Abstract
The contribution of seven known and nine predicted genes or operons associated with multidrug resistance to the susceptibility of Escherichia coli W3110 was assessed for 20 different classes of antimicrobial compounds that include antibiotics, antiseptics, detergents, and dyes. Strains were constructed with deletions for genes in the major facilitator superfamily, the resistance nodulation-cell division family, the small multidrug resistance family, the ATP-binding cassette family, and outer membrane factors. The agar dilution MICs of 35 compounds were determined for strains with deletions for multidrug resistance (MDR) pumps. Deletions in acrAB or tolC resulted in increased susceptibilities to the majority of compounds tested. The remaining MDR pump gene deletions resulted in increased susceptibilities to far fewer compounds. The results identify which MDR pumps contribute to intrinsic resistance under the conditions tested and supply practical information useful for designing sensitive assay strains for cell-based screening of antibacterial compounds.
Bacterial membrane transport systems function to take up essential nutrients, control cell homeostasis, export proteins, and control efflux of xenobiotic (including antibiotic) compounds (32). There are more than seven efflux systems in Escherichia coli that can export structurally unrelated antibiotics; these multidrug resistance efflux pump (MDR pump) systems contribute to intrinsic resistance for toxic compounds such as antibiotics, antiseptics, detergents, and dyes. They are of interest due to their unknown physiological roles (10), possible contribution to clinical resistance (20, 24), possible utility as antibacterial targets (20, 24), and potential value in cell-based screening for novel antibacterials (12).
In E. coli, seven different proton-dependent MDR pump systems have been identified in biological studies: AcrAB-TolC (22), EmrAB (21), MdfA (8), TehA (40), EmrE (33), AcrEF (15, 16), and EmrD (28). Others have been identified by comparative amino acid sequence analysis. All fall into three distinct families as compiled by Paulsen et al. (32): the major facilitator superfamily (MFS), the resistance nodulation-cell division (RND) family, and the small multidrug resistance (SMR) family. MFS members include emrD, mdfA, emrB, and predicted emrY. The RND family is comprised of at least acrB, acrF, predicted yhiV, and predicted acrD. The SMR family includes emrE (mvrC) and tehA, which is reported to have sequence identity to the SMR family (40).
In addition to proton-dependent systems, pumps that are ATP dependent have been identified in bacteria. One is LmrA, an ATP-binding cassette (ABC) MDR pump that has been identified in Lactococcus lactis that has homology to the eucaryotic P glycoprotein MDR1 (42). Putative ATP-dependent drug efflux pumps in E. coli have been identified by Paulsen et al. (31), including YhiG, MdlB, YbjZ, and MsbA.
Multidrug efflux systems in bacteria can consist of single gene products, such as NorA of Staphylococcus aureus or the multicomponent envelope translocases, such as MexA-MexB-OprM of Pseudomonas aeruginosa (19), that are found in gram-negative organisms and facilitate drug efflux across the gram-negative outer membrane (32). A well-studied example is the AcrA-AcrB-TolC MDR tripartite pump system of E. coli (9). This complex consists of an MDR pump, a membrane fusion protein (MFP), and an outer membrane factor (OMF). The MFP gene is located in an operon immediately upstream of the corresponding MDR pump gene. Examples of these gene pairs in E. coli are are emrAB, acrAB, acrEF, and the predicted yhiVU and emrKY. A second component is the outer membrane (OM) protein, such as the only known example in E. coli, TolC. It is the required third component for the AcrAB (9) and EmrAB (18) drug efflux systems.
Multidrug efflux pump systems and their substrates have, in part, been identified by experimental systems whereby the pump protein is overexpressed and substrates are identified by increased resistance to a panel of compounds. In a study of E. coli strains overexpressing acrEF (15), increased resistance was determined for compounds similar to the dyes, detergents, and antibiotic substrates of AcrAB. Strains overexpressing emrE (25, 44) identified the substrates methyl viologen, ethidium bromide, erythromycin, sulfadiazine, and tetraphenylphosphonium. Increased tehAB expression resulted in increased resistance to tetraphenylarsonium chloride, crystal violet, and proflavin (40). Overexpression of mdfA identified ethidium bromide, tetraphenylphosphonium, rhodamine, daunomycin, benzalkonium, rifampin, tetracycline, puromycin, chloramphenicol, erythromycin, some aminoglycosides, and fluoroquinolones (8).
Deletion of a multidrug pump sometimes results in increased susceptibility to the substrate of the pump. For example, E. coli overexpressing AcrAB demonstrates increased resistance to substrates such as bile salts, compared to an isogenic wild-type strain, and similarly, a strain with an acrAB deletion is more susceptible than the wild-type strain to the same compounds (23). However, although increased expression of AcrEF results in increased resistance to some AcrAB substrates, deletion of acrEF did not contribute to increased susceptibility to those substrates (15, 23). A comparison of the MICs for strains with deletions of different MDR pump genes can reveal the MDR pumps responsible for intrinsic resistance. Such a comparison can be useful to identify (i) potential physiological roles for a pump, (ii) contributors to clinical resistance, (iii) potential antibacterial targets, and (iv) MDR pumps that will result in susceptible strains useful for cell-based screening.
In this study, we determined which MDR pump genes, when deleted, result in strains with increased susceptibility to the approximately 35 compounds that have been previously reported as MDR pump substrates (8, 15, 16, 21, 22, 25, 28, 33, 40, 44). Predicted efflux genes whose substrate profiles are unknown were included in an attempt to functionally identify new MDR pump genes. Deletions of individual MDR pump genes and operons as well as deletions of entire families of MDR pump genes in one strain were studied. Sixteen genes or operons were selected and assessed for their contribution to intrinsic resistance to the compounds. All strains were tested side-by-side by an agar dilution MIC method so that the contribution of each MDR pump to susceptibility for each of the 35 compounds could be assessed. It was found that the major contributors to intrinsic resistance for the compounds tested were AcrAB and TolC.
MATERIALS AND METHODS
Bioinformatics methods.
Amino acid sequences of a set of multidrug resistance efflux pumps were identified which included members of the MFS, RND, and SMR families, and homology search was performed using BLASTP (2) against open reading frames of the E. coli MG1655 genome (5). The generated list was analyzed based on a set of criteria that included a probability score of <10−20.
Bacterial strains and media.
E. coli strains W3110 (13), MG1655 (5), JC7623 (4), DH5α (Life Technologies, Bethesda, Md.), and V355 (36) were used. Luria broth (LB), LB agar, and Mueller-Hinton agar were obtained from Difco (Detroit, Mich.). Antibiotics used for strain constructions and selection were obtained from Sigma (St. Louis, Mo.) and were used at the following concentrations in LB or LB agar: kanamycin, 40 μg/ml; ampicillin, 100 μg/ml; and chloramphenicol, 20 μg/ml.
Allelic exchange method for construction of deletion strains.
Methods for DNA recombinant techniques were done according to Ausubel et al. (3). DNA amplification was performed using PCR with Elongase Supermix (Life Technologies), according to the instructions of the manufacturer. Gene splicing by the overlap extension method of Horton et al. (11) using PCR was done to create DNA fragments for subsequent steps. Oligonucleotides were purchased from Life Technologies.
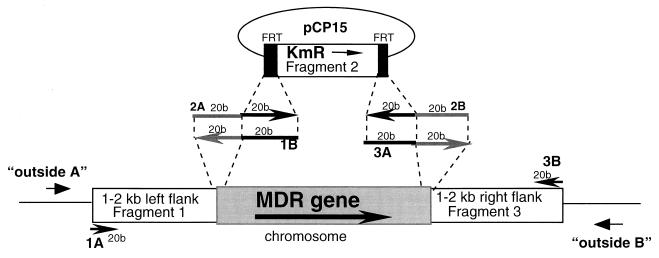
For each gene or tandem group of genes chosen for deletion, DNA fragments were produced that consisted of the Tn5 kanamycin resistance determinant (Kmr) flanked by the yeast 2μm Flp-recombination target (FRT) sites and further flanked by ca. 2 kb of MG1655 genomic DNA that normally flanks the designated gene to be disrupted. Briefly, the overlap extension method is as follows, and a schematic of primers and fragments generated is shown in Fig. 1. The Kmr-FRT DNA (Fig. 1, Fragment 1) was amplified from pCP15 (a kind gift from Wilfried Wackernagel) (7) using primers 2A and 2B (Fig. 1). The two chromosomal fragments 1 and 3 (Fig. 1) were amplified by PCR from MG1655 using primers 1A and 1B and 3A and 3B, respectively. All three DNA amplification products, fragments 1, 2, and 3 (Fig. 1), were purified from bands resolved by agarose gel electrophoresis using the Qiaex gel extraction purification method (Qiagen, Valencia, Calif.) per the instructions of the manufacturer. The three DNA amplification products were combined in an Elongase Supermix PCR mixture containing primers 1A and 3B, and the reaction was performed according to the manufacturer's instructions. Resulting amplified products of predicted sizes were purified, as described above, for subsequent manipulation.
FIG. 1.
