Abstract
Background
Inflammation is a complex physiological and pathological process. Although many types of inflammation are well characterized, their physiological functions are largely unknown. tRNA aspartic acid methyltransferase 1 (TRDMT1) has been implicated as a stress‐related protein, but its intrinsic biological role is unclear.
Methods
We constructed a Trdmt1 knockout rat and adopted the LPS‐induced sepsis model. Survival curve, histopathological examination, expression of inflammatory factors, and protein level of TLR4 pathway were analyzed.
Results
Trdmt1 deletion had no obvious impact on development and growth. Trdmt1 deletion slightly increased the mortality during aging. Our data showed that Trdmt1 strongly responded in LPS‐treated rats, and Trdmt1 knockout rats were vulnerable to LPS treatment with declined survival rate. We also observed more aggravated tissue damage and more cumulative functional cell degeneration in LPS‐treated knockout rats compared with control rats. Further studies showed upregulated TNF‐α level in liver, spleen, lung, and serum tissues, which may be explained by enhanced p65 and p38 phosphorylation.
Conclusions
Our data demonstrated that Trdmt1 plays a protective role in inflammation by regulating the TLR4‐NF‐κB/MAPK‐TNF‐α pathway. This work provides useful information to understand the TRDMT1 function in inflammation.
Keywords: inflammation, knockout rat, TLR4 pathway, TNF‐α, Trdmt1
Trdmt1 knockout rats were more vulnerable to LPS treatment with increased mortality and more aggravated tissue damage including noticeably thickened alveolar walls in lung tissues compared with WT. Elevated TNF‐α level production in liver, spleen, lung, and serum tissues were observed during LPS‐induced inflammation.
1. INTRODUCTION
Inflammation is a complex physiological and pathological process triggered by external and internal stimuli and conditions, such as infection, tissue injury, and aging. 1 , 2 Considerable achievements have been made in understanding the pathological process involved in the inflammatory response to infection or tissue injury. Dysregulation of inflammation during infection or tissue injury is a progressive and complex progress with unclear mechanism. Lipopolysaccharide (LPS), also known as endotoxin, is the main endogenous molecular component of the cell wall of Gram‐negative bacteria. 3 LPS is usually used as an exogenous stimulus of inflammatory responses. 4 LPS‐induced sepsis is a good tool to study inflammation progression. 5 , 6
tRNA aspartic acid methyltransferase 1 (TRDMT1), also known as DNMT2, was initially identified as a member of the DNA cytosine C5 methyltransferase (DNMT) family because it shares a conserved catalytic function domain with DNMT1. 7 TRDMT1 is highly conserved among vertebrates. 8 Evidence indicates that TRDMT1 acts as a tRNA methyltransferase rather than a DNA methyltransferase. 9 , 10 TRDMT1 methylates tRNAAsp‐GUC, tRNAGly‐GCC, tRNAVal‐AAC, tRNAGlu‐CUC, tRNAVal‐CAC, and tRNAGln‐CUG at the C5 position of C38 close to the anticodon. 11 , 12 This tRNA methylation function has been associated with tRNA stability and protein synthesis. 13 TRDMT1 has been implicated in regulation of brain development, myocardial hypertrophy, transgenerational inheritance, hematopoietic stem cell (HSC) steady state, human immunodeficiency virus (HIV) infection, and drug sensitivity. 14 , 15 , 16 , 17 , 18 , 19 , 20 Moreover, TRDMT1 plays an important role in resistance to stress, including oxidative stress, salt stress, and cellular senescence. 13 , 19 , 21 , 22 , 23 , 24 , 25 , 26 Even though TRDMT1 has been reported to be associated with longevity, the underlying mechanism is still unclear. Inflammation is a systemic and broad response to stress, and whether TRDMT1 plays a role in inflammation progression remains unknown. 26 , 27
Rats are indispensable laboratory animals to explore the human genome function by modeling disease processes. Rats differ from mice in terms of physiology, with some important traits that make rats a model of choice for complex human diseases. For example, rats share more immune characteristics with humans than mice do. 28 The complement levels in rats and humans are comparable, whereas in most inbred mice they are very low. 29 , 30 In this context, we used rats to construct a Trdmt1 gene knockout model and adopted LPS‐induced inflammation to study the function of Trdmt1 in inflammation progression. Our data showed that Trdmt1 deletion made rats more vulnerable to LPS treatment. The gene deletion was correlated with increased LPS‐induced death, and this phenotype can be largely explained by aggravated multiple organ damage with pronouncedly increased tumor necrosis factor‐alpha (TNF‐α) level. We further explored the underlying molecular mechanism of Trdmt1 participating in regulation of inflammation progression in LPS‐induced inflammation.
2. MATERIALS AND METHODS
2.1. Animal
All rats were purchased from Beijing Hfk Bioscience, and housed in a specific pathogen‐free (SPF) facility. Rats were under a 12/12‐hours light cycle with free access to food and water. All experiment procedures in this study were reviewed and approved by the Institutional Animal Care and Use Committee of the Institute of Laboratory Animal Sciences, Chinese Academy of Medical Sciences and Peking Union Medical College (MYW19004).
2.2. DNA construction and sgRNA preparation
The single‐guide RNA (sgRNA) expression plasmid was constructed as reported previously. 31 In brief, the synthesized paired oligos for sgRNAs were annealed and cloned into the pUC57‐sgRNA plasmid (#51132, Addgene). The oligos used in this study were purchased from Thermo Fisher and are listed in Table S1. The sgRNA expression plasmids were linearized by Dra I, purified by phenol‐chloroform, and used as template for transcription. The sgRNA transcription was performed using MEGAshortscript Kit (Ambion, AM1354) and purified using the MEGAclear Kit (Ambion, AM1908).
2.3. Generation of Trdmt1 knockout rat
The fertilized eggs were collected and prepared for microinjection as described previously. 32 In brief, the fertilized eggs were injected with a mixture of Cas9 protein and targeting sgRNAs with the concentration as reported previously. 33 The injected zygotes were transferred to prepared pseudopregnant Sprague–Dawley (SD) rats.
2.4. Rat genomic DNA extraction and genotyping
Genomic DNA of newborn pups was extracted from the tails of 7‐day‐old rat using phenol‐chloroform and recovered by alcohol precipitation. The primer pair Trdmt1‐P1 and Trdmt1‐P2 was used to detect the knockout brand. The primer pair Trdmt1‐P3 and Trdmt1‐P4 was used to detect the WT brand. The PCR conditions were as follows: 95°C for 30 s; 95°C for 30 s, 60°C for 30 s, and 72°C for 90 s for 30 cycles. Twenty microliters reaction mixture was run on a 1.5% agarose gel, and the GoodView Nucleic Acid Stain (HGV‐II, sbs)‐stained products were visualized using Gel Image System (Tanon‐1600). All primers used for PCR are listed in Table S2.
