6C). The presence of CsA prevented the effects of overexpression of IF1 or p53–CyPD complex on PTP (Fig. 6D). These observations indicated that CyPD was not a sufficient factor for the pore opening but necessary for IF1-induced PTP formation. Binding of IF1 to F-ATP synthase induced a conformational change that transmitted to the inner membrane subunits probably through the lateral stalk via CyPD. IF1 inhibited ATP hydrolysis but not ATP synthesis of the enzyme (37), which allowed ATP assay to detect the effect of IF1 overexpression on cell viability. Overexpression of IF1 slightly promoted cell death, which was dramatically facilitated by its combination with p53 and CyPD (Fig. 6E). IF1 and p53–CyPD complex showed synergistic effect on cell death (Fig. 6E). The effects of IF1 and p53–CyPD complex on cell death appeared to be contradictory to those on CRC. This discrepancy could be due to the possibility that the binding of Ca2+ to the pore weakened the impact of p53–CyPD overexpression on the conformational change of PTP.
Figure 6.
IF1 combined with p53–CyPD complex facilitates cell death.A, HEK293T cells were transfected with plasmids carrying the indicated genes for 24 h. Cell lysates were analyzed by WB. The blots are representative of at least three independent experiments. B, representative CRC traces of digitonin-permeabilized HEK293T cells transfected with EV (black) or ATPIF1 (red) or three plasmids carrying different genes ATPIF1, TP53, and PPIF (ATPIF1 + TP53 + PPIF) for 24 h. C and D, CRC/CRC0 ratio of digitonin-permeabilized HEK293T cells transfected with plasmids carrying the indicated genes normalized to those transfected with EV in the absence of CsA (C) or in the presence of CsA (D). CRC values of permeabilized HEK293T cells transfected with EV were set as one unit. Error bars indicated the mean ± SD of at least two independent experiments. ∗p < 0.05 versus EV, ∗∗p < 0.01 versus EV, ∗∗∗p < 0.001 versus EV, and ∗∗∗∗p < 0.0001 versus EV, Student’s t test. E, the effect of overexpression of indicated genes on cell viability in HEK293T cells revealed by ATP assay. Error bars indicated the mean ± SD of at least two independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, one-way ANOVA with Bonferroni post hoc test. CRC, calcium retention capacity; CsA, cyclosporine A; CyPD, cyclophilin D; EV, empty vector; HEK293T, human embryonic kidney 293T cell line; IF1, mitochondrial ATP synthase inhibitory factor 1; WB, Western blotting.
p53 interacts with CyPD in an IF1-dependent manner
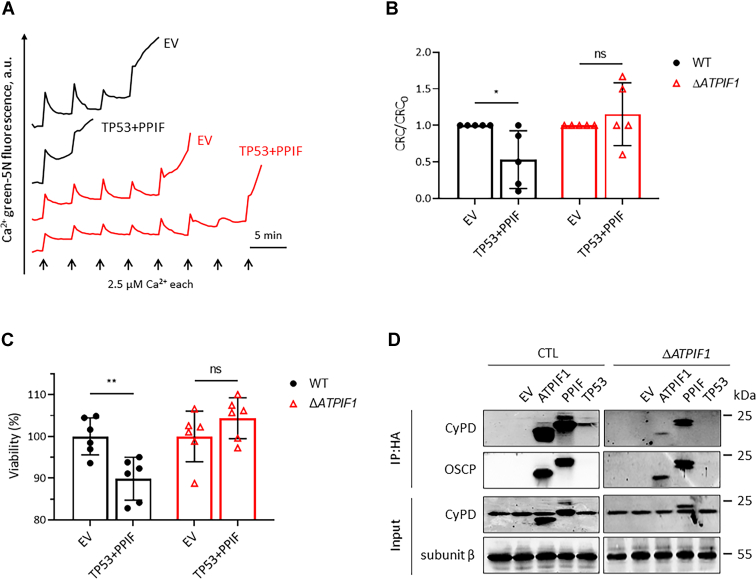
Whether the binding of IF1 to F-ATP synthase modulates the inducing effect of p53–CyPD complex on PTP was then investigated. In MIA PaCa-2 cells, overexpression of p53–CyPD complex dramatically sensitized the pore to Ca2+, which was completely abolished by the knockout of IF1 (Fig. 7, A and B). Cell death was facilitated by overexpression of p53–CyPD complex in WT MIA PaCa-2 cells, which was disappeared in ΔATPIF1 mutant (Fig. 7C). Consistent with the previous study (17), the interaction of p53 with CyPD was able to be detected in WT MIA PaCa-2 cells (Fig. 7D). Remarkably, knockout of IF1 prevented the interaction of p53 with CyPD (Fig. 7D). These observations suggested that IF1 was necessary for the p53–CyPD complex formation, probably because the binding of IF1 to F-ATP synthase caused a conformational change that favored the interaction of p53 with CyPD.
