Abstract
Peritoneal metastasis (PM) is the main site of gastric cancer (GC) distant metastasis and indicates an extremely poor prognosis and survival. Hypoxia is a common feature of peritoneal metastases and up-regulation of hypoxia inducible factor 1 alpha (HIF-1α) may be a potential driver in the occurrence of PM. Ferroptosis is a recently discovered form of regulated cell death and closely related to the occurrence and development of tumors. However, the underlying mechanism link HIF-1α to ferroptosis in PM of GC remains unknown. Here, lncRNA-microarrays and RNA library construction/lncRNA-seq results shown that lncRNA-PMAN was highly expressed in PM and significantly modulated by HIF-1α. Upregulation of PMAN is associated with poor prognosis and PM in patients with GC. PMAN was up-regulated by HIF-1α and improved the stability of SLC7A11 mRNA by promoting the cytoplasmic distribution of ELAVL1, which was identified in RNA-pulldown/mass spectrometry results. Accumulation of SLC7A11 increases the level of l-Glutathione (GSH) and inhibits the accumulation of reactive oxygen species (ROS) and irons in the GC cells. Finally protect GC cells against ferroptosis induced by Erastin and RSL3. Our findings have elucidated the effect of HIF-1α/PMAN/ELAVL1 in GC cells ferroptosis and provides theoretical support for the potential diagnostic biomarkers and therapeutic targets for PM in GC.
Keywords: Hypoxia, Ferroptosis, ROS, HIF-1α, lncRNA, ELAVL1 translocation, SLC7A11, Peritoneal tumor
Graphical abstract
Highlights
-
•
HIF-1⍺ mediates abnormally high expression of PMAN in PM from GC under hypoxia.
-
•
GC cells suppress ferroptosis by relieving ROS and irons accumulation through HIF-1⍺/PMAN under hypoxia.
-
•
Inhibition of ferroptosis may contributes to the development of PM from GC.
-
•
Increased cytoplasmic translocation of ELAVL1 is a key intermediate factor in PMAN inhibition of ferroptosis.
1. Introduction
Gastric cancer (GC) is one of the most common malignant tumors worldwide. Peritoneal metastasis (PM) occurs in 53–66% of patients with distant metastatic GC and frequently indicates an extremely poor prognosis [1,2]. However, the underlying molecular mechanism of PM remains poorly understood. Therefore, specific diagnostic biomarkers and therapeutic targets for PM have a great exploratory value.
Hypoxia is a major characteristic of the malignant tumor microenvironment and is directly related to tumorigenesis [3]. In the process of PM formation, due to the low oxygen partial pressure of ascites (around 50% less soluble oxygen than the blood [4]) and irregular tumor blood vessels [5], the microenvironment of the PM is indeed hypoxic [6]. Intratumoral hypoxia leads to the upregulation of hypoxia-inducible factor 1 alpha (HIF-1α). Accumulating evidence indicates that HIF-1α may be a potential driver of PM in GC [7,8]. However, the relationship between HIF-1α and PM formation remains unclear.
Studies have suggested that suppression of ferroptosis promotes tumor invasion and metastasis [9,10]. Notably, HIF-1α regulates iron metabolism-related gene expression through hypoxia response elements (HRE) [11] and promotes tumorigenesis by inhibiting ferroptosis [12,13]. Therefore, we reasoned that HIF-1α might promote the formation of PM by increasing the ferroptosis resistance of peritoneal disseminated tumors. Ferroptosis, a unique non-apoptotic form of cell death, is mainly caused by intracellular iron catalytic activity and lipid peroxidation and is characterized by the accumulation of reactive oxygen species (ROS) [14,15]. Erastin and RSL3 have been identified as ferroptosis stimulators by suppressing lipid hydroperoxidase glutathione peroxidase 4 (GPX4) or the cystine-glutamate exchanger system xc− (cystine/glutamate antiporter SLC7A11) [16,17]. SLC7A11 is a membrane channel transporter that alleviates ROS and iron accumulation induced by ferroptosis by increasing glutathione (GSH) synthesis in cells. It has been widely regarded as a protective factor for tumor cells to resist ferroptosis [18,19]. However, the molecular mechanisms by which HIF-1α affects ferroptosis remain unclear. We believe that exploring the mechanisms underlying HIF-1α/ferroptosis during hypoxia is of great significance for further understanding PM in GC.
Recent research showed that long non-coding RNA (lncRNA) plays an important role in ferroptosis [20,21]. By comparing the results of lncRNA-microarrays and RNA library construction/lncRNA-seq, we identified the key lncRNA-PMAN that regulates ferroptosis by enhancing the stability of SLC7A11 mRNA during hypoxia in GC cells and promotes the formation of peritoneal disseminated tumors in vitro. Our study elucidated that HIF-1α/lncRNA-PMAN inhibits the occurrence of ferroptosis and is expected to provide theoretical support for potential biomarkers and therapeutic targets for PM in GC.
2. Materials and methods
Detailed materials and methods are provided in supplemental experimental procedures (Supplementary file).
2.1. Patients and tissue samples
We collected 30 paired of GC and ANT samples, 3 paired of ANT, primary GC and PM from GC samples. All included patients underwent curative resection at ZhongNan Hospital of Wuhan University, and were identified as gastric adenocarcinoma by histopathology. In addition, all patients did not receive neoadjuvant chemotherapy or radiotherapy before surgical resection. Inform in writing that the consent of all participants has been obtained. This study was approved by the Research Ethics Committee of Wuhan University.
2.2. LncRNA microarrays and lncRNA library construction/lncRNA-seq
The lncRNA expression characteristics of 3 pairs GC/PM tissues were investigated by Arraystar Human LncRNA microarray V2.0, which contains 30,215 coding genes and 33,045 lncRNAs collected from several databases such as UCSC, Ensembl, RefSeq and the lncRNAs reported from literatures were also included. The microarray and data collection were conducted by KangChen Bio-tech (Shanghai, China). In addition, we investigated the lncRNA expression characteristics of 3 pairs of siHIF-1α transfected gastric cancer cells. The library construction and lncRNA-seq and data collection were conducted by HuaDa Gene Bio-tech (Shenzhen, China).
2.3. Animal experiments
We collected MGC-803 cell lines (1 × 10 [7] cells) that were infected by lentivirus with or without PMAN-OE. Then, suspended in 100 μl serum-free medium and subcutaneously/intraperitoneal injected into each flank of 4-week-old BALB/c nude female mice (six mice per group). We measured tumor volume and body weight each week, then mice were sacrificed after four weeks, and the maximum (L) and minimum (W) length and weight of the tumors were measured. Hematoxylin and eosin (H&E) staining was performed for tissue morphology evaluation.
For animal models of gastric subserosal injection, we collected MGC-803 cell lines (5 × 10 [6] cells) that were infected by lentivirus with or without PMAN-OE, and suspended in 40 μl serum-free medium (50% Matrigel). After that, nude mice (six mice per group) were anesthetized by intraperitoneal injection of 100 μl of pentobarbital (1%). After disinfection, the abdominal cavity was opened to expose the greater curvature of the stomach. The tumor suspension (40 μl) was implanted under the serosa of the greater curvature of the stomach of the nude mice through an insulin needle. Then, suturing the peritoneum and skin (absorbable surgical suture, Jinhuan Medical Co., ltd, China.), the incision was sterilized again and waited for recovery. The abdominal cavity was opened again after 35 days, and the incidence of abdominal tumor formation was observed. The animal experiments were approved by the Animal Experiment Center of Wuhan University.