2.5. Quantitative real‐time PCR
Total RNA from tissues and cells were extracted using TriQuick Reagent (Solarbio). Two micrograms of RNA samples was used for cDNA synthesis using PrimeScript RT reagent kit with a gDNA Eraser kit (Perfect Real Time) according the manufacturer's instructions. Real‐time PCR was performed with PerfectStart Green qPCR SuperMix (+Dye II) (Transgen) using QuantStudioT 3 (Applied Biosystems) as follows: 95°C for 30 s; 95°C for 5 s and 60°C for 30 s for annealing/extension for 40 cycles. All primers used for real‐time PCR are listed in Table S3.
2.6. LPS‐induced inflammation in rats
Eight‐week‐old SD rats were injected intraperitoneally with 5 or 1 mg/kg LPS (L4391, Sigma). Trdmt1 expression and animal survival rate were monitored. The rats treated with 1 mg/kg LPS were used for pathology and molecular detection.
2.7. Lung wet/dry (W/D) ratio
The isolated lung was rinsed with saline, and the excess water was absorbed with filter paper. Then, the lung was wrapped in tinfoil and weighed as W1. It was transferred to an incubator at 80°C for 24 hours. In the final hour, continuous measurement was taken 3 times and average recorded as W2. The lung W/D ratio was calculated as W1/W2.
2.8. Hematoxylin and eosin staining
Liver, spleen, lung, and kidney isolated from rats were preserved in 4% buffered formalin solution fixed, and then prepared for the paraffin sections (3–4 μm thickness) according to routine methods. All tissue sections were stained with hematoxylin and eosin (H&E) according to previous publications. 34 The histopathological changes of multiple tissues were observed and analyzed under Pannoramic 250 FLASH and CaseViewer 2.3 (3D HISTECH).
2.9. Lung injury score
Lung injury score was assessed using the following scoring system: (a) alveolar congestion, (b) alveolar hemorrhage, (c) neutrophil infiltration, (d) thickness of alveolar wall/hyaline membrane formation (absent = 0, minor = 1, moderate = 2, severe = 3, critical = 4). The total lung injury score was the sum of the 4 criteria. Twenty high‐power random fields (400× total magnification) were scored per rat.
2.10. ELISA
Serum from each rat was collected to measure the TNF‐α level by ELISA kit (RTA00, R&D) according to the manual. Briefly, after adding 50 μL of assay diluent to each well, the serum samples were diluted at 1:100, and then 50 μL standard, control, or samples were added to different precoated wells and maintained at room temperature for 2 hours, followed by 5 washes, and then 100 μL of conjugate was added to each well, and the samples were incubated at room temperature for 2 hours. Then, 100 μL Substrate Solution was added to each well, and the samples were incubated at room temperature for 30 minutes. Finally, 100 μL of Stop Solution was added to each well, and the absorbance of 450 nm was recorded by a microplate reader (Multiskan FC with Incubator 51 119 100, Thermo) within 30 minutes.
2.11. Alveolar macrophage (AM) isolation, culture, and treatment
SD rats were anesthetized with 2% pentobarbital sodium (3 mL/kg) via intraperitoneal injection. The abdominal cavity was opened, then the thoracic cavity was opened through the mediastinum, and a small incision (<2 mm) was made on the trachea posterior to the larynx. A 1‐inch 22 G catheter was inserted and secured with a silk braided suture with a square knot. Ice‐cold BAL buffer (Ca2+‐ and Mg2+‐free PBS + 1 mM EDTA) was filled in a 5 mL syringe attached to the catheter and instilled slowly into the lungs. The lavage fluid was aspirated and collected in a 15 mL conical tube placed on ice to full. BAL fluid was centrifuged at 250g at 4°C for 10 minutes, and the supernatant was removed. AM was washed 3 times with precooled RPMI‐1640 medium and incubated in a 37°C humidified incubator with 5% CO2. AM was treated with 100 ng/mL LPS and analyzed by quantitative real‐time PCR.
2.12. Peritoneal macrophage (PM) isolation, culture, and treatment
SD rats were anesthetized with 2% pentobarbital sodium (3 mL/kg) via intraperitoneal injection. The abdominal cavity was opened with a small incision. The abdominal cavity was washed with 10 mL PBS with a 25 G needle. The peritoneal lavage fluid was centrifuged at 250g at 4°C for 10 minutes, and the supernatant was removed. PM was washed 3 times with precooled RPMI‐1640 medium and incubated in a 37°C humidified incubator with 5% CO2. PM was treated with 100 ng/mL LPS and analyzed by quantitative real‐time PCR.
2.13. Western blot
Total proteins from tissue were extracted on ice and prepared in 1× loading buffer. Proteins were separated by 10% SDS‐PAGE and transferred to nitrocellulose (NC) membranes. The membranes were blocked with 5% skimmed milk at room temperature for 1 hour, followed by incubation with primary antibody overnight at 4°C. Then the membrane was incubated with HRP‐conjugated secondary antibody at room temperature for 1 hour. Antigen complexes were visualized with ECL and analyzed using Image Lab (Bio‐Rad). Antibodies are listed in Table S7.
2.14. Statistical analysis
All data were analyzed by the GraphPad Prism 8.0 software and presented as the mean ± standard error of mean (SEM). Data with 2 groups or multiple groups were analyzed by 2‐tailed Student's t test or 2‐way analysis of variance (ANOVA). The survival rate was analyzed by log‐rank nonparametric test and shown as Kaplan–Meier survival curve. P < 0.05 was defined as statistically significant.
3. RESULTS
3.1. Generation of Trdmt1 knockout rat
Trdmt1 is conserved among vertebrates, but the gene function is confusing and controversial. Here we use rat, an indispensable animal model for illustrating the human biological progress through genetic modification and phenotype networks, 35 , 36 to investigate the gene function of Trdmt1.
