Summary
Adult stem cells coordinate intrinsic and extrinsic, local and systemic, cues to maintain the proper balance between self-renewal and differentiation. However, the precise mechanisms stem cells use to integrate these signals remain elusive. Here, we show that Escargot (Esg), a member of the Snail family of transcription factors, regulates the maintenance of somatic cyst stem cells (CySCs) in the Drosophila testis by attenuating the activity of the pro-differentiation insulin receptor (InR) pathway. Esg positively regulates the expression of an antagonist of insulin signaling, ImpL2, while also attenuating the expression of InR. Furthermore, Esg-mediated repression of the InR pathway is required to suppress CySC loss in response to starvation. Given the conservation of Snail-family transcription factors, characterizing the mechanisms by which Esg regulates cell-fate decisions during homeostasis and a decline in nutrient availability is likely to provide insight into the metabolic regulation of stem cell behavior in other tissues and organisms.
Keywords: insulin, InR, ImpL2, Drosophila, escargot, cyst stem cells, metabolism, testis
Graphical abstract

Highlights
-
•
Escargot (Esg) is required for somatic cyst stem cell (CySC) maintenance
-
•
Esg attenuates the insulin receptor (InR) pathway in cyst stem cells (CySCs)
-
•
Esg stimulates expression of the insulin antagonist ImpL2 and represses InR
-
•
Esg-mediated InR suppression is needed for stem cell preservation under starvation
Sênos Demarco et al. show that the transcription factor Escargot controls somatic stem cell fate in the Drosophila testis. Mechanistically, Escargot suppresses insulin receptor (InR) signaling in stem cells to prevent differentiation. InR signaling suppression by Escargot is required for the preservation of stem cells during homeostasis and starvation.
Introduction
Adult stem cells provide an important reservoir of cells capable of self-renewing and differentiating into specialized cells that regenerate tissues and organs throughout life. Stem cells are maintained through both cell-autonomous mechanisms, as well as through microenvironmental or “niche”-derived cues. The Drosophila testis presents an ideal model system for characterizing mechanisms regulating stem cell behavior and the relationship to the surrounding microenvironment due to the available molecular and genetic tools, as well as the conservation of numerous signaling pathways (Demarco et al., 2014; Greenspan et al., 2015). In the testis, two stem cell populations—germline stem cells (GSCs) and somatic cyst stem cells (CySCs)—are maintained by both structural cues and molecular signals derived from a group of somatic cells that form the hub (Figure 1A). GSCs divide to self-renew and to generate a daughter gonialblast (GB), which undergoes four rounds of mitotic, transit-amplifying (TA) divisions prior to initiating spermatocyte-specific gene expression. Spermatocytes will then undergo meiosis to generate haploid spermatids that will eventually mature into sperm (Fuller, 1993; Hardy et al., 1979). CySCs, on the other hand, will divide to generate cyst cells (CCs) that can either become a new CySC or, together with another CC, encapsulate the GB and begin differentiating in concert with the developing germline cyst (Figure 1A). In this manner, CCs act in an analogous fashion to mammalian Sertoli cells to support differentiation of the developing germ line (Zoller and Schulz, 2012).
Figure 1.
escargot expression regulates early cyst cell number in the Drosophila testis
(A) Schematic of the Drosophila testis. Hub cells (green) are surrounded by germline stem cells (GSCs; pink) and somatic cyst stem cells (CySCs; blue). CySCs express high levels of Zfh1 and TJ. Upon division, GSCs self-renew and differentiate into a gonialblast (GB), which then undergoes four rounds of mitotic, transit amplifying (TA) divisions with incomplete cytokinesis. CySCs divide to generate daughter cells that can either become new CySCs or, upon InR/TOR signaling (red outline, p4EBP+), encapsulate the GB and start to differentiate. Cyst cells (CCs) will continue to elongate and co-differentiate with the developing spermatogonial cyst. Early CCs express TJ, and only toward the end of germline TA divisions will CCs start to express Eya (green outline in mid CCs, green filling in late CCs).
(B–C‴) Immunofluorescence (IF) images of testis tips from flies harboring a GFP-based “enhancer trap” reporting the expression of escargot (esg).
(B–B‴) Testes were stained with antibodies against Zfh1 (blue, CySCs and very early CCs), Fasciclin 3 (Fas3; green, hub), Eya (green, mid/late CCs), and GFP (red, esg-GFP).
(C–C‴) Testes were stained with an antibody against TJ (blue, CySCs and early CCs), Fas3, Eya, and GFP.
(B–C‴) Blue arrows point to Zfh1+/Eya− or TJ+/Eya−, and yellow arrows point to Zfh1+/Eya+ or TJ+/Eya+, respectively.
(D–F‴) Representative images of testis tips from animals harboring the c587-GAL4ts “driver” (see STAR Methods) that have developed at 18°C and were shifted to 29°C upon eclosion. Testes were stained with antibodies against Zfh1 (red, CySCs and very early CCs), TJ (blue, early CCs), Eya (green, mid/late CCs), and Fas3 (hub). Arrows point to the following: red, Zfh1HIGH CCs; blue, TJ+/Eya− CCs; yellow, TJ+/Eya+ CCs; and green, Eya+ CCs.
(D–D‴) Testes from c587-GAL4ts>w1118 outcross (control).
(E–E‴) Testes from c587-GAL4ts>UAS-esgRNAi.
(F–F‴) Testes from c587-GAL4ts>esgEP2009 (overexpression).
(G) Quantification of the number of TJ+/Eya− CCs per testis in animals in which GAL4/UAS gene expression was induced for 5 days at 29°C. Two-tailed t test used.
(H) Quantification of the number of Zfh1HIGH CCs per testis in animals induced for 10 days at 29°C. Two-tailed t test used.
(G and H) Data are represented as mean ± SD. All experiments were repeated at least 3 times in pools of at least 20 animals per condition. In all images, dotted green line circles the hub; scale bars, 20 μm.
Several evolutionarily conserved signaling pathways, including the bone morphogenic protein (BMP), Janus kinase (Jak)/signal transducer and activator of transcription (STAT), and Hedgehog (Hh) pathways, regulate GSC and/or CySC maintenance in the testis (Greenspan et al., 2015). Cadherin-based cell-cell adhesion is also important for the maintenance of both stem cell populations within the niche at the tip of the testis. In addition to responding to niche-derived factors, GSCs also have cell-autonomous mechanisms that regulate asymmetric cell division and, as such, contribute to maintenance of the germ line (Nelson et al., 2019). The intrinsic mechanisms promoting CySC maintenance, on the other hand, are less well understood. Recent work has demonstrated that epidermal growth factor receptor/MAP kinase (EGFR/MAPK) signaling and Hh activity can influence the competition of CySC-daughter cells to populate the niche (Amoyel et al., 2014, 2016a). Moreover, a somatic differentiation niche is established by a burst of Drosophila insulin-like peptide (Dilp) signaling, which is required for the onset of somatic CC differentiation. The fly homolog of the mammalian insulin-like growth factor binding protein IGFBP7, imaginal morphogenesis protein-late 2 (ImpL2), was shown to be expressed and secreted by CySCs to attenuate insulin signaling and prevent differentiation (Amoyel et al., 2016b).
In addition to homeostatic and developmental signals, stem cells in the testis respond to metabolic cues. For instance, in flies, protein deprivation (hereafter referred to as “starvation”) leads to loss of GSCs (Mcleod et al., 2010). However, not all GSCs are lost; instead, late CCs “sense” starvation conditions and promote the death of both late cyst and germ cells in order to preserve a reduced number of GSCs at the tip of the testis (Yang and Yamashita, 2015). Interestingly, CySCs appear to be spared during starvation (Yang and Yamashita, 2015), but the precise mechanisms that regulate CySC maintenance under these conditions are not known.
The transcription factor Escargot (Esg) is a member of the Snail family of transcription factors, which are involved in cell-fate decisions, migration, and regulation of the epithelial-mesenchymal transition (EMT) during development and tumorigenesis (Lye and Sanson, 2011; Barrallo-Gimeno and Nieto, 2005; Wu and Zhou, 2010; Wang et al., 2013). In Drosophila, esg is required for development (Whiteley et al., 1992) and is expressed in adult tissue stem cell populations, including in the intestine and testis (Micchelli and Perrimon, 2006; Kiger et al., 2000; (Voog et al., 2014)). Indeed, esg is expressed in the embryonic testis and is one of the first genes to be expressed in a sexually dimorphic manner (Voog et al., 2014). Our lab has shown that esg is expressed in germline and somatic cells at the tip of the testis (Figures 1B–1C‴) and that esg is required cell autonomously for the maintenance of both the hub and CySCs. Clonal analysis demonstrated that loss of esg in CySCs resulted in loss of stem cell identity and exit from the niche (Voog et al., 2014); however, the mechanism by which Esg regulates CySC fate was not determined.