Location of primers for (i) the design of the overlap extension method and (ii) for verification of allelic exchange. Three amplification products were constructed into one product by an overlap extension method for subsequent use in MDR pump gene replacement with the Kmr-FRT fragment. Primers 1A and 1B amplify Fragment 1 from the chromosome. Primers 2A and 2B amplify the Kmr-FRT fragment from pCP15. Primers 3A and 3B amplify Fragment 3 from the chromosome. Primers “outside A” and “outside B” amplify chromosomal DNA to verify those strains for which the Kmr-FRT fragment has replaced the MDR pump gene.
In the second step, the resulting overlap-extension DNA fragments for each gene were ligated to pBR322 (6) using T4 DNA ligase (Life Technologies). An exception was that the emrAB- and acrAB-related DNA fragments were ligated to pT7Blue (Novagen, Madison, Wis.) instead of pBR322. Ligated products were introduced into E. coli DH5α, and the correct plasmid recombinants were determined by restriction-digestion analysis.
The third step involved the introduction of plasmid recombinants into the recBC sbcBC strain JC7623 by CaCl2-mediated transformation with selection for colonies that were resistant to 40 μg of kanamycin/ml. In one case (emrAB), the recD strain V355 was used with linearized DNA. Both JC7623 and V355 are strains that facilitate allelic exchange by homologous recombination (30, 35) of incoming plasmid and chromosomal DNA, represented by fragments 1 and 3 (Fig. 1). Also, kanamycin-resistant colonies were screened for those that were sensitive to 100 μg of ampicillin (the antibiotic resistance marker for pBR322 and pT7Blue) per ml. Plasmids used for this step were ColE1 plasmids that are unstable in JC7623, and thus this strain facilitates loss of the plasmid backbone following an allelic exchange event (4). The kanamycin-resistant and ampicillin-sensitive colonies represent successful allelic exchange strains that had lost the plasmid DNA and contained the Kmr-FRT DNA, replacing the gene slated for deletion.
In the fourth step, verification of an allelic exchange for each gene was done by PCR (see Table 3). Analysis using oligonucleotides located outside primers 1A and 3B (Fig. 1, outside primers A and B) were used to amplify genomic DNA from wild-type and experimental samples. The calculation for the predicted size (bp) shown below in Table 3 was based on the following equation: predicted size = (distance in MG1655 contig between location of primers 1A and 3B) − (size of deleted gene or distance in MG1655 contig between regions of primers 1B and 3A homologous to MG1655) + (size of the Kmr-FRT fragment, 1,485 bp).
TABLE 3.
Location and evidence for MDR deletions in E. colia
| Operon | E. coli contig accession no. | Location of operon in contig (nt)b | Location of deletion in contig (nt) | Predicted amplification product (kbp)c
|
|
|---|---|---|---|---|---|
| WTd | Deletion strain | ||||
| tolC | AE000385 | 5769–7256 | 5810–7206 | 5.7 | 5.8 |
| yjcP | AE000481 | 9588–8122 | 8153–9502 | 5.8 | 5.9 |
| yohG | AE000303 | 3588–2392 | 2512–3490 | 5.4 | 5.9 |
| ylcB | AE000162 | 2377–3750 | 2402–3648 | 5.6 | 5.8 |
| acrAB | AE000152 | 9033–4668 | 4721–8937 | 8.5 | 5.7 |
| acrEF | AE000405 | 3660–7933 | 3585–7872 | 9.5 | 6.5 |
| yhiUV | AE000427 | 8349–12644 | 8432–12593 | 8.5 | 5.7 |
| acrD | AE000334 | 112–3225 | 139–3163 | 7.6 | 6.0 |
| yegMNO | AE000297 | 7262–14856 | 7317–14737 | 11.5 | 5.6 |
| emrD | AE000445 | 1091–2281 | 1114–2029 | 5.2 | 5.7 |
| mdfA | AE000186 | 5562–6794 | 5637–6720 | 5.2 | 5.5 |
| emrAB | AE000353 | 1902–4629 | 2018–4562 | 6.5 | 5.5 |
| emrKY | AE000325 | 2785–84 | 149–2716 | 6.7 | 5.5 |
| emrE | AE000160 | 1663–1995 | 1718–1974 | 4.5 | 5.7 |
| tehAB | AE000240 | 1932–3514 | 1990–3411 | 5.5 | 5.5 |
| ybjYZ | AE000189 | 8123–11208 | 8157–11189 | 7.1 | 5.5 |
Evidence for replacement of operon with the Kmr marker, based on predicted amplification product sites.
nt, nucleotides.
Amplification products were predicted based on location of primers located outside the DNA necessary for gene replacement (see Materials and Methods).
WT, wild type.
Finally, the Kmr-linked gene deletion strains were transduced to W3110 by P1 transduction using standard procedures (37) to create the group of W3110 isogenic strains listed below in Table 4.
TABLE 4.
Susceptibility of E. coli MDR gene deletion strains to toxic compounds
| W3110 pump deletion strain | Strain no. | MDR family | MIC (μg/ml)
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CAM | FLOR | CLO | PUR | ERM | METH | NOV | CPRO | NOR | NAL | RIF | FUS | SM | SUL | |||
| W3110 | HS414 | 12.5 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 3.125 | 5 | 400 | 1.95 | 2,000 | |
| ΔtolC::Kmr | HS151 | OMF | 6.25 | 0.78 | 0.78 | 1 | 1.56 | 1.56 | 20 | 0.78 | 0.0025 | 0.001 | 0.39 | 10 | 3.125 | 1.95 | 2,000 |
| ΔyjcP::Kmr | HS154 | OMF | 12.5 | 6.25 | 6.25 | >32 | 25 | 25 | >640 | 100 | 0.01 | 0.004 | 3.125 | 5 | 400 | 1.95 | 2,000 |
| ΔyohG::Kmr | HS157 | OMF | 12.5 | 6.25 | 3.12 | >32 | 25 | 50 | >640 | 100 | 0.01 | 0.004 | 3.125 | 5 | 400 | 1.95 | 2,000 |
| ΔylcB::Kmr | HS208 | OMF | 6.25 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.002 | 6.25 | 2.5 | 400 | 1.95 | 2,000 |
| ΔyjcP ΔyohG ΔylcB ΔtolC::Kmr | HS230 | OMF | 3.12 | 0.78 | 0.78 | 1 | 1.56 | 1.56 | 20 | 0.78 | 0.0025 | 0.001 | 0.39 | 10 | 3.125 | 1.95 | 2,000 |
| ΔacrAB::Kmr | HS832 | RND | 3.12 | 0.78 | 0.78 | 16 | 1.56 | 1.56 | 80 | 1.56 | 0.0025 | 0.004 | 1.56 | 2.5 | 3.125 | 1.95 | 2,000 |
| ΔacrEF::Kmr | HS212 | RND | 6.25 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 6.25 | 5 | 400 | 1.95 | 2,000 |
| ΔyhiUV::Kmr | HS193 | RND | 6.25 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 6.25 | 5 | 400 | 1.95 | 2,000 |
| ΔacrD::Kmr | HS215 | RND | 6.25 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 6.25 | 5 | 400 | 1.95 | 2,000 |
| ΔyegMNO::Kmr | HS199 | RND | 12.5 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.02 | 0.004 | 6.25 | 5 | 800 | 3.9 | 2,000 |
| ΔacrEF ΔacrAB ΔyhiUV ΔacrD ΔyegMNO::Kmr | HS276 | RND | 1.56 | 0.78 | 0.78 | 2 | 1.56 | 1.56 | 20 | 0.39 | 0.0025 | 0.001 | 1.56 | 2.5 | 3.125 | 1.95 | 2,000 |
| ΔemrD::Kmr | HS196 | MF | 6.25 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 3.125 | 2.5 | 400 | 1.95 | 2,000 |
| ΔmdfA::Kmr | HS221 | MF | 6.25 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 6.25 | 2.5 | 400 | 1.95 | 2,000 |
| ΔemrD ΔmdfA::Kmr | HS235 | MF | 6.25 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 3.125 | 2.5 | 400 | 1.95 | 2,000 |