2.4. Statistics analysis
All statistical analyses were performed with SPSS statistical software (version 22.0, USA) and GraphPad Prism software (version 9.0, USA). Two‐tailed Student's tests were used to compare the means quantitative data between groups, or when more than two groups were compared by one‐way analysis of variance. The associations between PMAN level in tumor tissues and clinicopathologic parameters were determined using the Chi-squared tests or Fisher's exact test. Kaplan–Meier method was used for survival analysis and drawing the survival curves, and difference among patients' subgroups (All GC patients were divided into a high- and low-expression group according to the median of the PMAN level) was calculated by log-rank test. The correlation between HIF-1α mRNA and PMAN or SLC7A11 mRNA was analyzed by Pearson's correlation coefficient analysis. Univariate and multivariate Cox-regression analyses were applied to identify the independent factors of prognosis. The results were expressed as the Mean ± SD from at least three independent experiments. In all cases, ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01.
3. Results
3.1. HIF-1α promoted the aberrant high expression of PMAN in GC with PM
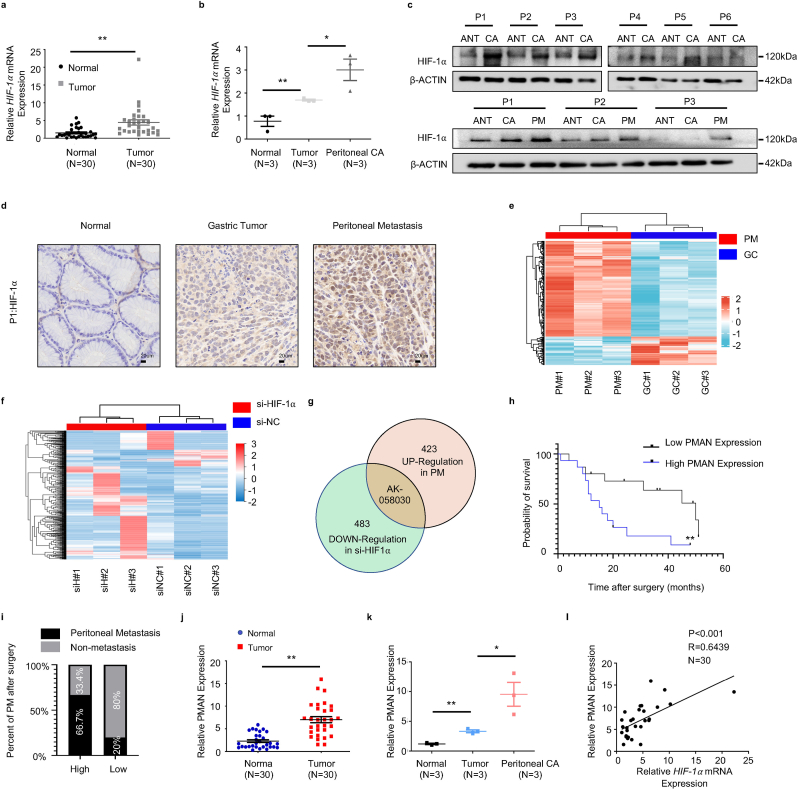
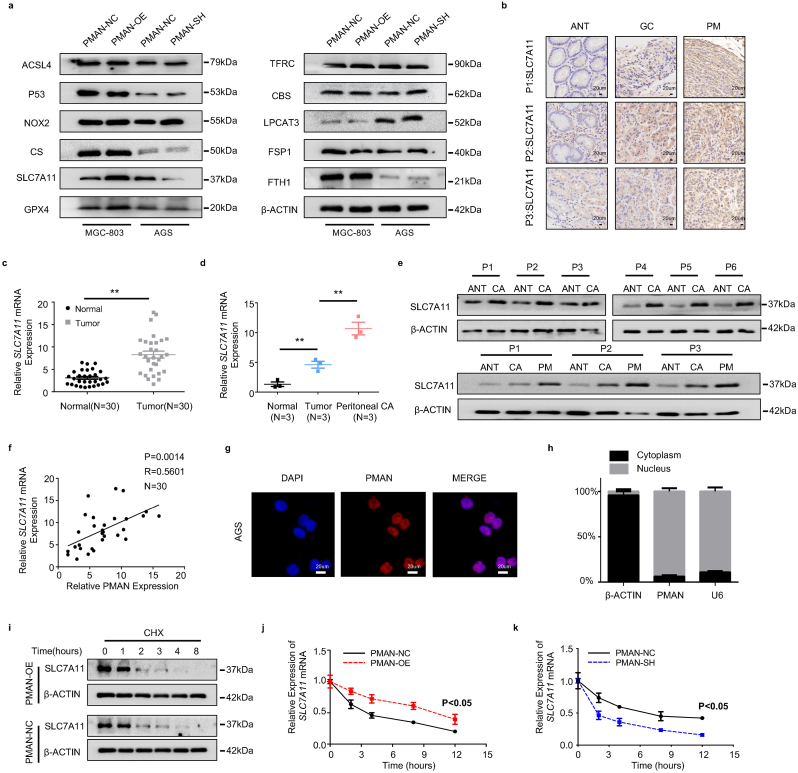
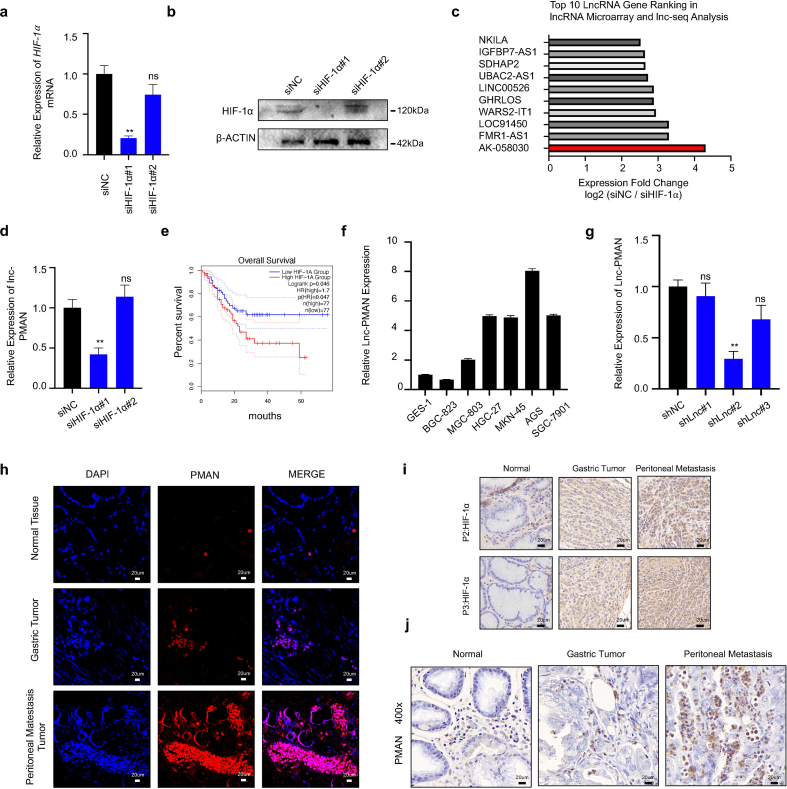
Previous studies [7,22] have shown that HIF-1α is a critical driver of PM malignancy. To further confirm the expression of HIF-1α in tumor tissues. We analyzed HIF-1α mRNA in GC and paired adjacent normal tissues (ANT) (n = 30) (Fig. 1.a) and samples of ANT, primary GC, and PM from GC (n = 3) (Fig. 1.b) by RT-qPCR. In addition, HIF-1α was detected in samples of GC and paired ANT (n = 6), samples of ANT, primary GC, and PM from GC (n = 3) by western blotting (WB) (Fig. 1.c) and immunohistochemistry (IHC) (Fig. 1.d, SupplementaryFig1.i). The results showed that HIF-1α mRNA and protein were significantly upregulated in PM compared to that in GC and ANT. Similarly, the results showed that the expression of HIF-1α was remarkably upregulated in GC compared to that in ANT. A multitude of studies [23,24] have reported that HIF-1α can participate in the occurrence and development of tumors by regulating aberrant lncRNA expression. To explore the PM-promoting lncRNAs and their activation by HIF-1α, lncRNA microarrays were performed in PM and matched primary GC (n = 3) (Fig. 1.e) [25] and lncRNA library construction/lncRNA-seq was performed in siHIF-1α AGS cells (n = 3) (Fig. 1.f). We combined the results from lncRNA microarray analyses and lncRNA library construction/lncRNA-seq, and found that LncRNA-AK058030 (SupplementaryTab.1) was highly expressed in PM and significantly modulated by HIF-1α (Fig. 1.g, SupplementaryFig1.c). We next verified the expressions of LncRNA-AK058030 in tumor tissues, and RT-qPCR results corresponded to lncRNA-seq analyses (Fig. 1.j-k). In addition, 66.7% of the high-expression LncRNA-AK058030 group had PM after surgery, and only 20% had PM after surgery in the low-expression LncRNA-AK058030 group (Fig. 1.i) (SupplementaryTab.2-3). LncRNA-AK058030 is not only highly expressed in PM from GC but also positively correlated with PM occurrence in patients with GC; therefore, we named LncRNA-AK058030 peritoneal metastasis associated long noncoding RNA (PMAN). In addition, we detected the endogenous expression levels of PMAN in various GC cells using RT-qPCR. These results were also in agreement with our previous study, wherein expression of PMAN, AGS, and MGC-803 cells were selected to complete the follow-up experiments (SupplementaryFig1.f). Notably, the correlation between PMAN and HIF-1α mRNA was determined by calculating the Pearson correlation coefficient, which indicated a positive correlation in tumors (R = 0.6439) (Fig. 1.l). Moreover, fluorescence in situ hybridization (FISH) and in situ hybridization (ISH) results demonstrated that PMAN was increased in PM from GC (SupplementaryFig1.h,j). Finally, we assessed patients who had relatively higher levels of PMAN associated with poor overall survival in GC through Kaplan-Meier analysis (Fig. 1.h), which indicated that PMAN might be a potential biomarker for PM.
Fig. 1.
PMAN is highly expressed in gastric cancer peritoneal metastasis and is highly correlated with the patient's prognosis.aHIF-1α mRNA expressions in 30 pairs GC samples. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. bHIF-1α mRNA expressions in 3 pairs GC peritoneal metastasis samples. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. c HIF-1α expression levels in 6 pairs GC samples and 3 pairs GC peritoneal metastasis samples. d Immunohistochemistry was used to analyze up-regulated HIF-1α expressions in paired GC peritoneal metastasis samples. Scale bar, 20 μm e Analysis of the expression of lncRNAs in GC peritoneal metastasis by lncRNA-microarrays. f Analysis of the expression of lncRNAs in HIF-1α knockdown AGS cells by lncRNA-seq. g AK-058030 is highly expressed in GC peritoneal metastasis samples and its expression decreases with knockdown of HIF-1α. h Kaplan-Meier curves for overall survival rates correlated with PMAN expression (high/low expression) in GC. Error bars, SD. i In the postoperative follow-up data, the relationship between the occurrence of PM and the expression of PMAN (high/low expression) in GC patients j PMAN expressions in 30 pairs GC samples. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. k PMAN expressions in 3 pairs GC peritoneal metastasis samples. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. l Correlations between HIF-1α mRNA levels and PMAN levels in GC. R-values and P-values were used via Pearson's correlation analysis. Mean ± SD are shown. Ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01.
3.2. PMAN is upregulated by HIF-1α during hypoxia
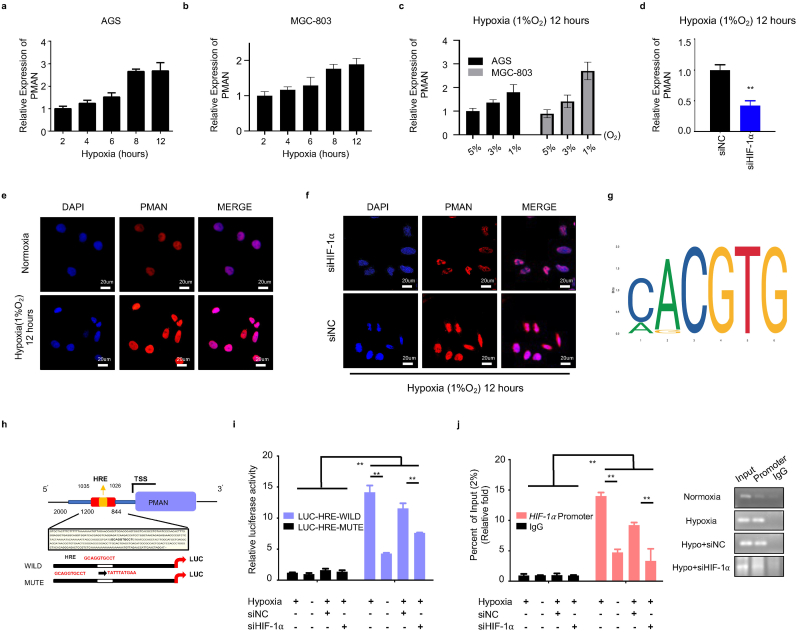
To elucidate the mechanism of PMAN activated by HIF-1α. A hypoxia model in vitro (1%O2, 5%CO2, 37 °C) was constructed, and we found that PMAN was upregulated in a time-dependent and concentration-dependent manner under hypoxia (Fig. 2.a-c). The FISH results also showed that compared with normoxia, a hypoxic environment can stimulate the expression of PMAN (Fig. 2.e). Next, siHIF-1α was transfected into GC cells under hypoxia in vitro, wherein siHIF-1α#1 was selected to complete the follow-up experiments according to RT-qPCR and WB results (SupplementaryFig1.a-b,d). Both the RT‐qPCR and FISH results suggested that siHIF-1α markedly decreased PMAN during hypoxia (Fig. 2.d,f). By analyzing the JASPAR database, we predicted the HRE sequences in the promoter regions of PMAN (Fig. 2.g). Two luciferase reporter plasmids containing the PMAN promoter (wild type, GCAGGTGCCT; mutant type, TATTTATGAA) were constructed and then transfected into AGS cells (Fig. 2.h). As expected, luciferase activity was significantly increased by hypoxia, whereas the HRE mutant exhibited markedly reduced luciferase activity. Moreover, siHIF-1α reversed the hypoxia-mediated promotion of WT-PMAN luciferase activity (Fig. 2.i). To further substantiate the direct binding of HIF-1α to the endogenous HRE site in the PMAN promoter, a chromatin immunoprecipitation (ChIP) assay was performed and showed the same results (Fig. 2.j). These results confirmed that HIF-1α could induce PMAN at the transcriptional level during hypoxia.
Fig. 2.