Here, we show that esg (Flybase: FBgn0287768) is required and sufficient to maintain a CySC-like state by repressing the pro-differentiation insulin signaling pathway. Overexpression of esg was sufficient to increase the number of CySC-like cells, increase ImpL2 (Flybase: FBgn0001257) expression, and reduce the expression and activity of the insulin receptor (InR) (Flybase: FBgn0283499). By contrast, loss of esg resulted in reduced ImpL2 and increased InR expression, which ultimately contributed to CySC loss. In addition, Esg activity and the suppression of the InR pathway are both required for the maintenance of CySCs and, consequently, preservation of the remaining GSCs under starvation conditions. Taken together, our data describe one mechanism by which a transcription factor controls stem cell maintenance through repression of a differentiation pathway and reveal that this mechanism can respond to changes in nutrient availability.
Results
Esg is necessary to support CySC maintenance and proliferation
CySCs and CCs can be visualized by immunofluorescence (IF) microscopy using a combination of antibodies against transcription factors that are expressed differentially throughout CC differentiation (Demarco et al., 2014). High levels of transcription repressor Zn finger homeodomain 1 (Zfh1) are observed in CySCs and very early CCs, while low levels are detectable until the mid CC stage. The Maf transcription factor traffic jam (TJ) is expressed in high levels in CySCs and early/mid CCs, while the differentiation factor eyes absent (Eya) is expressed in mid CCs and becomes highly expressed in late CCs, which are associated with spermatocyte cysts (Figures 1A and 1D–1D‴).
In order to further characterize the role of esg in regulating CySC behavior, the bipartite GAL4/UAS system was used to deplete and overexpress esg in early CCs (Brand and Perrimon, 1993). In order to bypass any developmental role of esg in the CC lineage, a ubiquitously expressed temperature-sensitive version of the GAL80 repressor (tub-GAL80ts) was used in combination with the CC “driver” c587-GAL4 to promote adult-only transgene expression (Mcguire et al., 2003) (for convenience, this system will be referred to as c587-GAL4ts). As expected, based on previous clonal analyses, when esg was depleted via RNAi, the number of early CCs was significantly reduced, as measured by the number of TJ+ CCs that do not express Eya (Amoyel et al., 2016b; Senos Demarco et al., 2020) (Figures 1E–E‴ and 1G). As another approach to quantify CySCs and very early CCs, the number of CCs at the very tip of the testis that expressed high levels of Zfh1 was quantified. Similar to the number of TJ+/Eya– CCs, the number of Zfh1HIGH CCs was also decreased when esg was depleted (Figures 1D′, 1E′, and 1H). A similar trend was obtained when esg was depleted using another, independent RNAi construct (Figures S1A, S1B, and S1D).
The number of proliferating CCs was assayed upon changes in esg expression as another approach to evaluate the effect of Esg on somatic stem cell activity. Consistent with the changes in early CC composition, the number of early CCs undergoing S phase decreased significantly when esg was depleted, as determined by the incorporation of the nucleotide analog 5-ethynyl-2'-deoxyuridine (EdU) (Figures S1E–S1H) (see STAR Methods). CC differentiation influences the adjacent germ cells; therefore, germline differentiation was also assayed. Spherical spectrosomes are found in GSCs and GBs, while highly branched fusomes, an endoplasmic reticulum (ER)-like organelle that branches through interconnected germ cells, are found in differentiating spermatogonial cysts (Figures 2A and 2A′). Moreover, GSCs and spermatogonia express high levels of the B-type nuclear lamina protein Lamin Dm0, while spermatocytes express the A-type Lamin C (Figures 2D, 2D′, 2G, and 2G′) (Chen et al., 2013; Hayashi et al., 2016). Consistent with a decrease in early CCs upon esg depletion, there was an apparent reduction in the region of the testis containing spermatogonia (Figures 2B, 2B′, 2E, 2E′, 2H, and 2H′). Because CySCs also participate in the transduction of signaling pathways required for GSC maintenance, the activities of the BMP and JAK/STAT pathways were analyzed (Figures 2J, 2J′, S2A, and S2A′). Depletion of esg in CCs resulted in the non-autonomous decrease in both pMad and nuclear accumulation of Stat92E, readouts of BMP and JAK/STAT signaling, respectively (Figures 2K, 2K′, S2B, and S2B′). Altogether, these data indicate that Esg is required for the maintenance of CySCs and early CCs and that it influences germline fate non-autonomously, consistent with our previously published results (Voog et al., 2014).
Figure 2.
esg levels in CCs non-autonomously control germline differentiation
(A–I′) Representative images of testis tips stained with antibodies against Vasa (red, germline), TJ (gray, CySCs and early CCs), Fas3 (green, hub), and proteins associated with germline differentiation.
(A–C′) Hts (green, fusome). Spherical spectrosomes are highlighted using white arrows, while branched fusomes are highlighted using yellow arrows.
(D–F′) LamDm0 (green, nuclear lamina), highly expressed in spermatogonia.
(G–I′) LamC (green, nuclear lamina), highly expressed in spermatocytes.
(D–I′) White arrows point to spermatogonia, while yellow arrows point to spermatocytes.
(J–L′) pMad stains (red) show BMP activity in GSCs (white arrows) and Eya+ CCs (yellow arrows). All experiments were repeated at least 3 times in pools of at least 20 animals per condition. In all images, dotted green line circles the hub; scale bars, 20 μm.
Esg is sufficient to support a CySC-like state
While loss of esg led to loss of CySCs and early CCs, overexpression of esg resulted in a significant increase in early CCs, including CySCs (Figures 1F–1H, S1C, and S1D). Consistent with these observations, the number of TJ+ early CCs that incorporated EdU or stained positive for phosphorylated histone H3 (pHH3) increased when esg was overexpressed, indicating that Esg is sufficient to drive somatic cell proliferation (Figures S1C, S1E, and S1I–K) (see STAR Methods). CCs overexpressing esg accumulated independently of germ cells (Figures 2C, 2C′, 2I, and 2I′). In addition, early CCs overexpressing esg were capable of supporting GSCs and early spermatogonia, as reflected by germline Lamin expression (Figures 2F, 2F′, 2I, and 2I′) and the accumulation of spherical and bar-bell-shaped fusomes (Figures 2C and 2C′).
Although an expansion of TJ+/Eya– CCs was clearly observed (Figures 1F and 1G), not all of those cells were Zfh1HIGH. Rather, Zfh1HIGH CCs were observed in “patches” throughout testis tips (Figures 1F and 1F′). As Zfh1 is a target of both JAK/STAT and Hh pathways (Michel et al., 2012; Leatherman and Dinardo, 2008) and both pathways are activated by the secretion of ligands from the hub, the difference in Zfh1 levels observed upon esg overexpression may be reflecting the activity of these pathways in CCs. As expected, when esg was overexpressed, STAT and Hh activities were high in CCs in the CySC position close to the hub (Figure S2A–S2C′, and S2E–S2G′). However, STAT nuclear accumulation and Ptc expression, a readout of Hh pathway activity, could also be observed in patches of CCs far away from the testis tip (Figures S2D, S2D′, S2H, and S2H′). Moreover, when esg was overexpressed in CCs throughout development (i.e., without tub-GAL80TS), a more dramatic accumulation of CySC-like cells was observed. Under these conditions, CCs far from the hub were also observed to stain positive for STAT and Hh activity (Figures S3A–S3D”), undergo mitosis (Figures S3E and S3F′), and support early spermatogonia (Figures S3G and S3H′). Taken together, these data suggest that esg overexpression is sufficient to induce a CySC/early CC fate, regardless of proximity to the hub.
Esg is required for the preservation of CySCs in response to changes in nutrient availability
Previous studies demonstrated that short-term protein deprivation results in loss of male GSCs, which is reversible upon a shift back to normal nutrient conditions (Mcleod et al., 2010; Yang and Yamashita, 2015). The number of late CCs also decreases; however, CySCs and very early CCs are preserved under these conditions (Figures 3A–3D) (Yang and Yamashita, 2015). Consistent with these observations, the activity of the somatic pro-differentiation InR/PI3K/Tor pathway also decreases, as assayed by measuring phosphorylation of the Tor target 4EBP (Figures 3E–3G) (see STAR Methods), indicating that CC differentiation is likely impaired.