| ΔemrAB::Kmr | HS833 | MF | 12.5 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 6.25 | 5 | 400 | 1.95 | 2,000 |
| ΔemrKY::Kmr | HS205 | MF | 6.25 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 6.25 | 5 | 400 | 1.95 | 2,000 |
| ΔemrKY | HS236 | MF | 6.25 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 6.25 | 5 | 400 | 1.95 | 2,000 |
| ΔemrKY ΔemrAB ΔemrD ΔmdfA::Kmr | HS275 | MF | 6.25 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 6.25 | 5 | 400 | 1.95 | 2,000 |
| ΔemrE::Kmr | HS218 | SMR | 6.25 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 6.25 | 2.5 | 400 | 1.95 | 2,000 |
| ΔtehAB::Kmr | HS202 | SMR | 12.5 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 400 | 0.01 | 0.004 | 6.25 | 10 | 400 | 1.95 | 2,000 |
| ΔemrE ΔtehAB::Kmr | HS238 | SMR | 6.25 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 6.25 | 5 | 400 | 1.95 | 2,000 |
| ΔybjYZ::Kmr | HS273 | ABC | 6.25 | 6.25 | 6.25 | >32 | 100 | 50 | >640 | 100 | 0.01 | 0.004 | 6.25 | 2.5 | 400 | 1.95 | 2,000 |
| MIC (μg/ml)
| |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TET | DOC | CHO | TDOC | SDS | OXAL | PROF | CV | ACR | EBR | TPP | R6G | TPAC | CTAB | DEQC | BENZ | DAUN | PLUM | VIOL | CCCP |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 800 | 1,600 | 1,280 | 1,600 | 200 | 160 | 62.5 | >80 | 40 | 50 | 26 |
| 0.156 | 200 | 1,600 | 400 | 50 | >3,200 | 6.25 | 0.78 | 3.12 | 3.12 | 6.25 | 2.5 | 1.56 | 25 | 25 | 2 | 0.625 | 10 | 50 | 1.6 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 50 | 25 | 100 | 400 | 800 | 1,280 | 400 | 100 | 160 | 31.2 | >80 | 40 | 50 | 13 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 50 | 25 | 100 | 400 | 800 | 1,280 | 400 | 100 | 160 | 31.2 | >80 | 40 | 50 | 13 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 400 | 1,600 | 1,280 | 1,600 | 200 | 160 | 62.5 | >80 | 80 | 50 | 26 |
| 0.156 | 200 | 1,600 | 400 | 50 | >3,200 | 12.5 | 0.78 | 3.12 | 3.12 | 6.25 | 2.5 | 3.125 | 25 | 2.5 | 2 | 0.625 | 10 | 50 | 1.6 |
| 0.156 | 1,600 | >12,800 | >12,800 | 100 | >3,200 | 12.5 | 0.78 | 3.12 | 3.12 | 6.25 | 2.5 | 3.125 | 2.5 | 2.5 | 2 | 0.625 | 10 | 50 | 13 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 400 | 1,600 | 1,280 | 1,600 | 200 | 160 | 62.5 | >80 | 80 | 50 | 13 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 400 | 1,600 | 1,280 | 1,600 | 200 | 160 | 62.5 | >80 | 80 | 50 | 13 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 400 | 1,600 | 1,280 | 1,600 | 200 | 160 | 62.5 | >80 | 80 | 50 | 13 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 800 | 1,600 | 1,280 | 1,600 | 200 | 160 | 62.5 | >80 | 80 | 50 | 52 |
| 0.156 | 1,600 | 6,400 | 400 | 100 | >3,200 | 6.25 | 0.78 | 3.12 | 1.56 | 6.25 | 2.5 | 1.56 | 25 | 2.5 | 2 | 0.625 | 10 | 25 | 26 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 400 | 1,600 | 1,280 | 1,600 | 200 | 160 | 62.5 | >80 | 80 | 50 | 13 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 200 | 1,600 | 1,280 | 1,600 | 100 | 160 | 15.6 | >80 | >80 | 50 | 13 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 200 | 1,600 | 1,280 | 1,600 | 100 | 160 | 15.6 | >80 | 80 | 50 | 13 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 800 | 1,600 | 1,280 | 1,600 | 200 | 160 | 62.5 | >80 | 40 | 50 | 13 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 400 | 1,600 | 1,280 | 1,600 | 200 | 160 | 62.5 | >80 | 80 | 50 | 13 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 400 | 1,600 | 1,280 | 1,600 | 200 | 160 | 62.5 | >80 | 80 | 50 | 13 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 200 | 200 | 1,600 | 1,280 | 1,600 | 100 | 160 | 15.6 | >80 | 80 | 50 | 13 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 50 | 200 | 1,600 | 1,280 | 1,600 | 100 | 160 | 62.5 | >80 | 80 | 6.25 | 13 |
| 1.25 | >3,200 | 12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 800 | 1,600 | 1,280 | 1,600 | 200 | 160 | 62.5 | >80 | 80 | 100 | 26 |
| 1.25 | >3,200 | >12,800 | >12,800 | >12,800 | >3,200 | 100 | 25 | 50 | 200 | 1,600 | 1,280 | 1,600 | 100 | 160 | 62.5 | >80 | 80 | 6.25 | 13 |
| 1.25 | >3,200 | >12,800 | 12,800 | >12,800 | >3,200 | 100 | 25 | 400 | 400 | 1,600 | 1,280 | 1,600 | 200 | 160 | 62.5 | >80 | 80 | 50 | 13 |
Values in bold differ from the W3110 control strain value by ≥4-fold.
Abbreviations: AMP, ampicillin; CAM, chloramphenicol; FLOR, florfenicol; CLO, clotrimazole; PUR, puromycin; ERM, erythromycin; METH, methotrexate; NOV, novobiocin; CIPRO, ciprofloxacin; NOR, norfloxacin; NAL, nalidixic acid; RIF, rifampin; FUS, fusidic acid; SM, streptomycin; SUL, sulfacetamide; TET, tetracycline; DOC, deoxycholate; CHO, sodium cholate; TDOC, sodium taurodeoxycholate; OXAL, sodium oxalate; PROF, proflavin; CV, crystal violet; ACR, acriflavin; EBR, ethidium bromide; TPP, tetraphenylphosphonium; R6G, rhodamine 6G; TPAC, tetraphenylarsonium chloride; DEQC, dequalinium chloride; BENZ, benzalkonium chloride; DAUN, daunomycin; PLUM, plumbagin; VIOL, methyl viologen.
Oligonucleotide sequences for 1A, 1B, 2A, 2B, 3A, and 3B (diagrammed in Fig. 1) used to construct each gene deletion by the overlap extension method are listed in Table 1 according to the respective MDR pump gene. This table also contains the outside A and outside B (Fig. 1) nucleotide sequences used to verify allelic exchanges.
TABLE 1.
Oligonucleotides used to construct and verify MDR deletion strains
| Gene(s) | Name | Oligonucleotide sequence (5′ to 3′) |
|---|---|---|
| tolC | 1A | AAAAAAAAGCGGCCGCTAGCTGAATATGACCGCACAGCAA |
| tolC | 1B | AACTTCAGAGCGCTTTTGAAGCTTGCTCAGGCCGATAAGAATGG |
| tolC | 2A | CCATTCTTATCGGCCTGAGCAAGCTTCAAAAGCGCTCTGAAGTT |
| tolC | 2B | GTGGTAGTGCGTGCGGATGTCCCGGGGATCTTGAAGTTCCTATT |
| tolC | 3A | AATAGGAACTTCAAGATCCCCGGGACATCCGCACGCACTACCAC |
| tolC | 3B | AAAAAAAAGCGGCCGCGGATCCGTGGATAAAACCGAGACATA |
| tolC | Outside A | AGCGGTATCAGGTTGGAAAC |
| tolC | Outside B | TACCTCGACCATGAAGGTGG |
| yjcP | 1A | AAAAAAAAGCGGCCGCGATCGATGCCTGCTGGTGATTCTGAT |
| yjcP | 1B | AACTTCAGAGCGCTTTTGAAGCTTCTTACGTACCAGGGCACAGC |
| yjcP | 2A | GCTGTGCCCTGGTACGTAAGAAGCTTCAAAAGCGCTCTGAAGTT |
| yjcP | 2B | GACGACGGGACCTGCCTGATCCCGGGGATCTTGAAGTTCCTATT |
| yjcP | 3A | AATAGGAACTTCAAGATCCCCGGGATCAGGCAGGTCCCGTCGTC |
| yjcP | 3B | AAAAAAAAGCGGCCGCGTCGACCGAACGGCAAGAAGTAGTGAC |
| yjcP | Outside A | TACAGGCACCAATCCACCAC |
| yjcP | Outside B | TCCGGTCAAAATCATGGTCG |
| yohG | 1A | AAAAAAAAGCGGCCGCGATCGATCCTTTAATCCGCTGACCATA |
| yohG | 1B | AACTTCAGAGCGCTTTTGAAGCTTCTCCATATCCGCCGAAAAGT |
| yohG | 2A | ACTTTTCGGCGGATATGGAGAAGCTTCAAAAGCGCTCTGAAGTT |
| yohG | 2B | GGATTCTGGCTTCGCTGACGCCCGGGGATCTTGAAGTTCCTATT |
| yohG | 3A | AATAGGAACTTCAAGATCCCCGGGCGTCAGCGAAGCCAGAATCC |
| yohG | 3B | AAAAAAAAGCGGCCGCGTCGACATAACGCCGTAAAACAAGAC |