Figure 7.
p53 interacts with CyPD in an IF1-dependent manner.A, representative CRC traces of digitonin-permeabilized WT (black) and ΔATPIF1 (red) MIA PaCa-2 cells transfected with C terminus HA-tagged plasmids carrying indicated genes for 24 h. B, CRC/CRC0 ratio of digitonin-permeabilized WT (black column) and ΔATPIF1 (red column) MIA PaCa-2 cells transfected with plasmids carrying the indicated genes normalized to those transfected with EV. CRC values of permeabilized WT (black) and ΔATPIF1 (red) MIA PaCa-2 cells transfected with EV were set as one unit, respectively. Error bars indicated the mean ± SD of five independent experiments. ∗p < 0.05, ns, two-way ANOVA with Bonferroni post hoc test. C, the effects of p53–CyPD complex overexpression on cell viability in WT and ΔATPIF1 MIA PaCa-2 cells revealed by ATP assay. Error bars indicated the mean ± SD of six independent experiments. ∗∗p < 0.01, ns, two-way ANOVA with Bonferroni post hoc test. D, WT and ΔATPIF1 MIA PaCa-2 cells were nontransfected or transfected with EV or with plasmids carrying ATPIF1 or PPIF or TP53 for 24 h. Protein was extracted for co-IP and WB. The blots are representative of three independent experiments. CRC, calcium retention capacity; CyPD, cyclophilin D; EV, empty vector; HA, hemagglutinin; IF1, mitochondrial ATP synthase inhibitory factor 1; IP, immunoprecipitation; ns, not significant; WB, Western blotting.
Discussion
This work reports a new role of IF1 in modulation of PTP. IF1, an intrinsic inhibitor of F-ATP synthase, inhibits ATP hydrolysis and has no direct effect on ATP synthesis in the presence of a proton motive force (18). Recent studies demonstrate that overexpressing IF1 inhibits both ATP hydrolysis and synthesis activities of F-ATP synthase (22). IF1 limits OXPHOS and thus promotes glycolysis, mediating metabolic switch in cancer cells (27). Besides regulating the activities of the enzyme, IF1 exerts additional roles in mitochondrial physiology (38). Inhibition of F-ATP synthase activity by IF1 increases mitochondrial membrane potential (ΔΨm) because of the inhibition of H+ translocation into matrix (26, 27). Mitochondrial hyperpolarization triggers generation of ROS, which mediates cellular proliferation and survival (26, 39, 40). Relative IF1 expression varies between different tissues and cell types (25) (Fig. 1, A and B). Overexpression of IF1 in HeLa cells instead downregulates the ΔΨm (25). MIA PaCa-2 cell lines displayed high basal expression of IF1 (Fig. 1, A and B). PDAC is the most lethal cancer, which is usually diagnosed at an advanced stage and is refractory to all currently available treatments (41). PDAC cancer cell line MIA PaCa-2 was used to assess the influence of IF1 overexpression on mitochondrial permeability transition and whether the decreased ΔΨm was due to the opening of PTP. IF1 binding to F-ATP synthase not only is regulated by the energetic state of mitochondria (33) but also depends on the molar ratio because the mass-action ratio controls the interaction between proteins (27). Overexpression of IF1 triggered the activation of caspase 3 (Figs. 1, E and F and S1), which was suppressed by the well-established PTP inhibitor CsA (Fig. 1, G and H). Overexpression of IF1 sensitized the PTP opening to Ca2+, which was abrogated by CsA (Fig. 2, A–E) or genetic ablation of CyPD (Fig. 4, B–G). Knockout of CyPD abolished the inducing effect of cell death by IF1 overexpression (Figs. 2, F and G and 4A). Genetic ablation of IF1 desensitized the PTP opening to Ca2+ (Fig. 3, A–D) and prevented cell death induced by tBHP, which displayed similar influence to that of CyPD (Fig. 3, E and F). These data suggested that promoted binding of IF1 to F-ATP synthase triggered mitochondrial permeability transition. Transient opening of the PTP participates in Ca2+/ROS homeostasis and chemoresistance (1), which may favor the malignance of PDAC with high expression of IF1. Next, the molecular mechanism through which IF1 favored PTP opening was then investigated.