Firstly, we examined the gene expression of Trdmt1 in different tissues of Sprague–Dawley (SD) rats by quantitative real‐time PCR. The data show that Trdmt1 was mainly expressed in liver and lung (Figure 1A). To study in vivo function, we constructed a Trdmt1 gene knockout rat using CRISPR/Cas9 system as described previously. 31 In brief, 2 sgRNAs were designed, one target site located in intron 1 and the other in intron 10, to produce fragment deletion from exon 2 to exon 10 (Figure 1B, Table S1). After sgRNA and Cas9 protein mixture microinjection and embryo transplantation, we obtained 7 rats, one with DNA fragment deletion detected by target primer pairs (Table S2). Then, we crossed this F0 rat with wild‐type (WT) SD rat and found the modification was germline transmittable (Figure 1C, Table S2). The target modification with a 28, 727 bp deletion was confirmed using Sanger sequencing. Further quantitative real‐time PCR results validated the expression of Trdmt1 knockout rat in various tissues (Figure 1D, Table S3). Moreover, The TRDMT1 protein deletion was further confirmed by western blot in lung tissue (Figure 1E).
FIGURE 1.
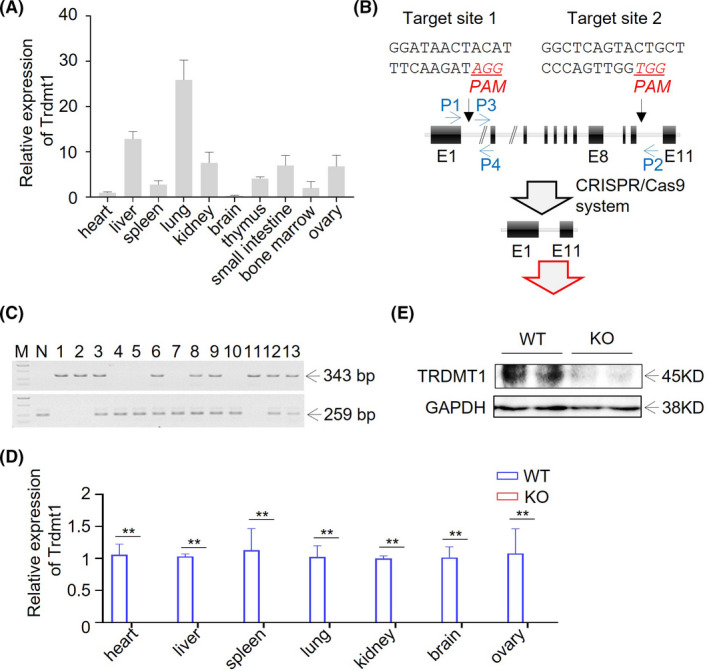
Generation of Trdmt1 gene knockout rat using CRISPR/Cas9. A, Relative expression of Trdmt1 in heart, liver, spleen, lung, kidney, brain, thymus, small intestine, bone marrow, and ovary tissues measured using real‐time PCR (n = 3). B, Schematic overview of CRISPR/Cas9‐mediated Trdmt1 gene deletion. The sgRNA targeting sites are shown. The protospacer adjacent motifs (PAMs) of sgRNA targeting sites are underlined in red and italic. Oligos used for sgRNAs preparation are listed in Table S1. Primers used for genotyping of Trdmt1 knockout rats are shown in blue and listed in Table S2. E, exon. C, PCR amplicon of Trdmt1 targeting locus. P1/P2 primer pair located outside of the sgRNA targeting sites and used for deleted DNA fragment detection. The P3/P4 primer pair located between the 2 sgRNA targeting sites and used for wild‐type DNA fragment detection. All primers used are listed in Table S2. M, marker for DL2, 000 (TaKaRa). N, negative control (wild type). D, The relative expression of Trdmt1 in heart, liver, spleen, lung, kidney, brain, and ovary tissues confirmed in Trdmt1 knockout rats using real‐time PCR (n = 3). E, Protein level of TRDMT1 in lung of Trdmt1 knockout rats. WT, wild‐type rats; KO, Trdmt1 knockout rats. Mean ± SEM; *P < 0.05, **P < 0.01
3.2. Loss of Trdmt1 slightly affected development and growth in rats
It has been reported that Trdmt1 knockout mice are viable and fertile. 10 Here, we monitored the development and growth of Trdmt1 knockout rats, including body weight change, organ coefficient, and lifespan. We found that the Trdmt1 knockout homozygous ratio accorded with Mendel's law (Table S4). The body weight changes in the absence of Trdmt1 during growth were comparable to those of WT in both female and male rats (Figure 2A). Organ coefficient analysis showed a slightly increased liver and spleen ratio in 8‐10‐week‐old knockout rats (Figure 2B). Moreover, we performed routine blood examination, with the results indicating normal blood cell count in Trdmt1 knockout rats (Table S5). In line with these findings, flow cytometry analysis revealed that the loss of Trdmt1 did not impair hematopoiesis, at least in 8‐10‐week‐old rats (Figure S1). To test possible compensations in Trdmt1 knockout rats, we measured the protein expression of DNMT1, DNMT3A, and NSUN2 (Figure 2C,D). DNMT1 and DNMT3A are members of the DNMT family. NSUN2 shares a similar function with TRDMT1 and methylates C34, C48, C49, and C50 of tRNA. 37 Surprisingly, the expression of NSUN2, but not other DNMTs, was significantly elevated in the Trdmt1 knockout rats (Figure 2C,D). Trdmt1 ablation had negligible impact on the growth and development of adult rats at steady state. Interestingly, we observed a minor but insignificant decrease of survival rate in Trdmt1 knockout rats (Figure 2E), which suggests that Trdmt1 deletion makes rats slightly more susceptible to aging.
FIGURE 2.
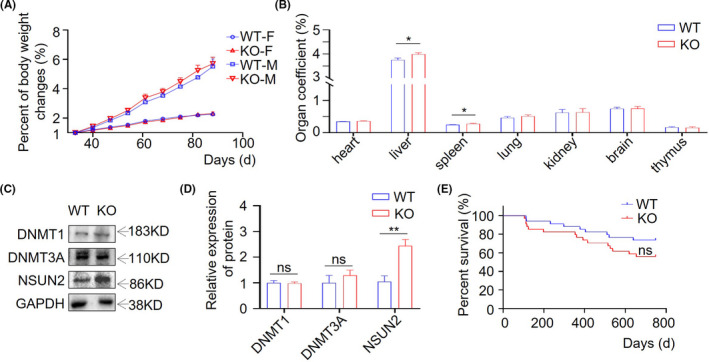
The effect of Trdmt1 deletion on rat development and survival. A, Body weight changes of Trdmt1 gene knockout male and female rats (n = 3). F, female; M, male. B, The organ coefficient of 8‐10‐week‐old Trdmt1 gene knockout rats (n ≥ 5). C‐D, Protein level of DNMT1, DNMT3A, and NSUN2 in liver analyzed by western blot of Trdmt1 knockout rats. Quantitative analysis of DNMT1, DNMT3A, and NSUN2 protein expression level was analyzed by Image Lab software and using GAPDH for normalization. E, Survival curves of Trdmt1 gene knockout rats (n = 34). WT, wild‐type rats; KO, Trdmt1 knockout rats. Mean ± SEM; ns, not significant; *P < 0.05
3.3. Trdmt1 deficiency resulted in increased mortality upon LPS treatment
Growing evidence illustrates that Trdmt1 plays a potential role in stress stimulation. 13 , 19 , 21 , 22 , 38 Here, we applied the LPS‐induced sepsis model to analyze Trdmt1 gene function. We first measured the expression of Trdmt1 in different tissues after administering SD rats with LPS. We observed significantly decreased Trdmt1 expression in liver, lung, kidney, and thymus (Figure 3A), suggesting a key role of Trdmt1 in LPS resistance. Then, we monitored the survival rate of Trdmt1 knockout rats in 5 mg/kg LPS‐induced sepsis model. Our data showed that the Trdmt1 knockout group experienced a dramatic increase in mortality (Figure 3B). At the same time, this phenotype recurred in cecal ligation and puncture (CLP)‐induced inflammation rat model (Figure S2a). 39 Thus, we came to the conclusion that Trdmt1 is essential to the stress response.