HIF-1α transcriptionally activates PMAN expression under hypoxia.a-b AGS cells (a) and MGC-803 cells (b) were cultured in hypoxia, RT-qPCR analyzes the expression level of PMAN at indicated timepoints. Mean ± SD is shown. c RT-qPCR analysis of PMAN expression of AGS cells and MGC-803 cells under different oxygen concentrations. Mean ± SD is shown. d RT-qPCR detects the expression level of PMAN in siHIF-1α-transfected AGS cells. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. e FISH detection of PMAN (red) in hypoxic or normoxic environment. The nucleus was stained with DAPI. Scale bar, 20 μm f FISH detection of PMAN (red) in HIF-1α knockdown AGS cells during hypoxia. The nucleus was stained with DAPI. Scale bar, 20 μm g The recognition sequences of HIF-1α (HRE) from the JASPAR database. h A potential HIF-1α binding site (HRE: GCAGGTGCCT) was found in the promoter sequence (0-2000 nt) of PMAN. i AGS cells were transfected with luciferase reporter plasmids containing PMAN promoter (wild/mute type) and further transfected with siHIF-1α and siNC in hypoxic or normoxic environment. The luciferase activity was measured and normalized. j ChIP assays with anti–HIF–1α were used to assess the specific binding between HIF-1α recognition sequence (HRE) of the PMAN promoter and HIF-1α during hypoxic or normoxic AGS cells transfected with siHIF-1α or siNC. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. Ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. PMAN could inhibited ferroptosis in GC cells
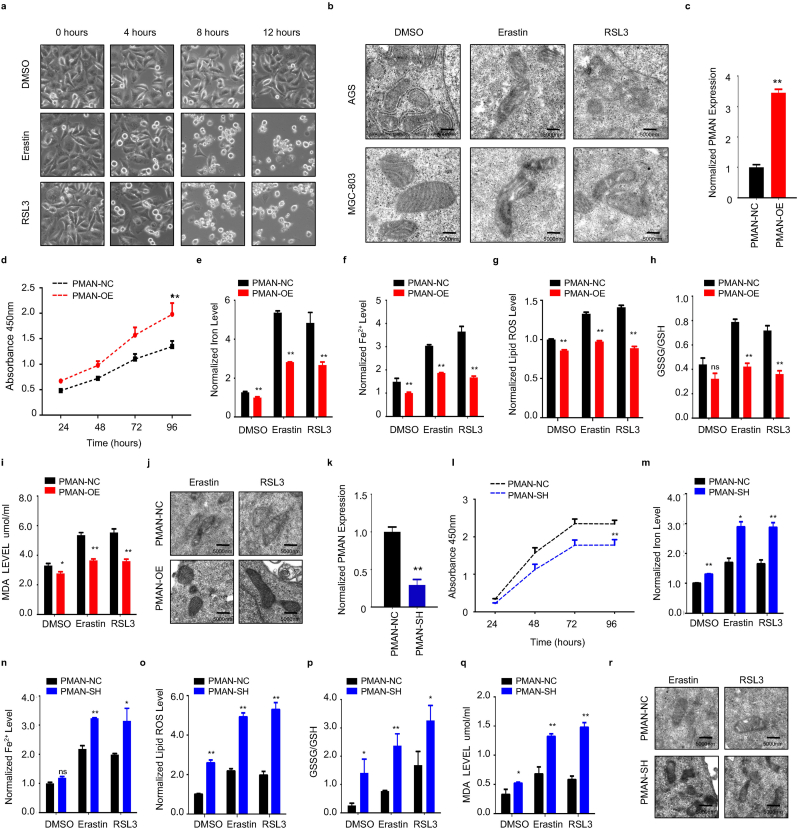
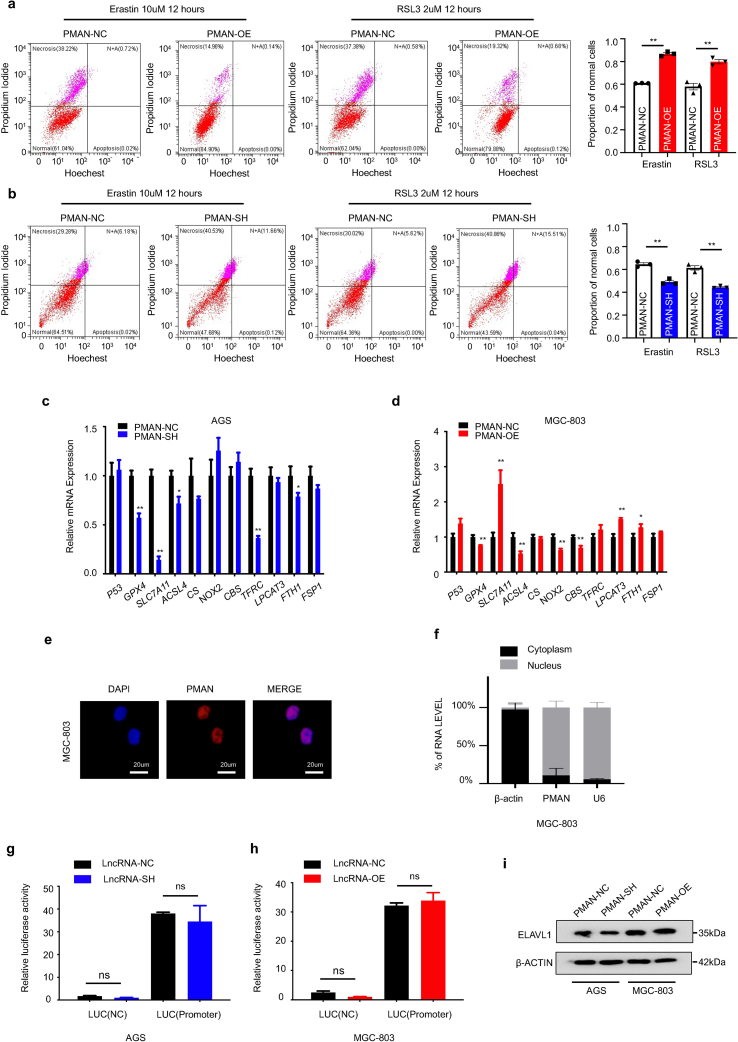
Recent studies [26] identified that HIF-1α might promote the progression of tumor growth by anti-ferroptosis. Thus, HIF-1α may play a protective role against ferroptosis. Therefore, we speculate that PMAN, which is activated by HIF-1α, is involved in ferroptosis. We first treated AGS cells with Erastin (10 μM) or RSL3 (2 μM) (12 h), stimulating drugs of ferroptosis, and then observed the cell morphology using an inverted microscope (Fig. 3.a). The cells lost their normal shape and became shrunken after treatment with Erastin or RSL3. Additionally, transmission electron microscopy (TEM) was performed to evaluate the changes in the mitochondria in AGS and MGC-803 cells after Erastin or RSL3 treatment (Fig. 3.b). These results showed that ferroptosis in the cell model was consistent with our expectations. Next, we constructed stable PMAN-OE (overexpression) MGC-803 cells using a lentivirus vector containing LV-PMAN (overexpression efficiency was confirmed by RT-qPCR) (Fig. 3.c). PMAN-OE increased the viability of MGC-803 cells in CCK-8 (Fig. 3.d) and reduced the proportion of MGC-803 cells damaged by Erastin or RSL3-induced ferroptosis stress (SupplementaryFig2.a). Furthermore, we found that PMAN-OE decreased the intracellular concentrations of total iron (FE), ferrous iron (FE2+), lipid ROS, glutathione disulfide (GSSG) to GSH ratio, and MDA in MGC-803 cells after treatment with Erastin or RSL3 (Fig. 3.e-i). TEM analysis revealed that the altered mitochondrial ultrastructure in MGC-803 cells was recovered after PMAN-OE (Fig. 3.j). These results indicated that PMAN-OE promoted resistance to ferroptosis in MGC-803 cells.
Fig. 3.
The overexpression of PMAN inhibits ferroptosis and knockdown of PMAN promotes ferroptosis in GC cells.a Cell morphology of AGS cells under an inverted microscope after treated with Erastin (10 μM) or RSL3 (2 μM) at indicated timepoints. b Transmission electron microscopy (TEM) was performed to evaluate the change in ultrastructural features such as mitochondria of AGS cells under Erastin or RSL3 treatment after 12 h. Scale bar, 5000 nm c RT-qPCR analysis was used to detect the expression of PMAN in MGC-803. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. d CCK-8 assay to analyze the effect of overexpression-PMAN on cell proliferation in MGC-803. e Level of total iron f level of ferrous iron g level of lipid ROS h the ratio of GSSG/GSH i level of MDA were detected in MGC-803 cells stably overexpressing PMAN under Erastin (10 μM) or RSL3 (2 μM) treatment after 12 h. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. j Transmission electron microscopy was performed to evaluate the change of mitochondrial ultrastructure in MGC-803 cells stably overexpressing PMAN after treated with Erastin (10 μM) or RSL3 (2 μM). Scale bar, 5000 nm k RT-qPCR analysis was used to detect the expression of PMAN in AGS cells. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. l CCK-8 assay to analyze the effect of knockdown-PMAN on cell proliferation in AGS cells. m Level of total iron n level of ferrous iron o level of lipid ROS p the ratio of GSSG/GSH q level of MDA were detected in AGS cells stably knockdown PMAN under Erastin (10 μM) or RSL3 (2 μM) treatment after 12 h. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. r Transmission electron microscopy was performed to evaluate the change of mitochondrial ultrastructure in AGS cells stably knockdown PMAN after treated with Erastin (10 μM) or RSL3 (2 μM). Scale bar, 5000 nm. Ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01.
To further validate the effect of PMAN on the ferroptosis of GC cells, AGS cells were transfected with lentiviral shRNA, wherein the knockdown efficiency of PMAN-SH was estimated, and sh-PMAN#2 was selected to complete the follow-up experiment according to RT-qPCR results (Fig. 3.k, SupplementaryFig1.g). The CCK-8 assay showed that PMAN-SH markedly decreased the proliferation of AGS cells (Fig. 3.l), and as we expected, knockdown of PMAN significantly increased the proportion of AGS cells damage under Erastin or RSL3-induced ferroptosis stress (SupplementaryFig2.b). We found that PMAN-SH increased the intracellular concentrations of FE, FE2+, lipid ROS, GSSG to GSH ratio, and MDA in AGS cells after treatment with Erastin or RSL3 (Fig. 3.m-q). TEM showed severely shriveled mitochondria in the PMAN-SH AGS cells (Fig. 3.r). Taken together, these results demonstrated that PMAN-SH increased the sensitivity of AGS cells to ferroptosis.
3.4. PMAN promoted mRNA stability of SLC7A11
Next, we sought to explore the mechanism by which PMAN regulates ferroptosis in GC cells. First, we filtered the protein and mRNA levels of ferroptosis-related genes (Fig. 4.a, SupplementaryFig2.c-d) in PMAN-SH AGS and PMAN-OE MGC-803 cells. The results showed that the expression of SLC7A11 mRNA and protein were significantly different between the two cell lines, while other ferroptosis-related genes such as GPX4, ACSL4, and FTH1 were significantly altered by PMAN-SH/OE at the mRNA level, but not at the protein level. IHC, WB, and RT-qPCR were conducted to detect SLC7A11 in samples of paired GC and ANT, and samples of ANT, primary GC, and PM from GC (n = 3) (Fig. 4.b-e). The results indicated that SLC7A11 was upregulated more in PM than in GC and ANT. Using the Pearson correlation coefficient, PMAN was found to be positively correlated with SLC7A11 mRNA levels in tumors (R = 0.5601) (Fig. 4.f). Based on this, we believe that PMAN and SLC7A11 mRNA/protein levels are positively correlated in GC tissues and GC cells. Next, we investigated the mechanisms underlying PMAN regulation of SLC7A11. FISH and nuclear/cytoplasmic RNA separation demonstrated that PMAN was mainly localized in the nucleus (Fig. 4.g-h, SupplementaryFig2.e-f). To determine whether PMAN altered the promoter activity of SLC7A11, a luciferase reporter plasmid containing the SLC7A11 promoter was constructed and transfected into AGS and MGC-803 cells. The results showed that neither PMAN-SH nor PMAN-OE affected luciferase activity of the SLC7A11 promoter (SupplementaryFig2.g-h). In addition, MGC‐803 cells were cultured with cycloheximide (CHX) (10 μM) for the indicated time, and PMAN-OE did not change the half-life of SLC7A11 (Fig. 4i). PMAN-SH AGS and PMAN-OE MGC-803 cells were exposed to actinomycin D (AD) (5 μg/ml) at the indicated time points and analyzed by RT-qPCR. Interestingly, the half-life of SLC7A11 mRNA was increased in PMAN-OE MGC-803 cells and reduced in PMAN-SH AGS cells (Fig. 4.j-k). We hypothesized that PMAN regulates SLC7A11 expression by influencing SLC7A11 mRNA stability.
Fig. 4.