IF1 binds to F-ATP synthase in a matrix pH-dependent manner (19, 20). IF1 overexpression promotes oligomerization of F-ATP synthase and the formation of mitochondrial cristae, thus, IF1 plays a role in mitochondrial biogenesis and morphology (25, 26). Overwhelming evidences support the hypothesis that F-ATP synthase contributes to the regulation and formation of PTP through electrophysiological analysis of purified enzymes (42, 43) and by site-directed mutagenesis (9, 10, 11, 44, 45, 46). The hypothesis that PTP forms from F-ATP synthase has been questioned by the studies based on the disruption of F-ATP synthase subunits (47, 48, 49, 50). In human mitochondria devoid of an assembled ATP synthase, the persistent observations of CsA-sensitive Ca2+ release and mitochondrial swelling in potassium chloride (KCl)-based buffer may be due to a distinct permeability pathway, which is provided by the adenine nucleotide translocator (ANT) (51, 52). Crosslinking and proteolysis experiments reveal that the central segment of IF1 binds to αβ subunits of F1 sector in a pH-dependent process and inhibits F-ATP synthase activity (53). Interestingly, the C-terminal region of IF1 simultaneously binds to the OSCP subunit of FO sector in a pH-independent process (53). This binding keeps IF1 anchored to F-ATP synthase both at acidic pH and alkaline pH (53). OSCP subunit acts as a flexible hinge by connection F1 and FO, indicating that OSCP is a hub of metabolic control (54). Binding of CyPD and the immunomodulator Bz-423 to OSCP subunit sensitizes the pore to Ca2+ and inhibits F-ATP synthase activity (5, 55). The tumor suppressor p53 interacts with OSCP subunit via p53–CypD axis, which plays an important role in mitochondrial physiology and tumor suppression activity (16, 17). IF1 appears to extend beyond what envisaged in the literature, and to know better about this fascinating little protein await more studies.
In this study, IF1 was found to interact with p53 and CyPD, as well as OSCP subunit, and either ablation of IF1 or ablation of CyPD affected the stable expression of p53 (Fig. 5). Overexpression of IF1 and p53–CyPD complex sensitized the pore opening to Ca2+, which was inhibited by the addition of CsA (Fig. 6, A–D). In addition, IF1 combined with p53–CyPD complex promoted cell death (Fig. 6E). These observations indicated that IF1 interacted with p53–CyPD complex resulting in a conformational change that transmitted to PTP via OSCP subunit. In the last, p53 appeared to interact with CyPD in an IF1-dependent manner (Fig. 7). Giorgio et al. (5) treated mitochondria with the membrane-permeable bifunctional reagent dimethyl 3,3-dithiobis-propionimidate prior to immunoprecipitation (IP) with anti–complex V antibody and found that CyPD associated to the lateral stalk of F-ATP synthase but not interacted with IF1 and ANT. These different observations might be due to the different techniques used. Some interactions could be missing because crosslinks with dimethyl 3,3-dithiobis-propionimidate is cleavable by DTT. Indeed, the interaction between CyPD and ANT exists, and ANT forms a CsA-sensitive channel, which acts as a distinct permeability pathway (51, 52). In this work, overexpression of IF1 probably further strengthened its interaction with CyPD.
IF1 is known to participate in the Warburg effect of cancer cells (27). This work identifies that IF1 played a role in modulation of PTP opening, which provides a new clue that PTP may be involved in regulating metabolic switch, cell fate, and chemoresistance in the development of lethal cancer (1).
Experimental procedures
Cell culture
HEK293T, MIA PaCa-2, HeLa, and HCT116 cells (Cell Bank, Chinese Academy of Sciences) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with fetal bovine serum (10% v/v) and 100 mg/ml penicillin and streptomycin. All the mentioned cell lines were cultured at 37 °C in a humidified atmosphere containing 5% CO2.
Plasmid DNA transfection
A polyethylenimine (PEI)-mediated transfection method was used in this work. For each well of cells seeded in a 6-well plate, 2 μg plasmids were diluted in 200 μl free DMEM, then 6 μl of PEI was added to the aforementioned solution, and mixed well. The mixture was incubated at room temperature for 15 min and added to each well. For cells cultured in a 10 cm dish, 10 μg plasmids were diluted in 1 ml free DMEM, then 30 μl PEI was added to the aforementioned solution, and mixed well. The mixture was incubated at room temperature for 15 min and added to 10 cm dish. Proteins were extracted, or cells were permeabilized 24 h after the transfection.