| yohG | Outside A | ACCAAGGTAAAGCAGGTAGC |
| yohG | Outside B | TCAACACCCCAGATGAGATG |
| ylcB | 1A | AAAAAAAAGCGGCCGCTAGCGTCCAGGTGCCCCAGATGCT |
| ylcB | 1B | AACTTCAGAGCGCTTTTGAAGCTTAATGGCAGAAGTTTACAAGG |
| ylcB | 2A | CCTTGTAAACTTCTGCCATTAAGCTTCAAAAGCGCTCTGAAGTT |
| ylcB | 2B | CAAATAAAGAACGCTCGGCACCCGGGGATCTTGAAGTTCCTATT |
| ylcB | 3A | AATAGGAACTTCAAGATCCCCGGGTGCCGAGCGTTCTTTATTTG |
| ylcB | 3B | AAAAAAAAGCGGCCGCGGATCCTCGCCCGCGACCTGACACTT |
| ylcB | Outside A | CCGCTACCTTCACCTTTTCG |
| ylcB | Outside B | CGGTGCCATCTTCGAAAATG |
| acrEF | 1A | AAAAAAAAGCGGCCGCGTCGACCATCAAAAGTCGCATCGTCT |
| acrEF | 1B | AACTTCAGAGCGCTTTTGAAGCTTCAGTTCGCTATCCTACAAAT |
| acrEF | 2A | ATTTGTAGGATAGCGAACTGAAGCTTCAAAAGCGCTCTGAAGTT |
| acrEF | 2B | TACGAAGAAGATTGCCAGCACCCGGGGATCTTGAAGTTCCTATT |
| acrEF | 3A | AATAGGAACTTCAAGATCCCCGGGTGCTGGCAATCTTCTTCGTA |
| acrEF | 3B | AAAAAAAAGCGGCCGCCACGTGAAACCTCGGAAAGCATAAC |
| acrEF | Outside A | TTGCCGACCCACCATATAAC |
| acrEF | Outside B | GATAGACCGACATGGTCATG |
| yhiUV | 1A | AAAAAAAAGCGGCCGCTAGCTATTGCCGGTTTGTTCTGTGT |
| yhiUV | 1B | AACTTCAGAGCGCTTTTGAAGCTTGTTTTCCGCCGATTTGTCAT |
| yhiUV | 2A | ATGACAAATCGGCGGAAAACAAGCTTCAAAAGCGCTCTGAAGTT |
| yhiUV | 2B | CCACTACAACGAAAAAGACCCCCGGGGATCTTGAAGTTCCTATT |
| yhiUV | 3A | AATAGGAACTTCAAGATCCCCGGGGGTCTTTTTCGTTGTAGTGG |
| yhiUV | 3B | AAAAAAAAGCGGCCGCGGATCCTTTTTCTTGAGGATGAGCAC |
| yhiUV | Outside A | GTGTATCAGTTTCCCGTTCG |
| yhiUV | Outside B | GTGTCAAGGACACGCTTTC |
| acrD | 1A | AAAAAAAAGCGGCCGCTAGCATGTGCCCAGAATAAGTAAC |
| acrD | 1B | AACTTCAGAGCGCTTTTGAAGCTTTGGGGCGATCAATAAAGAAA |
| acrD | 2A | TTTCTTTATTGATCGCCCCAAAGCTTCAAAAGCGCTCTGAAGTT |
| acrD | 2B | ACAAAGAACAGCGGCACGAACCCGGGGATCTTGAAGTTCCTATT |
| acrD | 3A | AATAGGAACTTCAAGATCCCCGGGTTCGTGCCGCTGTTCTTTGT |
| acrD | 3B | AAAAAAAAGCGGCCGCGGATCCGGTCGGCAGCGTTCACACAT |
| acrD | Outside A | GGGATGAAAATGGTGTTGGC |
| acrD | Outside B | GTTGAAATCACGATGCGGGG |
| yegM yegNO | 1A | AAAAAAAAGCGGCCGCTAGCCCAGTCTGGAAACGGCATCT |
| yegM yegNO | 1B | AACTTCAGAGCGCTTTTGAAGCTTGTCTGGAATCGTCTGAATGA |
| yegM yegNO | 2A | TCATTCAGACGATTCCAGACAAGCTTCAAAAGCGCTCTGAAGTT |
| yegM yegNO | 2B | CTCATTACCAGTCCGCCGACCCCGGGGATCTTGAAGTTCCTATT |
| yegM yegNO | 3A | AATAGGAACTTCAAGATCCCCGGGGTCGGCGGACTGGTAATGAG |
| yegM yegNO | 3B | AAAAAAAAGCGGCCGCTAGCCCAGAATGGGTCGCCGTGTA |
| yegM yegNO | Outside A | CATGAGGAGAATTATCCAGC |
| yegM yegNO | Outside B | TTTACCGGTGCCAGTAAACC |
| emrD | 1A | AAAAAAAAGCGGCCGCTAGCCCCCAGCATACCCAGCGACA |
| emrD | 1B | AACTTCAGAGCGCTTTTGAAGCTTGTTTCTTTGCCTTTTCATTA |
| emrD | 2A | TAATGAAAAGGCAAAGAAACAAGCTTCAAAAGCGCTCTGAAGTT |
| emrD | 2B | GAAACAGCATCCCGGCACCGCCCGGGGATCTTGAAGTTCCTATT |
| emrD | 3A | AATAGGAACTTCAAGATCCCCGGGCGGTGCCGGGATGCTGTTTC |
| emrD | 3B | AAAAAAAAGCGGCCGCGTCGACCCGGTCAGGAAGGAAAAGAT |
| emrD | Outside A | CCAGCACGTCATCAACATCC |
| emrD | Outside B | TCCGCAGTTACCAATGGAGT |
| mdfA | 1A | AAAAAAAAGCGGCCGCTAGCATCGTGATGGAAAATGTGGA |
| mdfA | 1B | AACTTCAGAGCGCTTTTGAAGCTTCGTAAAGCACCAGACAGAGA |
| mdfA | 2A | TCTCTGTCTGGTGCTTTACGAAGCTTCAAAAGCGCTCTGAAGTT |
| mdfA | 2B | GACAGCCACAAAATTCCGTTCCCGGGGATCTTGAAGTTCCTATT |
| mdfA | 3A | AATAGGAACTTCAAGATCCCCGGGAACGGAATTTTGTGGCTGTC |
| mdfA | 3B | AAAAAAAAGCGGCCGCTCCGGAGTTGCTGCTGATAGGTGTTG |
| mdfA | Outside A | AAGCGGGCTCAGTGACCATA |
| mdfA | Outside B | TTTATAGCCTGGGCACGCTG |
| emrKY | 1A | AAAAAAAAGCGGCCGCTAGCGAGCCATTTTCATCCGTCAT |
| emrKY | 1B | AACTTCAGAGCGCTTTTGAAGCTTAAACTACCGCCAATAAAGAA |
| emrKY | 2A | TTCTTTATTGGCGGTAGTTTAAGCTTCAAAAGCGCTCTGAAGTT |
| emrKY | 2B | AAAACCGTAAGTAAGATAAACCCGGGGATCTTGAAGTTCCTATT |
| emrKY | 3A | AATAGGAACTTCAAGATCCCCGGGTTTATCTTACTTACGGTTTT |
| emrKY | 3B | AAAAAAAAGCGGCCGCGGATCCGCCGTGTTTGTCGCCTATTA |
| emrKY | Outside A | AAGAATGTCCCCCATAACGC |
| emrKY | Outside B | ATTGGCAACCCTATCACTGC |
| emrE | 1A | AAAAAAAAGCGGCCGCTAGCACGCGCATTACTGGTTGGTT |
| emrE | 1B | AACTTCAGAGCGCTTTTGAAGCTTGTTGTACCAATGACCTCTGC |
| emrE | 2A | GCAGAGGTCATTGGTACAACAAGCTTCAAAAGCGCTCTGAAGTT |
| emrE | 2B | AATGTGGTGTGCTTCGTGACCCCGGGGATCTTGAAGTTCCTATT |
| emrE | 3A | AATAGGAACTTCAAGATCCCCGGGGTCACGAAGCACACCACATT |
| emrE | 3B | AAAAAAAAGCGGCCGCGGATCCTTTGTCACGTATTTTCAGTA |
| emrE | Outside A | GACATAATGCAGGCCTTCAC |
| emrE | Outside B | CCCCCAACCCAATTATTTGC |
| tehAB | 1A | AAAAAAAAGCGGCCGCTAGCAATCCTCGCTTTATGTCCTT |
| tehAB | 1B | AACTTCAGAGCGCTTTTGAAGCTTGTCCCCAACACAATACCAAA |
| tehAB | 2A | TTTGGTATTGTGTTGGGGACAAGCTTCAAAAGCGCTCTGAAGTT |
| tehAB | 2B | TTCATTGTATTTCACCCTCTCCCGGGGATCTTGAAGTTCCTATT |
| tehAB | 3A | AATAGGAACTTCAAGATCCCCAGAGGGTGAAATACAATGAA |
| tehAB | 3B | AAAAAAAAGCGGCCGCGGATCCTCTGTCCCCGCTTTTTGAAT |
| tehAB | Outside A | GGAGTGCGTATGTTTCCAGA |
| tehAB | Outside B | CTGATGTACCACTTACCGCA |
| ybjY ybjZ | 1A | AAAAAAAAGCGGCCGCGCTAGCTCGTATCGGGTTATTTTGCTCGG |
| ybjY ybjZ | 1B | TCAGAGCGCTTTTGAAGCTTCTTCTTCACGGTTTTCCGCT |
| ybjY ybjZ | 2A | AGCGGAAAACCGTGAAGAAGAAGCTTCAAAAGCGCTCTGA |
| ybjY ybjZ | 2B | TCTACTGGATCCAGTCGTGCCCCGGGGATCTTGAAGTTCC |
| ybjY ybjZ | 3A | GGAACTTCAAGATCCCCGGGGCACGACTGGATCCAGTAGA |
| ybjY ybjZ | 3B | AAAAAAAAGCGGCCGCGTCGACGGCACATCGCCCTGAACAATTC |
| ybjY ybjZ | Outside A | CAGGGACTTTCCGCAATGGTACA |
| ybjY ybjZ | Outside B | TTAGGTTAGCCGCATCGACCTGAC |
As opposed to the deletion strains described above, the acrAB and emrAB deletion strains were constructed differently. The acrAB and emrAB genes with flanking 2 kbp of DNA were amplified by PCR (primers from 5′ to 3′ are GCCCACCAGCAGTAGAGGGGAAGTAACAGA and GCCCACCAGCCTTCAGGACGGCAATCTGCT for acrAB and GCCCACCAGCGTGTTAAGAAGCCCATCAGT and GCCCACCAGCTCCGCAATTACTATCGTTTC for emrAB). These fragments were cloned into the pT7Blue cloning vector by blunt-end ligation using T4 DNA ligase. Another amplification was done using the recombinant plasmid as template with primers located near and directed away from the start and end of acrA or emrA (primers from 5′ to 3′ were CTTGTTGGGCCTGTTTGTCG and CTTCGCGCCCGTTACCATGATTTAGCCGCAAGAATGAAGA for acrAB and TGCTGGGGCTGGTGTGGTTT and GGTCCTGACTTTGGTCGCGACCCTATCGCTACGGCAATAA for emrAB). This product was blunt-end ligated to the purified Kmr-FRT SmaI/HindII fragment from pCP15 that was treated with T4 DNA polymerase. These products were introduced into strain DH5α via transformation, and recombinant clones were obtained. The resulting constructs were used for subsequent allelic exchange into E. coli JC7623 or V355 as described above. Verification of the allelic exchange was done as described above by PCR amplification using outside primers A and B that were from 5′ to 3′ CGCGGGTATTCAGAGACTCA and CAGACGCAAATCCGCGATGCG for acrAB and CATTCTGGCAAGCGAAGTGCGGTGATA and CCGCAAATTAAGCAATCAGCAACCGTT for emrAB.
