Abstract
In natural and engineered systems, most microorganisms would enter a state of dormancy termed as “viable but non-culturable” (VBNC) state when they are exposed to unpredictable environmental stress. One of the major advances in resuscitating from such a state is the discovery of a kind of bacterial cytokine protein called resuscitation-promoting factor (Rpf), which is secreted from Micrococcus luteus. In this study, the optimization of Rpf production was investigated by the response surface methodology (RSM). Results showed that an empirical quadratic model well predicted the Rpf yield, and the highest Rpf protein yield could be obtained at the optimal conditions of 59.56 mg L−1 IPTG, cell density 0.69, induction temperature 20.82 °C and culture time 7.72 h. Importantly, Phyre2 web portal characterized the structure of the Rpf domain to have a shared homology with lysozymes, and the highest lysozyme activity was at pH 5 and 50 °C. This study broadens the knowledge of Rpf production and provided potential strategies to apply Rpf as a bioactivator for environmental bioremediation.
A group of secreted proteins from M. luteus, recognized as resuscitation promoting factors (Rpf) can resuscitate the viable but non-culturable (VBNC) state bacteria which have the potential function of environmental bioremediation.
1. Introduction
Biological processes are known to be environmentally friendly and costeffective, and have been regarded as the most promising method for environmental bioremediation, such as wastewater treatment, soil pollution control and municipal solid waste disposal.1–3 Over the past several decades, numerous studies have optimized the biological processes for highly efficient pollutant degradation. However, in natural and engineered systems, most microorganisms live in unpredictable environments, such as physical (high/low temperatures), chemical stressors (highly-toxic pollutants), biocides, antibiotics, and typically experience conditions that are suboptimal for growth and reproduction.4,5 As a result, a common response of microorganisms to environmental stress is to enter a state of dormancy in which they do not grow on media usually employed to their detection, also termed as “viable but non-culturable” (VBNC) state.6–8 In particular, a variety of functional bacterial communities, which are the crucial populations of degrading target pollutants, reduced metabolic activity when exposed to unfavorable conditions due to entry into a VBNC state.9 For example, Escherichia coli and Vibrio spp. would enter into a VBNC state in adverse environments with long term cell viability but reduced metabolic activity.10
The VBNC state can only be an adaptive strategy if VBNC cells are resuscitated and able to reproduce. Therefore, the revival of VBNC cells has received considerable attention. In many cases, bacteria in the VBNC state are capable of exiting the dormant state in response to favorable changes in environmental conditions.5 In recent years, the discovery of an intracellular enzyme called resuscitation-promoting factor (Rpf), secreted from Micrococcus luteus, is considered as a major advance in the resuscitation of VBNC bacteria.11–14 It has been reported that Rpf could facilitate cell division and encourage the growth of a wide number of microorganisms at picomolar concentrations.15 For example, the genera of Actinobacteria, Rhizobium, Pseudomonas, Proteobacteria and Microbacterium, which contain numerous environmental microorganisms have been accepted to be sensitive to Rpf.16–18,58
Although the mechanism of Rpf function remains unknown, numerous investigators predicted that the peptidoglycan hydrolase activity of Rpf was correlated with its growth promotion and resuscitation activity. Keep et al.19 found that the highly conserved Rpf domain showed a structure similar to that of lysozyme, and it has been proved that Rpf cleaved peptidoglycan. Besides, Liu et al.20 showed that adding Rpf to the sequencing batch reactor (SBR) was conducive to increase microbial diversity, shorten the start-up process and significantly enhance the biological nutrient removal.
Due to the extensive application of Rpf in VBNC bacteria resuscitation as well as the enhanced degradation of target pollutants,7,20,21 a great need of Rpf has arisen for biological treatment. Fortunately, E. coli BL21 (DE3) is widely used as a host strain for the overproduction of recombinant proteins. There are a variety of factors influencing the expression of foreign proteins in E. coli, so it is crucial to optimize these factors to obtain higher expression of proteins. Recently, the response surface methodology (RSM), a method to design the experiments and build mathematical models, was applied to investigate the interactions among different parameters in complicated systems.
Numerous previous studies have proved the resuscitation of Rpf on VBNC state functional bacteria from the perspective of environmental bioremediation.21–24 Therefore, the main objective of this study is to explain the resuscitation mechanisms of Rpf more thoroughly and comprehensively from the structural analysis of the Rpf protein sequence. For this purpose, the Rpf protein sequence was subjected to a structural analysis using the Phyre2 portal, and experiments were conducted to ulteriorly investigate the peptidoglycan hydrolyzing activities of Rpf. More importantly, RSM was used to optimize the levels of the screened variables for increasing the Rpf production.
2. Materials and methods
2.1. Bacterial strain and expression system
E. coli BL21 (DE3) used as a host for gene expression has previously been reported.25,26 The rpf gene of M. luteus was ligated into a plasmid pET-28a vector and then transformed into E. coli, as described before.19–22 The plasmid pET-28a features a N-terminal 6×His tag for protein purification by metal affinity chromatography.27 The recombinant Rpf proteins were expressed as an intracellular enzyme from plasmid pET-28a by isopropyl-β-d-thiogalactopyranoside (IPTG) induction under the control of the strong T7 promoter.
2.2. Rpf protein overexpression, purification and electrophoretic analysis
E. coli cells were cultured overnight at 37 °C in SOB broth with 50 mg L−1 kanamycin. The overnight culture as an inoculum was transferred to a fresh SOB broth. When the density of cell reached between 0 and 1.7 at 600 nm (OD600), IPTG was added with different concentrations (0–80 mg L−1) for protein expression. During the induction phase, the medium was cultured at different temperatures (0–40 °C) at the required time interval (0–16 h). Different cultures were performed to investigate the effects of numerous parameters on the overexpression of the recombinant protein.
The cells were harvested via incubation broth, centrifuged and washed twice in phosphate buffered saline (PBS). The cell pellet was resuspended in a buffer solution containing 25 mM Tris–HCl and lysed using an ultrasonic disintegrator (Soniprep 150, Japan) on ice for 1 h. For the purification of overexpressed recombinant proteins, the supernatant was obtained by centrifugation, and then added into a nickel-nitrilotriacetic acid (Ni-NTA) column (Qiagen, Germany), which was equilibrated with binding buffer (0.5 M NaCl, 20 mM Tris–HCl, pH 7.6). The unabsorbed proteins were washed from the column with 10 volumes of buffer containing 10 mM imidazole. Rpf proteins were collected with 5 mL washing buffer (0.5 M NaCl, 100 mM imidazole and 20 mM Tris–HCl, pH 7.6). Afterwards, the proteins were concentrated to 2 mL fractions and dialyzed against 50 mm Na–phosphate buffer to eliminate imidazole. Samples were analyzed by 12.5% sodium dodecyl sulphate (SDS)–polyacrylamide gel electrophoresis (PAGE) using standard markers (Sangon Biotech, Shanghai, China). The concentration of the Rpf protein was determined by a modified Bradford Protein Assay Kit.27,28 Finally, the purified protein was diluted with an equal volume of glycerol and stored at −20 °C for further experiments.