Disruption of PPIF and ATPIF1 in MIA PaCa-2 cells
LentiCRISPR methods was used to disrupt PPIF (encoding CyPD) and ATPIF1 (encoding IF1) genes in MIA PaCa-2 cell line. The specific single-guide RNA (sgRNA) was constructed into a lentiviral expression vector lentiCRISPR v2. The specific sgRNA oligos were designed at the website https://www.benchling.com/crispr/. The target sequences are as follows: PPIF sgRNA-1: 5′-CACCGTGACGTCGCGCGCGCCCAGA-3’ (forward), 3′-AAACTCTGGGCGCGCGCGACGTCAC-5’ (reverse); PPIF sgRNA-2: 5′-CACCGCGGGAACCCGCTCGTGTACC-3’ (forward), 3′-AAACGGTACACGAGCGGGTTCCCGC-5’ (reverse); PPIF sgRNA-3: 5′-CACCGGTACACGAGCGGGTTCCCGG-3’ (forward), 3′-AAACCCGGGAACCCGCTCGTGTACC-5’ (reverse); ATPIF1 sgRNA-1: 5′-CACCGCCCGACCGTTGCCACGTATA-3’ (forward), 3′-AAACTATACGTGGCAACGGTCGGGC-5’ (reverse); ATPIF1 sgRNA-2: 5′-CACCGCGCGCCCCGGTCGACATTCT-3’ (forward), 3′-AAACAGAATGTCGACCGGGGCGCGC-5’ (reverse); and ATPIF1 sgRNA-3: 5′-CACCGAAAGATGGGATCCGCCCGCG-3’ (forward), 3′-AAACCGCGGGCGGATCCCATCTTTC-5’ (reverse). Each pair of oligos was annealed in a thermocycler (37 °C for 30 min and 95 °C for 5 min and then ramp down to 25 °C at 5 °C/min). The annealed oligos were cloned into the linearized lentiCRISPR v2 by BsmBI with a ligation kit (Ligation high version 2; Toyobo). For lentivirus production, psPAX2, pMD2.G, and plasmid DNA were diluted with free DMEM at 2:1:6 ratio and transfected to 50% confluence HEK293T cell with PEI-mediated transfection method. The culture medium containing lentivirus was collected and filtered with 0.45 μm filters. The lentivirus suspension was mixed with MIA PaCa-2 cell suspension supplemented with 10 μg/ml polybrene. The complete DMEM was refreshed after 24 h. MIA PaCa-2 cells were cultured for another 24 h. The selection was sustained with 2 μg/ml puromycin until the cells without lentivirus transduction were completely dead. Single cell was seeded into 96-well plate by limiting dilution method and cultured for 10 to 14 days to allow single cell–derived colony formation. Immunoblotting was used to validate the disruption of target genes.
Generation of hemagglutinin-tagged plasmids carrying target genes
Complementary DNA clones of ATPIF1, ATP5O, PPIF, and TP53 containing XhoI and EcoRI restriction sites were generated by a PCR-based method using the whole complementary DNA of HEK293T as template. The primers used for PCR are as follows: for ATPIF1: 5′-ATGCGAATTCATGGCAGTGACGGCGTTGGC-3’ (forward), 3′-ATGCCTCGAGATCATCATGTTTTAGCATTT-5’ (reverse); for ATP5O: 5′-AAGGGATCCGGAATTCATGGCTGCCCCAGCAGTG-3’ (forward), 3′-TAGATGCATGCTCGAGGACAATCTCCCGCATAGCCCTG-5’ (reverse); for PPIF: 5′-CAGTGTGCTGGAATTCATGCTGGCGCTGCGCTGC-3’ (forward), 3′-CGTAGGGGTACTCGAGGCTCAACTGGCCACAGTCTGTG-5’ (reverse); and for TP53: 5′-CAGTGTGCTGGAATTATGGAGGAGCCGCAGTCAG-3’ (forward), 3′-CGTAGGGGTACTCGAGTCTGAGTCAGGCCCTTCTGTC-5’ (reverse). The PCR products were cloned into a C terminus hemagglutinin (HA)-tagged pCDNA3.1 vector linearized by XhoI and EcoRI restriction enzymes using a ligation high kit (Toyobo). The insertion of target gene was confirmed using DNA sequencing.
Preparation of isolated mitochondria from HEK293T cells
HEK293T cells were collected and washed once with cold Dulbecco’s PBS by centrifugation at 600g for 5 min. The cell pellet was resuspended with several milliliters of mitochondrial isolation buffer containing 250 mM sucrose, 10 mM Tris–HCl, 0.1 mM EGTA, and pH 7.4. The homogenate suspension was centrifuged at 600g for 10 min. The supernatant was collected and centrifuged at 12,000g for 10 min. The pellet was resuspended in 1 ml isolation buffer and centrifuged at 12,000g for 10 min. The mitochondrial pellet was resuspended in 100 to 200 μl isolation buffer. Bicinchoninic acid assay was used to determine mitochondrial protein concentration.
Cell permeabilization
Human cells were harvested by a centrifugation at 600g for 5 min. Cells were resuspended and washed with a KCl-based buffer containing 130 mM KCl, 10 mM Mops–Tris, 1 mM Pi, 0.1 mM EGTA, and pH 7.4. The cell pellet was resuspended in a KCl-based buffer containing 130 mM KCl, 10 mM Mops–Tris, 1 mM Pi, 1 mM EGTA, and pH 7.4 supplemented with 0.1 mM digitonin at a density of 2 × 107 cells × ml−1. The incubation with digitonin took about 10 min. The cell permeabilization by digitonin was terminated by addition of 15 ml of a KCl-based buffer containing 130 mM KCl, 10 mM Mops–Tris, 1 mM Pi, 0.1 mM EGTA, and pH 7.4. Cell pellet was collected after a centrifugation at 600g for 5 min and washed once with 5 ml of the same KCl buffer by centrifugation at 600g for 5 min. The cell pellet was resuspended in a sucrose-based buffer (250 mM sucrose, 10 mM Tris–Mops, 10 μM EGTA, pH 7.4) for CRC assay.