Figure 3.
esg is required for early CC maintenance under starvation conditions
(A–B″) Representative images of testis tips stained with antibodies against TJ (blue, CySCs and early CCs), Eya (green, mid/late CCs), and Fas3 (green, hub). Arrows point to the following: blue, TJ+/Eya− CCs; yellow, TJ+/Eya+; and green, Eya+. Gray insets show Vasa stains (early germ cells) in testis niche. (A)–(A″) show a testis from control (w1118) flies fed a regular diet for 14 days, while control flies in (B)–(B″) were starved in parallel.
(C) Quantification of the number of GSCs per testis.
(D) Quantification of the number of TJ+/Eya− CCs per testis.
(C and D) Two-tailed t test used.
(E–F′) Images of testis tips stained for the InR/PI3K/TOR target p4EBP (red), TJ (early CCs, white), Fas3 (hub, green), and 4′,6-diamidino-2-phenylindole (DAPI; nuclei, blue). (E) and (E′) show a testis from a fly fed a regular diet, while in (F) and (F′), the fly was starved.
(G) Quantification of the p4EBP signal intensity in differentiating CCs (see STAR Methods). Two-tailed t test used.
(H and I) Quantification of the number of GSCs (H) or TJ+/Eya− CCs (I) in testes from c587-GAL4ts>UAS-TdTomato or c587-GAL4ts>UAS-esgRNAi fed either a regular diet or starved for 14 days or those that were switched back to a regular diet for 7 days (after the initial 14-day starvation). Two-tailed t test used.
(C, D, and G–I) Data are represented as mean ± SD. All experiments were repeated at least 3 times in pools of at least 20 animals per condition. In all images, dotted green line circles the hub; scale bars, 20 μm.
When CySCs and early CCs are depleted of esg in flies that are protein deprived, there was a significant decrease in the number of TJ+/Eya− early CCs when compared with testes from control flies fed a similar diet. In addition, the number of TJ+/Eya− early CCs was significantly lower than the number resulting from esg depletion under normal nutrient conditions (Figure 3H). The decrease in early CCs resulting from esg depletion impacted GSC maintenance as well, leading to a further reduction in GSCs when compared with control flies subjected to protein deprivation or depletion of esg in flies fed a normal diet (Figure 3I). Importantly, depletion of esg prevented complete recovery of CySCs and GSCs when flies were shifted back to normal nutrient conditions (Figure 3I). These data suggest that Esg plays an important role in in the maintenance of CySCs during acute metabolic stress in addition to a previously documented role in CySC maintenance under homeostatic conditions. Furthermore, the presence of Esg is important for stem cell regeneration when nutrient conditions improve.
Esg controls the expression of ImpL2 and InR
In order to determine how Esg controls CySC maintenance and the onset of CC differentiation, putative targets of esg in the Drosophila testis were determined through the use of DNA adenine methylase identification (DamID) technology, which allows for mapping of transcription-factor binding to DNA in vivo (Southall and Brand, 2009, Van Steensel and Henikoff, 2000). Similar to our previous work in the intestine (Loza-Coll et al., 2014), a Dam:esg fusion transgene was expressed in the testis in order to methylate adenine nucleotides surrounding Esg-binding sites, and genomic DNA from testes was extracted and processed in order to determine “peaks” of Esg-binding (Table S1) (Choksi et al., 2006; see STAR Methods). Interestingly, Esg was found to bind to sites within, and in close proximity to, the ImpL2 and InR transcription units. As noted above, ImpL2 and InR have been demonstrated to regulate CySC maintenance and CC differentiation, respectively (Amoyel et al., 2016b) (Figures 4A and 4B). Given the important roles that ImpL2 and InR also play in metabolic signaling, we wanted to determine whether these two genes are physiologically relevant Esg targets. ImpL2 is expressed in CySCs to attenuate insulin signaling, while InR is expressed in CySCs and CCs, and is required for the onset of CC differentiation (Amoyel et al., 2016b) (Figures 4C–4D‴). Because Esg is required for CySC maintenance, we hypothesized that Esg may positively regulate ImpL2 expression and repress InR expression in order to modulate InR signaling, thereby influencing CySC maintenance and differentiation in a coordinated fashion. In order to test whether Esg is sufficient to regulate the expression of these two genes, Drosophila S2 cells stably expressing either an N-terminal localization and affinity purification (NLAP)-tagged Esg fusion protein (NLAP-Esg) or a control (NLAP) were established (as in Kyriakakis et al., 2008; Voog et al., 2014), and gene expression levels were determined by microarray in response to NLAP-Esg overexpression, relative to control (Table S2) (see STAR Methods). While InR levels were significantly reduced upon expression of NLAP-esg (∼2-fold decrease compared with NLAP alone), the expression of ImpL2 was significantly upregulated (∼2-fold increase compared with NLAP alone) (Figure 4E; Table S2), suggesting that Esg expression is sufficient to drive changes in ImpL2 and InR expression. In order to test whether Esg is required for the expression of ImpL2 in testes in vivo, esg was depleted for 2 days in adult CCs, a time point prior to significant changes in cell composition, and quantitative reverse-transcription PCR (qRT-PCR) was performed (see STAR Methods). Upon esg depletion, the levels of ImpL2 decreased significantly (Figure 4F), suggesting that Esg acts in early CCs to positively regulate ImpL2 expression. By contrast, the depletion of esg led to increased InR mRNA expression levels (Figure 4G), consistent with what was observed in the DamID and microarray datasets (Tables S1 and S2).
Figure 4.
Esg regulates expression of ImpL2 and InR
(A and B) Average plots of Dam-Esg occupancy from three independent biological sample pools. Gene loci for ImpL2 (A) and InR (B) are represented in blue, with exons represented in dark red. Light gray boxes display additional genes. Dam-Esg occupancy is displayed below in green. Significant binding sites are represented by dark gray rectangles under the plot. Y axis represent the log2 ratio of Dam-Esg over Dam alone (average for each GATC fragment; see STAR Methods).
(C and D‴) IF images of testis tips from flies harboring a GFP-based “enhancer trap” reporting the expression of esg.
(C–C‴) Testes were stained with antibodies against TJ (blue, CySCs and very early CCs), Fas3 (Fas3, blue, hub), ImpL2 (green puncta), and GFP (red, esg-GFP).
(D–D‴) Testes were stained with an antibody against TJ (blue, CySCs and early CCs), Fas3, InR (green, membrane bound), and GFP.
(C–D‴) White arrows point to signal in cells in the CySC position.
(D) Pink arrows point to signal in CCs. Dotted green line circles the hub; scale bars, 20 μm; experiments were repeated at least 3 times in pools of at least 20 animals per condition.
(E) Representation of the fold change in gene expression between NLAP-Esg and NLAP only for InR and ImpL2. Two-tailed t test used.
(F and G) qRT-PCR quantification of ImpL2 mRNA (F) or InR mRNA (G) normalized to Act5c mRNA. Each dot represents a pool of 150 biological samples (i.e., whole testes) of each condition (control and esgRNAi). Depletion of esg was achieved by inducing the RNAi response in CCs with the c587-GAL4ts driver for 2 days (at which point no significant changes to CC number were observed). Two-tailed t test used.
(F and G) Data are represented as mean ± SD.
CySC maintenance is regulated by the interaction between esg and ImpL2
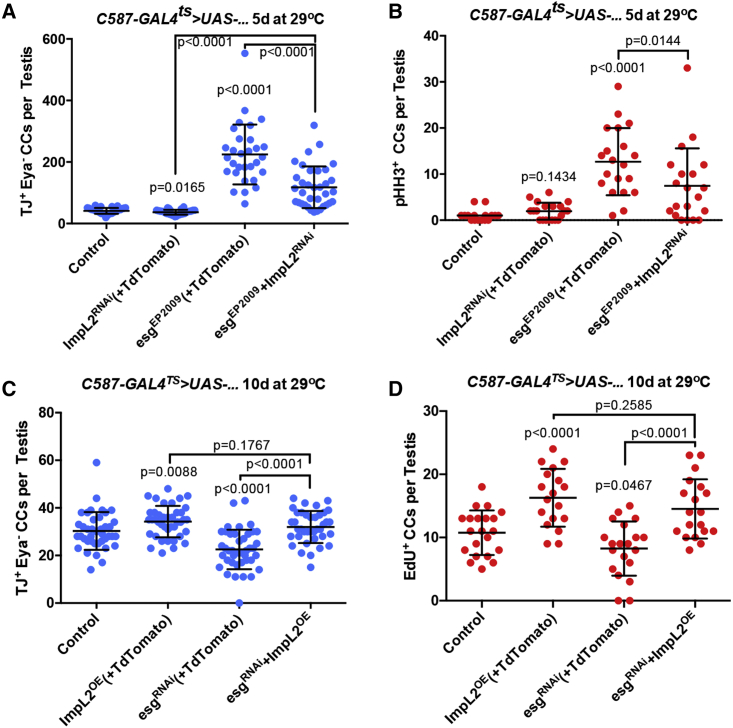
In order to test whether esg and ImpL2 interact genetically, the number of early CCs was quantified upon manipulation of ImpL2 and esg, both individually and in combination. Consistent with previous results demonstrating a role for ImpL2 in CySCs (Amoyel et al., 2016b), conditional RNAi-mediated depletion of ImpL2 with c587-GAL4ts resulted in a modest, but significant, decrease in the number of TJ+/Eya− CCs (Figures 5A and S4A–S4B″), while overexpression of ImpL2 led to a modest increase in early CC number (Figures 5C and S4D–S4D″). Strikingly, the increase in TJ+/Eya− CCs observed with the overexpression of esg (with “UAS-TdTomato” to control for UAS levels; see STAR Methods) (Figure 1G) was significantly suppressed when ImpL2 was depleted (Figures 5A, S4C–S4C″). A similar trend was observed when assaying the number of mitotic CCs per testis (Figure 5B), indicating that the ability of Esg to promote an increase in CySC number is dependent on ImpL2. Consistent with these findings, overexpression of ImpL2 was sufficient to rescue the decrease in CC number resulting from esg depletion (Figures 5C and S4E–S4E″). The number of EdU+ CCs also increased, demonstrating that ImpL2 acts downstream of Esg to regulate early CC activity (Figure 5D). Taken together, these data strongly suggest that esg acts upstream of ImpL2 to maintain CySCs in the testis.