Because Kmr-marked deletions were flanked by FRT sites, it was possible to remove the Kmr-FRT DNA directly from the chromosome, leaving a deletion marked by a single FRT site by the method of Cherepanov and Wackernagel (7). Briefly, the pCP20 plasmid (7) that is temperature sensitive for replication and contains the yeast 2μm Flp recombinase was introduced into Kmr-FRT-marked deletion strains. Introduction of the plasmid by transformation was performed using selection for chloramphenicol resistance at 30°C. The plasmid expressing Flp recombinase facilitated the FRT site-specific recombination and subsequent loss of the chromosomal Kmr-FRT cassette. Culturing of the strain at 42°C in the absence of antibiotic selection produced strains cured for pCP20. The resulting strains were chloramphenicol sensitive and kanamycin sensitive. Then another Kmr-marked MDR pump deletion was introduced by P1 transduction, and again the Kmr determinant was removed. Reiteration of this method resulted in multiple unmarked gene deletions in one strain. Verification of the loss of the Kmr marker was done by PCR using outside primers as described above.
Susceptibility testing.
Agar dilution antimicrobial susceptibility tests were performed as outlined in the NCCLS standard M7-A4 (29), using Mueller-Hinton agar with the modification that concentrations of the initial compounds (shown below) may differ from those recommended. Experiments were repeated at least twice. The inoculum E. coli cultures consisted of 2 × 104 bacteria/2-μl spot on Mueller-Hinton agar media containing serial dilutions of the compounds listed below. The plates were incubated for 16 to 20 h at 37°C. The lowest concentration of antibiotic that completely inhibited growth was identified as the MIC. A ≥4-fold difference in susceptibility of the deletion strain versus the isogenic W3110 parent strain was considered significant. The following compound stock solutions were prepared in water unless indicated otherwise, and the compound class is listed in parentheses: clotrimazole (imidazole; Sigma) at 0.5 mg/ml in 50% ethanol; ampicillin (β-lactam; Sigma) at 100 mg/ml; chloramphenicol (Sigma) at 25 mg/ml in ethanol; florfenicol (Schering-Plough Research Institute) at 12.5 mg/ml in ethanol; puromycin (lipophilic basic compound; Sigma) at 100 mg/ml; methotrexate (Calbiochem-Novobiochem, La Jolla, Calif.) at 10 mg/ml in alkaline water; erythromycin (macrolide; Sigma) at 100 mg/ml in methanol; novobiocin (Sigma) at 50 mg/ml in 50% methanol; fusidic acid sodium salt (steroidal antibiotic; Aldrich Chemical Co., Inc., Milwaukee, Wis.) at 100 mg/ml in methanol; tetracycline (Sigma) at 10 mg/ml in methanol; ciprofloxacin (fluoroquinolone) at 10 mg/ml; norfloxacin (fluoroquinolone; Sigma) at 10 mg/ml in dimethyl sulfoxide; nalidixic acid sodium salt (quinolone precursor; Aldrich) at 50 mg/ml; rifampin (Sigma) at 20 mg/ml in methanol; streptomycin sulfate (Sigma) at 500 mg/ml; sulfacetamide (Sigma) at 500 mg/ml; sodium dodecyl sulfate (detergent; Sigma) at 200 mg/ml; deoxycholate sodium salt (bile salt; Sigma) at 100 mg/ml; sodium cholate (bile salt; Sigma) at 100 mg/ml; sodium taurodeoxycholate (bile salt; Sigma) at 100 mg/ml; sodium oxalate (dicarboxylic acid; Aldrich) at 25 mg/ml; proflavin (intercalator; Sigma) at 100 mg/ml; crystal violet (intercalator; Aldrich) at 50 mg/ml; acriflavin (intercalator; Sigma) at 200 mg/ml; ethidium bromide (intercalator; Sigma) at 10 mg/ml; cetyltrimethylammonium bromide (quaternary amino compound; Aldrich) at 100 mg/ml; dequalinium chloride (quaternary amino compound; Aldrich) at 25 mg/ml; benzalkonium chloride (quaternary amino compound; Sigma) at 500 mg/ml; tetraphenylphosphonium chloride (lipophilic quaternary amino compound; Aldrich) at 100 mg/ml; tetraphenylarsonium chloride (lipophilic quaternary amino compound; Aldrich) at 100 mg/ml; rhodamine 6G (lipophilic quaternary amino compound; Sigma) at 20 mg/ml; daunomycin (redox cycling drug; Sigma) at 10 mg/ml; plumbagin (redox cycling drug; Sigma) at 20 mg/ml in methanol; methyl viologen (redox cycling drug; Sigma) at 100 mg/ml; and carbonyl cyanide-chlorophenyl hydrazone (CCCP) (uncoupler of proton motive force; Sigma) at 8 mM in 50% methanol–20% dimethyl sulfoxide.
RESULTS
Identification of MDR pump genes.
In order to conduct a comprehensive survey on the effect of the loss of MDR pump genes on susceptibility to toxic compounds, genes encoding 25 E. coli multidrug efflux pumps, OMFs, and an MFP were deleted (Table 2). These genes include known and predicted MDR genes as well as predicted OMF and MFP elements that were identified by bioinformatics analysis as described in Materials and Methods. Specifically, for the OMF class of genes, OprM—the P. aeruginosa OM component of the MexAB multidrug resistance pump (19)—identified YlcB (45% identity and 62% amino acid similarity over 455 residues), YjcP (26% identity and 47% similarity over 466 residues), and YohG (27% identity and 45% similarity over 345 residues). For additional MF family proteins, the Neisseria gonorrhoeae MtrC lipoprotein open reading frame (32) was used to identify yegM (32% identity and 50% similarity over 378 residues). The gene yegM is located immediately upstream, in operon-like form, of two genes that show sequence similarity to the hypothetical RND family efflux pumps identified by Paulsen et al. (31): yegN (30% identity and 53% similarity over 1,027 residues with acrD) and yegO (29% identity and 52% similarity over 1,031 residues with acrD). MtrC was homologous to YbjY (28% identity and 45% sequence similarity over 332 residues), which is proximal to ybjZ, a gene that encodes a putative ABC MDR pump. YbjZ is a putative ABC drug efflux transporter (31). Although most bacterial MDR pumps identified are proton motive force-dependent efflux genes, the identification of ybjY immediately upstream of a putative ABC transporter prompted us to include this putative multidrug resistance-like pump in the study.
TABLE 2.