IP and Western blotting
Cells were lysed in co-IP lysis buffer containing 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 10% glycerophosphate, and a cocktail of proteinase inhibitors. After incubation on ice for 30 min, cell lysate was harvested by centrifugation at 12,000g for 20 min at 4 °C. The cleared cell lysate was incubated with anti-HA agarose beads (Abmart) at 4 °C overnight. The beads were washed at least three times in co-IP lysis buffer and boiled in 2× loading buffer for 10 min. Proteins were separated by SDS-PAGE gel electrophoreses and transferred onto nitrocellulose membranes (Millipore). The membranes were blocked with 5% (w/v) nonfat dry milk and incubated with the corresponding primary antibody at 4 °C overnight. The immunoblot bands were detected with the Odyssey system (LI-COR Biosciences).
Mitochondrial Ca2+ retention capacity
Human permeabilized cells were suspended in a sucrose-based assay buffer containing 250 mM sucrose, 10 mM Tris–Mops, 10 μM EGTA, 1 mM Pi, 0.5 μM Calcium green-5N, 5 mM succinate, 2 μM rotenone, and pH 7.4 at a cell density of 1 × 107 cells × ml−1. The cell suspension was added to a black 96-well plate (0.2 ml per well). The concentration of external Ca2+ was monitored by Calcium Green-5N fluorescence (excitation/emission: 485 nm/538 nm) with a Tecan microplate reader.
Cell viability assay
Cells were seeded in 96-well plates (100 μl/well) at a cell density of 1 × 105 cells × ml−1 1 day before treatment. After 24 h, different doses of tBHP were added as indicated in the figure legends. For overexpression, the PEI-mediated transfection method was used. After 24 h, 10 μl of CellTiter-Glo 2.0 assay (Promega) was added to each well. The plates were incubated at room temperature for 10 min. Luminescence was recorded with a Tecan microplate reader.
Statistics
All the data were presented as mean ± SD of at least three independent experiments or as representative blots and traces. p Values indicated in the figures are calculated with GraphPad (GraphPad Software, Inc). One-way ANOVA was used to analyze the difference between more than two groups. Two-way ANOVA was used to determine the interaction between two independent variables. ∗p < 0.05 was considered statistically significant.
Data availability
All data are contained within the article and supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The author declares no conflicts of interest with the contents of this article.
Acknowledgments
This work was sponsored by the Shanghai Sailing Program (grant no.: 20YF1453200).
Author contributions
L. G. conceptualization, methodology, software, validation, investigation, data curation, writing–review & editing, visualization, and funding acquisition.
Edited by Phyllis Hanson
Supporting information
References
- 1.Guo L. Mitochondria and the permeability transition pore in cancer metabolic reprogramming. Biochem. Pharmacol. 2021;188:114537. doi: 10.1016/j.bcp.2021.114537. [DOI] [PubMed] [Google Scholar]
- 2.Gonnern C.P., Halestrap A.P. Chaotropic agents and increased matrix volume enhance binding of mitochondrial cyclophilin to the inner mitochondrial membrane and sensitize the mitochondrial permeability transition to [Ca2+] Biochemistry. 1996;35:8172–8180. doi: 10.1021/bi9525177. [DOI] [PubMed] [Google Scholar]
- 3.Giorgio V., Soriano M.E., Basso E., Bisetto E., Lippe G., Forte M.A., Bernardi P. Cyclophilin D in mitochondrial pathophysiology. Biochim. Biophys. Acta. 2010;1797:1113–1118. doi: 10.1016/j.bbabio.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicolli A., Basso E., Petronilli V., Wenger R.M., Bernardi P. Interactions of cyclophilin with the mitochondrial inner membrane and regulation of the permeability transition pore , a Cyclosporin A-sensitive channel. J. Biol. Chem. 1996;271:2185–2192. doi: 10.1074/jbc.271.4.2185. [DOI] [PubMed] [Google Scholar]
- 5.Giorgio V., Bisetto E., Soriano M.E., Dabbeni-Sala F., Basso E., Petronilli V., Forte M.A., Bernardi P., Lippe G. Cyclophilin D modulates mitochondrial F0F1-ATP synthase by interacting with the lateral stalk of the complex. J. Biol. Chem. 2009;284:33982–33988. doi: 10.1074/jbc.M109.020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giorgio V., von Stockum S., Antoniel M., Fabbro A., Fogolari F., Forte M., Glick G.D., Petronilli V., Zoratti M., Szabo I., Lippe G., Bernardi P. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5887–5892. doi: 10.1073/pnas.1217823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giorgio V., Fogolari F., Lippe G., Bernardi P. OSCP subunit of mitochondrial ATP synthase : Role in regulation of enzyme function and of its transition to a pore. Br. J. Pharmacol. 2019;176:4247–4257. doi: 10.1111/bph.14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carraro M., Checchetto V., Sartori G., Kucharczyk R., di Rago J.-P., Minervini G., Franchin C., Arrigoni G., Giorgio V., Petronilli V., Tosatto S.C.E., Lippe G., Szabó I., Bernardi P. High-conductance channel formation in yeast Mitochondria is mediated by F-ATP synthase e and g subunits. Cell. Physiol. Biochem. 2018;50:1840–1855. doi: 10.1159/000494864. [DOI] [PubMed] [Google Scholar]