Figure 5.
esg positively interacts with ImpL2 to maintain CySCs
(A) Quantification of the number of TJ+/Eya− CCs per testis at 5 days of induction. Two-tailed t test used.
(B) Quantification of pHH3+ CCs per testis at 5 days of induction. Two-tailed t test used.
(C) Quantification of the number of TJ+/Eya− CCs per testis at 10 days of induction. Two-tailed t test used.
(D) Quantification of EdU+ CCs per testis at 10 days of induction. Two-tailed t test used.
All data are represented as mean ± SD. All experiments were repeated at least 3 times in pools of at least 20 animals per condition. Note that UAS-TdTomato was used to balance the number of UAS elements, as described in STAR Methods. See Figure S3 for representative images used in the displayed quantifications.
esg and InR interact genetically to regulate CC differentiation
The relationship between esg and InR was also tested using a combination of loss- and gain-of-function genetic analyses. The constitutive activation of the InR was achieved by overexpressing a construct in which a single amino acid (A1325D, “InRCA”) was replaced in the transmembrane domain to enhance ligand-independent activation, mimicking the human V938D mutation. Expression of InRCA resulted in a very modest decrease in the number of TJ+/Eya− CCs (Figures 6A and S5A–S5B″), while the RNAi-mediated depletion of InR resulted in a very modest increase in early CC number (Figures 6C and S5D–S5D″), consistent with the role of InR in priming somatic cells for CC differentiation (Amoyel et al., 2016b). The increases in TJ+/Eya− and pHH3+ CCs observed with esg overexpression (Figures 1G and S1K) were significantly suppressed when InRCA was expressed simultaneously (Figures 6A, 6B, and S5C–S5C″). Furthermore, depletion of esg resulted in a decrease in TJ+/Eya− CCs and EdU+ CCs (Figures 1G and S1H), which was suppressed by co-depletion of InR and esg (Figures 6C, 6D, and S5E–S5E″). Together, these data support a model in which the ability of Esg to regulate CySC maintenance and proliferation could be due to enhanced expression of ImpL2 and repression of InR, in order to suppress InR signaling and block the differentiation of CySCs.
Figure 6.
esg negatively interacts with InR to prevent CySC differentiation
(A) Quantification of the number of TJ+/Eya− CCs per testis at 5 days of induction. Two-tailed t test used.
(B) Quantification of pHH3+ CCs per testis at 5 days of induction. Two-tailed t test used.
(C) Quantification of the number of TJ+/Eya− CCs per testis at 10 days of induction. Two-tailed t test used.
(D) Quantification of EdU+ CCs per testis at 10 days of induction. Two-tailed t test used.
All data are represented as mean ± SD. All experiments were repeated at least 3 times in pools of at least 20 animals per condition. Note that UAS-TdTomato was used to balance the number of UAS elements, as in Figure 4. See Figure S4 for representative images used in the displayed quantifications.
Inhibition of InR signaling suppresses loss of stem cells due to esg depletion
Upon binding to a Dilp, InR activates the phosphoinositide 3-kinase/target of rapamycin (PI3K/TOR) pathway, which is required for the initial onset of CC differentiation (Amoyel et al., 2016b). Therefore, we hypothesized that Esg expression would be sufficient to inhibit InR-mediated activation of TOR signaling in CCs. To assay changes in TOR signaling, the levels of phosphorylation of 4EBP, a highly conserved target of TOR, were assayed in vivo via IF (Gingras et al., 1999; Miron et al., 2001). Under homeostatic conditions, a ring of p4EBP+ CCs in the initial stages of differentiation is observed roughly 1-cell diameter away from the hub (Figures S6A and S6A′), consistent with the onset of InR/TOR activation (Amoyel et al., 2016b). However, no p4EBP signal was detected in CCs upon conditional overexpression of esg with c587-GAL4ts (Figures S6B and S6B′). These results suggest that InR/PI3K/TOR signaling is strongly reduced upon Esg expression and that Esg may be sufficient to repress InR/TOR signaling in CCs. Conversely, when esg was depleted in early CCs, strong p4EBP staining was observed in somatic cells immediately adjacent to the hub (Figures S6C and S6C′), further supporting our model that Esg expression maintains low InR/PI3K/TOR activity in these cells to support CySC maintenance by blocking differentiation.
To test whether the increase in InR/PI3K/TOR activity upon depletion of esg mediates loss of CySCs through differentiation, flies were fed food with or without rapamycin, a potent TOR inhibitor (Sabatini, 2017). Similar to the suppression of early CC loss and cell division observed when ImpL2 was overexpressed (Figures 5C and 5D) or when InR was depleted (Figures 6C and 6D), rapamycin treatment also rescued the CC phenotypes caused by the depletion of esg (Figures S6D and S6E), indicating that TOR activity contributes to loss of CySCs caused by the depletion of esg.
Suppression of the InR pathway is required for the preservation of CySCs during starvation
As data suggest that Esg negatively regulates the InR pathway in CySCs under homeostatic conditions and Esg activity is required for the preservation of CySCs under starvation conditions (Figure 3H), we next sought to determine whether the Esg targets ImpL2 and InR were also involved in the preservation of CySCs during starvation. Strikingly, both esg and ImpL2 mRNA levels were upregulated in testes from starved flies, while InR mRNA was slightly downregulated (Figure 7A). These data suggest that the expression of esg and its target genes ImpL2 and InR are responding to changes in the overall metabolic state of the organism. Similar to esg (Figures 3H and 3I), ImpL2 was required cell autonomously for the preservation of CySCs upon starvation (Figure 7B) and to prevent a further decrease in GSCs upon starvation (Figure 7C). Moreover, ImpL2 depletion in CySCs and early CCs prevented the recovery of GSCs upon shifting from starvation back to normal nutrient conditions (Figure 7C). Ectopic activation of the InR pathway in CySCs and early CCs, achieved via expression of InRCA in early CCs (Figures 7B and 7C), led to a decline in TJ+/EyA− early CCs and GSCs and prevented the recovery of GSCs upon shifting back to a regular diet (Figures 7B and 7C). Hence, Esg activity and the suppression of the InR pathway are required in CySCs for preservation of both somatic and GSCs in the testis during metabolic stress.
Figure 7.
Suppression of the InR pathway is required for stem cell maintenance under starvation conditions
(A) qRT-PCR quantification of esg, ImpL2, and InR mRNA levels normalized to Act5c mRNA from testes from control flies (w1118) fed a regular diet or starved for 14 days. Each dot represents a pool of 150 biological samples (i.e., whole testes) of each condition. Two-tailed t test used.
(B and C) Quantification of the number of TJ+/Eya− CCs (B) and GSCs (C) in testes from c587-GAL4ts>UAS-TdTomato (control), c587-GAL4ts>UAS-ImpL2RNAi, or c587-GAL4ts>UAS-InRCA fed either a regular diet or starved for 14 days or those that were switched back to a regular diet for 7 days (after the initial 14-day starvation). Two-tailed t test used.
All data are represented as mean ± SD. Experiments were repeated at least 3 times in pools of at least 20 animals per condition.