2.3. Statistical experimental design
Response surface methodology was employed for the statistical analysis of the experimental data using the Design-Expert 12.0 software (Statease, Minneapolis, MN, USA). The Central Composite Design (CCD) was utilized to obtain the optimum conditions for maximizing the Rpf protein yield. The effects of the main operating parameters including IPTG concentration, induced cell density, induction temperature and induction culture time were investigated. Table 1 shows the factors and their levels. According to the design, the optimization process of the protein was carried out at five levels (−2, −1, 0, +1, and +2), which required a total of thirty runs to extend the design domain. The CCD and experimental results of RSM are shown in Table 2. All the runs were performed in triplicate to acquire the average value as the response. In order to obtain the optimum combinations of independent parameters, a quadratic model, which included all the independent variables, was fitted to predict the response value. Five experimental runs were replicated at the center point of the design to confirm any differences in the estimation.29 At the same time, the changes in the protein concentration with and without optimization were analyzed by fluorescence.30,31
Experimental range and levels coded for ANOVA.
| Factor | Unit | Range and levels | ||||
|---|---|---|---|---|---|---|
| −2 | −1 | 0 | +1 | +2 | ||
| IPTG concentration | mg L−1 | 0 | 20 | 40 | 60 | 80 |
| Induced cell density | — | −0.3 | 0.2 | 0.7 | 1.2 | 1.7 |
| Induction temperature | °C | 0 | 10 | 20 | 30 | 40 |
| Induction culture time | h | 0 | 4 | 8 | 12 | 16 |
Central composite design and experimental results of response surface methodology.
| Run | A: IPTG concentration (mg L−1) | B: induced cell density | C: induction temperature (°C) | D: induction culture time (h) | Protein yield (mg mL−1) | |
|---|---|---|---|---|---|---|
| Actual value | Predicted value | |||||
| 1 | 20 | 0.2 | 10 | 4 | 0.035 ± 0.003 | 0.031 |
| 2 | 20 | 1.2 | 10 | 4 | 0.165 ± 0.012 | 0.17 |
| 3 | 20 | 1.2 | 30 | 4 | 0.150 ± 0.002 | 0.15 |
| 4 | 60 | 1.2 | 10 | 4 | 0.180 ± 0.005 | 0.18 |
| 5 | 60 | 0.2 | 10 | 12 | 0.080 ± 0.004 | 0.076 |
| 6 | 40 | 0.7 | 20 | 16 | 0.385 ± 0.004 | 0.38 |
| 7 | 40 | 0.7 | 20 | 8 | 0.517 ± 0.009 | 0.52 |
| 8 | 60 | 0.2 | 30 | 4 | 0.195 ± 0.011 | 0.19 |
| 9 | 40 | 0.7 | 20 | 8 | 0.517 ± 0.009 | 0.52 |
| 10 | 40 | 0.7 | 20 | 8 | 0.517 ± 0.009 | 0.52 |
| 11 | 20 | 0.2 | 30 | 4 | 0.180 ± 0.005 | 0.19 |
| 12 | 40 | 1.7 | 20 | 8 | 0.225 ± 0.005 | 0.22 |
| 13 | 40 | 0.7 | 0 | 8 | 0.170 ± 0.005 | 0.17 |
| 14 | 0 | 0.7 | 20 | 8 | 0.080 ± 0.004 | 0.077 |
| 15 | 20 | 0.2 | 10 | 12 | 0.090 ± 0.009 | 0.097 |
| 16 | 60 | 1.2 | 30 | 12 | 0.120 ± 0.005 | 0.12 |
| 17 | 20 | 1.2 | 10 | 12 | 0.145 ± 0.007 | 0.14 |
| 18 | 60 | 1.2 | 10 | 12 | 0.155 ± 0.004 | 0.16 |
| 19 | 40 | 0.7 | 20 | 8 | 0.517 ± 0.009 | 0.52 |
| 20 | 40 | −0.3 | 20 | 8 | 0.009 ± 0.054 | 0.00575 |
| 21 | 60 | 0.2 | 10 | 4 | 0.085 ± 0.0124 | 0.092 |
| 22 | 20 | 0.2 | 30 | 12 | 0.210 ± 0.030 | 0.21 |
| 23 | 60 | 1.2 | 30 | 4 | 0.145 ± 0.002 | 0.15 |
| 24 | 40 | 0.7 | 40 | 8 | 0.120 ± 0.009 | 0.12 |
| 25 | 40 | 0.7 | 20 | 8 | 0.517 ± 0.009 | 0.52 |
| 26 | 80 | 0.7 | 20 | 8 | 0.120 ± 0.005 | 0.12 |
| 27 | 40 | 0.7 | 20 | 8 | 0.517 ± 0.009 | 0.52 |
| 28 | 60 | 0.2 | 30 | 12 | 0.130 ± 0.007 | 0.14 |
| 29 | 20 | 1.2 | 30 | 12 | 0.075 ± 0.002 | 0.082 |
| 30 | 40 | 0.7 | 20 | 0 | 0.000 ± 0.000 | −0.00325 |
2.4. Structure assay of Rpf protein
Comparative homology modeling has become an efficient and easy method for predicting the unknown three-dimensional structure of a protein based on sequence alignment.32 In view of the structural similarity between Rpf and lysozyme-like proteins, the primary amino acid sequence of Rpf was submitted to the Phyre2 portal (http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index) to predict its three-dimensional protein structure by template searching and modeling.33 Among many models generated, the model with the highest confidence was selected as the best one for evaluation. Finally, the structure was confirmed through Verify3D.
2.5. Determination of the peptidoglycan hydrolases activity of Rpf
Rpf was added into 100 mM of sodium citrate buffer (pH 4.8) containing 1 mg mL−1 4-nitrophenyl N-acetyl-β-d-glucosaminide (NP-GlcNAc, Sigma-Aldrich) to identify the activities of peptidoglycan hydrolase, while the control group added with distilled water instead of enzymes.34 The enzymatic reaction was carried out for 30 min and ionization of p-nitrophenylates were initiated by the addition of 140 mM sodium carbonate, and the absorbance of this ion at 405 nm was monitored by a spectrophotometer (TU-1810, Purkinje, China).35
The muralytic enzyme of Rpf was further investigated by zymography.36,37 Purified Rpf was tested renaturing PAGE electrophoresis through 12% polyacrylamide separating gels (without SDS in the gel) containing 0.1% (wt/vol) of M. luteus as substrate.38 Gels were incubated at 37 °C for 16 h with 200 mL renaturing buffer (100 mmol L−1 Tris–HCl, 10 mL L−1, Triton X-100, pH 7.0) after electrophoresis. Finally, gels were stained for 30 min utilizing 1 g L−1 methylene blue and 0.1 g L−1 KOH, and then destained in distilled water. The appearance of clear bands indicated that proteins possess cell lytic activity.