PMAN affected mRNA stability of SLC7A11 to enhance the expression of SLC7A11.a The expression level of ferroptosis-related proteins in the PMAN knockdown AGS cells and the PMAN overexpression MGC-803 cells. b Immunohistochemistry was used to analyze SLC7A11 expressions in 3 pairs GC peritoneal metastasis samples. Scale bar, 20 μm cSLC7A11 mRNA expressions in 30 pairs GC samples. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. dSLC7A11 mRNA expressions in 3 pairs GC peritoneal metastasis samples. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. e SLC7A11 expression levels in 6 pairs GC samples and 3 pairs GC peritoneal metastasis samples. f Correlations between SLC7A11 mRNA levels and PMAN levels in GC. R-values and P-values were used via Pearson's correlation analysis. g The location of PMAN (red) was detected by FISH assay in AGS cells. The nucleus was stained with DAPI. Scale bar, 20 μm h RNA nuclear-cytoplasmic fractionation assay showed the expression level of PMAN in the subcellular fractions of AGS cells by RT-qPCR. U6 was treated as a nuclear control while β-actin was a cytoplasmic control. Mean ± SD is shown. i SLC7A11 was detected by western blotting in PMAN-overexpression MGC-803 cells after treated with CHX (10 μM). j-kSLC7A11 mRNA decay line chart of PMAN-overexpression MGC-803 cells (j) and PMAN knockdown AGS cells (k) after treated with actinomycin D (5 μg/ml). Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. Ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
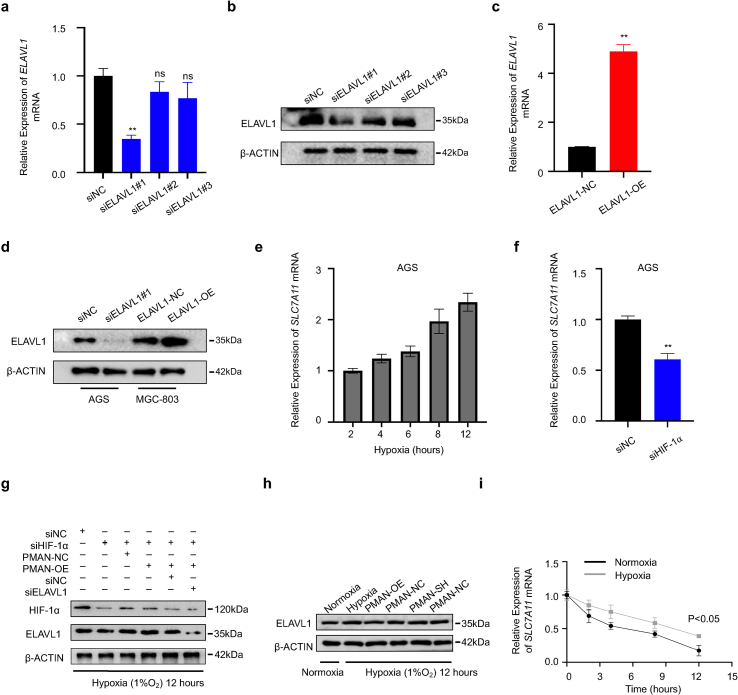
3.5. PMAN promoted ELAVL1 cytoplasmic translocation by direct bound to ELAVL1
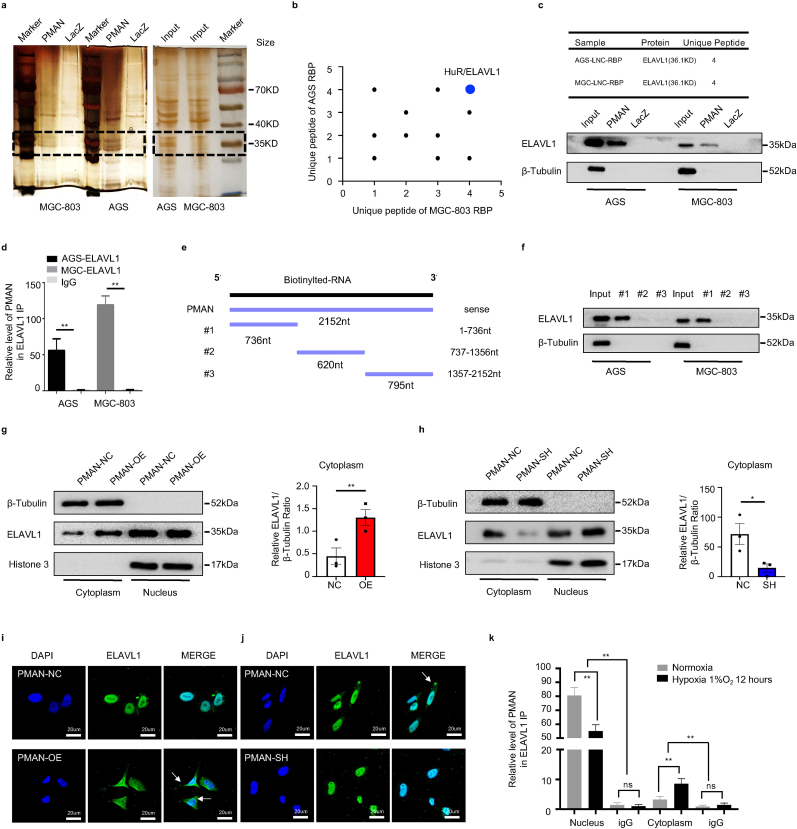
Next, we investigated how PMAN in the nucleus regulates the mRNA stability of SLC7A11 in the cytoplasm. Studies [21,27] prompted us that lncRNAs usually exert their regulatory function by binding to proteins (RBPs); therefore, we reasoned that there was a major protein between PMAN and SLC7A11 mRNA. To probe the potential RBPs, RNA pull-down/silver staining was performed, and we found that the protein band present in PMAN was located at approximately 37 kD (Fig. 5.a). Subsequently, mass spectrometry was performed, and ELAVL1 caught our attention because numerous ELAVL1 binding sites (AREs GUUU/AUUU) were found in the PMAN (RBPDB http://rbpdb.ccbr.utoronto.ca/) (SupplementaryTab.4), which was confirmed by WB (Fig. 5.b-c). To further strengthen the above results, we performed RNA immunoprecipitation (RIP) and found significant PMAN enrichment in the anti-ELAVL1 IP compared with the anti-IgG IP (>50-fold) (Fig. 5.d). Next, we constructed three different PMAN deletion fragments based on the distribution of ELAVL1 binding sites (Fig. 5.e, SupplementaryTab.5), and RNA pull-down assay was performed. The results demonstrated that ELAVL1 interacted with fragment #1 of PMAN rather than with other fragments (Fig. 5.f). We then measured the expression of ELAVL1 by WB and found that neither PMAN-SH nor PMAN-OE altered ELAVL1 expression (SupplementaryFig2.i). Notably, ELAVL1 is an important RNA-binding protein that regulates mRNA stability by cytoplasmic translocation [28,29]; therefore, we reasoned that ELAVL1 cytoplasmic translocation was associated with PMAN. First, we extracted the nuclear and cytoplasmic proteins from PMAN-OE MGC-803 and PMAN-SH AGS cells. As expected, PMAN-OE showed that the enrichment of ELAVL1 increased in the cytoplasm, and PMAN-SH showed the opposite result (Fig. 5.g-h). The immunofluorescence (IF) results also supported the above conclusion (Fig. 5.i-j). Furthermore, we isolated the nuclear and cytoplasmic contents of the MGC-803 cells. Under hypoxic and normoxic conditions, the results of the RIP experiment showed that the direct binding of PMAN and ELAVL1 was mostly located in the nucleus. Moreover, compared with IgG, the enrichment level of PMAN in the nucleus was reduced under hypoxia (∼55-fold) compared with normoxia (>80-fold), which we speculate is related to the promotion of ELAVL1 cytoplasmic translocation by hypoxia (Fig. 5.k). Based on this, we confirmed that PMAN could induce ELAVL1 cytoplasmic translocation by direct binding.
Fig. 5.
PMAN promotes ELAVL1 cytoplasmic translocation by direct bound to ELAVL1a RNA pull‐down proteins were precipitated in AGS cells and MGC-803 cells were detected by silver staining. b Proteins identification by LC-MS/MS in pulldowns of biotinylated PMAN from two independent experiences, two technical replicates. c Western blotting analysis of PMAN-binding protein ELAVL1 in AGS cells and MGC-803 cells. LacZ was treated as a negative control d RIP assay was performed with anti-ELAVL1 antibodies and control antibodies (IgG) in AGS cells and MGC-803 cells. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. e Three different PMAN deletion fragments were constructed based on the distribution of ELAVL1 specific binding sequence (GUUU). f The RNA binding protein ELAVL1 in three different PMAN deletion fragments were detected by western blotting in AGS cells and MGC-803 cells. g-h Protein nuclear-cytoplasmic fractionation assay showed the expression level of ELAVL1 in the subcellular fractions of PMAN-overexpression MGC-803 cells (g) and PMAN knockdown AGS cells (h) by western blotting and protein quantification on ImageJ. Histone H3 was treated as a nuclear control while β-tubulin was a cytoplasmic control. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. i-j IF showed ELAVL1 cytoplasmic fluorescence intensity (green) in PMAN-overexpression MGC-803 cells (i) and PMAN knockdown AGS cells (j). Scale bar, 20 μm k RIP assay was performed with anti-ELAVL1 antibodies and control antibodies (IgG) in cytoplasmic and nuclear components of MGC-803 cells during hypoxia and normoxia. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. Ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. ELAVL1 was the key RBP in the regulation of SLC7A11 mRNA stability by PMAN
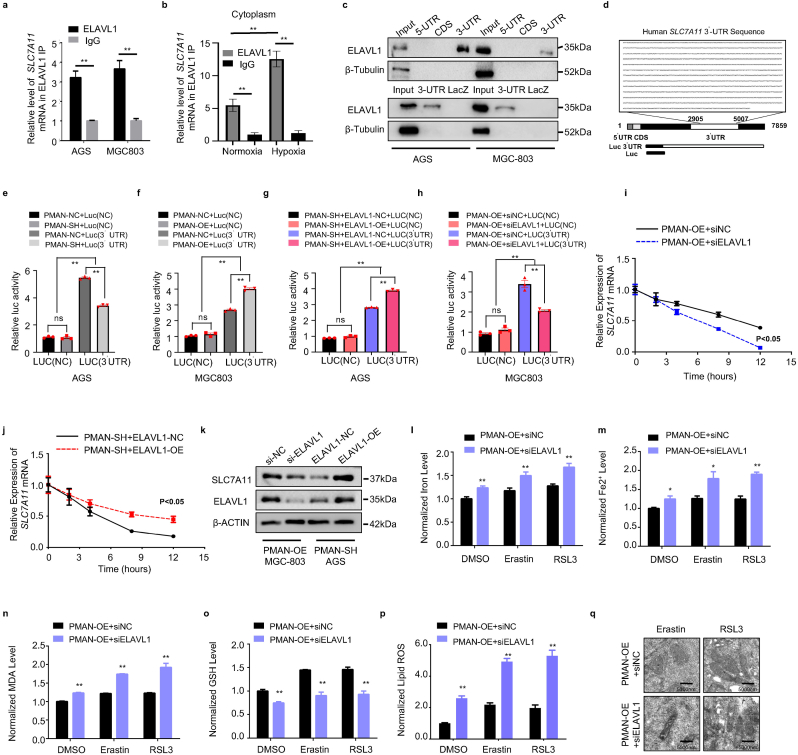
We verified that the increased cytoplasmic translocation of ELAVL1 was the key intermediate protein for PMAN-regulating SLC7A11 mRNA. RIP was performed and results showed a remarkable SLC7A11 mRNA enrichment in the anti-ELAVL1 IP compared with anti-IgG IP (>5-fold) (Fig. 6.a), in addition, by extracting the cytoplasmic components of MGC-803 cells treated with hypoxia or normoxia, we found that hypoxia increased the enrichment level of SLC7A11 mRNA in the cytoplasm ELAVL1 IP compared to normoxia by RIP results (Fig. 6.b). ELAVL1 is known to regulate mRNA stability of target genes by directly binding to AREs in the 3′-UTR [30]. Therefore, we speculated that ELAVL1 increases SLC7A11 mRNA stability by directly binding to AREs. As expected, a wide distribution of AREs was found in the SLC7A11 mRNA 3′-UTR (SupplementaryTab.6). To verify the results from RBPDB (http://rbpdb.ccbr.utoronto.ca/), proteins pulled down by SLC7A11 mRNA 5′-UTR/CDS/3′-UTR/LacZ specific probes were analyzed by WB. The results confirmed that SLC7A11 mRNA 3′-UTR was significantly enriched in ELAVL1 (Fig. 6.c). Then, we transfected the luciferase reporter plasmid containing the SLC7A11 mRNA 3′-UTR (Fig. 6.d) (SupplementaryTab.7) into cells and found that PMAN-OE increased luciferase activity, while PMAN-SH repressed luciferase activity (Fig. 6.e-f). In addition, siELAVL1 suppressed the luciferase activity induced by PMAN-OE, and ELAVL1-OE showed significantly improved luciferase activity, which was inhibited by PMAN-SH (Fig. 6.g-h); siELAVL1 or ELAVL1-OE efficiency was estimated by RT-qPCR and WB, wherein siELAVL1#1 was selected to complete the follow-up experiments (SupplementaryFig3.a-d). Next, an AD assay was performed and the results revealed that siELAVL1 in PMAN-OE MGC-803 cells shortened the half-life of SLC7A11 mRNA. Similarly, ELAVL1-OE in PMAN-SH AGS cells increased the half-life of SLC7A11 mRNA (Fig. 6.i-j). In addition, WB results indicated that ELAVL1 markedly affected SLC7A11 (Fig. 6.k). siELAVL1 in PMAN-OE MGC-803 cells significantly increased FE, FE2+, lipid ROS, and MDA levels, while the intracellular GSH level was also reduced, and mitochondria were shrunken in TEM (Fig. 6.l-q). We demonstrated that ELAVL1 could directly bind to the AREs of SLC7A11 mRNA 3′-UTR, thereby improving the mRNA stability and expression of SLC7A11.