Discussion
The results presented here advance our understanding of how a member of the Snail family of transcription factors, Esg, controls somatic stem cell behavior in the Drosophila testis. Previous results indicated that esg is expressed at the tip of the testis (Figures 1B–1C‴), where it is required autonomously for maintaining CySCs and hub cell fates (Voog et al., 2014). Here, we provide a mechanism by which Esg serves to support CySC maintenance, as assayed through combinations of early CC markers and markers of cell proliferation (Figures 1, S1, 2, S2, and S3). In addition to playing an essential role during tissue homeostasis, Esg is also required for somatic stem cell maintenance under metabolic challenges, such as starvation (Figures 3A–3I). DamID was used to provide insight into putative targets of Esg in the testis, which revealed that Esg bound close to control regions of two genes involved in the insulin signaling pathway, ImpL2 and InR (Figures 4A and 4B). Subsequent assays suggested that Esg, indeed, could enhance expression of ImpL2 while repressing expression of InR (Figures 4E–4G). Genetically, esg was found to positively interact with ImpL2 to maintain CySCs and early CCs (Figures 5A–5D and S4A–S4E″) and to negatively interact with InR to prevent CySC loss through differentiation (Figures 6A–6D and S5A–S5E″). Moreover, esg expression was found to be sufficient to modulate InR-mediated TOR activity in early CCs (Figures S6A–S6C′), which contributes to the loss of early CC as a consequence of esg depletion (Figures S6D and S6E). Finally, Esg-mediated suppression of the InR pathway in CySCs is required for the maintenance of CySCs under starvation conditions, which in turn is important for the preservation of the remaining GSCs (Figures 7A–7C).
Esg has been characterized to act as both an enhancer and repressor of gene expression, depending on the cellular context (Voog et al., 2014; Loza-Coll et al., 2014; Tanaka-Matakatsu et al., 1996; Korzelius et al., 2014; Miao and Hayashi, 2016). Our data here suggest that Esg promotes the expression of ImpL2 while suppressing InR expression in early CCs. One explanation for the ability of Esg to play such a dual role is its ability to interact with multiple binding partners, including the repressor C-terminal binding protein (CtBP) (Voog et al., 2014). CtBP has also been shown to be required for CySC maintenance (Leatherman and Dinardo, 2008). It will be interesting to investigate whether Esg acts through CtBP to downregulate InR and what additional Esg-binding partners are important for the positive regulation of ImpL2 (Loza-Coll and Jones, 2016). Moreover, given the fact that Esg putatively controls the expression of many other genes (Tables S1 and S2), future studies will aim to address whether these genes are also important in regulating CySC behavior in an Esg-dependent manner.
Esg has been shown to repress differentiation in another stem cell population in Drosophila, the intestinal stem cells (ISCs). In the intestine, Esg represses Notch signaling, in part, by repressing Amun, a negative regulator of the Notch pathway, which prevents ISC loss due to differentiation (Loza-Coll et al., 2014). Esg also regulates the fate of intestinal progenitor cells, as loss of esg resulted in the accumulation of secretory enteroendocrine cells, at the expense of absorptive enterocytes (Loza-Coll et al., 2014). In addition, Esg also represses expression of the POU-domain transcription factor Pdm1, which is enriched in enterocytes, to maintain stemness (Korzelius et al., 2014). Interestingly, mammalian Snai1 has also been shown to be required for the maintenance of ISCs and the regulation of lineage choice in the mouse intestinal epithelium (Horvay et al., 2015). In neuroblasts, the neural stem/progenitor cells in flies, Esg, together with family members Snail and Worniu, is required for the proper expression of genes involved in cell division and for specifying asymmetric divisions (Ashraf and Ip, 2001). Hence, Snail-family members act to maintain stem cell populations by promoting proliferation and asymmetric outcomes to stem cell divisions while also repressing differentiation.
Our data also suggest an intriguing role for Esg in responding to metabolic changes at the organismal level and integrating a cellular response within the testis. Although the InR/PI3K/TOR pathway has been shown to regulate CySC differentiation (Amoyel et al., 2016b), the effects of different diets on CySC maintenance have not been explored thoroughly. During starvation, germ cells and differentiating CCs undergo cell death in order to preserve a smaller population of GSCs at the tip of the testis (Yang and Yamashita, 2015); importantly, CySC maintenance is required for preservation of the GSCs that remain in the testis niche (Figures 3A–3D). Here we find that CySCs rely on Esg activity and the suppression of the InR pathway for maintenance (Figures 3E–3H, 7A, and 7B). Upon depletion of esg or ImpL2, or the overexpression of constitutively active InR, all of which ectopically activate the InR pathway in CySCs, the number of GSCs was reduced further in testes from starved flies (Figures 3I and 7C). These data suggest a model in which the Esg-mediated repression of the InR pathway in CySCs is a strategy employed to maintain CySCs and to act as a “safeguard” to prevent a loss of CySCs and an exacerbated loss of GSCs upon metabolic stress. Of note, the tumor necrosis factor (TNF) homolog Eiger and JNK signaling have been implicated in the regulation of CCs in starved animals (Chang et al., 2020). As such, one interesting possibility is that the TNF-JNK pathway could be interacting with Esg to preserve CySCs during starvation.
In sum, our work advances the understanding of how Esg acts to maintain somatic stem cells in the Drosophila testis and provides a model for how Snail-family members may control stem cell behavior across systems. Through the regulation of the InR pathway, Esg integrates homeostatic, developmental, and metabolic cues for the maintenance of stem cells and tissue homeostasis in the testis.
Limitations of the study
Because RNAi-based knockdowns were performed in the genetic-relationship experiments, likely resulting in partial loss of function, a potential caveat is that esg and ImpL2/InR could be acting in parallel pathways to control CySC maintenance. However, additional evidence presented in this work, including the Dam:Esg target mapping and measuring levels of ImpL2 and InR upon up/downregulation of esg, together with the genetic epistasis analyses, support a model in which esg acts upstream of ImpL2 and InR to control CySC maintenance.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse monoclonal anti-Fasciclin 3 (7G10) (1:50) | Developmental studies hybridoma bank (DSHB) | RRID: AB_528238 |
| mouse monoclonal anti-Eyes absent (10H6) (1:10) | DSHB | RRID: AB_528232 |
| rat anti-shg/DE-Cadherin (DCAD2) (1:20) | DSHB | RRID: AB_528120 |
| mouse anti-Hts (1B1) (1:10) | DSHB | RRID: AB_528070 |
| mouse anti-Ptc (Apa 1.3) (1:500) | DSHB | RRID: AB_528441 |
| mouse anti-LamDm0 (ADL101) (1:50) | DSHB | RRID: AB_528332 |
| mouse anti-LamC (LC28.26) (1:50) | DSHB | RRID: AB_528339 |
| rabbit anti-Vasa (d260) (1:100) | Santa Cruz Biotechnology | Cat#sc-30210; RRID: AB_793874 |
| rabbit anti-pSmad1/5 (41D10) (1:100) | Cell Signaling Technology | Cat#9516T; RRID: AB_491015 |
| chicken anti-Green Fluorescent Protein (GFP-1020) (1:1000) | Aves Labs | Cat#GFP-1020; RRID: AB_10000240 |
| rabbit anti-P-4E-BP1 (Thr37/46 236B4, 2855S) (1:500) | Cell Signaling Technology | Cat#2855; RRID: AB_560835 |
| rabbit anti-Zfh1 (1:500) | Ruth Lehmann (New York University) | N/A |
| rabbit anti-Stat92E (1:800) | Denise Montell (UCSB) | N/A |
| guinea pig anti-Traffic Jam (1:100) | Dorothea Godt (U. Toronto, Canada) | RRID: AB_2568583 |
| Mouse monoclonal anti-phospho-Histone H3 (Ser10) (6G3) | Cell Signaling Technology | Cat#9706; RRID: 331748 |
| Chemicals, peptides, and recombinant proteins | ||