3. Results and discussion
3.1. Production and purification of Rpf
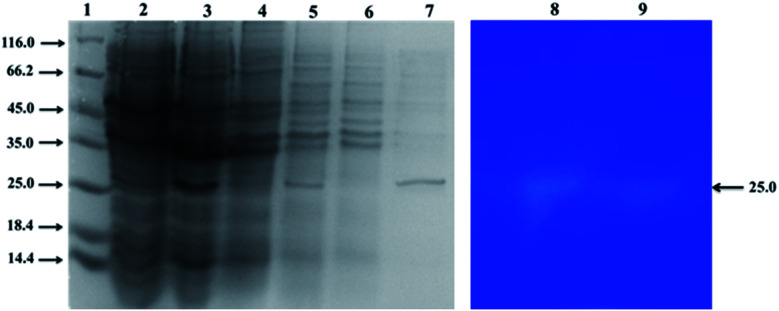
The recombinant Rpf protein was expressed in E. coli BL21 (DE3) and different components were analyzed on SDS-PAGE gels. As presented in Fig. 1, Rpf had a higher expression level than those without induction in the cultures that were employed IPTG as the inducer of foreign gene expression (Fig. 1, lane 2–3). Importantly, proteins mainly existed in the supernatant in the water-soluble form and hardly formed inclusion bodies (Fig. 1, lane 4–5). Upon affinity purification of Rpf, the target protein was absorbed in the column, with a limited amount of protein penetrating (Fig. 1, lane 5–6). After elution, a distinct band was observed, as shown in Fig. 1, lane 7, which corresponded to a protein with an apparent molecular weight of 25–27 kDa. The purified protein was equivalent to the recombinant Rpf protein (27 kDa).39 Therefore, Rpf proteins obtained by affinity chromatography could be selected for further experiments.
Fig. 1. Protein profiles and zymograms. Lane 1: molecular weight high range; lane 2: uninduced; lane 3: induced; lane 4: the precipitation of the cell lysate of E. coli BL21 (DE3); lanes 5: the supernatant of the cell lysate of E. coli BL21 (DE3); lane 6: unbound washed fractions; lane 7: eluted fractions; lane 8 and 9: zymograms against M. luteus.
3.2. The optimized conditions and validation of the models
In order to optimize the production of recombinant proteins, effects of the IPTG concentration (X1), induced cell density (X2), induction temperature (X3) and induction culture time (X4) were selected as independent variables. The experimental runs and the corresponding results are illustrated in Table 2. According to the experimental data, the following second-order polynomial equation was found to demonstrate the relationships between significant variables and protein production via the multiple regression analysis. The calculated regression equation is given below:
| Y1 = 0.52 + 0.005X1 + 0.023X2 + 0.007083X3 + 0.027X4 + 0.005625X1X2 − 0.005625X1X3 − 0.006875X1X4 − 0.036X2X3 − 0.01X2X4 − 0.00875X3X4 − 0.11X12 − 0.1X22 − 0.094X32 − 0.082X42 | 1 |
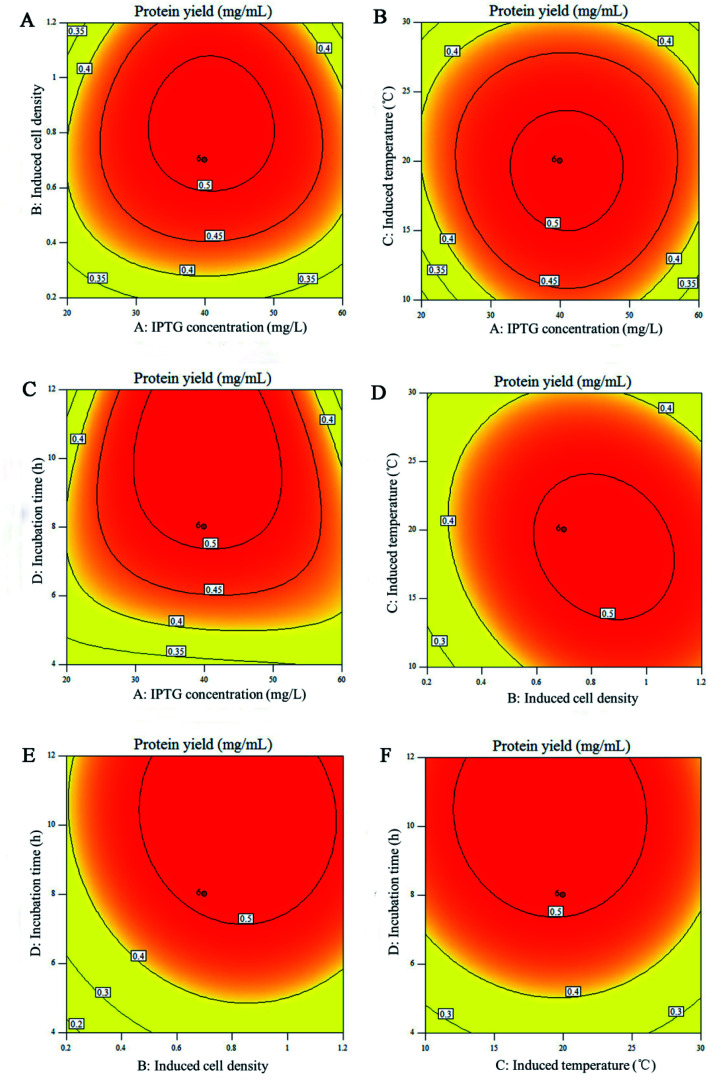
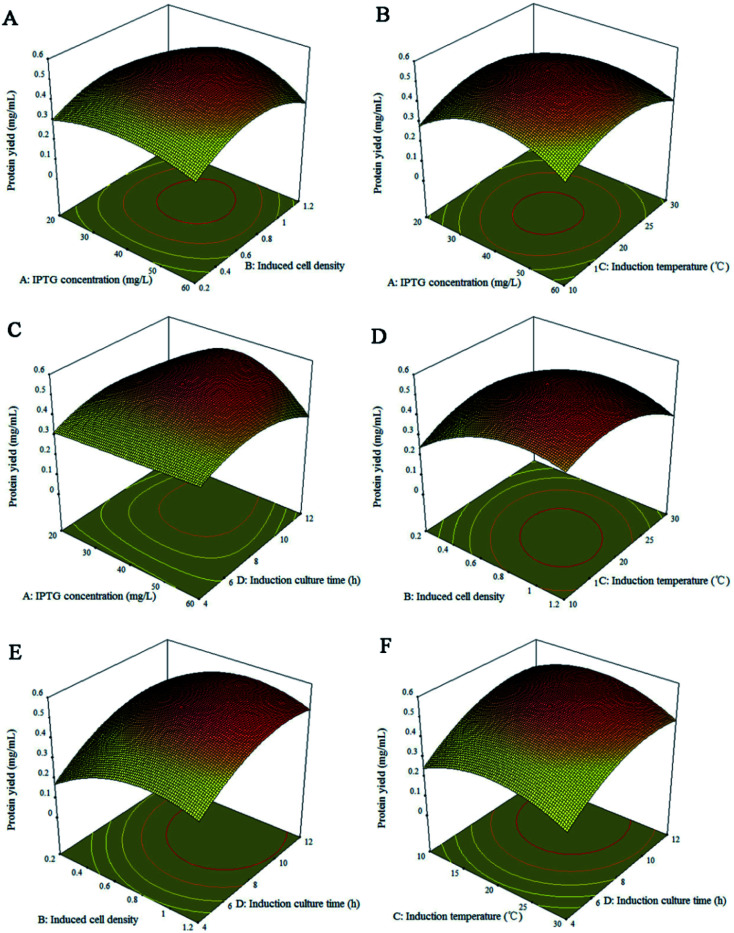
Eqn (1) showed that the positive coefficients for X1, X2, X3 and X4 had favorable effects on protein expression. In addition, the significance of parameter affecting the protein production would be determined by the variance (ANOVA) using the Design Expert program.29 As presented in Table 3, the model fit was verified by the coefficient of determination (R2 = 0.907) with highly significant level (P < 0.001), which implies that the predicted value is correlated with the experimental value. Therefore, the model can reliably predict the response for the optimization process. Furthermore, IPTG concentration (X1), induced cell density (X2), induction temperature (X3) and induction culture time (X4) have no significant effect on protein yield, as well as the interaction between the two factors is not significant. Among them, although the difference is not notable, induction culture time (X4) is the most influential factor affecting protein production, and the interaction effect of induced cell density (X2) and induction temperature (X3) is slightly stronger than other parameters. Likewise, Su et al.40 reported that the concentration of Lithium l-lactate, initial pH and induction time significantly affect the protein production from M. luteus, which was consistent with the result of RSM in this study. From the coefficient of the interaction term, it also can be observed that the IPTG concentration (X1) and induced cell density (X2) are synergistic effects, the other five factors are antagonistic effects, which should be suppressed in the optimization process.