Known and predicted E. coli MDR pump proteins
| MDR component | Assigned family | GenBank contig accession no. (and ORF no. in contig)b | Reference or source | Genetic contexta |
|---|---|---|---|---|
| TolC | OMF | AE000385_0008 | 9 | tolC |
| YjcP | OMF | AE000481_0007 | This study; homology to P. aeruginosa OprM | yjcP |
| YohG | OMF | AE000303_0004 | This study; homology to P. aeruginosa OprM | yohG |
| YlcB | OMF | AE000162_0003 | This study; homology to P. aeruginosa OprM | ylcB |
| AcrB | RND | AE000152_0008 | 22 | acrAB |
| AcrF | RND | AE000405_0007 | 16 | acrEF |
| YhiU | RND | AE000427_0010 | 32 | yhiUV |
| AcrD | RND | AE000334_0001 | 27 | acrD |
| YegN | RND | AE000297_0008 | 31 | yegMNO |
| YegO | RND | AE000297_0009 | 31 | yegMNO |
| EmrD | MF | AE000445_0002 | 28 | emrD |
| MdfA | MF | AE000186_0007 | 8 | mdfA |
| EmrB | MF | AE000353_0005 | 21 | emrAB |
| EmrY | MF | AE000325_0001 | 32 | emrKY |
| EmrE | SMR | AE000160_0004 | 33 | emrE |
| TehA | SMR | AE000240_0004 | 40 | tehAB |
| YbjZ | ABC | AE000189_0008 | 31 | ybjYZ |
| AcrA | MFP | AE000152_0007 | 22 | acrAB |
| AcrE | MFP | AE000405_0006 | 16 | acrEF |
| YhiV | MFP | AE000427_0011 | 32 | yhiUV |
| YegM | MFP | AE000297_0007 | This study; homology to N. gonorrhoeae MtrC | yegMNO |
| EmrA | MFP | AE000353_0004 | 21 | emrKY |
| EmrK | MFP | AE000325_0002 | 32 | emrKY |
| YbjY | MFP | AE000189_0007 | This study; homology to N. gonorrhoeae MtrC | ybjYZ |
| TehB | NAc | AE000240_0005 | 40 | tehAB |
The location of the MDR genes that are contiguous to and oriented in the same transcriptional direction as other MDR genes.
Last four digits shown correspond to the open reading frame (ORF) in the contig.
NA, not applicable.
Construction of E. coli strains with deletions for multidrug efflux genes.
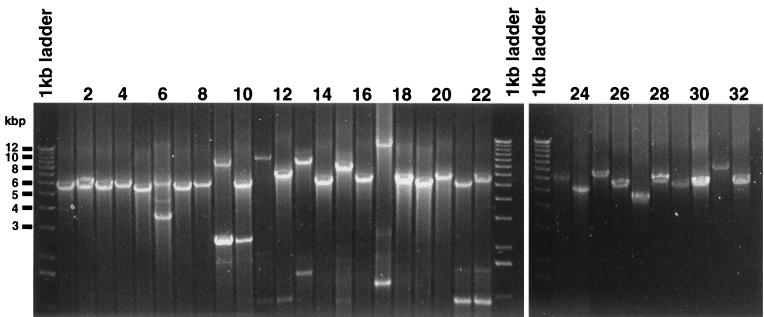
Contiguous MDR pump genes oriented in the same direction can constitute a multidrug efflux operon (Table 2, genetic context of MDR pump genes). We chose to delete the contiguous MDR pump genes, with the rationale that such putative operon gene products may associate to form multicomponent MDR efflux systems. Moreover, all of the MFP genes were immediately upstream of a pump (except for yhiUV, which has the MFP located downstream), which is a configuration that is seen in the best-studied E. coli pumps, acrAB and emrAB. Therefore, 16 unique deletions representing 25 individual gene deletions were made, and the specific genes deleted are shown in Table 2. The corresponding nucleotide deletion locations in the E. coli contigs (5) are shown in Table 3. E. coli W3110 strains with deletions for MDR pump genes or operons had a chromosomal deletion replaced by the Tn5 kanamycin resistance determinant, which was itself flanked by the yeast 2μm Flp recombination target FRT sites (Kmr-FRT fragment). In addition to the 16 deletion strains, 5 other strains (Table 4; HS230, HS276, HS235, HS236, HS275, and HS238) were constructed that had multiple operon or gene deletions. These strains had deletions for operons or genes of the same OMF, MFS, RND, or SMR MDR pump families. They were constructed by initially deleting the chromosomal Kmr-FRT fragment in vivo using the method of Cherepanov and Wackernagel (7). A new Kmr-FRT-marked MDR pump deletion locus of the same MDR pump family was introduced by P1 transduction. This marker was removed as described above, and another marked deletion was introduced by P1 transduction. This iterative process was repeated such that many MDR pump deletions were introduced into a single strain. The construction of all strains listed in Table 4 is described in Materials and Methods. All deletion strains were verified as correct by analyzing the chromosomal locus by PCR amplification using oligonucleotide primers located outside the region of DNA used to make the deletion constructs. The outside A and B oligonucleotides used for PCR for each deletion strain are shown in Table 1, and the corresponding wild-type and mutant predicted PCR product sizes are depicted in Table 3. The PCR product size experimental data for the wild-type and mutant strains are seen in Fig. 2. The predicted sizes in Table 3 are the same as the experimental PCR product sizes seen in Fig. 2, providing strong evidence that the deletion strains are correct.
FIG. 2.
Evidence for successful allelic exchange of the Kmr-FRT determinant for the genes or operons used in this study, as assessed by ethidium bromide-stained PCR products following electrophoresis in a 0.7% agarose gel. PCRs using oligonucleotides located outside the operons deleted (outside primers in Table 1) were used with genomic DNA templates of the wild type (W3110) or of the specific mutant Kmr strain. PCR products were amplified using tolC primers with genomic W3110 (lane 1) and with HS151 (lane 2); yjcP primers with W3110 (lane 3) and HS154 (lane 4); yohG primers with W3110 (lane 5) and HS157 (lane 6); ylcB primers with W3110 (lane 7) and HS208 (lane 8); acrAB primers with W3110 (lane 9) and HS832 (lane 10); acrEF primers with W3110 (lane 11) and HS212 (lane 12); yhiUV primers with W3110 (lane 13) and HS193 (lane 14); acrD primers with W3110 (lane 15) and HS215 (lane 16); b2074yegNO primers with W3110 (lane 17) and HS199 (lane 18); emrD primers with W3110 (lane 19) and HS196 (lane 20); mdfA primers with W3110 (lane 21) and HS221 (lane 22); emrAB primers with W3110 (lane 23) and HS833 (lane 24); emrKY primers with W3110 (lane 25) and HS205 (lane 26); emrE primers with W3110 (lane 27) and HS218 (lane 28); tehAB primers with W3110 (lane 29) and HS202 (lane 30); and b0878ybjC primers with W3110 (lane 31) and HS273 (lane 32). The two tehAB 5.5-kbp bands seen for both wild-type and mutant strains in lanes 29 and 30 do not distinguish mutant from wild-type alleles. The wild-type product was distinguished from the ΔtehAB::Kmr PCR product upon digestion with HindII; the ΔtehAB::Kmr PCR product seen in lane 30 was digested to two fragments of size 2 and 3.5 kbp, whereas the wild-type product seen in lane 29 remained a 5.5-kbp undigested product as expected (data not shown).
Susceptibility testing results.
MICs of 35 compounds were determined for the newly constructed deletion strains. The agar dilution MIC results for E. coli strain W3110 and each of its 22 isogenic null mutants are shown in Table 4. Results were considered significant when there was a ≥4-fold difference between deletion strains and the wild-type control, E. coli W3110 (Table 4). The data are grouped by gene families, e.g., OMF and MDR pump families. For the OMF family, a deletion in tolC (strain HS151) resulted in a strain with increased susceptibility to 29 of the 35 compounds tested; single deletions in yjcP (strain HS154) or yohG (strain HS157) resulted in strains with increased susceptibility to puromycin, acriflavin, and tetraphenylarsonium chloride. In contrast, a strain with a deletion in another family member, ylcB (strain HS208), showed no difference versus W3110 for any of the compounds tested. Strain HS230, with all four genes in this OMF class deleted, showed a susceptibility profile comparable to that of the single tolC deletion strain HS151.
Another group of genes deleted are members of the RND family: a deletion in acrAB resulted in a strain with increased susceptibility to 25 of the 35 compounds (Table 4, compare strains HS832 and W3110). Strains containing single deletions of either acrEF, yhiUV, acrD, or yegMNO (strains HS212, HS193, HS215, and HS199) demonstrated no alteration in susceptibility compared to W3110 for all compounds tested. Twenty-four of these compounds resulted in increased susceptibility when tested on strains with a deletion in tolC. When all five deletions of the RND class were combined in one strain (strain HS276), there were eight compounds to which the strain showed increased susceptibility above that of the acrAB-deleted strain. For these eight compounds, susceptibility appeared to be dependent on multiple RND efflux systems.
Single, double, and quadruple deletions of the MF family were constructed. Strains with single deletions in emrD, emrAB, and emrKY (strains HSHS196, HS833, and HS205) showed no alternations in susceptibility to the 35 compounds when compared to W3110. In contrast, a strain with a single deletion of mdfA (strain HS221) resulted in increased susceptibility to ethidium bromide and benzalkonium chloride. Strains with two pump deletions were also created. A strain with emrD and mdfA deleted (strain HS235) had the same susceptibility profile as a single mdfA deletion. The strain with deletions for emrAB and emrKY (strain HS236) had susceptibility data similar to W3110. A deletion of all four MF family loci resulted in MICs comparable to those for the strain with a deletion for mdfA alone (compare strain HS275 to strain HS221), suggesting that among the four members there is no additive or synergistic effect of multiple deletion in this family.