| Rapamycin | TSZCHEM | Cat#R1017; CAS#53123-88-9 |
| Trizol | Life Technologies | Cat#15596026 |
| Linear Poly-Acrylamide | Sigma | Cat#56575 |
| DNase Q1 | Promega | Cat# M610A |
| iScriptkit | Biorad | Cat#170-8841 |
| Sso Advanced SYBR Green | Biorad | Cat#1725-264 |
| Critical commercial assays | ||
| Click-iT™ Plus EdU Cell Proliferation Kit for Imaging, Alexa Fluor™ 555 dye | Invitrogen/ThermoFisher | Cat# C10638 |
| Deposited data | ||
| Dam:Esg raw array data | This paper | GEO: GSE157685 |
| Experimental models: Cell lines | ||
| D. melanogaster: Cell line S2: S2- NLAP | (Voog et al., 2014) | N/A |
| D. melanogaster: Cell line S2: S2- NLAP:Esg | (Voog et al., 2014) | N/A |
| D. melanogaster: Cell line S2: S2- NLAP:EsgG387E | (Voog et al., 2014) | N/A |
| Experimental models: Organisms/strains | ||
| D. melanogaster: tub-Gal80TS | Bloomington Drosophila Stock Center | BDSC:7108 |
| D. melanogaster: UAS-InRRNAi [JF01482] | Bloomington Drosophila Stock Center |
BDSC:31037 |
| UAS-esgRNAi[HMS02538] | Bloomington Drosophila Stock Center |
BDSC:42846 |
| D. melanogaster: D. melanogaster: UAS-InRA1325D (“InRCA”) | Bloomington Drosophila Stock Center |
BDSC:8440 |
| D. melanogaster: UAS-IVS-myr::TdTomato | Bloomington Drosophila Stock Center |
BDSC:32221 |
| D. melanogaster: UAS-ImpL2RNAi[GD6004] | Vienna Drosophila RNAi Center | v30930 |
| D. melanogaster: UAS-esgRNAi[GD1437] | Vienna Drosophila RNAi Center | v9793 |
| D. melanogaster: UAS-ImpL2, UAS-ImpL2 | Kyoto Stock Center | DGRC:117649 |
| D. melanogaster: ptc-GFP (enhancer trap) | Carnegie collection, through A. Spradling and M. Buszczak (Carnegie Institution of Washington) | CB02030 |
| D. melanogaster: UAS-esgNLAP | (Voog et al., 2014) | N/A |
| D. melanogaster: esg-GFP | Lynn Cooley, Yale | P01986 |
| D. melanogaster: esgEP2009 | Berkeley Drosophila Genome Project, ((Rørth et al., 1998); (Abdelilah-Seyfried et al., 2000)) | N/A |
| D. melanogaster: w1118 | D. Walker, UCLA | FBal0018186 |
| D. melanogaster: c587-Gal4 | T. Xie (Stowers Institute of Biomedical Research) | N/A |
| D. melanogaster: UAS-Dam-esg | (Loza-Coll et al., 2014) | N/A |
| D. melanogaster: UAS-Dam | (Loza-Coll et al., 2014) | N/A |
| Oligonucleotides | ||
| Primer Act5C For: TTGTCTGGGCAAGAGGATCAG | (Senos Demarco et al., 2020) | N/A |
| Primer Act5C Rev: ACCACTCGCACTTGCACTTTC | (Senos Demarco et al., 2020) | N/A |
| InR Fwd: GCACCATTATAACCGGAACC | This paper | N/A |
| InR Rev: TTAATTCATCCATGACGTGAGC | This paper | N/A |
| ImpL2 Fwd: GCCGATACCTTCGTGTATCC | This paper | N/A |
| ImpL2 Rev: TTTCCGTCGTCAATCCAATAG | This paper | N/A |
| Esg Fwd: CGCCAGACAATCAATCGTAAGC | (Loza-Coll et al., 2014) | N/A |
| Esg Rev: TGTGTACGCGAAAAAGTAGTGG | (Loza-Coll et al., 2014) | N/A |
| Software and algorithms | ||
| Prism v6.0 | Graphpad | N/A |
| Illustrator CC 2015 | Adobe | N/A |
| Photoshop CC 2015 | Adobe | N/A |
| Image J v2.0.0 | Wayne Rasband, NIH | http://imagej.nih.gov/ij |
| CFX ManagerTM | Biorad | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, D. Leanne Jones (leanne.jones@ucsf.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Male flies were raised on a standard cornmeal and molasses diet (“regular diet”) with no more than 25 flies per vial. For TOR inhibition, Rapamycin (final concentration - 400μM) was mixed with regular molten Drosophila media and poured into vials. Eclosed flies of the specific genotypes were transferred to Rapamycin-containing food vials and transferred every 2-3 days. Starvation diet was achieved by maintaining adult flies in vials containing 10% ultrapure sucrose, 1% agar, and transferred every 2-3 days. Specific fly strains are listed in the key resources table.
Method details
Tissue-specific genetic manipulation
To express transgenes in CySCs and early CCs using c587-GAL4ts (c587-GAL4/Y;tub-GAL80ts/+;+/+), crosses were performed and maintained at 18°C until eclosion. Males were then shifted after eclosion to 29°C to induce the expression of UAS-driven transgenes. Flies maintained at 29°C were transferred onto new food every 2-3 days and were dissected after 5 or 10 days as stated. Control flies were the progeny of outcrosses from the GAL4 driver line to w1118 flies. When determining the genetic interaction between two UAS-based constructs, UAS-levels were taken into consideration – UAS-TdTomato was incorporated into every cross using a single UAS-element (i.e., UAS-InRCA or UAS-esgEP2009) but not incorporated in crosses using 2 UAS-elements (i.e., UAS-InRCA + UAS-esgEP2009). The control used for these epistatic analyses was the progeny of the cross between c587-GAL4ts and UAS-TdTomato.
Immunostaining
Testes from adult flies were dissected and fixed in a 4% paraformaldehyde solution for 30 minutes. Samples were washed 15 minutes twice with 0.1% Triton X-100 in PBS (PBS-T) with 0.3% sodium deoxycholate, then washed once for 10 minutes with PBS-T. Testes were blocked with a 3% bovine serum albumin (BSA) solution in PBS-T and incubated overnight at 4°C in primary antibodies diluted in the block solution. Samples were then washed for 10 minutes three times with PBS-T, incubated in Alexa-conjugated secondary antibody (1:500, Invitrogen) with 3% BSA in PBS-T, washed 10 minutes three times, and mounted in vectashield with DAPI (Vector Labs). Primary antibodies and concentrations used are listed in the key resources table. Samples were imaged with a Carl Zeiss LSM 780 or 880 Confocal microscope, or Axio Vert.A1 inverted light microscope with a 40x water-immersion or 63x oil-immersion objective. Digital images were processed using ZEN digital imaging (version 4.1, Zeiss), Image J (v2.0.0, Wayne Rasband, National Institute of Health, http://imagej.nih.gov/ij), and Adobe Illustrator software.
EdU incorporation assays
EdU incorporation was done using the Click-iT EdU Imaging kit (Invitrogen). Testes were dissected in 1X Ringer’s buffer (NaCl 155mM, KCl 5mM, CaCl2 2mM, MgCl2 1mM, NaH2PO4 2mM, HEPES 10mM, Glucose 10mM) and incubated in a 30μM EdU/1X Ringer’s buffer solution during 30 min. Testes were fixed 20min in 4% formaldehyde, washed twice 5min in 3%BSA/1XPBS, and incubated 20min in 1X PBS 0.3% Triton X-100 (PBST) 0.3% Sodium Deoxycholate. Testes were then incubated for 30min with the Click-iT reaction cocktail, rinsed and subsequently blocked in 3% BSA/PBST. Samples were then subjected to the regular IF protocol.
Characterization of CySC and early cyst cells
Quantification of early cyst cells
Zfh1HIGH CCs were counted as CCs located approximately within two rows of CC nuclei away from the hub (a clear decrease in Zfh1 intensity is observed beyond this limit). Since CySCs are the only dividing cells in the CC lineage, all cells within this region were counted. For a more precise definition of early progenitor CCs (including CySCs), a similar approach as to the one described in (Amoyel et al., 2016b) and (Senos Demarco et al., 2020) was used – CCs were co-stained with TJ (which marks early CCs including CySCs) and Eya (which marks late CCs), and counted only CCs that expressed TJ but not Eya.
CC proliferation
To determine the proliferative capacity of CCs, the number of Edu+ CC (as determined by those cells that co-stained positive for Edu and TJ, but absent for Eya) per testis was obtained in different experimental conditions. Since the expansion of mitotic cells was so dramatic in c587-GAL4ts>esgEP2009 conditions, EdU staining proved rather hard to be quantified because a large portion of CCs would stain positive. Instead, we used pHH3, as it is a more specific marker of mitosis and therefore would yield a smaller but still relevant and significant number of dividing CCs.
p4EBP measurements
To determine the signal intensity of p4EBP stains, all images within an experiment were acquired at the same laser power and exposure and analyzed with Image J. A 32x32 circle was drawn to fit within the cell limits of cyst cells located approximately in the second row (where p4EBP signal is highest). Three individual measurements (i.e., three different cyst cells) were performed per testis.