ANOVA for response surface quadratic model.
| Factors | Statistics | ||||
|---|---|---|---|---|---|
| Sum of squares | df | Mean square | F-Value | P-Value | |
| Model | 0.77 | 14 | 0.055 | 10.48 | < 0.0001 |
| A-IPTG concentration | 0.0006 | 1 | 0.0006 | 0.11 | 0.7394 |
| B-induced cell density | 0.013 | 1 | 0.013 | 2.52 | 0.1333 |
| C-induction temperature | 0.001204 | 1 | 0.001204 | 0.23 | 0.638 |
| D-induction culture time | 0.017 | 1 | 0.017 | 3.27 | 0.0907 |
| AB | 0.0005063 | 1 | 0.0005063 | 0.097 | 0.7598 |
| AC | 0.0005063 | 1 | 0.0005063 | 0.097 | 0.7598 |
| AD | 0.0007563 | 1 | 0.0007563 | 0.14 | 0.7089 |
| BC | 0.021 | 1 | 0.021 | 4.03 | 0.0632 |
| BD | 0.0016 | 1 | 0.0016 | 0.31 | 0.5881 |
| CD | 0.001225 | 1 | 0.001225 | 0.23 | 0.6352 |
| A2 | 0.3 | 1 | 0.3 | 57.97 | < 0.0001 |
| B2 | 0.28 | 1 | 0.28 | 53.37 | < 0.0001 |
| C2 | 0.24 | 1 | 0.24 | 46.22 | < 0.0001 |
| D2 | 0.18 | 1 | 0.18 | 35.26 | < 0.0001 |
| Residual | 0.078 | 15 | 0.005223 | ||
| Lack of fit | 0.078 | 10 | 0.007834 | ||
| Pure error | 0 | 5 | 0 | ||
| Cor total | 0.84 | 29 | |||
| R 2 = 0.9073 | Adj R2 = 0.8207 | ||||
The experimental variables and their interactive effects on the expression process are also graphically displayed on the corresponding 2D contour plots and 3D response surface plots. Fig. 2 and 3 explained the correlation between response and experimental levels for each variable. Each figure shows the interaction between two variables while keeping the other variables at their central levels.41 The response surface plots and their corresponding counter plots showed that the induction culture time (X4) was the most influential factor for increasing the protein production, which is in accordance with the data summarized in Table 3. The interaction between the four influencing factors presented in Fig. 2 and 3 are consistent with the results in Table 3. Fig. 2D and 3D present the interaction effect of induced cell density (X2) and induction temperature (X3) on the Rpf preparation under a fixed cell density of 0.7. With the increase in the induced cell density, the yield of Rpf first increased and then decreased rapidly with the Rpf yield reaching 0.52 mg mL−1 at the optimum induction temperature. Likewise, the other five responses had similar tendency towards Rpf expression. At first, the Rpf yield increased with the elevated induction time, cell density, induction temperature, IPTG concentration, and then began to decrease. To find the optimal conditions of Rpf expression, numerical optimization was accomplished by the response optimizer. The highest Rpf protein yield was obtained at the optimal conditions of 59.56 mg L−1 IPTG, cell density 0.69, induction temperature 20.82 °C and culture time 7.72 h.
Fig. 2. Contour plots for yield of Rpf. (A) IPTG concentration and induced cell density; (B) IPTG concentration and induction temperature; (C) IPTG concentration and culture time; (D) induced cell density and induction temperature; (E) induced cell density and culture time; (F) induction temperature and culture time. Remaining variables were fixed at coded zero level.
Fig. 3. Response surface plots for yield of Rpf. (A) IPTG concentration and induced cell density; (B) IPTG concentration and induction temperature; (C) IPTG concentration and culture time; (D) induced cell density and induction temperature; (E) induced cell density and culture time; (F) induction temperature and culture time. Remaining variables were fixed at coded zero level.
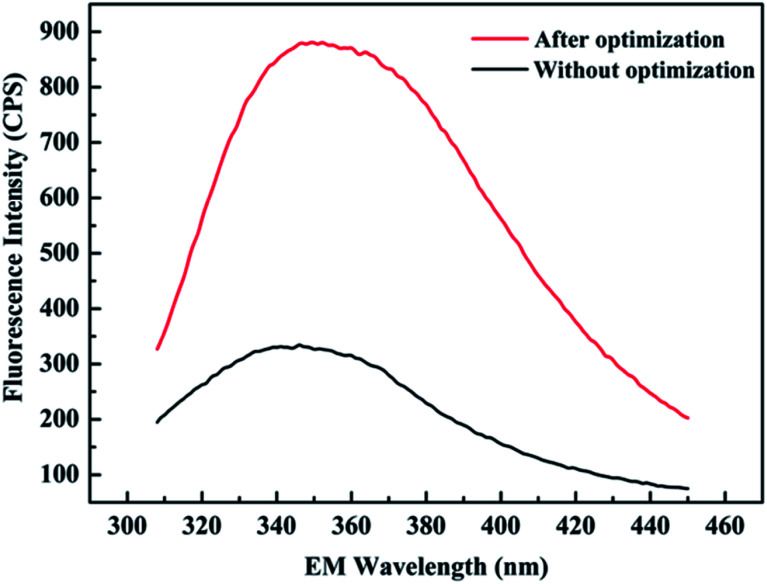
To verify the accuracy of the experimental results, the above-mentioned optimum conditions were replicated five times. Similarity between the predicted (0.42 mg mL−1) and actual (0.348 mg mL−1) values reached as high as 82.9%, it could be concluded that the optimal conditions would be effectively applied to the actual protein production process. In addition, the emission fluorescence spectra of protein concentration with and without optimization are shown in Fig. 4. For each sample, it was apparent that the washing buffer containing recombinant protein had the maximum fluorescence intensity at 350 nm (excitation at 288 nm), which was consistent with that of tryptophan. It can be seen that there is a sharp attenuation of the fluorescence intensities in the spectra without optimization, while the position and shape of the emission peak did not change significantly, indicating that the protein conformation of Rpf has not changed. Moreover, a maximum protein yield of 3-fold increase in fluorescence was discovered with optimization, which suggests that a higher efficiency for protein expression was discovered compared with the protein yields obtained from other groups.