Fig. 6.
PMAN improves SLC7A11 mRNA stability through ELAVL1.a RIP assay was performed with anti-ELAVL1 antibodies and control antibodies (IgG) in AGS cells and MGC-803 cells. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. b RIP assay was performed with anti-ELAVL1 antibodies and control antibodies (IgG) in cytoplasmic components of MGC-803 cells during normoxia and hypoxia. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. c RNA pulldown assay was performed with biotinylated SLC7A11 mRNA 5′-UTR CDS 3′-UTR in AGS cells and MGC-803 cells. LacZ was treated as a negative control. d Sequence of human SLC7A11 mRNA 3′UTR from UCSC database. e-f Relative luciferase activity of reporter plasmids containing SLC7A11 mRNA 3′UTR in stably PMAN knockdown AGS cells (e) and PMAN-overexpression MGC-803 cells (f) The luciferase activity was measured and normalized. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. g-h Relative luciferase activity of reporter plasmids containing SLC7A11 mRNA 3′UTR in stably PMAN knockdown AGS cells co-transfected with ELAVL1-over expression or negative control (NC) (g) and PMAN-overexpression MGC-803 cells co-transfected with siELAVL1 or negative control (siNC) (h).The luciferase activity was measured and normalized. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. i-jSLC7A11 mRNA decay line chart of PMAN-overexpression MGC-803 cells co-transfected with siELAVL1 or negative control (siNC) (i) and PMAN knockdown AGS cells co-transfected with ELAVL1-over expression or negative control (NC) (j) after treated with actinomycin D (5 μg/ml). Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. k SLC7A11 expression in PMAN-overexpression MGC-803 cells co-transfected with siELAVL1 or negative control (siNC) and PMAN knockdown AGS cells co-transfected with ELAVL1-over expression or negative control (NC). l Level of total iron m level of ferrous iron n level of MDA o level of GSH p level of lipid ROS were detected in PMAN-overexpression MGC-803 cells co-transfected with siELAVL1 or negative control (siNC) under Erastin (10 μM) or RSL3 (2 μM) treatment after 12 h. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. q Transmission electron microscopy was performed to evaluate the change of mitochondrial ultrastructure in PMAN-overexpression MGC-803 cells co-transfected with siELAVL1 or negative control (siNC) under Erastin (10 μM) or RSL3 (2 μM) treatment after 12 h. Scale bar, 5000 nm. Ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01.
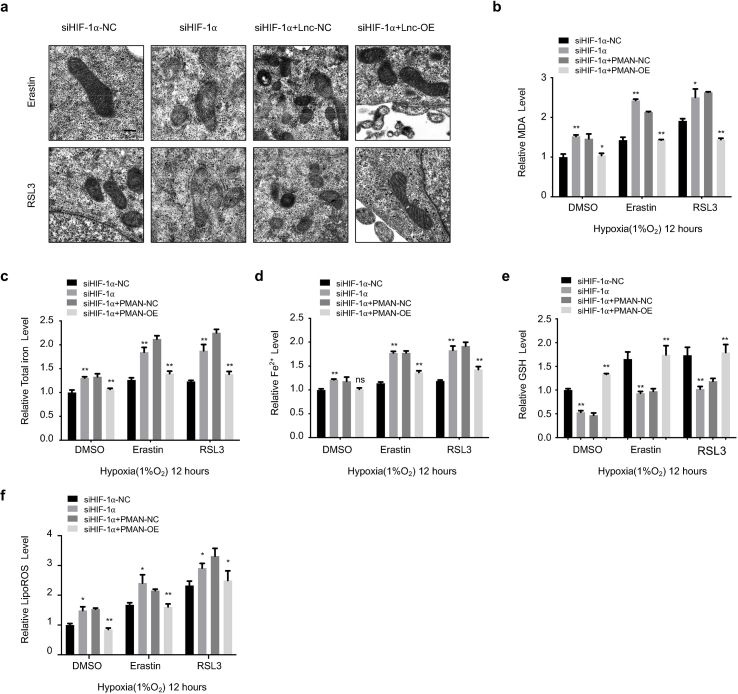
3.7. PMAN was the key lncRNA for HIF-1α to regulate ELAVL1 cytoplasmic translocation and expression of SLC7A11 in hypoxia
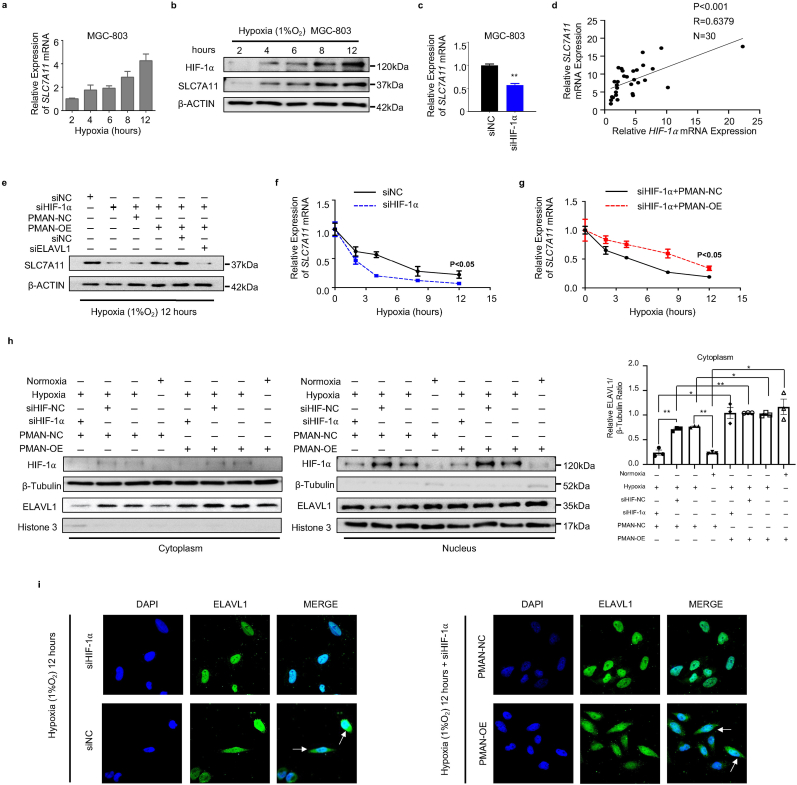
MGC-803 and AGS cells were cultured under hypoxic conditions in vitro at various time points. RT-qPCR and WB results indicated that HIF-1α increased SLC7A11 expression (Fig. 7.a-b, SupplementaryFig3.e). In addition, siHIF-1α markedly decreased SLC7A11 mRNA levels in GC cells (Fig. 7.c, SupplementaryFig3.f). Additionally, the correlation between SLC7A11 mRNA and HIF-1α mRNA was calculated and indicated a positive correlation in the tumors (R = 0.6379) (Fig. 7.d). To further validate that HIF-1α regulates SLC7A11 expression by activating PMAN during hypoxia, WB showed that PMAN restored SLC7A11 expression, which was decreased by siHIF-1α. Meanwhile, siELAVL1 significantly suppressed SLC7A11 expression, which was restored by PMAN (Fig. 7.e SupplementaryFig3.g). When cells were exposed to AD in hypoxia and normoxia, the results showed that hypoxia increased SLC7A11 mRNA stability, siHIF-1α under hypoxia accelerated the decay rate of SLC7A11 mRNA, and PMAN-OE alleviated the decay rate (Fig. 7.f-g SupplementaryFig3.i). Thus, we showed that the regulation of SLC7A11 mRNA by HIF-1α was dependent on PMAN/ELAVL1. Next, to explore the regulatory mechanism of HIF-1α in ELAVL1 cytoplasmic translocation, we confirmed that PMAN-OE/SH under hypoxia did not change the expression of ELAVL1 (SupplementaryFig3.h). Notably, WB results confirmed that hypoxia increased the cytoplasmic level of ELAVL1, which was dependent on HIF-1α, and PMAN-OE inverted levels of ELAVL1 in the cytoplasm after siHIF-1α (Fig. 7.h). As expected, the IF results verified the above conclusion (Fig. 7.i). Finally, we found that siHIF-1α reduced GSH levels and increased the concentrations of MDA, FE, FE2+, and lipid ROS in MGC-803 cells under hypoxia. However, PMAN-OE treatment reversed these results. Moreover, mitochondrial alterations supported the conclusions of our study (SupplementaryFig4.a-f). In summary, we demonstrated that HIF-1α/PMAN enhances ferroptosis resistance in gastric cancer cells by promoting cytoplasmic translocation of ELAVL1 and expression of SLC7A11 under hypoxia.
Fig. 7.