FIGURE 3.
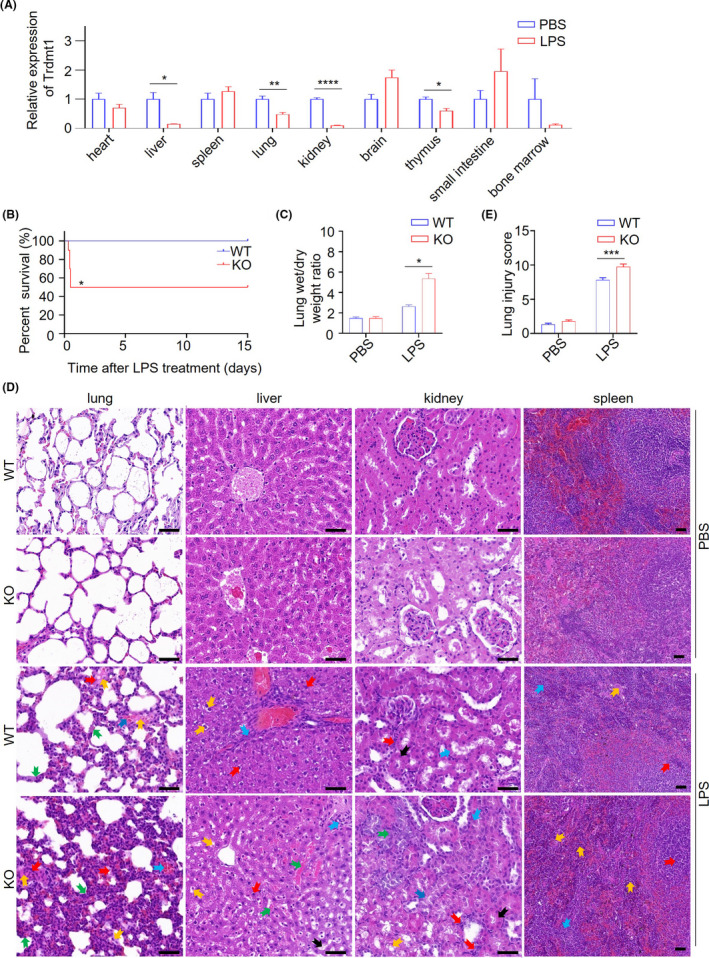
Trdmt1 deletion leads to impaired response after LPS treatment in rats. A, The relative expression of Trdmt1 in LPS‐treated rats in heart, liver, spleen, lung, kidney, brain, thymus, small intestine, and bone marrow tissues (n = 3). B, Survival curve of Trdmt1 knockout rats after 5 mg/kg LPS treatment (n = 10). C, Lung wet/dry weight ratio of Trdmt1 knockout rats after 5 mg/kg LPS treatment (n ≥ 3). D, Representative images of HE staining of lung, liver, kidney, and spleen from Trdmt1 knockout rats with 1 mg/kg LPS or PBS treatment. In liver, labels indicate vacuolization of liver lobules (yellow arrow), infiltration of inflammatory cells (blue arrow), nuclei color change (red arrow), bleeding (green arrow), and hepatocyte degeneration (black arrow) in LPS‐treated groups; in lung tissue, labels indicate alveolar congestion (yellow arrow), alveolar hemorrhage (blue arrow), neutrophil infiltration (red arrow), and thickness of alveolar wall/hyaline membrane formation (green arrow) in LPS‐treated groups; in spleen tissue, labels indicate extramedullary hematopoietic foci (yellow arrow), splenic corpuscle in red pulp (blue arrow), and marginal zone thinning (red arrow) in LPS‐treated groups; in kidney, labels indicate vacuolization (yellow arrow), dilatation of renal tubules (blue arrow), neutrophil infiltration (red arrow), degeneration of renal tubules (green arrow), and tubular thickening (black arrow) in LPS‐treated groups. Scale bars, 50 μm. (E) Lung injury score of Trdmt1 knockout rats after 1 mg/kg LPS treatment (n = 20). WT, wild‐type rats; KO, Trdmt1 knockout rats. Mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001
3.4. Trdmt1 deletion aggravated multiple organ damage in inflammation
From the routine blood test, we did not observe a significant difference in blood cell function between wild‐type and knockout rats after LPS treatment (Table S5). It has been reported that LPS‐induced damage mainly occurs in liver, lung, spleen, and kidney. 40 , 41 , 42 , 43 , 44 On autopsy, we observed obviously increased scattered bleeding points and flaky bleeding foci on the fresh lung surface in Trdmt1 knockout rats (Figure S2b). We also observed increased lung wet/dry weight (W/D) ratio in Trdmt1 knockout rats (5.37 ± 1.41) compared with negative control (2.64 ± 0.26) (Figure 3C). On further pathological section examination, we observed an increase in the frequency of liver lobule vacuolation and hepatocyte degeneration, and lung injury score increased considerably (Figure 3E). Moreover, extramedullary hematopoietic foci increased in red pulp, and dilation and degeneration of renal tubules was more remarkable (Figure 3D). Collectively, these data reflect more severe damage of these tissues in Trdmt1 knockout rats than their littermates. Then we explored the protein expression of DNMT1, DNMT3A, and NSUN2 in knockout rats and control rats with LPS treatment (Figure S2c‐f). Our data showed decreased expression of DNMT1, DNMT3A, and NSUN2 in LPS‐treated gene knockout rats and control rats.
