Abstract
Animal models of human diseases play a critical role in medical research. Pigs are anatomically and physiologically more like humans than are small rodents such as mice, making pigs an attractive option for modeling human diseases. Advances in recent years in genetic engineering have facilitated the rapid rise of pig models for use in studies of human disease. In the present review, we summarize the current status of pig models for human cardiovascular, metabolic, neurodegenerative, and various genetic diseases. We also discuss areas that need to be improved. Animal models of human diseases play a critical role in medical research. Advances in recent years in genetic engineering have facilitated the rapid rise of pig models for use in studies of human disease. In the present review, we summarize the current status of pig models for human cardiovascular, metabolic, neurodegenerative, various genetic diseases and xenotransplantation.
Keywords: animal model, gene‐editing, human disease, pig
Animal models of human diseases play an important role in the research of medical field. In the current review, we attempt to summarize the advances of the pig models for human cardiovascular diseases, metabolic diseases, neurodegenerative diseases, and some other genetic diseases. We also discuss the areas that need to be improved.
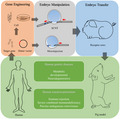
1. INTRODUCTION
Research on human disease pathogenesis is critical for progress in therapeutic medicine. Insufficient sample acquisition, environmental conditions, and ethics often impede studies to examine human disease directly, and therefore animal models are crucial for gaining in vivo insight into disease etiology and pathogenesis. Mice and other small rodents have long been important model animals for basic research, and have contributed greatly to our understanding of human disease pathogenesis. However, the limitations of rodent models are many. For example, metabolic rate is influenced by body size, and their small size leads to difficulties in performing surgery and using organs (Table 1). Considerable differences exist between rodents and humans in the regulatory networks controlling the activity of the immune system, metabolic functions, and responses to stress. 1 , 2 For example, age‐associated fasting blood glucose exhibits differential trends between mice and monkeys/humans. 3 Importantly, more than 80% of potential therapeutics fail in human trials despite showing safety and efficacy in mice. 4
TABLE 1.
| Species | Average body length (cm) | Average body weight (kg) | Average age (year) | Pregnancy length (day) | Offspring per litter | Heart as % of body weight | Brain as % of body weight |
|---|---|---|---|---|---|---|---|
| Homo sapiens | 170 | 40–100 | 72 | 280 | 1–2 | 0.5 | 2.1 |
| Mus musculus | 8–10 | 0.03–0.05 | 1–2 | 18–21 | 3–12 | 0.5 | 1.42 |
| Rattus norvegicus | 17 | 0.2–0.6 | 1–2 | 20–23 | 6–12 | 0.38 | 0.29 |
| Oryctolagus cuniculus | 48 | 3.5–7.5 | 5–12 | 23–34 | 3–9 | 0.3 | 0.4 |
| Sus scrofa | 125 | 40–120 | 20 | 114 | 10 | 0.6 | 0.5 |
| Ovis aries | 90 | 80–100 | 10–15 | 142–155 | 1–2 | 0.27 | 0.12 |
| Bos taurus | 220 | 500–900 | 20–23 | 270 | 1 | 0.03 | 0.08 |
| Canis familiaris | 75 | 10–25 | 12–15 | 58–67 | 5–6 | 0.85 | 0.59 |
| Rhesus monkey | 50 | 8–10 | 20 | 150 | 1–2 | 0.36 | 0.9 |
Pigs are one of the most common domestic animals in the world. Compared to other livestock and primates, pigs have a rapid growth rate, short generation intervals, large litter sizes, and standardized breeding techniques. These advantages, combined with comparable human and pig body sizes, anatomical and physiological characteristics, diets, and genome (Table 1), 5 have driven a gradual rise in the use of pigs as animal models for human diseases.
Similar body and organ sizes between pigs and humans will likely hasten the translation of pig studies (in comparison to mouse studies) to the clinic. Even before the advent of transgenic and gene‐editing technology, pig models enabled important advances in human heart, bone, metabolism, and even genetic diseases, to name a few. For example, pig models of acute myocardial infarction (MI) were generated by permanently ligating the trunk near one‐third of the apex after the first branch or by inflating an angioplasty balloon in the mid‐left anterior descending artery, 12 , 13 , 14 , 15 , 16 facilitating testing, and development of MI therapies for use in humans. Similarly, bone and cartilage models have been generated through surgically‐induced lesions in pigs for the development of biomaterials. 17 , 18 As both humans and pigs are monogastric omnivores, diet modification has been a fruitful approach for creating pig models of human metabolic disease. A high‐fat diet (HFD) induces obesity and metabolic syndrome and has been used in pigs to research the renal disease and nonalcoholic fatty liver disease. 19 , 20 To obtain genetic disease models, ENU chemical mutagenesis has been used to induce a set of point mutations that frequently mimic the subtlety and heterogeneity of human genetic lesions. 21 For example, microphthalmia‐associated transcription factor (MITF +/l247s) mutants mimic Waardenburg’s syndrome type II, dual oxidase 2 (DUOX 2 D409G/D409G) mutants mimic congenital hypothyroidism, SRY‐box transcription factor 10 (SOX 10 +/R109W) mutants mimic Mondini dysplasia, and mutants with a 2 bp CC insertion in the melanocortin receptor 1 (MC1R) mimic albinism. 22 , 23 , 24 , 25 These genetic models are heritable and require no special diet or surgical intervention to obtain experimental animals (Table 2).
TABLE 2.