HIF-1α to regulate ELAVL1 cytoplasmic translocation and the mRNA stability of SLC7A11 in hypoxic environment by PMAN.a MGC-803 cells were cultured in hypoxia, RT-qPCR analyzes the expression level of SLC7A11 mRNA at indicated timepoints. Mean ± SD is shown. b MGC-803 cells were cultured in hypoxia, western blotting analyzes the expression level of SLC7A11 at indicated timepoints. c RT-qPCR detects the expression level of SLC7A11 mRNA in siHIF-1α-transfected MGC-803 cells. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. d Correlations between HIF-1α mRNA levels and SLC7A11 mRNA levels in GC. R-values and P-values were used via Pearson's correlation analysis. e The expression of SLC7A11 in siHIF-1⍺ or siNC transfected MGC-803 cells, PMAN-overexpression MGC-803 cells co-transfected with siHIF-1⍺ or siNC and transfected siHIF-1⍺ or siNC in PMAN-overexpression MGC-803 cells co-transfected with siELAVL1 or siNC after 24 h under hypoxia. fSLC7A11 mRNA decay line chart in MGC-803 cells transfected with siHIF-1⍺ or siNC during hypoxia. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. gSLC7A11 mRNA decay line chart of PMAN-overexpression MGC-803 cells co-transfected with siHIF-1⍺ or siNC during hypoxia. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. h Protein nuclear-cytoplasmic fractionation assay showed the expression level of ELAVL1 in the subcellular fractions of PMAN-NC MGC-803 cells under hypoxia or normoxia and PMAN-NC MGC-803 cells transfected with siHIF-1⍺ or siNC under hypoxia by western blotting. Similarly, the expression level of ELAVL1 in the subcellular fractions of PMAN-overexpression MGC-803 cells under hypoxia or normoxia and PMAN-overexpression MGC-803 cells transfected with siHIF-1⍺ or siNC under hypoxia. Protein quantification on ImageJ and Histone H3 was treated as a nuclear control while β-tubulin was a cytoplasmic control. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. i IF showed ELAVL1 cytoplasmic fluorescence intensity (green) in transfected siHIF-1⍺ or siNC MGC-803 cells and PMAN-overexpression or PMAN-NC MGC-803 cells transfected with siHIF-1⍺. The nucleus was stained with DAPI. Scale bar, 20 μm. Ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
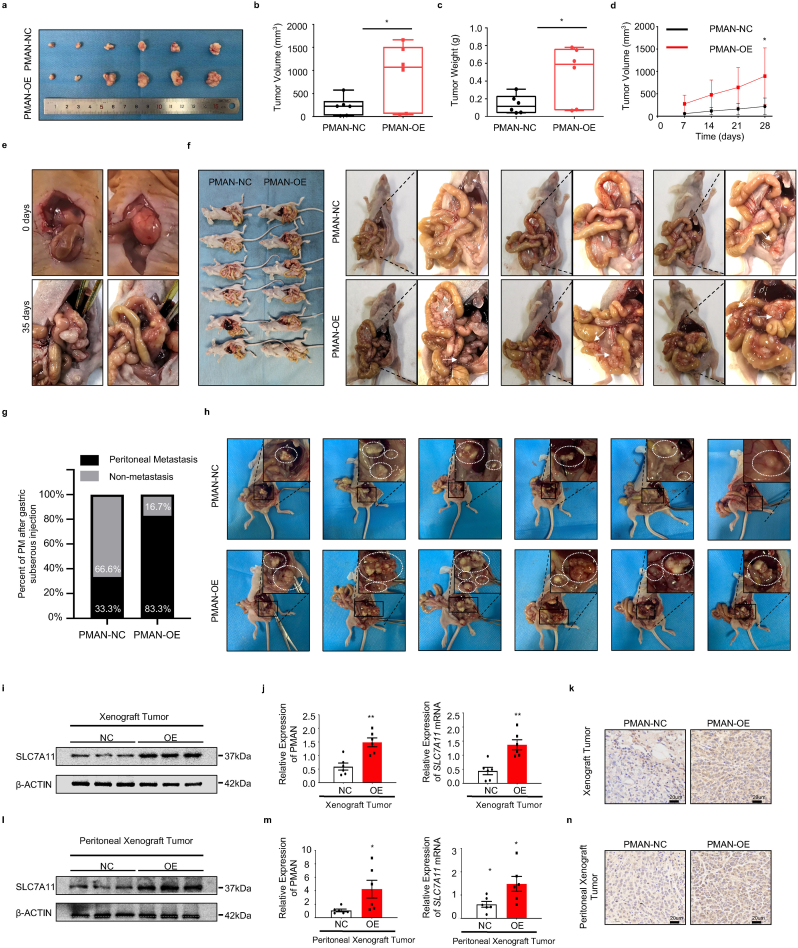
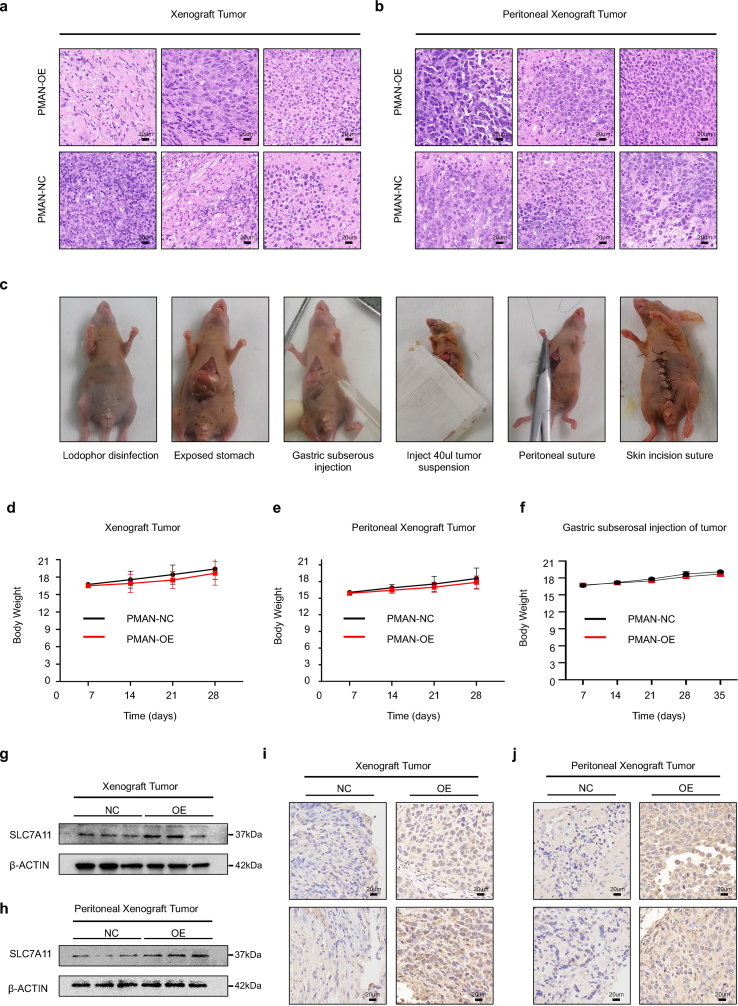
3.8. PMAN promoted the occurrence and development of tumors in vivo
To validate the role of PMAN in tumor progression and PM promotion in vivo, we constructed stable PMAN-OE/NC in MGC-803 cells (1 × 10 [7]cells), subcutaneously or intraperitoneally injected into each flank of 4-week-old BALB/c nude female mice (n = 6). The volume of the subcutaneous tumor and the body weight were recorded (SupplementaryFig5.d-e). After four weeks, we found that the size and volume of subcutaneous xenograft tumors and abdominal disseminated tumors in the PMAN-OE group increased significantly compared to those in the NC group (Fig. 8.a-d,h). The results of RT‐qPCR, WB, and IHC suggested that PMAN-OE markedly increased SLC7A11 levels in vivo (Fig. 8.i-n, SupplementaryFig5.g-j). To further demonstrate that PMAN can promote gastric cancer peritoneal metastasis, we collected and suspended MGC-803 cells (5 × 10 [6]cells) with stable PMAN-OE/NC in 40 μl of serum-free medium (50% Matrigel), subserosal injection at the greater curvature of the stomach (Fig. 8.e, SupplementaryFig5.c), and recorded the body weight changes in nude mice every week (SupplementaryFig5.f). After 35 days, the mice were sacrificed (n = 6), and we found that the PMAN-OE group not only had a higher incidence of intraperitoneal metastases, but also a wider range of dissemination (Fig. 8.f-g). Finally, hematoxylin and eosin (H&E) staining was performed to evaluate mouse tumor tissue morphology evaluation (SupplementaryFig5.a-b). Taken together, we demonstrated that PMAN promotes the occurrence and development of tumors in vivo.
Fig. 8.
PMAN promotes tumor occurrence and development in vivo.a-d Subcutaneous tumors of nude mice injected with (1 × 10 [7] cells) PMAN-overexpression or PMAN-NC MGC-803 cells after 4 weeks. Tumor image (a) was monitored, volumes (b), weights (c) and weekly volume (d) were recorded (n = 6). Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. e After subserosal injection in nude mice, the stomach of day 0 and day 35 was monitored f 5 weeks after subserosal injection of the greater curvature of the stomach of nude mice (n = 6), the abdominal cavity was opened to observe intraperitoneal metastases g The proportion of peritoneal metastases in the PMAN-OE group and PMAN-NC group after 5 weeks of subserosal injection in the greater curvature of the stomach in nude mice (n = 6). h Intraperitoneal injection of tumor of nude mice injected with (1 × 10 [7] cells) PMAN-overexpression or PMAN-NC MGC-803 cells after four weeks. Intraperitoneal tumor image (h) was monitored. (n = 6). i-k The expression of SLC7A11 in subcutaneous tumors from PMAN-overexpression group and PMAN-NC group. SLC7A11 level (i), PMAN and SLC7A11 mRNA level (j), immunohistochemistry (k). Scale bar, 20 μm. l-n The expression of SLC7A11 in intraperitoneal tumors from PMAN-overexpression group and PMAN-NC group. SLC7A11 level (l), PMAN and SLC7A11 mRNA level (m) and immunohistochemistry (n). Scale bar, 20 μm. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. Ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01.
4. Discussion
Hypoxia is a key feature of the tumor microenvironment, and tumor cells must activate the HIF-signaling pathway for adaption to hypoxia [31,32]. In this study, we determined that high HIF-1α expression in PM tissues may be caused by low partial pressure of oxygen (pO2 of 40–50 mmHg) in the abdominal cavity [33]. HIF-1α, a master transcriptional regulator of hypoxia, participates in tumorigenesis and angiogenesis by regulating the activation of hypoxia-related genes [34,35]. Regrettably, owing to technical limitations, our study lacked ferroptosis detection in PM, which is one of the shortcomings of our study. This is also a direction worthy of further exploration. The identification of specific markers of ferroptosis in clinical specimens, such as apoptosis, will help us to further apply ferroptosis in tumor treatment and efficacy evaluation. Here, PMAN was confirmed to be activated by HIF-1α and highly expressed in the PM. HIF-1α/PMAN improves the survival of tumor cells during ferroptosis.
Ferroptosis is a novel form of regulated cell death. Accumulation of lipid peroxidation and intracellular FE/FE2+ are key determinants of ferroptosis [36]. Our findings support that PMAN has the ability to alleviate ferroptosis in tumor cells, and HIF-1α is shown to be a key protective factor for tumor cells to resist ferroptosis under hypoxia. Furthermore, we found that the potential molecular mechanism of PMAN ferroptosis in GC cells is mediated by SLC7A11. The xCT system, which are heterodimers comprising the heavy subunit SLC3A2 and light subunit SLC7A11, imports cystine for GSH biosynthesis in GC cells and defends against damage caused by oxidative stress [37,38]. Owing to its unique function SLC7A11 has been widely confirmed as an important target for inducing ferroptosis [39]. Here, we demonstrated that HIF-1α/PMAN increases the expression of SLC7A11 at the post-transcriptional level by promoting the stability of SLC7A11 mRNA, thereby inhibiting ferroptosis.
RBPs are critical regulators of gene expression that affect the localization, translation, and stability of target genes [40,41]. We identified ELAVL1/HuR as the key RBP in the regulation of SLC7A11 expression by PMAN. Abnormal ELAVL1 directly binds to AREs in the mRNA 3′-UTR, which improves the mRNA translation of target genes. Involvement of ELAVL1 in various malignancies has been widely shown [42]. Our results demonstrate that ELAVL1 improves the stability of SLC7A11 mRNA, thereby enhancing its protein expression. In general, the functional activity of ELAVL1 is regulated by dynamic subcellular localization [28]. Under normal physiological conditions, ELAVL1 is mainly located in the nucleus, but when subjected to internal and/or external stimuli, ELAVL1 can be transferred to the cytoplasm, where it stabilizes and increases the translation of target mRNA. Changes in ELAVL1 expression or subcellular localization may affect its function [43]. Our results showed that PMAN directly binds to ELAVL1 and causes an obvious increase in cytoplasmic ELAVL1 expression. Unfortunately, the molecular mechanisms by which PMAN affects ELAVL1 subcellular localization are not fully understood. The reason is that we tried to synthesize wild/mut-type lncRNA probes for PMAN-AREs, however, AREs have a considerable amount of distribution on PMAN and SLC7A11 mRNA and if the mutated AREs are greater than 50% it may lead to changes in the structure of PMAN and lose their meaning. On the contrary, if a small number of AREs are mutated, the experimental results show that there is no significant difference compared with wild type. We speculate that PMAN may act as a scaffold during cytoplasmic distribution of ELAVL1 or PMAN may regulate ELAVL1 subcellular localization by posttranslational modification of crystal structures such as RRM1, RRM2, and HNS [44,45].