- 9.Giorgio V., Burchell V., Schiavone M., Bassot C., Minervini G., Petronilli V., Argenton F., Forte M., Tosatto S., Lippe G., Bernardi P. Ca 2+ binding to F-ATP synthase β subunit triggers the mitochondrial permeability transition. EMBO Rep. 2017;18:1065–1076. doi: 10.15252/embr.201643354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo L., Carraro M., Carrer A., Minervini G., Urbani A., Masgras I., Tosatto S.C.E., Szabò I., Bernardi P., Lippe G. Arg-8 of yeast subunit e contributes to the stability of F-ATP synthase dimers and to the generation of the full-conductance mitochondrial megachannel. J. Biol. Chem. 2019;294:10987–10997. doi: 10.1074/jbc.RA119.008775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo L., Carraro M., Sartori G., Minervini G., Eriksson O., Petronilli V., Bernardi P. Arginine 107 of yeast ATP synthase subunit g mediates sensitivity of the mitochondrial permeability transition to phenylglyoxal. J. Biol. Chem. 2018;293:14632–14645. doi: 10.1074/jbc.RA118.004495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinke G., Zhou L., Sazanov L.A. Cryo-EM structure of the entire mammalian F-type ATP synthase. Nat. Struct. Mol. Biol. 2020;27:1077–1085. doi: 10.1038/s41594-020-0503-8. [DOI] [PubMed] [Google Scholar]
- 13.Riley T., Sontag E., Chen P., Levine A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 14.Chipuk J.E., Maurer U., Green D.R., Schuler M. Pharmacologic activation of p53 elicits Bax-dependent apoptosis in the absence of transcription. Cancer Cell. 2003;4:371–381. doi: 10.1016/s1535-6108(03)00272-1. [DOI] [PubMed] [Google Scholar]
- 15.Chipuk J.E., Kuwana T., Bouchier-hayes L., Droin N.M., Newmeyer D.D., Schuler M., Green D.R. Direct activation of bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Am. Assoc. Adv. Sci. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 16.Bergeaud M., Mathieu L., Guillaume A., Moll U.M., Mignotte B., Le Floch N., Vayssière J.L., Rincheval V. Mitochondrial p53 mediates a transcription-independent regulation of cell respiration and interacts with the mitochondrial F1F0-ATP synthase. Cell Cycle. 2013;12:3781–3793. doi: 10.4161/cc.25870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaseva A.V., Marchenko N.D., Ji K., Tsirka S.E., Holzmann S., Moll U.M. P53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149:1536–1548. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pullman M.E., Monroy G.C. A naturally occurring inhibitor of mitochondrial adenosine triphosphatase occurring adenosine inhibitor of mitochondrial triphosphatase. J. Biol. Chem. 1963;238:3762–3769. [PubMed] [Google Scholar]
- 19.Cabezon E., Butler P.J.G., Runswick M.J., Walker J.E. Modulation of the oligomerization state of the bovine F1-ATPase inhibitor protein, IF1, by pH. J. Biol. Chem. 2000;275:25460–25464. doi: 10.1074/jbc.M003859200. [DOI] [PubMed] [Google Scholar]
- 20.Walker J.E. The regulation of catalysis in ATP synthase. Curr. Opin. Struct. Biol. 1994;4:912–918. doi: 10.1016/0959-440x(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 21.Walker J.E. The ATP synthase: The understood, the uncertain and the unknown. Biochem. Soc. Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 22.García-Bermúdez J., Sánchez-Aragó M., Soldevilla B., del Arco A., Nuevo-Tapioles C., Cuezva J.M. PKA phosphorylates the ATPase inhibitory factor 1 and inactivates its capacity to bind and inhibit the mitochondrial H+-ATP synthase. Cell Rep. 2015;12:2143–2155. doi: 10.1016/j.celrep.2015.08.052. [DOI] [PubMed] [Google Scholar]
- 23.Cabezón E., Arechaga I., Butler P.J.G., Walker J.E. Dimerization of bovine F1-ATPase by binding the inhibitor protein, IF1. J. Biol. Chem. 2000;275:28353–28355. doi: 10.1074/jbc.C000427200. [DOI] [PubMed] [Google Scholar]
- 24.García J.J., Morales-Ríos E., Cortés-Hernández P., Rodríguez-Zavala J.S. The inhibitor protein (IF1) promotes dimerization of the mitochondrial F1F0-ATP synthase. Biochemistry. 2006;45:12695–12703. doi: 10.1021/bi060339j. [DOI] [PubMed] [Google Scholar]