3.5. Knockout of Trdmt1 dramatically elevated the TNF‐α level in sepsis model
Giving that changes in inflammatory factors, including TNF‐α, IL‐1β, IL‐6, IL‐10, and iNOS, are a major characteristic of the sepsis model, 45 , 46 we measured the inflammatory cytokines after LPS treatment. We observed changes in inflammatory cytokines at different timepoints after LPS treatment (S3). Remarkably, the TNF‐α level was strikingly elevated in both mRNA and protein levels in the sepsis model when Trdmt1 was deleted (Figure 4A‐D). In addition, the expression of IL‐1β was elevated at 1.5 hours after LPS treatment (Figure S3a‐c), while IL‐6 level increased at the 2‐hour timepoint (Figure S3a‐c). Decreased IL‐10 and iNOS levels were observed at the 2‐hour timepoint after LPS treatment in Trdmt1 knockout rats (Figure S3a‐c). We further analyzed the TNF‐α level of alveolar macrophage (AM) and peritoneal macrophage (PM) treated with 100 ng/mL LPS derived from Trdmt1 knockout and control rats. As expected, a marked escalation of TNF‐α mRNA level was also detected in AM and PM derived from Trdmt1 knockout rats (Figure 4E‐F). Our results underscore a critical role of Trdmt1 in regulating the levels of inflammatory cytokines, especially TNF‐α in the sepsis model. The increased release of pro‐inflammatory factors and decreased release of anti‐inflammatory factors might contribute to the enhanced aggravated tissue damage in LPS‐treated Trdmt1 knockout rats.
FIGURE 4.
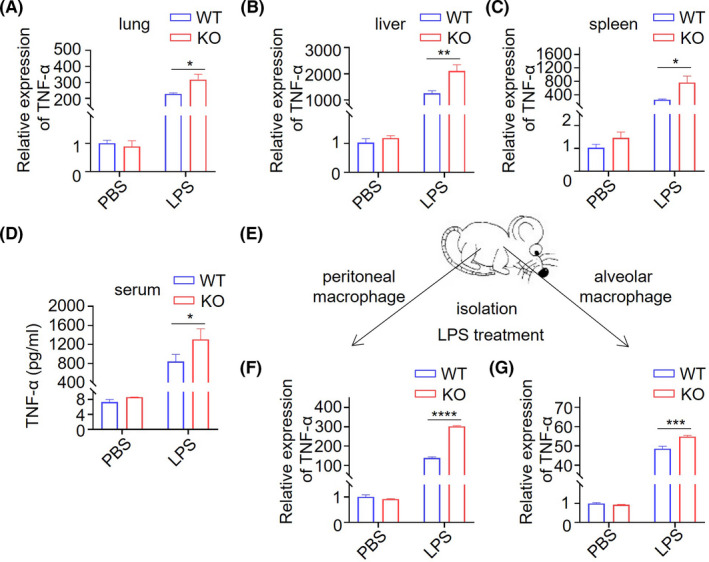
Trdmt1 deletion leads to impaired inflammatory factor response after LPS treatment. A‐C, The relative expression of TNF‐α in lung (A), liver (B), and spleen (C) of 1 mg/kg LPS‐treated Trdmt1 knockout rats. D, The TNF‐α protein level in serum of 1 mg/kg LPS‐treated Trdmt1 knockout rats. E‐H, Alveolar macrophage and peritoneal macrophage derived from Trdmt1 knockout rats were cultured and treated with 100 ng/mL LPS. The relative expression of TNF‐α of peritoneal macrophage (F), and alveolar macrophage (G) was determined. WT, wild‐type rats; KO, Trdmt1 knockout rats. Mean ± SEM; *P < 0.05, ***P < 0.001, ****P < 0.0001
3.6. Trdmt1 knockout enhanced the activation of TLR4 pathway after LPS treatment
Considering that TLR4, but not other TLR pathways, is commonly activated in sepsis, 45 , 47 we performed an in‐depth analysis of the expression of TLR4 signal pathway protein. The protein level of TLR4, p‐p65, and p‐p38, but not p‐ERK, was significantly increased in liver of the LPS‐treated group upon Trdmt1 ablation compared with WT (Figure 5A‐J). In addition, enhanced TLR4 pathway activation was also observed in spleen after Trdmt1 knockout (Figure S4). Taken together, our findings indicate a protective role of Trdmt1 in LPS‐induced inflammation through regulation of the TLR4 signaling pathway.
FIGURE 5.
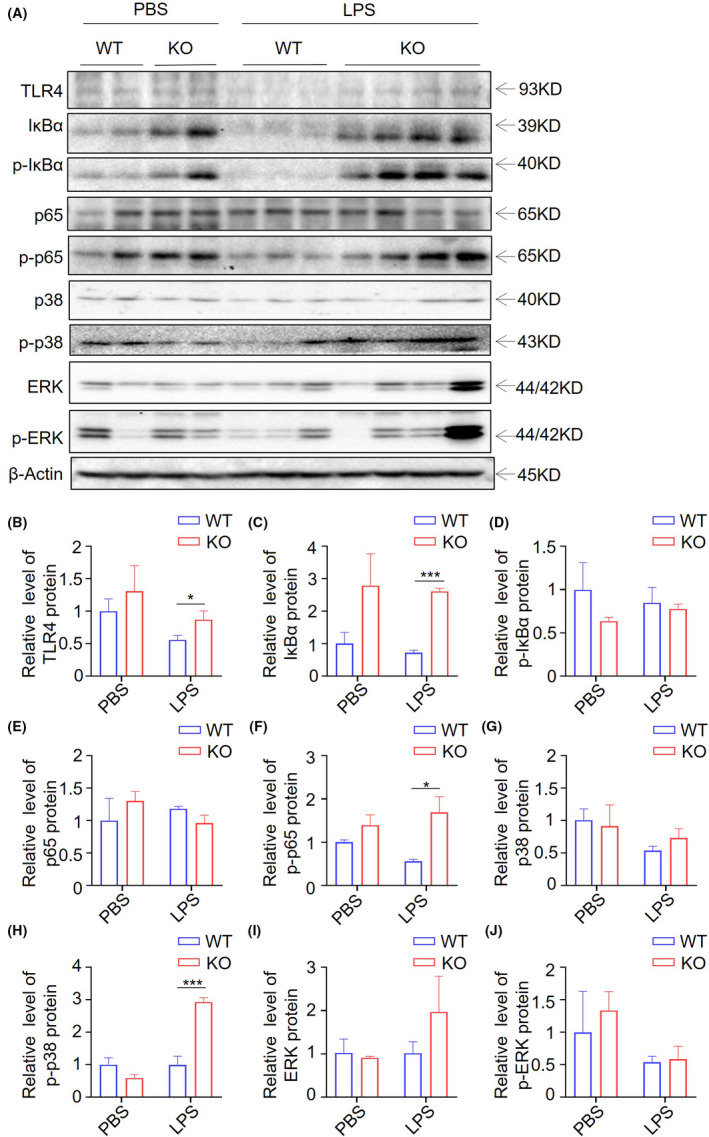
TLR4 pathway activation contributed to LPS‐induced mortality of Trdmt1 knockout rats. A, Protein level of TLR4, IκBα, p‐IκBα, p65, p‐p65, p38, p‐p38, ERK, and p‐ERK in liver analyzed by western blot in LPS‐ or PBS‐treated Trdmt1 knockout rats. B‐J, Quantitative analysis of TLR4 (B), IκBα (C), p‐IκBα (D), p65 (E), p‐p65 (F), p38 (G), p‐p38 (H), ERK (I), and p‐ERK (J) protein expression level analyzed by Image Lab software and using β‐Actin for normalization. WT, wild‐type rats; KO, Trdmt1 knockout rats. Mean ± SEM; *P < 0.05, **P < 0.01, ****P < 0.0001
4. DISCUSSION
For a long time, the function of TRDMT1 has been unclear. It was originally identified from Schizosaccharomyces pombe as DNMT2 because it shares a catalytic function domain with de novo DNA methyltransferase DNMT1. 7 Increasing evidence has indicated that TRDMT1 functions as a transfer RNA methyltransferase instead of a DNA methyltransferase. 10 Though TRDMT1 depletion in zebrafish resulted in developmental perturbations, in most vertebrates TRDMT1 disruption causes no phenotypic defects. 14 , 17 For example, Trdmt1‐mutant mice were shown to be viable and fertile without visible abnormal development. 10