To date, there is still no effective treatment for PM in GC, and the 5-year survival rate of patients with PM in GC remains low [46]. Inducing ferroptosis in tumor cells by inhibiting SLC7A11 or GPX4 is an emerging concept in tumor therapy [15,47]. Our data showed that HIF-1α/PMAN makes gastric cancer cells more resistant to ferroptosis induced by Erastin or RSL3 and may be closely related to the ratio of ELAVL1 nuclear and cytoplasmic distribution. This suggests that targeting HIF-1α/PMAN/ELAVL1 may be the Achilles heel of cancer cells and might provide theoretical support for the development of drugs related to tumor ferroptosis.
5. Conclusion
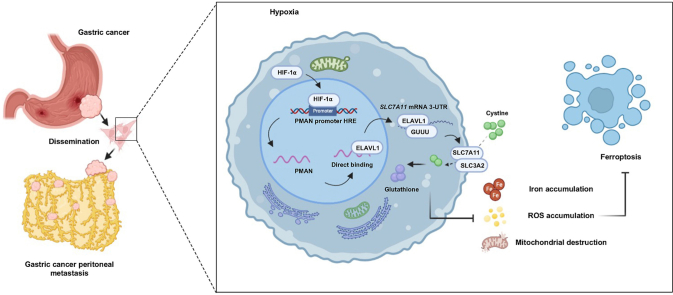
In summary, our current findings reveal that under hypoxia, HIF-1α could act as a protective factor against ferroptosis in GC cells. HIF-1α activates PMAN at the transcriptional level, which greatly improves the output of ELAVL1 in the cytoplasm. ELAVL1 directly combines with the AREs of SLC7A11 mRNA 3′-UTR and improves the stability of SLC7A11 mRNA, thereby increasing the expression of SLC7A11 and reducing the accumulation of ROS and iron in ferroptosis, ultimately promoting the proliferation and development of tumor cells (Fig. 9).
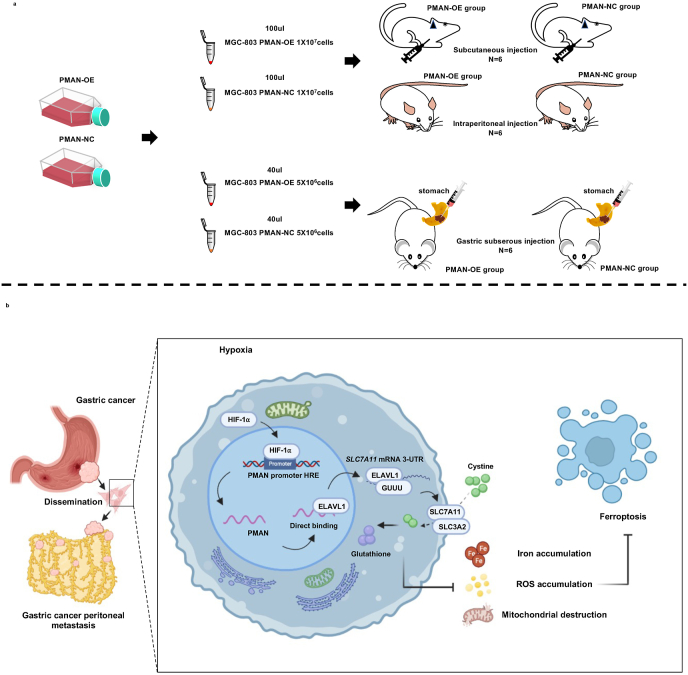
Fig. 9.
Schematic diagrams of mechanism diagram and animal experiments.a We collected MGC-803 cells (1 × 10 [7] cells) that were infected by lentivirus with or without PMAN overexpression (six mice per group).Then, suspended in 100 μl serum-free medium and subcutaneously injected and intraperitoneal injected into each flank of 4-week-old BALB/c nude female mice. For the construction of intraperitoneal metastatic orthotopic tumors, we collected MGC-803 cells (5 × 10 [6] cells) that were infected by lentivirus with or without PMAN overexpression (six mice per group).Then, suspended in 40 μl serum-free medium (50% Matrigel) and gastric subserosal injection into each flank of 4-week-old BALB/c nude female mice.b Schematic diagram of PMAN regulating ferroptosis through SLC7A11 during hypoxia. The expression of HIF-1⍺ is up-regulated under hypoxia and as a transcription factor to activates the expression of PMAN by combining with the promoter sequence HRE of PMAN. PMAN directly binds to ELAVL1 and improves the cytoplasmic translocation of ELAVL1. ELAVL1 directly binds to the (GUUU) site of the mRNA of SLC7A11 to improve the stability of mRNA and enhance the expression of SLC7A11. Tumor cells can transport Cys (cystine) into the cell and reduce it to cysteine through the overexpression of SLC7A11, and participate in the synthesis of glutathione (GSH). Reducing the accumulation of ROS and iron in tumor cells and the destruction of mitochondria. Thereby inhibit the occurrence of ferroptosis and promote the growth of tumor cells.
Ethics approval and consent to participate
All experimental protocols were approved by the Ethics Committee of Wuhan University School of Medicine. Paired specimens of peritoneal metastases, gastric cancer, and adjacent normal tissues were collected from patients undergoing surgical resection with informed consent from all subjects. All patients did not receive anticancer therapy before surgery. Animal experiments were approved by the Animal Experiment Center of Wuhan University.
Consent for publication
All authors agree to submit the article for publication.
Availability of data and materials
The datasets analyzed during the current study are temporarily unpublished for research reasons, but are available from the respective authors upon reasonable request.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81872376) and Translational Medicine and Interdisciplinary Research of Zhongnan Hospital of Wuhan University (ZNJC201913).
Authors’ contributions
ZL, JS and YG are the experimental designer and executor of the experimental research of this study, completes data analysis, writing the first draft of the paper; SH, RD, PZ and GH participates in experimental design and analysis of experimental results; LH, JZ and XZ participates in experimental design, data analysis, thesis writing and revision. BX and SW conceived the project and supervised research. All authors read and approved the final manuscript.
Declaration of competing interest
No competing interests.
Acknowledgements
The authors also thank the Medical Science Research Center of Zhongnan Hospital of Wuhan University for providing equipment and excellent technical support for the experiments, and we would like to thank Editage (www.editage.cn) for English language editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102312.
Contributor Information
Shuyi Wang, Email: shuyiwang@whu.edu.cn.
Bin Xiong, Email: binxiong1961@whu.edu.cn.
Abbreviations
- PM
Peritoneal metastasis
- GC
Gastric cancer
- ANT
Adjacent normal tissues
- siRNA
Small interfering RNA
- Fe
Total iron
- Fe2+
Ferrous iron
- MDA
Malondialdehyde
- lipid ROS
Lipid reactive oxygen species
- GSH
glutathione
- GSSG
l-Glutathione
- lncRNA
Long non-coding RNA
- IHC
Immunohistochemistry
- IF
immunofluorescence
- FISH
Fluorescence in situ hybridization
- HRE
Hypoxia-response element
- AREs
AU-rich elements
- TEM
Transmission electron microscopy
- AD
Actinomycin D
- CHX
Cycloheximide;
- RIP
Immunoprecipitation
- ChIP
Chromatin immunoprecipitation
- H&E
Hematoxylin and eosin
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
Supplementary Fig. 1; The knockdown efficiency of siHIF-1⍺ and the expression of PMAN in gastric cancer peritoneal metastases.a,b The expression level of HIF-1α mRNA was detected by RT-qPCR (a) and protein was analyzed by western blotting (b) in AGS cells transfected with siHIF-1⍺#1, siHIF-1⍺#2 or siNC during hypoxia. c Ranking of the top ten lncRNA genes associated with HIF-1α, expression as assessed by lncRNA microarray analysis and lncRNA-seq results. d RT-qPCR detects the expression of PMAN in AGS cells transfected with siHIF-1⍺#1, siHIF-1⍺#2 or siNC during hypoxia. According to the results,siHIF-1⍺#1 was selected for follow-up experiments. e Kaplan-Meier curves for overall survival rates correlated with HIF-1α expression in STAD from GEPIA database. f RT-qPCR detects the expression of PMAN in GC cells. According to the expression level of PMAN and cell state, AGS and MGC-803 cells were selected for follow-up experiments. g The expression of PMAN after transfected with shPMAN#1, shPMAN#2, shPMAN#3 or shNC were detected by RT-qPCR. According to the results shPMAN#2 was selected for follow-up experiments. h Fish showed the expression of PMAN (red) in 3 pairs GC peritoneal metastasis samples. The nucleus was stained with DAPI. Scale bar, 20 μm i Immunohistochemistry was used to analyze up-regulated HIF-1α expressions in paired GC peritoneal metastasis samples. Scale bar, 20 μm j ISH showed the expression of PMAN in 3 pairs GC peritoneal metastasis samples. Scale bar, 20 μm. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. Ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01.
figs2.
Supplementary Fig. 2; l Ferroptosis-related mRNA expression and PMAN have no effect on SLC7A11 promoter activity in the nucleus.a-b The ratio of necrotic cells (Propidium Iodide), apoptotic cells (Hoechest) and normal cells was measured by flow cytometry for PMAN-OE MGC-803 cells (a) and PMAN-SH AGS cells (b) under Erastin (10 μM) or RSL3 (2 μM) treatment after 12 h. Quantify the proportion of normal cells. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. c-d The mRNA expression level of ferroptosis-related genes in the PMAN knockdown AGS cells (c) and the PMAN overexpression MGC-803 cells (d). Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. e The location of PMAN (red) was detected by FISH assay in MGC-803 cells. The nucleus was stained with DAPI. Scale bar, 20 μm f RNA nuclear-cytoplasmic fractionation assay showed the expression level of PMAN in the subcellular fractions of MGC-803 cells by RT-qPCR. U6 was treated as a nuclear control while β-actin was a cytoplasmic control. Mean ± SD is shown. g-h PMAN knockdown AGS cells was transfected with luciferase reporter plasmids containing SLC7A11 promoter or NC (g) and PMAN overexpression MGC-803 cells was transfected with luciferase reporter plasmids containing SLC7A11 promoter or NC (h). The luciferase activity was measured and normalized. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. i ELAVL1 expression in PMAN knockdown AGS cells and PMAN overexpression MGC-803 cells. Ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01.
figs3.
Supplementary Fig. 3; The efficiency of siELAVL1/OE-ELAVL1 and the expression of SLC7A11 under hypoxia.a-b The expression of ELAVL1 mRNA in AGS cells after transfected with siELAVL1#1,siELAVL1#2,siELAVL1#3 or siNC. The ELAVL1 mRNA were detected by RT-qPCR (a) and protein were analyzed by western blotting (b). According to the results siELAVL1#1 was selected for follow-up experiments. c The expression of ELAVL1 mRNA in MGC-803 cells after transfected with ELAVL1-overexpression or ELAVL1-NC. d Western blotting showed the SLC7A11 expression in AGS cells after transfected with siELAVL1 or siNC and MGC-803 cells after transfected with ELAVL1-overexpression or ELAVL1-NC. e AGS cells were cultured in hypoxia, RT-qPCR analyzes the expression level of SLC7A11 mRNA at indicated timepoints. f RT-qPCR detects the expression level of SLC7A11 mRNA in siHIF-1α-transfected AGS cells. g The HIF-1α and ELAVL1 expression were detected by western blotting. h The ELAVL1 expression in hypoxia or normoxia and PMAN overexpression/NC MGC-803 cells and PMAN knockdown/NC AGS cells. iSLC7A11 mRNA decay line chart in MGC-803 cells under hypoxia and normoxia. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. Ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01.
figs4.