- 25.Campanella M., Casswell E., Chong S., Farah Z., Wieckowski M.R., Abramov A.Y., Tinker A., Duchen M.R. Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab. 2008;8:13–25. doi: 10.1016/j.cmet.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Formentini L., Sánchez-Aragó M., Sánchez-Cenizo L., Cuezva J.M.C. The mitochondrial ATPase inhibitory factor 1 triggers a ROS-mediated retrograde prosurvival and proliferative response. Mol. Cell. 2012;45:731–742. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Sánchez-Cenizo L., Formentini L., Aldea M., Ortega Á.D., García-Huerta P., Sánchez-Aragó M., Cuezva J.M. Up-regulation of the ATPase inhibitory Factor 1 (IF1) of the mitochondrial H+-ATP synthase in human tumors mediates the metabolic shift of cancer cells to a warburg phenotype. J. Biol. Chem. 2010;285:25308–25313. doi: 10.1074/jbc.M110.146480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-outschoorn U.E., Peiris-Pagés M., Pestell R.G., Sotgia F., Lisanti M.P. Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 2017;14:11–31. doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- 29.Ward P.S., Thompson C.B. Metabolic reprogramming : A cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu M., Nguyen N.D., Huang Y., Lin D., Fujimoto T.N., Molkentine J.M., Deorukhkar A., Kang Y., Anthony San Lucas F., Fernandes C.J., Koay E.J., Gupta S., Ying H., Koong A.C., Herman J.M., et al. Mitochondrial fusion exploits a therapeutic vulnerability of pancreatic cancer. JCI Insight. 2019;5 doi: 10.1172/jci.insight.126915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masoud R., Reyes-Castellanos G., Lac S., Garcia J., Dou S., Shintu L., Abdel Hadi N., Gicquel T., El Kaoutari A., Diémé B., Tranchida F., Cormareche L., Borge L., Gayet O., Pasquier E., et al. Targeting mitochondrial complex I overcomes chemoresistance in high OXPHOS pancreatic cancer. Cell Rep. Med. 2020;1:100143. doi: 10.1016/j.xcrm.2020.100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piret J.P., Arnould T., Fuks B., Chatelain P., Remacle J., Michiels C. Mitochondria permeability transition-dependent tert-butyl hydroperoxide-induced apoptosis in hepatoma HepG2 cells. Biochem. Pharmacol. 2004;67:611–620. doi: 10.1016/j.bcp.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 33.Gledhill J.R., Montgomery M.G., Leslie A.G.W., Walker J.E. How the regulatory protein, IF1, inhibits F1-ATPase from bovine mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15671–15676. doi: 10.1073/pnas.0707326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson G.C., Bason J.V., Montgomery M.G., Fearnley I.M., Mueller D.M., Leslie A.G.W., Walker J.E. The structure of F₁-ATPase from Saccharomyces cerevisiae inhibited by its regulatory protein IF₁. Open Biol. 2013;3:120164. doi: 10.1098/rsob.120164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cabezón E., Montgomery M.G., Leslie A.G.W., Walker J.E. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat. Struct. Biol. 2003;10:744–750. doi: 10.1038/nsb966. [DOI] [PubMed] [Google Scholar]
- 36.Giorgio V., Guo L., Bassot C., Petronilli V., Bernardi P. Calcium and regulation of the mitochondrial permeability transition. Cell Calcium. 2018;70:56–63. doi: 10.1016/j.ceca.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 37.Burwick N.R., Wahl M.L., Fang J., Zhong Z., Moser T.L., Li B., Capaldi R.A., Kenan D.J., Pizzo S.V. An inhibitor of the F1 subunit of ATP synthase (IF1) modulates the activity of angiostatin on the endothelial cell surface. J. Biol. Chem. 2005;280:1740–1745. doi: 10.1074/jbc.M405947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernardi P., Rasola A., Forte M., Lippe G. The mitochondrial permeability transition pore: Channel formation by F-ATP synthase, integration in signal transduction, and role in pathophysiology. Physiol. Rev. 2015;95:1111–1155. doi: 10.1152/physrev.00001.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuyama S., Xu Q., Velours J., Reed J.C. The mitochondrial F0F1-ATPase proton pump is required for function of the proapoptotic protein bax in yeast and mammalian cells. Mol. Cell. 1998;1:327–336. doi: 10.1016/s1097-2765(00)80033-7. [DOI] [PubMed] [Google Scholar]
- 40.Santamaría G., Martínez-Diez M., Fabregat I., Cuezva J.M. Efficient execution of cell death in non-glycolytic cells requires the generation of ROS controlled by the activity of mitochondrial H+-ATP synthase. Carcinogenesis. 2006;27:925–935. doi: 10.1093/carcin/bgi315. [DOI] [PubMed] [Google Scholar]
- 41.Meslar E. Pancreatic adenocarcinoma. JAAPA. 2020;33:50–51. doi: 10.1097/01.JAA.0000718300.59420.6c. [DOI] [PubMed] [Google Scholar]