RNA extraction and quantitative RT-PCR
One hundred and fifty testes per condition after dissection were frozen at −80°C in fresh Trizol buffer (Trizol Life Technologies, 15596026; 5μg Linear Poly-Acrylamide Sigma 56575, 100ηg of tRNA). Total RNA was extracted pooling testes samples, followed by 5 rounds of freezing (liquid nitrogen)/thawing at 37°C in a water bath. Then 5 Vortex rounds at RT for 30’’, letting stand at RT for 5 min to disrupt all RNA-protein complexes. Finally, RNA was isolated by phenol/chloroform extraction. Purified RNA was treated with DNase Q1 (Promega, M610A). RNA (1μg) from testes dissected from males was reverse-transcribed using the iScriptkit (Bio-Rad, 170-8841). Standard qPCRs were carried out on a Bio-Rad CFX96/C1000 Touch system (Bio-Rad), using Sso Advanced SYBR Green (Bio-Rad, 1725-264). The following primer sequences were used: Act5c Fwd: TTGTCTGGGCAAGAGGATCAG; Act5c Rev: ACCACTCGCACTTGCACTTTC; InR Fwd: GCACCATTATAACCGGAACC; InR Rev: TTAATTCATCCATGACGTGAGC; ImpL2 Fwd: GCCGATACCTTCGTGTATCC; ImpL2 Rev: TTTCCGTCGTCAATCCAATAG; Esg Fwd: CGCCAGACAATCAATCGTAAGC; Esg Rev: TGTGTACGCGAAAAAGTAGTGG. Cycling conditions were as follows: 95°C for 30s; 95°C for 5s then 55°C for 30s, cycled 40 times. All calculated gene expression values were normalized to the value of the loading control gene, Actin5c. In Figure 3, testes from 2-day-old control or esgRNAi flies (genotypes: c587-GAL4/Y; tub-GAL80ts/+; +/+ or c587-GAL4/Y; tub-GAL80ts/UAS-esgRNAi-TRiP/+; +/+) were used. In Figure 6, recently eclosed w1118 males were maintained for 14 days on either regular diet or under starvation conditions at 25°C prior to dissection and mRNA extraction.
DamID
Flies carrying a UAS-Dam-esg transgene were generated as previously described (Loza-Coll et al., 2014). Tissue from UAS-Dam-esg and UAS-Dam control adult fly testes (∼12,000 flies) was collected and immediately preserved on dry ice and kept at −80°C. Genomic DNA was then isolated and amplified as described in (Choksi et al., 2006). Samples were labelled and hybridized to a whole genome 2.1 million feature tiling array with 50–75 mer oligonucleotides spaced at approximately 55 bp intervals. Arrays were scanned and intensities extracted (Nimblegen systems). Three biological replicates (with one dye-swap) were performed. Log2 mean ratios of each spot were median normalized. The data were binned to GATC fragments (DNA fragments between each GATC site) before a peak finding algorithm with false discovery rate (FDR) was used to identify significant binding sites (https://github.com/tonysouthall/Peak_calling_DamID) (Estacio-Gómez et al., 2020). A summary of the statistically significant results is presented on Table S1. Raw data from the arrays were submitted to Gene Expression Omnibus (http://ncbi.nlm.nih.gov/geo) with the access number GEO: GSE157685.
S2 cell line generation and analysis
S2 cells were transfected and stabilized with vectors derived from the N-terminal localization and affinity purification (NLAP) vector, in which the IgG binding domain was replaced with GFP (Kyriakakis et al., 2008): NLAP alone (“control”), an NLAP:Esg fusion (as seen in (Voog et al., 2014)), or an NLAP:EsgG387E fusion (in which DNA binding is disrupted due to a missense mutation in the third of the five zinc finger domains of Esg). All vectors contain a metallothionine promoter that allows a Cu2+-inducible expression in S2 cells. RNA was then extracted from S2 cells that have been treated with 350nM CuSO4 16 hours prior to induce gene expression. Cells were centrifuged in 15 mL conical tubes at 4°C and 1400rpm for 5 min (Eppendorf 5810R). The supernatant was removed and resuspended in cold PBS (600μL per 10cm dish of cells). Cells were then divided into 200μL aliquots, and RNA was extracted with the RNease Mini Kit (Qiagen) according to manufacturer’s instructions. Finally, the RNA was eluted from columns with 2 x 30μL RNAse free water and stored at −80°C.
Microarray analysis of gene expression
100ng of RNA was used per cell line. The hybridization reaction to the GeneChip Drosophila Genome 2.0 Array (Affymetrix) was performed by the Functional Genomics Core facility at the Salk Institute, covering 18,500 transcripts. For each cell line, three biological replicates were analyzed and the mean signal intensities were calculated. Genomics Suite software (Partek) was used to analyze the raw microarray data. Briefly, the fold changes of signal intensities were calculated between the two cell types used (NLAP and NLAP:Esg), looking for at least a 2-fold increase or decrease in signal between the two cell types with a p value <0.005 based on ANOVA tests. In total, 1369 genes experienced a statistically significant change of greater than 2-fold between the NLAP and NLAP:Esg cell lines, and an approximate equal number of up- (49%) and down-regulated (51%) genes were found. Further analyses compared the NLAP:Esg to the NLAP:EsgG387E arrays for evidence of a more direct DNA-binding effect of Esg on the regulation of gene expression. All of the results are presented on Table S2.
Quantification and statistical analysis
All quantitative experiments were evaluated for statistical significance using the software Graphpad Prism v6.0, after verifying the normality of values and equivalence of variances. For stem cell counts and replicative cell counts, means with standard deviations are displayed, and the statistical differences between mutant or RNAi-treated samples and controls were addressed using a Student’s two-tailed t-test. Results presented in figures as mean +/- SD. Individual statistical tests (with specific p values) are listed and noted in figures and their associated figure legends. DamID and microarray detailed statistics are described in their respective method details section.
Acknowledgments
The authors thank T. Xie, L. Cooley, D. Walker, R. Lehmann, D. Godt, the Vienna Drosophila RNAi Center (VDRC), Kyoto Drosophila Genetic Resource Center (DGRC), Bloomington Drosophila Stock Center (BDSC), and the Developmental Studies Hybridoma Bank (DSHB) for reagents, the BSCRC/MCDB microscopy core at UCLA, and the Jones laboratory for comments on the manuscript. A.H.B. was funded by the Royal Society Darwin Trust Research Professorship, a Wellcome Trust Senior Investigator Award (103792), and a Wellcome Trust Program grant (092545) and acknowledges core funding to the Gurdon Institute from the Wellcome Trust (092096) and CRUK (C6946/A14492). T.D.S. was funded by the Wellcome Trust Investigator grant (104567). R.S.D. and D.L.J. were funded by the Eli & Edythe Broad Center of Regenerative Medicine & Stem Cell Research, and D.L.J was funded by the NIH (AG040288, AG052732, and GM135767).
Author contributions
R.S.D. designed, performed, and analyzed experiments and wrote the manuscript. B.J.S. designed, performed, and analyzed experiments. A.M.T. performed experiments. S.L.S. designed, performed, and analyzed the microarray dataset. J.V., T.D.S., and A.H.B. designed, analyzed, and performed the DamID-related experiments. D.L.J. designed and analyzed experiments and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: April 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110679.
Supplemental information
List of genes with significant methylated-DNA peaks (putative binding sites) of the Dam-Esg fusion protein (see Methods for more details).
List of genes with significant changes in gene expression (increase or decrease) upon NLAP-Esg expression in S2 cells, as compared to NLAP alone or NLAP-EsgG387E (point mutation in the Zn-finger, which blocks binding to DNA). Fold change and p-values displayed (see Methods for more details).