Fig. 4. Emission fluorescence spectra of the protein yield with and without optimization. Conditions without optimization: IPTG = 100 mg L−1, cell density = 0.4, induction temperature = 20 °C, culture time = 7 h.
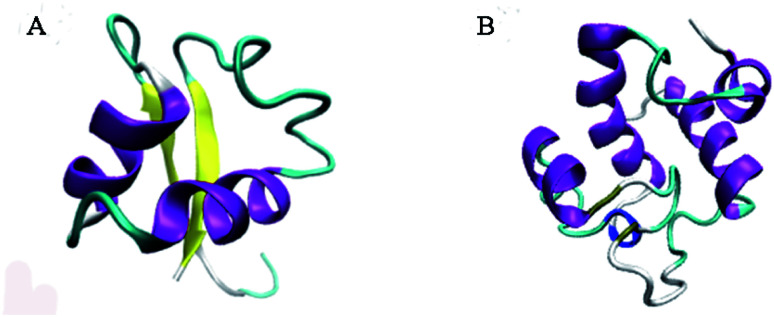
3.3. Structure prediction
Phyre2 portal and Verify3D were used to analyze the molecular model of Rpf. As illustrated in Fig. 5, the intensive model of Rpf was predicted using the “c5fimA” model as the most suitable protein template, with 99.8% confidence (Fig. 5A). Moreover, the predicted model demonstrated 48% identity with the “d1xsfa1” model adopting a lysozyme-like fold (Fig. 5B), which suggests that Rpf has the same domain architecture of lysozyme. The indication that Rpf might be a muralytic enzyme was provided by its weak sequence similarity to the c-type lysozymes.42 This was also suggested by the frequent occurrence in the Rpf-like proteins of domains such as LysM,43 which are characteristic of cell wall-associated proteins.44 Besides, Rpf from M. luteus, the founder member of this protein family, is indeed a muralytic enzyme, as revealed by its activity in zymograms containing M. luteus cell walls.15 Interestingly, NMR and X-ray diffraction were used to establish that the conserved domain of Rpf was structurally similar to the analogous domain of lysozyme, and the conserved domain of the Rpf contributed to resuscitate the growth of bacteria.45 These results suggest that Rpf had a great possibility to resuscitate bacteria in the VBNC state by the mechanism of lysozyme activity, thus greatly affecting the potential function of bacterial floras under adverse environment.
Fig. 5. Structure of Rpf and“d1xsfa1” model. (A) Prediction of the Rpf structure generated by the Phyre2 server; (B) “d1xsfa1” model belong to the lysozyme-like superfamily.
3.4. Peptidoglycan hydrolase activity of Rpf and zymograms
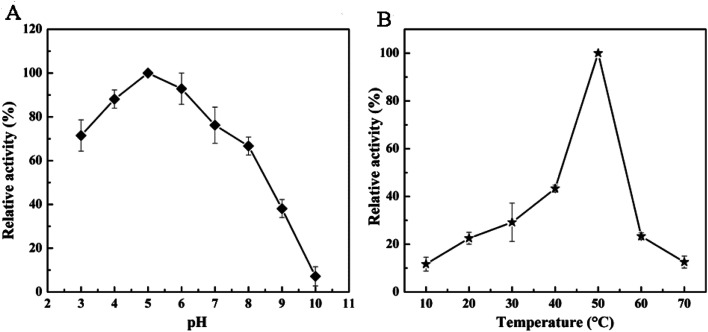
Rpf obtained from E. coli BL21 (DE3) was analyzed by zymography. As shown from Fig. 1, lane 8–9, the protein of 25 kDa was able to present a distinct clearance band with lytic activity against M. luteus, which meant Rpf was able to solubilize the peptidoglycan substrate.46 In order to further investigate the biochemical properties of Rpf, 4-nitrophenyl N-acetyl-β-d-glucosaminide (NP-GlcNAc) as a substrate was broken down in the enzyme activity assays. Fig. 6 shows the change of p-nitrophenylate under different conditions, which were performed by spectrophotometric assays at 405 nm. Results indicated that the optimal pH of the recombinant enzyme was 5.0. The enzyme exhibited 80% activity at acidic pH (3.0–4.0), while 60% of the activity was lost at pH 9.0 and 85% at pH 10.0 (Fig. 6A). Moreover, Fig. 6B shows the change in the peptidoglycan hydrolase activity among different temperatures, and it was found that the peptidoglycan hydrolase activity reached a maximum value at 50 °C and reduced greater than 70% at temperatures above 60 °C. Similarly, a previous study demonstrated that the protein secreted by Pediococcus acidilactici ATCC 8042 was stable at pH 5.0–7.0 and 45–55 °C, while 70% of the activity was lost at a temperature higher than 70 °C.
Fig. 6. Effects of pH (A) and temperature (B) on the peptidoglycan hydrolases activity of Rpf.
The obtained results suggest that Rpf has peptidoglycan hydrolase activity, which was consistent with the previous reports that the lytic activity of the Rpf protein was clearly confirmed by a decrease in the absorbance of the cell wall suspension.47 In addition, lysis zones were also observed in the polyacrylamide gel plate containing M. luteus cell walls treated with the recombinant Rpf protein.48 Moreover, Vollmer et al.49 indicated that bacterial peptidoglycan hydrolases participated in the bacterial cell wall growth and the separation of daughter cells during cell division, which could further explain the influence of Rpf on the abundance of bacterial communities.
3.5. Implication of bacterial cytokine protein Rpf for environmental bioremediation
In previous studies, it has been demonstrated that Rpf have positive effects on many biological processes (Table 4), including phenol/biphenyl biodegradation, polychlorinated biphenyl (PCB) biodegradation, dye decolorization, cellulose degradation in composting, etc. However, the prerequisite for Rpf to resuscitate the potential contaminant degraders is to optimize the Rpf preparation method. In the previous studies, culture supernatant containing Rpf from M. luteus was used to resuscitate and cultivate VBNC bacteria, but the content of Rpf in the supernatant is low and a large amount of supernatant was needed in practice.39,50,51 In recent years, recombinant Rpf was used to resuscitate and stimulate bacterial populations in pollutant-contaminated environments.11,18,52,53 However, to date, limited studies have systematically investigated the optimal conditions for the preparation of recombinant Rpf. This study revealed that bacterial cytokine protein Rpf can be obtained from recombinant E. coli by RSM, and the protein yield Rpf can be achieved when the IPTG concentration, cell density, induction temperature and culture time were 59.56 mg L−1, 0.69, 20.82 °C and 7.72 h, respectively. Importantly, the obtained Rpf protein shared homology with lysozymes. The role of Rpf in the resuscitation and stimulation of VBNC bacteria can be explained by different models, such as signaling molecules (cytokines), inducing cell remodeling and cleaving cell's wall.52 However, the mechanism of Rpf protein is still lacking. With the help of optimized Rpf protein production, more in-depth studies can be conducted to elaborate the role of Rpf in promoting the bacterial growth and resuscitation. Therefore, bacterial cytokine protein Rpf can be used as a potential bioactivator for environmental bioremediation.