Two members of the SMR family were deleted and analyzed. A strain with a deletion of emrE (strain HS218) displayed increased susceptibility to acriflavin, ethidium bromide, and methyl viologen, while a strain with a deletion of tehAB (a possible SMR family member) (strain HS202) resulted in increased resistance to novobiocin. Deletions of both genes in one strain (strain HS238) showed the same susceptibility profiles as single deletions of emrE and tehAB for the compounds tested, with the exception of novobiocin, for which the profile was unchanged from wild type.
A putative ABC transporter and the MF component located immediately upstream were also deleted. The strain with deletion of ybjYZ (strain HS273) showed no differences in MICs of any compound compared to MICs for W3110.
DISCUSSION
E. coli strains were constructed with null mutations in efflux pump genes. Efflux genes were deleted singly or by groups according to the class of pump (RND, MF, SMR, ABC) or MDR multicomplex component (OM). The susceptibility profiles for each of the isogenic E. coli strains were determined using 35 compounds, including antibiotics, antiseptics, detergents, and dyes that have been previously identified by other investigators as substrates for the known MDR pumps. This study was designed to systematically identify the contribution of efflux pump genes for intrinsic susceptibility to broad classes of compounds. By and large, deletions of tolC (OM family) and acrAB (RND family) resulted in the greatest increase in susceptibility for most of the compounds tested. They each individually accounted for increased susceptibility to ≥25 of the 35 compounds (Table 4).
The data support the concept that TolC functions with and without AcrAB in its contribution to intrinsic resistance. Multicomponent gram-negative pump systems are comprised of a pump, an MFP, and an OMF. Two examples are AcrA(MFP)-AcrB(pump)-TolC(OMF) of E. coli (9) and the MexA(MFP)-MexB(pump)-OprM(OMF) of P. aeruginosa (19). The acrAB and tolC mutant strains have overlapping substrate susceptibility profiles, supporting their interaction as a tripartite pump system. None of the other OM components showed similar overlapping profiles with other strains with deletions for different classes of pumps. For example, yjcP and yohG mutants showed increased sensitivities to two compounds for which no mutant strains other than acrAB showed increased susceptibility. On the other hand, it was evident that TolC also functions independently of AcrA-AcrB. This is supported by (i) the strain deleted for tolC showed hypersusceptibility to more compounds than any of the single-locus-deletion strains tested; (ii) for 7 of 24 compounds, the MIC for the tolC strain was lower than that for the acrAB-deletion strain; and (iii) the tolC strain was susceptible to five more compounds than was the acrAB-deletion strain.
This study identifies those genes (tolC, acrAB, yjcP, yohG, mdfA, and emrE) that contribute to intrinsic resistance for the compounds studied here. First, consistent with previous studies (9, 18, 43), the absence of tolC in E. coli accounts for increased susceptibility to many classes of compounds (29 of the 35 compounds tested), and it is known that acrAB mutant strains are more sensitive to antibiotics, detergents, and dyes (22). Second, we have found that in addition to TolC, two putative members of the OM family also affect intrinsic resistance. Deletion of yjcP and yohG resulted in a fourfold increase in susceptibility to puromycin, acriflavin, and tetraphenylarsonium chloride. These data support the idea that these two genes are OM components. Third, the mdfA null mutant displayed increased susceptibility to two (ethidium bromide and benzalkonium chloride) of the 12 compounds (the other compounds were chloramphenicol, erythromycin, tetraphenylphosphonium, puromycin, tetracycline, daunomycin, rhodamine 6G, rifampin, ciprofloxacin, and norfloxacin) identified by Edgar and Bibi (8). Interestingly, the two compounds for which the null mutant displayed increased susceptibility are those for which the strain overexpressing mdfA displayed the highest increases in resistance compared to the wild-type control (8). Finally, it is known that strains that overexpress emrE demonstrate increased resistance to ethidium bromide and methyl viologen. Here, the emrE null mutant displayed increased susceptibility to these two compounds and also to acriflavin.
In contrast to the deletion strains mentioned above, there are others for which no increase was seen in susceptibility. They are strains in which individual deletions are in emrAB, emrD, tehAB, acrEF, acrD, ylcB, yhiUV, yegMNO, and ybjYZ. First, although E. coli strains overexpressing emrAB are resistant to nalidixic acid and CCCP (21) (compounds tested in this study), the emrAB-deletion strain was not found to exhibit susceptibility to these compounds. Second, whereas an emrD mutant has been shown to be sensitive to CCCP by the efficiency-of-plating method used by Naroditskaya et al. (28), such a result was not identified here. However, it is plausible that the discrepancy in results can be attributed to the use here of a less sensitive agar dilution MIC method compared to the more sensitive efficiency-of-plating method used by Naroditskaya et al. (28). Third, the tehAB deletion had little effect on susceptibility and in fact showed increased resistance to novobiocin, a phenomenon that may be related to the fact that strains overexpressing tehAB are more susceptible to methyl viologen and dequalinium chloride (40). Fourth, the data here are consistent with studies indicating that single acrEF and acrD null mutants have not been shown to increase susceptibility in E. coli K-12 (23). However, for acrD, Rosenberg and Nikaido have recently shown that a deletion in acrD decreased MICs of aminoglycosides (amikacin, gentamicin, neomycin, kanamycin, and tobramycin) by a factor of 2 to 8 (34). The aminoglycosides kanamycin and neomycin could not be tested here because the Tn5 Kmr marker that confers resistance to these two compounds is present in all strains. More work is needed to identify the contributions of the MDR pumps for susceptibility to both aminoglycosides and also to other compounds not studied here. Finally, for the putative pumps ylcB, yhiUV, yegMNO, and ybjYZ, we were unable to identify a functional contribution to intrinsic resistance.
There can be a number of reasons why strains with deletions for single pumps do not show susceptibility to those substrates for which strains overexpressing the same pump genes show resistance. MDR pumps, such as EmrAB, AcrEF, EmrD and TehAB (8, 15, 25, 40, 44), have been identified by their ability to confer resistance to compounds when overexpressed in bacteria. In this study, strains with deletions for acrAB and emrE had increased susceptibility to specific compounds for which overexpression reportedly confers resistance. However, strains with deletions for emrAB, acrEF, emrD, and tehAB did not. There are a number of possible explanations for this. First, for those MDR pumps where no clear effect on intrinsic resistance was seen, it is possible that the set of growth conditions employed here is not sufficient to detect their contribution. Other growth conditions may be necessary. Second, since pumps have overlapping substrate profiles (e.g., acrAB and acrEF [25]), an MDR pump that is active in the cell can mask the effect of the deletion of another MDR pump. An example may be the AcrA-AcrB-TolC pump, which is the primary efflux system in E. coli under the conditions tested. A deletion in acrAB or tolC would be necessary to identify the contribution of other pumps with overlapping substrate profiles. This was in fact shown to be the case when emrAB was identified as contributing to bile salt susceptibility in an acrAB mutant background by Thanassi et al. (39). This may also be the case for emrAB, acrEF, emrD, and tehAB and others. Third, these MDR pumps are normally poorly expressed and therefore a deletion may have little effect—an idea previously suggested by Ma et al. for acrEF (23). Fourth, a corollary to the previous explanation may be that the true inducer of the pump (such as bile salts, as postulated for AcrAB) is not present, and so the pump is not expressed in the presence of its alternative pump substrates. An example of this explanation is found for two Bacillus pumps. Bacillus subtilis bmr and blt have similar substrate specificities (e.g., to rhodamine) when overexpressed but have different inducers. Rhodamine is the inducer of bmr but not of blt (1). Whereas rhodamine both induces and is effluxed by bmr, it does not induce expression of blt and thus is not effluxed. Perhaps emrAB, acrEF, emrD, and tehAB are like blt, where the inducer is not present in the list of compounds used in this study. One possible way to identify inducers of these pumps is to treat E. coli with many structurally unrelated compounds and look for increased expression of these genes.
The effect on susceptibility for strains with multiple pump knockouts was assessed only with MDR pumps of the same family (OM, RND, MF, and SMR) in an initial effort to identify possible new combinations of deletions that may lead to a susceptible phenotype. No additive or synergistic effects on susceptibility were seen for the strains with deletions for all genes comprising the OM, MF, or SMR families. However, the strain with deletions for genes in the RND family led to increased susceptibility above levels seen for the acrAB-deletion strain alone, indicating that another MDR pump in this family contributes to intrinsic resistance. A recent report by Lee et al. (17) indicated that simultaneous expression of single-component (MdfA or CmlA) and multicomponent (AcrAB) MDR pump genes results in resistance to antibiotics that is greater than the additive resistance of each MDR pump expressed singly. Perhaps for the RND family the AcrD single-component pump contributes to resistance with AcrAB. The strains constructed in this study are a resource that can be used to construct other novel strains to assess the interplay among efflux pumps for intrinsic resistance.