Supplementary Fig. 4; Si–HIF-1α increases the sensitivity of GC cells to ferroptosis, and PMAN could rescue this situation.a-f Ultrastructure of cell mitochondria. Scale bar, 5000 nm (a), level of MDA (b), level of total iron (c), level of ferrous iron (d), level of GSH (e) and level of lipid ROS were analyzed in MGC-803 cells transfected with siHIF-1α/siNC or PMAN overexpression/NC MGC-803 cells co-transfected with siHIF-1⍺ after treated with Erastin (10 μM) or RSL3 (2 μM) under hypoxia (1%O2,12 h). Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. Ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01.
figs5.
Supplementary Fig. 5; PMAN promotes tumor occurrence and development in vivo.a-b (H&E) staining was performed for tissue from subcutaneous tumors (a) and intraperitoneal tumors in nude mice (b). Scale bar, 20 μm c Process for establishing an animal model of peritoneal metastasis in nude mice by gastric subserosal injection. d-f The body weight of nude mice in the subcutaneous tumor group (d) and intraperitoneal tumor group (e) and gastric subserous injection group (f) were recorded once a week for 4 or 5 weeks (n = 6). Mean ± SD is shown. g,i The expression of SLC7A11 in subcutaneous tumors from PMAN-overexpression group and PMAN-NC group. SLC7A11 level (g), immunohistochemistry (i). Scale bar, 20 μm h,j The expression of SLC7A11 in intraperitoneal tumors from PMAN-overexpression group and PMAN-NC group. SLC7A11 level (h), and immunohistochemistry (j). Scale bar, 20 μm.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Fujitani K., Yang H.K., Mizusawa J., Kim Y.W., Terashima M., Han S.U., et al. Gastrectomy plus chemotherapy versus chemotherapy alone for advanced gastric cancer with a single non-curable factor (REGATTA): a phase 3, randomised controlled trial. Lancet Oncol. 2016;17(3):309–318. doi: 10.1016/S1470-2045(15)00553-7. [DOI] [PubMed] [Google Scholar]
- 3.Choudhry H., Harris A.L. Advances in hypoxia-inducible factor biology. Cell Metabol. 2018;27(2):281–298. doi: 10.1016/j.cmet.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Kim K.S., Sengupta S., Berk M., Kwak Y.G., Escobar P.F., Belinson J., et al. Hypoxia enhances lysophosphatidic acid responsiveness in ovarian cancer cells and lysophosphatidic acid induces ovarian tumor metastasis in vivo. Cancer Res. 2006;66(16):7983–7990. doi: 10.1158/0008-5472.CAN-05-4381. [DOI] [PubMed] [Google Scholar]
- 5.Mikula-Pietrasik J., Uruski P., Tykarski A., Ksiazek K. The peritoneal "soil" for a cancerous "seed": a comprehensive review of the pathogenesis of intraperitoneal cancer metastases. Cell. Mol. Life Sci. 2018;75(3):509–525. doi: 10.1007/s00018-017-2663-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Natarajan S., Foreman K.M., Soriano M.I., Rossen N.S., Shehade H., Fregoso D.R., et al. Collagen remodeling in the hypoxic tumor-mesothelial niche promotes ovarian cancer metastasis. Cancer Res. 2019;79(9):2271–2284. doi: 10.1158/0008-5472.CAN-18-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun C., Li X., Guo E., Li N., Zhou B., Lu H., et al. MCP-1/CCR-2 axis in adipocytes and cancer cell respectively facilitates ovarian cancer peritoneal metastasis. Oncogene. 2020;39(8):1681–1695. doi: 10.1038/s41388-019-1090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Che X., Yu Y., Cheng Y., Bai M., Yang Z., et al. Hypoxia-autophagy axis induces VEGFA by peritoneal mesothelial cells to promote gastric cancer peritoneal metastasis through an integrin alpha5-fibronectin pathway. J. Exp. Clin. Cancer Res. 2020;39(1):221. doi: 10.1186/s13046-020-01703-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng D.W., Lei Q., Zhu J.Y., Fan J.X., Li C.X., Li C., et al. Switching apoptosis to ferroptosis: metal-organic network for high-efficiency anticancer therapy. Nano Lett. 2017;17(1):284–291. doi: 10.1021/acs.nanolett.6b04060. [DOI] [PubMed] [Google Scholar]
- 10.Li D., Li Y. The interaction between ferroptosis and lipid metabolism in cancer. Signal Transduct. Targeted Ther. 2020;5(1):108. doi: 10.1038/s41392-020-00216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah Y.M., Xie L. Hypoxia-inducible factors link iron homeostasis and erythropoiesis. Gastroenterology. 2014;146(3):630–642. doi: 10.1053/j.gastro.2013.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z., Jiang L., Chew S.H., Hirayama T., Sekido Y., Toyokuni S. Carbonic anhydrase 9 confers resistance to ferroptosis/apoptosis in malignant mesothelioma under hypoxia. Redox Biol. 2019;26:101297. doi: 10.1016/j.redox.2019.101297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu J., Li T., Yang Y., Jiang L., Wang W., Fu L., et al. Activatable nanomedicine for overcoming hypoxia-induced resistance to chemotherapy and inhibiting tumor growth by inducing collaborative apoptosis and ferroptosis in solid tumors. Biomaterials. 2021;268:120537. doi: 10.1016/j.biomaterials.2020.120537. [DOI] [PubMed] [Google Scholar]
- 14.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang C., Zhang X., Yang M., Dong X. Recent progress in ferroptosis inducers for cancer therapy. Adv. Mater. 2019;31(51) doi: 10.1002/adma.201904197. [DOI] [PubMed] [Google Scholar]
- 16.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei G., Zhang Y., Koppula P., Liu X., Zhang J., Lin S.H., et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020;30(2):146–162. doi: 10.1038/s41422-019-0263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T., Jiang L., Tavana O., Gu W. The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res. 2019;79(8):1913–1924. doi: 10.1158/0008-5472.CAN-18-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z., Chen X., Liu N., Shi Y., Liu Y., Ouyang L., et al. A nuclear long non-coding RNA LINC00618 accelerates ferroptosis in a manner dependent upon apoptosis. Mol. Ther. 2021;29(1):263–274. doi: 10.1016/j.ymthe.2020.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M., Mao C., Ouyang L., Liu Y., Lai W., Liu N., et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019;26(11):2329–2343. doi: 10.1038/s41418-019-0304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiyozumi Y., Iwatsuki M., Kurashige J., Ogata Y., Yamashita K., Koga Y., et al. PLOD2 as a potential regulator of peritoneal dissemination in gastric cancer. Int. J. Cancer. 2018;143(5):1202–1211. doi: 10.1002/ijc.31410. [DOI] [PubMed] [Google Scholar]
- 23.Liang Y., Song X., Li Y., Chen B., Zhao W., Wang L., et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol. Cancer. 2020;19(1):85. doi: 10.1186/s12943-020-01206-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Chen F., Chen J., Yang L., Liu J., Zhang X., Zhang Y., et al. Extracellular vesicle-packaged HIF-1αlpha-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat. Cell Biol. 2019;21(4):498–510. doi: 10.1038/s41556-019-0299-0. [DOI] [PubMed] [Google Scholar]
- 25.Fang Y., Huang S., Han L., Wang S., Xiong B. Comprehensive analysis of peritoneal metastasis sequencing data to identify LINC00924 as a prognostic biomarker in gastric cancer. Cancer Manag. Res. 2021;13:5599–5611. doi: 10.2147/CMAR.S318704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang M., Chen P., Liu J., Zhu S., Kroemer G., Klionsky D.J., et al. Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci. Adv. 2019;5(7) doi: 10.1126/sciadv.aaw2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H., Jiang Y., Xu X., Su X., Liu Y., Ma Y., et al. Inducible degradation of lncRNA Sros1 promotes IFN-gamma-mediated activation of innate immune responses by stabilizing Stat1 mRNA. Nat. Immunol. 2019;20(12):1621–1630. doi: 10.1038/s41590-019-0542-7. [DOI] [PubMed] [Google Scholar]
- 28.Xiao L., Li X.X., Chung H.K., Kalakonda S., Cai J.Z., Cao S., et al. RNA-binding protein HuR regulates paneth cell function by altering membrane localization of TLR2 via post-transcriptional control of CNPY3. Gastroenterology. 2019;157(3):731–743. doi: 10.1053/j.gastro.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berkovits B.D., Mayr C. Alternative 3' UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522(7556):363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lal S., Burkhart R.A., Beeharry N., Bhattacharjee V., Londin E.R., Cozzitorto J.A., et al. HuR posttranscriptionally regulates WEE1: implications for the DNA damage response in pancreatic cancer cells. Cancer Res. 2014;74(4):1128–1140. doi: 10.1158/0008-5472.CAN-13-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jing X., Yang F., Shao C., Wei K., Xie M., Shen H., et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer. 2019;18(1):157. doi: 10.1186/s12943-019-1089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilkes D.M., Semenza G.L., Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat. Rev. Cancer. 2014;14(6):430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renvall S., Niinikoski J. Kinetics of oxygen in peritoneal cavity. Effects of chemical peritonitis and intraperitoneally administered colloids in rats. J. Surg. Res. 1980;28(2):132–139. doi: 10.1016/0022-4804(80)90156-0. [DOI] [PubMed] [Google Scholar]
- 34.Semenza G.L. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Invest. 2013;123(9):3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semenza G.L. Pharmacologic targeting of hypoxia-inducible factors. Annu. Rev. Pharmacol. Toxicol. 2019;59:379–403. doi: 10.1146/annurev-pharmtox-010818-021637. [DOI] [PubMed] [Google Scholar]
- 36.Zheng J., Conrad M. The metabolic underpinnings of ferroptosis. Cell Metabol. 2020;32(6):920–937. doi: 10.1016/j.cmet.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 37.Koppula P., Zhuang L., Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2021;12(8):599–620. doi: 10.1007/s13238-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daher B., Parks S.K., Durivault J., Cormerais Y., Baidarjad H., Tambutte E., et al. Genetic ablation of the cystine transporter xCT in PDAC cells inhibits mTORC1, growth, survival, and tumor formation via nutrient and oxidative stresses. Cancer Res. 2019;79(15):3877–3890. doi: 10.1158/0008-5472.CAN-18-3855. [DOI] [PubMed] [Google Scholar]
- 39.Lang X., Green M.D., Wang W., Yu J., Choi J.E., Jiang L., et al. Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Cancer Discov. 2019;9(12):1673–1685. doi: 10.1158/2159-8290.CD-19-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatterji P., Rustgi A.K. RNA binding proteins in intestinal epithelial biology and colorectal cancer. Trends Mol. Med. 2018;24(5):490–506. doi: 10.1016/j.molmed.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jankowsky E., Harris M.E. Specificity and nonspecificity in RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 2015;16(9):533–544. doi: 10.1038/nrm4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siddiqui N., Borden K.L. mRNA export and cancer. Wiley Interdiscip. Rev. RNA. 2012;3(1):13–25. doi: 10.1002/wrna.101. [DOI] [PubMed] [Google Scholar]
- 43.Ibrahim H., Lee Y.J., Curthoys N.P. Renal response to metabolic acidosis: role of mRNA stabilization. Kidney Int. 2008;73(1):11–18. doi: 10.1038/sj.ki.5002581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan X.C., Steitz J.A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. U. S. A. 1998;95(26):15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brennan C.M., Gallouzi I.E., Steitz J.A. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 2000;151(1):1–14. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rawicz-Pruszynski K., van Sandick J.W., Mielko J., Cisel B., Polkowski W.P. Current challenges in gastric cancer surgery: European perspective. Surg. Oncol. 2018;27(4):650–656. doi: 10.1016/j.suronc.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 47.Mou Y., Wang J., Wu J., He D., Zhang C., Duan C., et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J. Hematol. Oncol. 2019;12(1):34. doi: 10.1186/s13045-019-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are temporarily unpublished for research reasons, but are available from the respective authors upon reasonable request.