The application of bacterial cytokine Rpf protein in environmental bioremediation.
| Type of Rpf | Rpf preparation | Application | Performance | Reference |
|---|---|---|---|---|
| Culture supernatant containing Rpf (SRpf) from M. luteus | The strain of M. luteus was incubated in a lactate minimal medium (LMM) on a rotary shaker (160 rpm, 30 °C) for 36 h, and then was inoculated to fresh LMM and incubated under the same culture condition until a stationary phase was achieved. Finally, the culture supernatant was obtained by centrifugation and filtration | Biphenyl degradation | Biphenyl at concentration of 1500 mg L−1 was almost completely degraded in 24 h using SRpf at a dosage of 15% (v/v) | 51 and 54 |
| Recombinant protein RpfSm (a truncated form of Rpf from M. luteus) | The strain of E. coli was first grown to an OD600nm = 0.65–0.8 and induced with 1 mM IPTG for 2 h at room temperature | Resuscitate and stimulate gram-positive bacteria | Addition of the recombinant Rpf protein (15 μg mL−1) resulted dispersion of cell aggregates and emergence of solitary cells | 55 |
| Purified recombinant Rpf protein from M. luteus | The E. coli BL21 (DE3) cells were cultured with 50 μg mL−1 kanamycin and induced by 100 μg mL−1 IPTG. Then, the culture supernatant obtained by centrifugation and sonication was applied to the Ni-NTA-agarose column. Finally, the eluate was dialyzed against 25 mM Tris–HCl at 4 °C for 17–24 h | Salt-tolerant phenol degradation | Rpf at a dosage of 1% (v/v) accelerated the start-up process during sludge domestication with higher concentrations of phenol (1500 mg L−1) and NaCl (30 g L−1) | 17 |
| Purified recombinant Rpf protein from M. luteus | The E. coli BL21 (DE3) cells were cultured with 50 μg mL−1 kanamycin and induced by 100 μg mL−1 IPTG. Then, the culture supernatant obtained by centrifugation and sonicatation, and was applied to the Ni-NTA-agarose column. Finally, the eluate was dialyzed against 25 mM Tris–HCl at 4 °C for 17–24 h with the concentration of 0.7218 g L−1 | Cellulose-degrading of the bacterial community in composting | The activity of filter paper cellulose and carboxymethyl cellulase increased 0.1028 IU mL−1 and 0.0.1282 IU mL−1 in the treatment group with 0.25% Rpf addition (v/v) | 21 |
| Purified recombinant Rpf protein from M. luteus | The E. coli BL21 (DE3) cells were cultured with 50 μg mL−1 kanamycin and induced by 100 μg mL−1 IPTG. Then, the culture supernatant obtained by centrifugation and sonication, and was applied to the Ni-NTA-agarose column. Finally, the eluate was dialyzed against 25 mM Tris–HCl at 4 °C for 17–24 h | Treatment of high-saline phenolic wastewater in MBR system | Phenol removal of sludge with Rpf addition (1%, v/v) was more than twice as that without Rpf in the MBR system | 18 |
| Culture supernatant containing Rpf (SRpf) from M. luteus | The strain of M. luteus was incubated in lactate minimal medium (LMM) on a rotary shaker (160 rpm, 30 °C) for 36 h, and then was inoculated to fresh LMM and incubated under the same culture condition until stationary phase. Finally, the culture supernatant was obtained by centrifugation and filtration | Biological nutrient removal in SBR process | PO43−-P removal efficiency increased by over 12% and total nitrogen removal efficiency increased by over 8% in the treatment reactor acclimated with SRpf addition (10%, v/v) | 20 and 54 |
| Purified recombinant Rpf protein from M. luteus | The E. coli BL21 (DE3) cells were cultured with 50 μg mL−1 kanamycin and induced by 100 μg mL−1 IPTG. Then, the culture supernatant obtained by centrifugation and sonication, and was applied to the Ni-NTA-agarose column. Finally, the eluate was dialyzed against 25 mM Tris–HCl at 4 °C for 17–24 h | Nitrogen removal | Strain SSPR1 resuscitated by Rpf (3%, v/v) showed high NH4+ removal efficiency and the removal efficiency reached 72.3% after 72 h | 23 |
| Extracellular organic matter (EOM) from M. luteus | The strain of M. luteus was incubated in lactate minimal medium (LMM) on a rotary shaker (160 rpm, 30 °C) for 36 h, and then was inoculated to fresh LMM and incubated under the same culture condition until stationary phase was obtained. Finally, the culture supernatant was obtained by centrifugation and filtration | Biphenyl degradation | Under a concentration of 3,500 mg L−1 biphenyl, biphenyl degradation efficiency reached 60.8% at a dosage of 10% EOM (v/v) | 50 and 54 |
| Crude Rpf protein from M. luteus | The E. coli BL21 (DE3) cells were grown in a 2×YT medium broth with 50 μg mL−1 kanamycin and 100 μg mL−1 IPTG. After centrifugation and sonication, the supernatant was then filtered through a 0.22 mm filter | Biodegradation of polychlorinated biphenyls (PCBs) | In soil microcosms containing 50 mg kg−1 Aroclor 1242 and inoculated with VBNC TG9T cells, after 49 d of supplementation with Rpf 20% (v/v), degradation efficiency of PCB reached 34.2% | 56 |
| Crude Rpf protein from M. luteus | The E. coli BL21 (DE3) cells were cultured with 50 μg mL−1 kanamycin and induced by 100 μg mL−1 IPTG. Then, the culture supernatant obtained by centrifugation and sonication, and was applied to the Ni-NTA-agarose column. Finally, the eluate was dialyzed against 25 mM Tris–HCl at 4 °C for 17–24 h | Degradation of reactive blue 19 | The strain Bacillus sp. JF4 resuscitated by Rpf addition (0–1.6%, v/v) could effectively decolorize RB19 | 57 |
4. Conclusions
The obtained results indicated that RSM well explored the relationships between Rpf protein production and influencing factors. The optimum conditions for obtaining Rpf protein were 59.56 mg L−1 IPTG, cell density of 0.69, induction temperature of 20.82 °C and culture time of 7.72 h. The molecular model demonstrated that Rpf had a lysozyme-like structure,which exhibits the highest lysozyme activity at pH 5.0 and 50 °C. This study provided potential strategies to apply Rpf as a bioactivator for environmental bioremediation.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We gratefully acknowledge the financial support provided by the National Natural Science Foundation of China (Grant No. 41701354), the Natural Science Foundation of Zhejiang Province of China (Grant No. LY21D010006), the Project of Jinhua Science and Technology Department, China (Grant No. 2021-4-345), the Key R&D Program of Zhejiang Province (Grant No. 2020C03080 and 2021C03021).