Mutations leading to increased expression of efflux genes can contribute to increased resistance to antibiotics, such as quinolone resistance in norA mutants of S. aureus (14). Conversely, inhibition of MDR pumps can lead to increased susceptibility (24), such as is seen for the 5′-methoxyhydnocarpin inhibition of NorA (38). The notion of inhibiting efflux pumps in order to potentiate the efficacy of antibiotics was described for S. aureus by Stermitz et al. (38) when S. aureus had increased susceptibility to berberine and NorA substrates in the presence of the NorA inhibitor, 5′-methoxyhydnocarpin. Lomovskaya et al. (20) and Hsieh et al. (12) reported that the potential effect of inhibiting MDR pumps in combination with fluoroquinolone treatment would result in increased fluoroquinolone susceptibility for P. aeruginosa and S. aureus, respectively. Moreover, other studies suggest that a pump inhibitor combined with an antimicrobial would decrease the likelihood of the emergence of strains clinically resistant to fluoroquinolones (20, 24). This study suggests that an inhibitor specific for the dominant efflux system identified here, AcrA-AcrB-TolC, would result in an increase in susceptibility to a broad spectrum of toxic compounds including antibiotics, detergents, and dyes. Such an inhibitor could potentially be used in conjunction with current antimicrobials that are substrates for the AcrA-AcrB-TolC pump. The strains generated in this study could also be used to identify those E. coli MDR pumps that contribute to resistance to any given antimicrobial, and subsequent identification of a compound that inhibits that MDR pump could be used with the particular antimicrobial for effective cotherapy.
Efflux pump deletion strains can yield sensitive bacterial strains that are useful for cell-based screening of compound libraries to identify novel new antimicrobials (12). The susceptible strains can be used to detect lower concentrations of antibiotics in compound libraries than those for wild-type E. coli strains. Of the 16 deletion strains studied here, the most sensitive strains for cell-based screening are the acrAB or tolC deletion strains. This study shows that highly susceptible E. coli strains may be obtained for cell-based screening if other efflux pump genes are disrupted in combination with an acrAB or tolC deletion. Conceivably, non-MDR pump alleles that affect susceptibility, such as the E. coli lpxA, firA, or rfa alleles that increase sensitivity to multiple antibiotics (41), can also be used in combination with the efflux pump mutants to produce strains hypersusceptible to broad classes of compounds. Systematic deletions of MDR pumps, similar to those in this study, in organisms other than E. coli can be performed to generate gram-negative, gram-positive, or fungal-sensitive strains for cell-based antimicrobial screens.
Use of bioinformatics, genetics, and susceptibility testing will continue to identify new MDR pumps and additional substrates. For example, MDR pumps such as YdhE, a homolog of the NorM MDR pump from Vibrio parahaemolyticus, can be added for study (26), and newer substrates such as the recently identified substrate indole for the AcrEF pump can be tested (15). Thus, the systematic approach used in this study is an initial step to determine the relative contributions to intrinsic resistance of many efflux pumps. Specifically, this approach identified AcrAB-TolC as the dominant MDR pump among the 16 MDR pumps evaluated for the 20 different classes of compounds assessed.
REFERENCES
- 1.Ahmed M, Lyass L, Markham P N, Taylor S S, Vazquez-Laslop N, Neyfak A. Two highly similar multidrug transporters of Bacillus subtilis whose expression is differentially regulated. J Bacteriol. 1995;177:3904–3910. doi: 10.1128/jb.177.14.3904-3910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1989. [Google Scholar]
- 4.Bassett C L, Kushner S R. Exonucleases I, III, and V are required for stability of ColEl-related plasmids in Escherichia coli. J Bacteriol. 1984;157:661–664. doi: 10.1128/jb.157.2.661-664.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett G R, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W, Crose J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 7.Cherepanov P P, Wackernagel W. Gene disruption in Escherichia coli TcR and KmR cassettes with the option of FLP-catalyzed excision of the antibiotic-resistant determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 8.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. . (Erratum, 179:5654.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fralick J A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George A M. Multidrug resistance in enteric and other gram-negative bacteria. FEMS Microbiol Lett. 1996;139:1–10. doi: 10.1111/j.1574-6968.1996.tb08172.x. [DOI] [PubMed] [Google Scholar]
- 11.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh P C, Siegel S A, Rogers B, Davis D, Lewis K. Bacteria lacking a multidrug pump: a sensitive tool for drug discovery. Proc Natl Acad Sci USA. 1998;95:6602–6606. doi: 10.1073/pnas.95.12.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen K. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaatz G W, Seo S M, Ruble C A. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1086–1094. doi: 10.1128/aac.37.5.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawamura-Sato K, Shibayama K, Horii T, Iimuma Y, Arakawa Y, Ohta M. Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol Lett. 1999;179:345–352. doi: 10.1111/j.1574-6968.1999.tb08748.x. [DOI] [PubMed] [Google Scholar]
- 16.Klein J R, Henrich B, Plapp R. Molecular analysis and nucleotide sequence of the envCD operon of Escherichia coli. Mol Gen Genet. 1991;230:230–240. doi: 10.1007/BF00290673. [DOI] [PubMed] [Google Scholar]
- 17.Lee A, Mao W, Warren M S, Mistry A, Hoshino K, Okumura R, Ishida H, Lomovskaya O. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J Bacteriol. 2000;182:3142–3150. doi: 10.1128/jb.182.11.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis K. Multidrug resistance efflux. In: Broome-Smith J K, Baumberg S, Sterling C J, Ward F B, editors. Transport of molecules across biological membranes. Cambridge, United Kingdom: Cambridge University Press; 1999. pp. 15–40. [Google Scholar]
- 19.Li X Z, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomovskaya O, Lee A, Hoshino K, Ishida H, Mistry A, Warren M S, Boyer E, Chamberland S, Lee V J. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:1340–1346. doi: 10.1128/aac.43.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lomovskaya O, Lewis K. emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci USA. 1992;89:8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 23.Ma D, Cook D N, Hearst J E, Nikaido H. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 24.Markham P N, Neyfakh A A. Inhibition of the multidrug transporter NorA prevents emergence of norfloxacin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:2673–2674. doi: 10.1128/aac.40.11.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morimyo M, Hongo E, Hama-Inaba H, Machida I. Cloning and characterization of the mvrC gene of Escherichia coli K-12 which confers resistance against methyl viologen toxicity. Nucleic Acids Res. 1992;20:3159–3165. doi: 10.1093/nar/20.12.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother. 1998;42:1778–1782. doi: 10.1128/aac.42.7.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura H. Novel acriflavin resistance genes, acrC and acrD, in Escherichia coli K-12. J Bacteriol. 1979;139:8–12. doi: 10.1128/jb.139.1.8-12.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naroditskaya V, Schlosser M J, Fang N Y, Lewis K. An E. coli gene emrD is involved in adaptation to low energy shock. Biochem Biophys Res Commun. 1993;196:803–809. doi: 10.1006/bbrc.1993.2320. [DOI] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. Methods for dilution antibacterial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 30.Oden K L, DeVeaux L C, Vibat C R T, Cronan J E, Jr, Gennis R B. Genomic replacement in Escherichia coli K-12 using covalently closed circular plasmid DNA. Gene. 1990;96:29–36. doi: 10.1016/0378-1119(90)90337-q. [DOI] [PubMed] [Google Scholar]
- 31.Paulsen I, Sliwinski M, Saier M J. Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J Mol Biol. 1998;277:573–592. doi: 10.1006/jmbi.1998.1609. [DOI] [PubMed] [Google Scholar]
- 32.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purewal A S. Nucleotide sequence of the ethidium efflux gene from Escherichia coli. FEMS Microbiol Lett. 1991;66:229–231. doi: 10.1016/0378-1097(91)90338-b. [DOI] [PubMed] [Google Scholar]
- 34.Rosenberg E Y, Nikaido H. AcrD of Escherichia coli is an aminoglycoside pump. J Bacteriol. 2000;182:1754–1756. doi: 10.1128/jb.182.6.1754-1756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell C B, Thaler D S, Dahlquist F W. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J Bacteriol. 1989;171:2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shevell D, Abou-Zamzam A, Demple B, Walker G. Construction of an Escherichia coli K-12 ada deletion by gene replacement in a recD strain reveals a second methyltransferase that repairs alkylated DNA. J Bacteriol. 1988;170:3294–3296. doi: 10.1128/jb.170.7.3294-3296.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 38.Stermitz F R, Lorenz P, Tawara J N, Zenewicz L A, Lewis K. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc Natl Acad Sci USA. 2000;97:1433–1437. doi: 10.1073/pnas.030540597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thanassi D G, Cheng L W, Nikaido H. Active efflux of bile salts by Escherichia coli. J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner R J, Taylor D E, Weiner J H. Expression of Escherichia coli TehA gives resistance to antiseptics and disinfectants similar to that conferred by multidrug resistance efflux pumps. Antimicrob Agents Chemother. 1997;41:440–444. doi: 10.1128/aac.41.2.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaara M. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium. Antimicrob Agents Chemother. 1993;37:2255–2260. doi: 10.1128/aac.37.11.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Veen H W, Venema K, Bolhuis H, Oussenko I, Kok J, Poolman B, Driessen A J, Konings W N. Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR1. Proc Natl Acad Sci USA. 1996;93:10668–10672. doi: 10.1073/pnas.93.20.10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitney E N. The tolC locus in Escherichia coli K12. Genetics. 1971;67:39–53. doi: 10.1093/genetics/67.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yerushalmi H, Lebendiker M, Schuldiner S. EmrE, an Escherichia coli 12-kDA multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem. 1995;270:6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]