In this work, we constructed Trdmt1 knockout rats and analyzed the phenotype of Trdmt1 knockout rats, which displayed virtually normal growth, development, reproduction, and hematopoiesis. We concluded that Trdmt1 plays a negligible role in homeostasis under normal condition. We also assessed the possible compensations in Trdmt1 knockout rats and observed elevated NSUN2 expression. TRDMT1 and NSUN2 are the only 2 discovered (cytosine‐5) tRNA methyltransferases to date, and they appear to play a role in tRNA stability and protein synthesis. Although lack of either TRDMT1 or NSUN2 alone has no detectable effects on mouse viability, double knockout showed a systemic lethal interaction. 48 This indicated that the function of TRDMT1 could be compensated in part by NSUN2 in Trdmt1 knockout rats.
Emerging evidence has suggested that Trdmt1 is strongly correlated with stress stimulus. 13 , 19 , 49 Lin et al. reported that Trdmt1 overexpression in Drosophila melanogaster could extend its lifespan. 27 In another report Li et al. found that gene expression of Dnmt1 and Dnmt2 (Trdmt1) was decreased in aging rats. 25 These clues link the Trdmt1 to aging, but little is known about the underlying mechanism. Here, we found the survival rate of Trdmt1‐deficient rats slightly but insignificantly decreased compared with their negative control, suggesting that Trdmt1‐deficient rats are vulnerable to environment or stress.
Trdmt1 played a protective role against heat‐shock‐stress‐induced tRNA cleavage by transfer RNA methylation. 13 Further study demonstrated that Trdmt1 deletion led to increased tRNA fragmentation (tRFs) that competed with endogenous small interfering RNAs (siRNAs) to interact with Dicer‐2. 49 In another report, Trdmt1 participated in RNA processing during cellular stress by shuttling from the nucleus into the cytoplasm to interact with stress granules and RNA processing bodies. 19 Inflammation is a systemic and broad stress response. However, whether Trdmt1 functions in inflammation progression remains unknown. In the present study, we concentrated on the relationship between Trdmt1 and inflammation. LPS has toxic effects and is widely used as a potent stimulus of inflammation, 4 In addition, dysregulation of inflammation to LPS treatment can result in sepsis, 5 , 6 which places a substantial burden on the public owing to its high morbidity and mortality. It should be pointed out that LPS‐induced inflammation is easy to control and reproducible. Surprisingly, Trdmt1 knockout significantly decreased the survival rate of rats and caused more aggravated multiple organ damage in both LPS‐treated model and CLP model. Why? We analyzed the expression of NSUN2 and DNMTs family members. The expression of DNMT1 and DNMT3A is comparable in both Trdmt1 knockout and control rats. The expression of DNMT1 was repressed in both WT and Trdmt1 knockout rats after LPS treatment. This is consistent with a previous report. 50 The expression of NSUN2 in LPS‐treated Trdmt1 knockout and WT rats is comparable, but the elevated NSUN2 expression in LPS‐treated Trdmt1 knockout rats was repressed.
Inflammation imbalance represents the basis of the sepsis pathogenesis and is sustained throughout the process of sepsis. 51 Infectious agents induce acute responses, including macrophages activation and pro‐inflammatory cytokine release, further forming a cytokine storm. 52 This process is mediated by diverse pattern‐recognition receptors (PRRs), including the most‐studied Toll‐like receptors (TLRs), through damage‐associated molecular patterns (DAMPs) or pathogen‐associated molecular patterns (PAMPs). 53 TLRs interact with their ligands and activate several signaling pathways, including mitogen‐activated protein kinase (MAPK) and nuclear factor‐κB (NF‐κB), to produce inflammatory cytokines such as interleukin (IL)‐1, IL‐6, and tumor necrosis factor‐α (TNF‐α). 45 , 54 , 55 , 56 , 57 We observed increased release of pro‐inflammatory factors and decreased release of anti‐inflammatory factors release at different timepoints. More importantly, TNF‐α level was remarkably elevated in the Trdmt1 knockout rat liver, lung, and spleen tissues as well as in AM and PM. Moreover, in K562 cells, we found that overexpression of Trdmt1 resulted in lower level of TNF‐α after LPS treatment (data not shown). This phenotype was in accordance with our results presented in this study. Further, we explored the underlying mechanism and found increased phosphorylation levels of NF‐κB and p38 in Trdmt1 knockout rats. This indicates enhanced TLR4 signal pathway activation. Even though the direct relationship between TRDMT1 and TLR4 signal remains unclear, our data highlight Trdmt1 as playing a protective role in the inflammation process through the TLR4‐NF‐κB/MAPK‐TNF‐α signaling pathway.