References
- Guo M. Jiang Y. Xie J. Cao Q. Zhang Q. Mabruk A. Chen C. J. Environ. Sci. 2022;115:55–64. doi: 10.1016/j.jes.2021.07.010. [DOI] [PubMed] [Google Scholar]
- Ji B. Zhu L. Wang S. Liu Y. J. Environ. Manage. 2021;282:111955. doi: 10.1016/j.jenvman.2021.111955. [DOI] [PubMed] [Google Scholar]
- Ma R. Yan X. Mi X. Wu Y. Qian J. Zhang Q. Chen G.-H. Chem. Eng. J. 2021;425:131457. doi: 10.1016/j.cej.2021.131457. [DOI] [Google Scholar]
- Xie M. Q. Xu L. N. Zhang R. Zhou Y. Xiao Y. Y. Su X. M. Shen C. F. Sun F. Q. Hashmi M. Z. Lin H. J. Chen J. R. Appl. Environ. Microbiol. 2021;87:e01110–01121. doi: 10.1128/AEM.01110-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon J. T. Jones S. E. Nat. Rev. Microbiol. 2011;9:119–130. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- Flemming H.-C. Water Res. 2020;173:115576. doi: 10.1016/j.watres.2020.115576. [DOI] [PubMed] [Google Scholar]
- Ke Q. Zhang Y. G. Wu X. Su X. M. Wang Y. Y. Lin H. J. Mei R. W. Zhang Y. Hashmi M. Z. Chen C. J. Chen J. R. J. Environ. Manage. 2018;222:185–189. doi: 10.1016/j.jenvman.2018.05.061. [DOI] [PubMed] [Google Scholar]
- Chen S. Li X. Wang Y. Zeng J. Ye C. Li X. Guo L. Zhang S. Yu X. Water Res. 2018;142:279–288. doi: 10.1016/j.watres.2018.05.055. [DOI] [PubMed] [Google Scholar]
- Ayrapetyan M. Oliver J. D. Curr. Opin. Food Sci. 2016;8:127–133. doi: 10.1016/j.cofs.2016.04.010. [DOI] [Google Scholar]
- Orruño M. Kaberdin V. R. Arana I. World J. Microbiol. Biotechnol. 2017;33:33–45. doi: 10.1007/s11274-017-2218-5. [DOI] [PubMed] [Google Scholar]
- Su X. M. Li S. Xie M. Q. Tao L. Q. Zhou Y. Xiao Y. Y. Lin H. J. Chen J. R. Sun F. Q. Chemosphere. 2021;263:128283. doi: 10.1016/j.chemosphere.2020.128283. [DOI] [PubMed] [Google Scholar]
- Wang Y. Y. Wang H. L. Wang X. M. Xiao Y. Y. Zhou Y. Su X. M. Cai J. F. Sun F. Q. Sci. Total Environ. 2020;730:139034. doi: 10.1016/j.scitotenv.2020.139034. [DOI] [PubMed] [Google Scholar]
- Pinto D. Santos M. A. Chambel L. Crit. Rev. Microbiol. 2015;41:61–76. doi: 10.3109/1040841X.2013.794127. [DOI] [PubMed] [Google Scholar]
- Mukamolova G. V. Turapov O. A. Kazarian K. Telkov M. Kaprelyants A. S. Kell D. B. Young M. Mol. Microbiol. 2002;46:611–621. doi: 10.1046/j.1365-2958.2002.03183.x. [DOI] [PubMed] [Google Scholar]
- Mukamolova G. V. Murzin A. G. Salina E. G. Demina G. R. Kell D. B. Kaprelyants A. S. Young M. Mol. Microbiol. 2006;59:84–98. doi: 10.1111/j.1365-2958.2005.04930.x. [DOI] [PubMed] [Google Scholar]
- Nikitushkin V. D. Demina G. R. Shleeva M. O. Guryanova S. V. Ruggiero A. Berisio R. Kaprelyants A. S. FEBS J. 2015;282:2500–2511. doi: 10.1111/febs.13292. [DOI] [PubMed] [Google Scholar]
- Sun J. T. Pan L. L. Zhu L. Z. Sci. Total Environ. 2018;613:54–61. doi: 10.1016/j.scitotenv.2017.09.063. [DOI] [PubMed] [Google Scholar]
- Su X. M. Wang Y. Y. Xue B. B. Hashmi M. Z. Lin H. J. Chen J. R. Wang Z. Mei R. W. Sun F. Q. Chem. Eng. J. 2019;357:715–723. doi: 10.1016/j.cej.2018.09.197. [DOI] [Google Scholar]
- Keep N. H. Ward J. M. Cohen-Gonsaud M. Henderson B. Trends Microbiol. 2006;14:271–276. doi: 10.1016/j.tim.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Liu Y. D. Su X. M. Lu L. Ding L. X. Shen C. F. Environ. Sci. Pollut. Res. 2016;23:4498–4508. doi: 10.1007/s11356-015-5603-3. [DOI] [PubMed] [Google Scholar]
- Su X. M. Zhang S. Mei R. W. Zhang Y. Hashmi M. Z. Liu J. J. Lin H. J. Ding L. X. Sun F. Q. Microb. Biotechnol. 2018;11:527–536. doi: 10.1111/1751-7915.13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X. M. Wang Y. Y. Xue B. B. Zhang Y. G. Mei R. W. Zhang Y. Hashmi M. Z. Lin H. J. Chen J. R. Sun F. Q. Bioresour. Technol. 2018;261:394–402. doi: 10.1016/j.biortech.2018.04.048. [DOI] [PubMed] [Google Scholar]
- Su X. M. Xue B. B. Wang Y. Y. Hashmi M. Z. Lin H. J. Chen J. R. Mei R. W. Wang Z. Sun F. Q. Ecotoxicol. Environ. Saf. 2019;179:188–197. doi: 10.1016/j.ecoenv.2019.04.067. [DOI] [PubMed] [Google Scholar]
- Su X. M. Zhou M. Hu P. Xiao Y. Y. Wang Z. Mei R. W. Hashmi M. Z. Lin H. J. Chen J. R. Sun F. Q. Chemosphere. 2019;232:76–86. doi: 10.1016/j.chemosphere.2019.05.195. [DOI] [PubMed] [Google Scholar]
- Kilikian B. V. Suarez I. D. Liria C. W. Gombert A. K. Process Biochem. 2000;35:1019–1025. doi: 10.1016/S0032-9592(00)00137-0. [DOI] [Google Scholar]
- Acero-Navarro K. E. Jiménez-Ramírez M. Villalobos M. A. Vargas-Martínez R. Perales-Vela H. V. Velasco-García R. Protein Expression Purif. 2018;142:53–61. doi: 10.1016/j.pep.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Allers T. Barak S. Liddell S. Wardell K. Mevarech M. Appl. Environ. Microbiol. 2010;76:1759–1769. doi: 10.1128/AEM.02670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill M. A. Denos M. Reed D. Pest Manage. Sci. 2018;74:705–714. doi: 10.1002/ps.4766. [DOI] [PubMed] [Google Scholar]
- Ghafoori S. Mowla A. Jahani R. Mehrvar M. Chan P. K. J. Environ. Manage. 2015;150:128–137. doi: 10.1016/j.jenvman.2014.11.011. [DOI] [PubMed] [Google Scholar]