Data and code availability
-
•
The dataset supporting the DamID experiments has been deposited at Gene Expression Omnibus and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microarray and microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abdelilah-Seyfried S., Chan Y.M., Zeng C., Justice N.J., Younger-Shepherd S., Sharp L.E., Barbel S., Meadows S.A., Jan L.Y., Jan Y.N. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics. 2000;155:733–752. doi: 10.1093/genetics/155.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoyel M., Anderson J., Suisse A., Glasner J., Bach E.A. Socs36e controls Niche competition by repressing Mapk signaling in the Drosophila testis. PLoS Genet. 2016;12:E1005815. doi: 10.1371/journal.pgen.1005815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoyel M., Hillion K.H., Margolis S.R., Bach E.A. Somatic stem cell differentiation is regulated by Pi3k/Tor signaling in response to local cues. Development. 2016;143:3914–3925. doi: 10.1242/dev.139782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoyel M., Simons B.D., Bach E.A. Neutral competition of stem cells is skewed by proliferative changes downstream of Hh and Hpo. EMBO J. 2014;33:2295–2313. doi: 10.15252/embj.201387500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf S.I., Ip Y.T. The Snail protein family regulates neuroblast expression of inscuteable and string, genes involved in asymmetry and cell division in Drosophila. Development. 2001;128:4757–4767. doi: 10.1242/dev.128.23.4757. [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A., Nieto M.A. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chang Y.C., Tu H., Huang T.W., Xu B.W., Pi H. Upregulated Tnf/Eiger signaling mediates stem cell recovery and tissue homeostasis during nutrient resupply in Drosophila testis. Sci. Rep. 2020;10:11674. doi: 10.1038/s41598-020-68313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Chen X., Zheng Y. The nuclear lamina regulates germline stem cell niche organization via modulation of Egfr signaling. Cell Stem Cell. 2013;13:73–86. doi: 10.1016/j.stem.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi S.P., Southall T.D., Bossing T., Edoff K., De Wit E., Fischer B.E., Van Steensel B., Micklem G., Brand A.H. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Demarco R.S., Eikenes A.H., Haglund K., Jones D.L. Investigating spermatogenesis in Drosophila melanogaster. Methods. 2014;68:218–227. doi: 10.1016/j.ymeth.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estacio-Gómez A., Hassan A., Walmsley E., Le L.W., Southall T.D. Dynamic neurotransmitter specific transcription factor expression profiles during Drosophila development. Biol. Open. 2020;9 doi: 10.1242/bio.052928. bio052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller M.T. In: The Development of Drosophila. Bate M., Arias A.M., editors. Cold Spring Harbor Press; 1993. Spermatogenesis. [Google Scholar]
- Gingras A.C., Gygi S.P., Raught B., Polakiewicz R.D., Abraham R.T., Hoekstra M.F., Aebersold R., Sonenberg N. Regulation of 4e-Bp1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan L.J., De Cuevas M., Matunis E. Genetics of gonadal stem cell renewal. Annu. Rev. Cell Dev. Biol. 2015;31:291–315. doi: 10.1146/annurev-cellbio-100913-013344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.W., Tokuyasu K.T., Lindsley D.L., Garavito M. The germinal proliferation center in the testis of Drosophila melanogaster. J. Ultrastruct. Res. 1979;69:180–190. doi: 10.1016/s0022-5320(79)90108-4. [DOI] [PubMed] [Google Scholar]
- Hayashi D., Tanabe K., Katsube H., Inoue Y.H. B-type nuclear Lamin and the nuclear pore complex Nup107-160 influences maintenance of the spindle envelope required for cytokinesis in Drosophila male meiosis. Biol. Open. 2016;5:1011–1021. doi: 10.1242/bio.017566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvay K., Jarde T., Casagranda F., Perreau V.M., Haigh K., Nefzger C.M., Akhtar R., Gridley T., Berx G., Haigh J.J., et al. Snai1 regulates cell lineage allocation and stem cell maintenance in the mouse intestinal epithelium. EMBO J. 2015;34:1319–1335. doi: 10.15252/embj.201490881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger A.A., White-Cooper H., Fuller M.T. Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature. 2000;407:750–754. doi: 10.1038/35037606. [DOI] [PubMed] [Google Scholar]
- Korzelius J., Naumann S.K., Loza-Coll M.A., Chan J.S., Dutta D., Oberheim J., Glasser C., Southall T.D., Brand A.H., Jones D.L., Edgar B.A. Escargot maintains stemness and suppresses differentiation in Drosophila intestinal stem cells. EMBO J. 2014;33:2967–2982. doi: 10.15252/embj.201489072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakakis P., Tipping M., Abed L., Veraksa A. Tandem affinity purification in Drosophila: the advantages of the Gs-tap system. Fly (Austin) 2008;2:229–235. doi: 10.4161/fly.6669. [DOI] [PubMed] [Google Scholar]
- Leatherman J.L., Dinardo S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008;3:44–54. doi: 10.1016/j.stem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza-Coll M.A., Jones D.L. Simultaneous control of stemness and differentiation by the transcription factor escargot in adult stem cells: how can we tease them apart? Fly (Austin) 2016;10:53–59. doi: 10.1080/19336934.2016.1176650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza-Coll M.A., Southall T.D., Sandall S.L., Brand A.H., Jones D.L. Regulation of Drosophila intestinal stem cell maintenance and differentiation by the transcription factor escargot. EMBO J. 2014;33:2983–2996. doi: 10.15252/embj.201489050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye C.M., Sanson B. Tension and epithelial morphogenesis in Drosophila early embryos. Curr. Top Dev. Biol. 2011;95:145–187. doi: 10.1016/B978-0-12-385065-2.00005-0. [DOI] [PubMed] [Google Scholar]
- Mcguire S.E., Le P.T., Osborn A.J., Matsumoto K., Davis R.L. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Mcleod C.J., Wang L., Wong C., Jones D.L. Stem cell dynamics in response to nutrient availability. Curr. Biol. 2010;20:2100–2105. doi: 10.1016/j.cub.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao G., Hayashi S. Escargot controls the sequential specification of two tracheal tip cell types by suppressing Fgf signaling in Drosophila. Development. 2016;143:4261–4271. doi: 10.1242/dev.133322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli C.A., Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Michel M., Kupinski A.P., Raabe I., Bokel C. Hh signalling is essential for somatic stem cell maintenance in the Drosophila testis niche. Development. 2012;139:2663–2669. doi: 10.1242/dev.075242. [DOI] [PubMed] [Google Scholar]
- Miron M., Verdu J., Lachance P.E., Birnbaum M.J., Lasko P.F., Sonenberg N. The translational inhibitor 4e-bp is an effector of Pi(3)K/Akt signalling and cell growth in Drosophila. Nat. Cell Biol. 2001;3:596–601. doi: 10.1038/35078571. [DOI] [PubMed] [Google Scholar]
- Nelson J.O., Chen C., Yamashita Y.M. Germline stem cell homeostasis. Curr. Top Dev. Biol. 2019;135:203–244. doi: 10.1016/bs.ctdb.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Rørth P., Szabo K., Bailey A., Laverty T., Rehm J., Rubin G.M., Weigmann K., Milán M., Benes V., Ansorge W., Cohen S.M. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- Sabatini D.M. Twenty-five years of Mtor: uncovering the link from nutrients to growth. Proc. Natl. Acad. Sci. U S A. 2017;114:11818–11825. doi: 10.1073/pnas.1716173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senos Demarco R., Uyemura B.S., Jones D.L. Egfr signaling stimulates autophagy to regulate stem cell maintenance and lipid homeostasis in the Drosophila testis. Cell Rep. 2020;30:1101–1116.e5. doi: 10.1016/j.celrep.2019.12.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall T.D., Brand A.H. Neural stem cell transcriptional networks highlight genes essential for nervous system development. EMBO J. 2009;28:3799–3807. doi: 10.1038/emboj.2009.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M., Uemura T., Oda H., Takeichi M., Hayashi S. Cadherin-mediated cell adhesion and cell motility in Drosophila trachea regulated by the transcription factor escargot. Development. 1996;122:3697–3705. doi: 10.1242/dev.122.12.3697. [DOI] [PubMed] [Google Scholar]
- Van Steensel B., Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered Dam methyltransferase. Nat. Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- Voog J., Sandall S.L., Hime G.R., Resende L.P., Loza-Coll M., Aslanian A., Yates J.R., 3rd, Hunter T., Fuller M.T., Jones D.L. Escargot restricts niche cell to stem cell conversion in the Drosophila testis. Cell Rep. 2014;7:722–734. doi: 10.1016/j.celrep.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shi J., Chai K., Ying X., Zhou B.P. The role of Snail in Emt and tumorigenesis. Curr. Cancer Drug Targets. 2013;13:963–972. doi: 10.2174/15680096113136660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M., Noguchi P.D., Sensabaugh S.M., Odenwald W.F., Kassis J.A. The Drosophila gene escargot encodes a zinc finger Motif found in snail-related genes. Mech. Dev. 1992;36:117–127. doi: 10.1016/0925-4773(92)90063-p. [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhou B.P. Snail: more than Emt. Cell Adh. Migr. 2010;4:199–203. doi: 10.4161/cam.4.2.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yamashita Y.M. The regulated elimination of transit-amplifying cells preserves tissue homeostasis during protein starvation in Drosophila testis. Development. 2015;142:1756–1766. doi: 10.1242/dev.122663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller R., Schulz C. The Drosophila cyst stem cell lineage: partners behind the scenes? Spermatogenesis. 2012;2:145–157. doi: 10.4161/spmg.21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes with significant methylated-DNA peaks (putative binding sites) of the Dam-Esg fusion protein (see Methods for more details).
List of genes with significant changes in gene expression (increase or decrease) upon NLAP-Esg expression in S2 cells, as compared to NLAP alone or NLAP-EsgG387E (point mutation in the Zn-finger, which blocks binding to DNA). Fold change and p-values displayed (see Methods for more details).
Data Availability Statement
-
•
The dataset supporting the DamID experiments has been deposited at Gene Expression Omnibus and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microarray and microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.