- 42.Mnatsakanyan N., Llaguno M.C., Yang Y., Yan Y., Weber J., Sigworth F.J., Jonas E.A. A mitochondrial megachannel resides in monomeric F1FO ATP synthase. Nat. Commun. 2019;10:5823. doi: 10.1038/s41467-019-13766-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urbani A., Giorgio V., Carrer A., Franchin C., Arrigoni G., Jiko C., Abe K., Maeda S., Shinzawa-Itoh K., Bogers J.F.M., McMillan D.G.G., Gerle C., Szabò I., Bernardi P. Purified F-ATP synthase forms a Ca2+-dependent high-conductance channel matching the mitochondrial permeability transition pore. Nat. Commun. 2019;10:4341. doi: 10.1038/s41467-019-12331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alavian K.N., Beutner G., Lazrove E., Sacchetti S., Park H.-A., Licznerski P., Li H., Nabili P., Hockensmith K., Graham M., Porter G.A., Jonas E.A. An uncoupling channel within the c-subunit ring of the F1FO ATP synthase is the mitochondrial permeability transition pore. Proc. Natl. Acad. Sci. U. S. A. 2014;111:10580–10585. doi: 10.1073/pnas.1401591111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antoniel M., Jones K., Antonucci S., Spolaore B., Fogolari F., Petronilli V., Giorgio V., Carraro M., Di Lisa F., Forte M., Szabó I., Lippe G., Bernardi P. The unique histidine in OSCP subunit of F-ATP synthase mediates inhibition of the permeability transition pore by acidic pH. EMBO Rep. 2017;19:257–268. doi: 10.15252/embr.201744705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carraro M., Jones K., Sartori G., Schiavone M., Antonucci S., Kucharczyk R., di Rago J.P., Franchin C., Arrigoni G., Forte M., Bernardi P. The unique cysteine of F-ATP synthase OSCP subunit participates in modulation of the permeability transition pore. Cell Rep. 2020;32:108095. doi: 10.1016/j.celrep.2020.108095. [DOI] [PubMed] [Google Scholar]
- 47.He J., Ford H.C., Carroll J., Ding S., Fearnley I.M., Walker J.E. Persistence of the mitochondrial permeability transition in the absence of subunit c of human ATP synthase. Proc. Natl. Acad. Sci. U. S. A. 2017;114:3409–3414. doi: 10.1073/pnas.1702357114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He J., Carroll J., Ding S., Fearnley I.M., Walker J.E. Permeability transition in human mitochondria persists in the absence of peripheral stalk subunits of ATP synthase. Proc. Natl. Acad. Sci. U. S. A. 2017;114:9086–9091. doi: 10.1073/pnas.1711201114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carroll J., He J., Ding S., Fearnley I.M., Walker J.E. Persistence of the permeability transition pore in human mitochondria devoid of an assembled ATP synthase. Proc. Natl. Acad. Sci. U. S. A. 2019;116:12816–12821. doi: 10.1073/pnas.1904005116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker J.E., Carroll J., He J. Reply to BERNARDI : The mitochondrial permeability transition pore and the ATP synthase. Proc. Natl. Acad. Sci. U. S. A. 2020;117:2745–2746. doi: 10.1073/pnas.1921409117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neginskaya M.A., Solesio M.E., Berezhnaya E.V., Amodeo G.F., Mnatsakanyan N., Jonas E.A., Pavlov E.V. ATP synthase C-subunit-deficient mitochondria have a small cyclosporine A-sensitive channel, but lack the permeability transition pore. Cell Rep. 2019;26:11–17.e2. doi: 10.1016/j.celrep.2018.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carrer A., Tommasin L., Šileikytė J., Ciscato F., Filadi R., Urbani A., Forte M., Rasola A., Szabò I., Carraro M., Bernardi P. Defining the molecular mechanisms of the mitochondrial permeability transition through genetic manipulation of F-ATP synthase. Nat. Commun. 2021;12:1–12. doi: 10.1038/s41467-021-25161-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zanotti F., Raho G., Gaballo A., Papa S. Inhibitory and anchoring domains in the ATPase inhibitor protein IF 1 of bovine heart mitochondrial ATP synthase. J. Bioenerg. Biomembr. 2004;36:447–457. doi: 10.1023/B:JOBB.0000047327.68173.9b. [DOI] [PubMed] [Google Scholar]
- 54.Murphy B.J., Klusch N., Langer J., Mills D.J., Yildiz Ö., Kühlbrandt W. Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F1-Fo coupling. Science. 2019;364 doi: 10.1126/science.aaw9128. [DOI] [PubMed] [Google Scholar]
- 55.Johnson K.M., Chen X., Boitano A., Swenson L., Opipari A.W., Glick G.D. Identification and validation of the mitochondrial F1F0-ATPase as the molecular target of the immunomodulatory benzodiazepine Bz-423. Chem. Biol. 2005;12:485–496. doi: 10.1016/j.chembiol.2005.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and supporting information.