In summary, we proved for the first time the function of Trdmt1 in inflammation. Specifically, Trdmt1 was required to survive inflammation stress through the NF‐κB/MAPK‐TNF‐α signaling pathway. Indeed, our work provides useful information to extend our understanding of Trdmt1 function under stress conditions, especially inflammation. This work may be helpful for patients with sepsis in clinic treatment.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Y.M. conceived the project and supervised the research. X.Q. and Z.L. designed all the experiments, performed the experiments, and analyzed the data with the assistance of X.Z., L.Y., W.K., W.C., W.D., L.L., D.L., L.G., and L.Z. Y.M., X.Q., and Z.L. wrote the manuscript, data analysis, and paper discussion.
Supporting information
Data S1
ACKNOWLEDGMENTS
The present work was supported in part by the CAMS Innovation Fund for Medical Sciences (CIFMS) (2021‐I2M‐1‐024 and 2021‐I2M‐1‐034), Beijing Municipal Natural Science Foundation (M21004), National Natural Science Foundation of China (31970508), and 111 Project of the Ministry of Education (B20095).
Li Z, Qi X, Zhang X, et al. TRDMT1 exhibited protective effects against LPS‐induced inflammation in rats through TLR4‐NF‐κB/MAPK‐TNF‐α pathway. Anim Models Exp Med. 2022;5:172–182. doi: 10.1002/ame2.12221
Funding information
CAMS Innovation Fund for Medical Sciences (CIFMS) (2021‐I2M‐1‐024 and 2021‐I2M‐1‐034), and Beijing Municipal Natural Science Foundation (M21004), National Natural Science Foundation of China (31970508), and 111 Project of the Ministry of Education (B20095).
Zhengguang Li and Xiaolong Qi contributed equally to this work.
REFERENCES
- 1. Zhang X, Meng X, Chen Y, Leng SX, Zhang H. The biology of aging and cancer: frailty, inflammation, and immunity. Cancer J. 2017;23(4):201‐205. [DOI] [PubMed] [Google Scholar]
- 2. Bektas A, Schurman SH, Sen R, Ferrucci L. Aging, inflammation and the environment. Exp Gerontol. 2018;105:10‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ciesielska A, Matyjek M, Kwiatkowska K. TLR4 and CD14 trafficking and its influence on LPS‐induced pro‐inflammatory signaling. Cell Mol Life Sci. 2021;78(4):1233‐1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fritsche KL. The science of fatty acids and inflammation. Adv Nutr. 2015;6(3):293s‐301s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Płóciennikowska A, Hromada‐Judycka A, Borzęcka K, Kwiatkowska K. Co‐operation of TLR4 and raft proteins in LPS‐induced pro‐inflammatory signaling. Cell Mol Life Sci. 2015;72(3):557‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuzmich NN, Sivak KV, Chubarev VN, Porozov YB, Savateeva‐Lyubimova TN, Peri F. TLR4 signaling pathway modulators as potential therapeutics in inflammation and sepsis. Vaccines (Basel). 2017;5(4):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilkinson CR, Bartlett R, Nurse P, Bird AP. The fission yeast gene pmt1+ encodes a DNA methyltransferase homologue. Nucleic Acids Res. 1995;23(2):203‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19(2):81‐92. [DOI] [PubMed] [Google Scholar]
- 9. Raddatz G, Guzzardo PM, Olova N, et al. Dnmt2‐dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci USA. 2013;110(21):8627‐8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goll MG, Kirpekar F, Maggert KA, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311(5759):395‐398. [DOI] [PubMed] [Google Scholar]
- 11. Wnuk M, Slipek P, Dziedzic M, Lewinska A. The roles of host 5‐methylcytosine RNA methyltransferases during viral infections. Int J Mol Sci. 2020;21(21):8176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li H, Zhu D, Wu J, et al. New substrates and determinants for tRNA recognition of RNA methyltransferase DNMT2/TRDMT1. RNA Biol. 2021;18(12):2531‐2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schaefer M, Pollex T, Hanna K, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress‐induced cleavage. Genes Dev. 2010;24(15):1590‐1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rai K, Chidester S, Zavala CV, et al. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007;21(3):261‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ghanbarian H, Wagner N, Polo B, et al. Dnmt2/Trdmt1 as mediator of RNA polymerase II transcriptional activity in cardiac growth. PLoS One. 2016;11(6):e0156953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Y, Zhang X, Shi J, et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non‐coding RNAs. Nat Cell Biol. 2018;20(5):535‐540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tuorto F, Herbst F, Alerasool N, et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015;34(18):2350‐2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dev RR, Ganji R, Singh SP, Mahalingam S, Banerjee S, Khosla S. Cytosine methylation by DNMT2 facilitates stability and survival of HIV‐1 RNA in the host cell during infection. Biochem J. 2017;474(12):2009‐2026. [DOI] [PubMed] [Google Scholar]
- 19. Thiagarajan D, Dev RR, Khosla S. The DNA methyltranferase Dnmt2 participates in RNA processing during cellular stress. Epigenetics. 2011;6(1):103‐113. [DOI] [PubMed] [Google Scholar]
- 20. Cheng JX, Chen L, Li Y, et al. RNA cytosine methylation and methyltransferases mediate chromatin organization and 5‐azacytidine response and resistance in leukaemia. Nat Commun. 2018;9(1):1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fisher O, Siman‐Tov R, Ankri S. Pleiotropic phenotype in Entamoeba histolytica overexpressing DNA methyltransferase (Ehmeth). Mol Biochem Parasitol. 2006;147(1):48‐54. [DOI] [PubMed] [Google Scholar]
- 22. Mytych J, Lewinska A, Bielak‐Zmijewska A, Grabowska W, Zebrowski J, Wnuk M. Nanodiamond‐mediated impairment of nucleolar activity is accompanied by oxidative stress and DNMT2 upregulation in human cervical carcinoma cells. Chem Biol Interact. 2014;220:51‐63. [DOI] [PubMed] [Google Scholar]
- 23. Bloniarz D, Adamczyk‐Grochala J, Lewinska A, Wnuk M. The lack of functional DNMT2/TRDMT1 gene modulates cancer cell responses during drug‐induced senescence. Aging (Albany NY). 2021;13(12):15833‐15874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lewinska A, Adamczyk‐Grochala J, Kwasniewicz E, et al. Reduced levels of methyltransferase DNMT2 sensitize human fibroblasts to oxidative stress and DNA damage that is accompanied by changes in proliferation‐related miRNA expression. Redox Biol. 2018;14:20‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Q, Cai M, Wang J, et al. Decreased ovarian function and autophagy gene methylation in aging rats. J Ovarian Res. 2020;13(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lewinska A, Adamczyk‐Grochala J, Kwasniewicz E, Wnuk M. Downregulation of methyltransferase Dnmt2 results in condition‐dependent telomere shortening and senescence or apoptosis in mouse fibroblasts. J Cell Physiol. 2017;232(12):3714‐3726. [DOI] [PubMed] [Google Scholar]