- Keerati-u-rai M. Miriani M. Iametti S. Bonomi F. Corredig M. Colloids Surf., B. 2012;93:41–48. doi: 10.1016/j.colsurfb.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Mendonca A. Rocha A. C. Duarte A. C. Santos E. B. H. Anal. Chim. Acta. 2013;788:99–107. doi: 10.1016/j.aca.2013.05.051. [DOI] [PubMed] [Google Scholar]
- Fischer D. Curr. Opin. Struct. Biol. 2006;16:178–182. doi: 10.1016/j.sbi.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Kelley L. A. Mezulis S. Yates C. M. Wass M. N. Sternberg M. J. E. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cano I. Campos-Gómez M. Contreras-Cruz M. Serrano-Maldonado C. E. González-Canto A. Peña-Montes C. Rodríguez-Sanoja R. Sánchez S. Farrés A. Appl. Microbiol. Biotechnol. 2015;99:8563–8573. doi: 10.1007/s00253-015-6593-2. [DOI] [PubMed] [Google Scholar]
- Serrano-Maldonado C. E. García-Cano I. González-Canto A. Ruiz-May E. Elizalde-Contreras J. M. Quirasco M. J. Mol. Microbiol. Biotechnol. 2018;28:14–27. doi: 10.1159/000486757. [DOI] [PubMed] [Google Scholar]
- García-Cano I. Serrano-Maldonado C. E. Olvera-García M. Delgado-Arciniega E. Peña-Montes C. Mendoza-Hernández G. Quirasco M. LWT-Food Sci. Technol. 2014;59:26–34. doi: 10.1016/j.lwt.2014.04.059. [DOI] [Google Scholar]
- Serrano-Maldonado C. E. Quirasco M. J. Biotechnol. 2018;283:28–36. doi: 10.1016/j.jbiotec.2018.06.346. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S. Choudhury J. D. Gachhui R. Mukherjee J. Int. J. Biol. Macromol. 2018;109:1140–1146. doi: 10.1016/j.ijbiomac.2017.11.106. [DOI] [PubMed] [Google Scholar]
- Mukamolova G. V. Kaprelyants A. S. Young D. I. Young M. Kell D. B. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8916–8921. doi: 10.1073/pnas.95.15.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X. M. Liu Y. D. Hu J. X. Ding L. X. Shen C. F. SpringerPlus. 2014;3:117. doi: 10.1186/2193-1801-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar M. Rasul M. G. Ashwath N. Energy Convers. Manage. 2018;156:103–112. doi: 10.1016/j.enconman.2017.11.004. [DOI] [Google Scholar]
- Cohen-Gonsaud M. Keep N. H. Davies A. P. Ward J. Henderson B. Labesse G. Trends Biochem. Sci. 2004;29:1–4. doi: 10.1016/j.tibs.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Bateman A. Bycroft M. J. Mol. Biol. 2000;299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- Ravagnani A. Finan C. L. Young M. BMC Genomics. 2005;6:45. doi: 10.1186/1471-2164-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Gonsaud M. Barthe P. Bagneris C. Henderson B. Ward J. Roumestand C. Keep N. H. Nat. Struct. Mol. Biol. 2005;12:270–273. doi: 10.1038/nsmb905. [DOI] [PubMed] [Google Scholar]
- Escobar C. A. Cross T. A. Anal. Biochem. 2018;543:162–166. doi: 10.1016/j.ab.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana B. D. Mizrahi V. FEMS Immunol. Med. Microbiol. 2010;58:39–50. doi: 10.1111/j.1574-695X.2009.00606.x. [DOI] [PubMed] [Google Scholar]
- Telkov M. V. Demina G. R. Voloshin S. A. Salina E. G. Dudik T. V. Stekhanova T. N. Mukamolova G. V. Kazaryan K. A. Goncharenko A. V. Young M. Kaprelyants A. S. Biochemistry (Mosc.) 2006;71:414–422. doi: 10.1134/S0006297906040092. [DOI] [PubMed] [Google Scholar]
- Vollmer W. Joris B. Charlier P. Foster S. FEMS Microbiol. Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- Su X. M. Zhang Q. Hu J. X. Hashmi M. Z. Ding L. X. Shen C. F. Appl. Microbiol. Biotechnol. 2015;99:1989–2000. doi: 10.1007/s00253-014-6108-6. [DOI] [PubMed] [Google Scholar]
- Su X. M. Shen H. Yao X. Y. Ding L. X. Yu C. N. Shen C. F. Bioresour. Technol. 2013;146:27–34. doi: 10.1016/j.biortech.2013.07.028. [DOI] [PubMed] [Google Scholar]
- Cai J. F. Pan A. D. Li Y. L. Xiao Y. Y. Zhou Y. Chen C. J. Sun F. Q. Su X. M. Chemosphere. 2021;263:127922. doi: 10.1016/j.chemosphere.2020.127922. [DOI] [PubMed] [Google Scholar]
- Su X. M. Li S. Cai J. F. Xiao Y. Y. Tao L. Q. Hashmi M. Z. Lin H. J. Chen J. R. Mei R. W. Sun F. Q. Sci. Total Environ. 2019;688:917–925. doi: 10.1016/j.scitotenv.2019.06.364. [DOI] [PubMed] [Google Scholar]
- Kaprelyants A. S. Kell D. B. J. Appl. Bacteriol. 1992;72:410–422. doi: 10.1111/j.1365-2672.1992.tb01854.x. [DOI] [Google Scholar]
- Nikitushkin V. D. Demina G. R. Kaprelyants A. S. Biochemistry. 2017;81:1719–1734. doi: 10.1134/S0006297916130095. [DOI] [PubMed] [Google Scholar]
- Ye Z. Li H. X. Jia Y. Y. Fan J. H. Wan J. X. Guo L. Su X. M. Zhang Y. Wu W. M. Shen C. F. Environ. Pollut. 2020;263:114488. doi: 10.1016/j.envpol.2020.114488. [DOI] [PubMed] [Google Scholar]
- Cai J. F. Liu J. L. Pan A. D. Liu J. F. Wang Y. Y. Liu J. B. Sun F. Q. Lin H. J. Chen J. R. Su X. M. Water Sci. Technol. 2020;81:1159–1169. doi: 10.2166/wst.2020.201. [DOI] [PubMed] [Google Scholar]
- Li Y. L. Zhang W. J. Dai Y. Su X. M. Wu D. Sun F. Q. Mei R. W. Chen J. R. Lin H. J. Science of The Total Environment. 2022:150975. doi: 10.1016/j.scitotenv.2021.150975. [DOI] [PubMed] [Google Scholar]