- 27. Lin MJ, Tang LY, Reddy MN, Shen CK. DNA methyltransferase gene dDnmt2 and longevity of drosophila. J Biol Chem. 2005;280(2):861‐864. [DOI] [PubMed] [Google Scholar]
- 28. Wildner G. Are rats more human than mice? Immunobiology. 2019;224(1):172‐176. [DOI] [PubMed] [Google Scholar]
- 29. Ménoret S, Ouisse LH, Tesson L, et al. In vivo analysis of human immune responses in immunodeficient rats. Transplantation. 2020;104(4):715‐723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chenouard V, Remy S, Tesson L, et al. Advances in genome editing and application to the generation of genetically modified rat models. Front Genet. 2021;12:615491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma Y, Shen B, Zhang X, et al. Heritable multiplex genetic engineering in rats using CRISPR/Cas9. PLoS One. 2014;9(3):e89413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma Y, Zhang L, Huang X. Building Cre knockin rat lines using CRISPR/Cas9. Methods Mol Biol. 2017;1642:37‐52. [DOI] [PubMed] [Google Scholar]
- 33. Ma Y, Chen W, Zhang X, et al. Increasing the efficiency of CRISPR/Cas9‐mediated precise genome editing in rats by inhibiting NHEJ and using Cas9 protein. RNA Biol. 2016;13(7):605‐612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu D, Ma Y, Zhang W, et al. Knockdown of cytochrome P450 2E1 inhibits oxidative stress and apoptosis in the cTnT(R141W) dilated cardiomyopathy transgenic mice. Hypertension. 2012;60(1):81‐89. [DOI] [PubMed] [Google Scholar]
- 35. Yang X, Lu D, Zhang X, et al. Knockout of ISCA1 causes early embryonic death in rats. Animal Model Exp Med. 2019;2(1):18‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dong W, Zhang L, Sun C, et al. Knock in of a hexanucleotide repeat expansion in the C9orf72 gene induces ALS in rats. Animal Model Exp Med. 2020;3(3):237‐244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chellamuthu A, Gray SG. The RNA methyltransferase NSUN2 and its potential roles in cancer. Cell. 2020;9(8):1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Singh D, Yadav R, Kaushik S, Wadhwa N, Kapoor S, Kapoor M. Transcriptome analysis of ppdnmt2 and identification of superoxide dismutase as a novel interactor of DNMT2 in the moss Physcomitrella patens . Front Plant Sci. 2020;11:1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dhaya I, Griton M, Konsman JP. Magnetic resonance imaging under isoflurane anesthesia alters cortical cyclooxygenase‐2 expression and glial cell morphology during sepsis‐associated neurological dysfunction in rats. Animal Model Exp Med. 2021;4(3):249‐260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Woźnica EA, Inglot M, Woźnica RK, Łysenko L. Liver dysfunction in sepsis. Adv Clin Exp Med. 2018;27(4):547‐551. [DOI] [PubMed] [Google Scholar]
- 41. Lelubre C, Vincent JL. Mechanisms and treatment of organ failure in sepsis. Nat Rev Nephrol. 2018;14(7):417‐427. [DOI] [PubMed] [Google Scholar]
- 42. Boomer JS, To K, Chang KC, et al. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA. 2011;306(23):2594‐2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cheng KT, Xiong S, Ye Z, et al. Caspase‐11‐mediated endothelial pyroptosis underlies endotoxemia‐induced lung injury. J Clin Invest. 2017;127(11):4124‐4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bagshaw SM, Lapinsky S, Dial S, et al. Acute kidney injury in septic shock: clinical outcomes and impact of duration of hypotension prior to initiation of antimicrobial therapy. Intensive Care Med. 2009;35(5):871‐881. [DOI] [PubMed] [Google Scholar]
- 45. Bosmann M, Ward PA. The inflammatory response in sepsis. Trends Immunol. 2013;34(3):129‐136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Regmi B, Shah MK. Possible implications of animal models for the assessment of visceral pain. Animal Model Exp Med. 2020;3(3):215‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hu J, Xu J, Feng X, Li Y, Hua F, Xu G. Differential expression of the TLR4 gene in pan‐cancer and its related mechanism. Front Cell Dev Biol. 2021;9:700661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tuorto F, Liebers R, Musch T, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19(9):900‐905. [DOI] [PubMed] [Google Scholar]
- 49. Durdevic Z, Mobin MB, Hanna K, Lyko F, Schaefer M. The RNA methyltransferase Dnmt2 is required for efficient Dicer‐2‐dependent siRNA pathway activity in drosophila. Cell Rep. 2013;4(5):931‐937. [DOI] [PubMed] [Google Scholar]
- 50. Cai L, Zhan M, Li Q, Li D, Xu Q. DNA methyltransferase DNMT1 inhibits lipopolysaccharide‐induced inflammatory response in human dental pulp cells involving the methylation changes of IL‐6 and TRAF6. Mol Med Rep. 2020;21(2):959‐968. [DOI] [PubMed] [Google Scholar]
- 51. Huang M, Cai S, Su J. The pathogenesis of sepsis and potential therapeutic targets. Int J Mol Sci. 2019;20(21):5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805‐820. [DOI] [PubMed] [Google Scholar]
- 53. Raymond SL, Holden DC, Mira JC, et al. Microbial recognition and danger signals in sepsis and trauma. Biochim Biophys Acta Mol Basis Dis. 2017;1863(10 Pt B):2564‐2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kumar V. Toll‐like receptors in sepsis‐associated cytokine storm and their endogenous negative regulators as future immunomodulatory targets. Int Immunopharmacol. 2020;89(Pt B):107087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chi H, Barry SP, Roth RJ, et al. Dynamic regulation of pro‐ and anti‐inflammatory cytokines by MAPK phosphatase 1 (MKP‐1) in innate immune responses. Proc Natl Acad Sci USA. 2006;103(7):2274‐2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Moynagh PN. TLR signalling and activation of IRFs: revisiting old friends from the NF‐kappaB pathway. Trends Immunol. 2005;26(9):469‐476. [DOI] [PubMed] [Google Scholar]
- 57. Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42(2):145‐151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1