Pig models by surgery, HFD and ENU mutagenesis
| Human disease | Method | Phenotype | References |
|---|---|---|---|
| Myocardial infarction | Permanent ligation of the trunk near one‐third of the apex after the first branch | Mir‐590‐3p suppresses proliferation and migration of cardiac fibroblasts | 13 |
| Myocardial infarction | Inflated angioplasty balloon in the mid‐left anterior descending artery for 90‐min | Reduction of apoptosis by Cortical bone stem cells | 16 |
| Myocardial infarction | 90‐min occlusion of the left anterior coronary artery | Improvement of cardiomyocyte proliferation by microrna‐199a | 48 |
| Meniscal lesions | 4 mm defect created in the medial meniscus by surgery | Reduced the chondral lesions by tissue‐engineered construct | 17 |
| Cartilage lesions | 6 mm created on the femoral condyles of stifle joints by surgery | Repaired by living hyaline cartilaginous graft | 18 |
| Renal disease | High‐fat diet | Diabetic changes and glomerulomegaly | 20 |
| Nonalcoholic fatty liver disease | High‐fat diet | Selenoproteins against damage induced by high‐fat diet | 19 |
| Waardenburg’s syndrome type II | ENU mutagenesis | Hearing loss, white coat color and MITF +/L247S | 22 |
| Congenital hypothyroidism | ENU mutagenesis | Anemia, immunodeficiency and DUOX 2 D409G/D409G | 23 |
| Mondini dysplasia | ENU mutagenesis | Inner ear mondini malformation and SOX 10 +/R109W | 22 |
| Albinism | ENU mutagenesis | White coat color and 2 bp CC insertion in the MC1R | 25 |
With the development of transgenic and gene‐editing technology, genetically engineered pig models are greatly expanding our understanding of human disease pathogenesis while aiding the development of novel treatments. Existing pig models comprise a wide range of human diseases, including cardiovascular diseases, diabetes, neurodegenerative diseases, genetic diseases, and cancer. Our review will focus on important genetically engineered pig models of human diseases in current use, generated using novel approaches, such as the combined technologies of microinjection (MI), somatic cell nuclear transfer (SCNT), and embryo transfer. We include helpful references for the construction of pig models and the research of human diseases.
2. CURRENT PIG MODELS OF HUMAN DISEASE
2.1. Metabolic diseases
Metabolic diseases are diseases that disrupt the normal metabolic process and are generally affected by both genetics and environments. Common metabolic diseases include obesity, hyperglycemia, hyperlipidemia, hypertension, hyperuricemia, fatty liver, cardiovascular disease, and cerebrovascular disease.
2.1.1. Diabetes
Diabetes mellitus (DM) is a group of metabolic disorders characterized by high blood sugar. Prolonged high blood glucose can damage the kidneys, heart, eyes, and nervous system. The three main classifications of DM are type I, type II, and gestational diabetes, although rarer forms of diabetes caused by mutations in specific genes also occur. Although type I and type II diabetes can appear in individuals without any family history of diabetes, they still show a highly heritable and generally involve insulin (INS) deficiency (type I) or insulin resistance (type II). 26 As insulin is secreted by the pancreatic islet cells, pigs—with a pancreas similar in size, shape, and blood circulation to the human pancreas—have become an attractive diabetes model. INS is believed to play a central role in insulin‐dependent diabetes, permanent neonatal diabetes, type 10 juvenile mature diabetes, and hyperinsulinemia. Mutations 27 and deletions 28 of INS were achieved in pigs using transgenic and gene editing techniques, providing invaluable models for studying the onset of diabetes and insulin supplement therapy. These pig models are often improved by insulin treatment and can be used for the research of insulin supplementation and islet transplantation. Type II diabetes is mainly caused by insufficient insulin secretion and excessive insulin resistance. In 2010, Renner et al. 29 generated transgenic pigs expressing a dominant‐negative GIP (glucose‐dependent insulinotropic polypeptide) receptor (GIPR[dn]) in pancreatic islets, demonstrating an essential role of GIP 30 for insulin secretion, the proliferation of β‐cells, and physiological expansion of β‐cell mass. As patients with type II diabetes show significant insulin resistance to exogenous GIP, these pigs are good models to study the role of GIP in glucose homeostasis and pancreatic development. IAPP can induce oxidative stress and further promote the production of amyloid deposits. Its deposition is considered to be one of the major causes of type II diabetes. Zou et al. 31 successfully established an IAPP gene humanized pig model, which exhibited symptoms of human type II diabetes, such as increased glucose tolerance. These pigs are suitable models for research into islet amyloid deposits in type II diabetes. In addition to the two main types of diabetes, Umeyama et al. 32 generated cloned pigs with a mutation in human hepatocyte nuclear factor 1α (HNF‐1α), which has been reported to cause type III maturity‐onset diabetes of the young (MODY3). 33 Although the majority of cloned MODY3 pigs died two weeks after birth, the viable pigs, showed high blood glucose levels and proved useful for studying the disease.
Following the development of gene‐editing technology, researchers also pay attention to models with multiple gene modifications. In 2015, Kong et al. 34 developed knock‐in pigs using the polycistronic system, which contains an expression cassette of 11‐β‐hydroxysteroid dehydrogenase 1 (11β‐HSD1) and another expression cassette of human islet amyloid polypeptide (HIAPP) and C/EBP homologous protein (CHOP). 11β‐HSD1 is important in insulin resistance when hIAPP and CHOP can induce β cell apoptosis in the pancreas. These pigs showed diabetic phenotypes such as hepatic insulin resistance and pancreatic cell apoptosis, which modeled type II diabetes better than some pigs with single‐gene modifications. Similarly, Zhang et al. 35 engineered pigs to carry three knock‐in risk genes, glucose‐dependent insulinotropic polypeptide receptor (GIPR dn ), human islet amyloid polypeptide (hIAPP), and Patatin‐like phospholipase domain‐containing three variant rs738409 C>G p.I148M (PNPLA3 I148M ), resulting in glucose and lipid metabolism disorders, abnormal fat development and liver necrosis, ideal for research on non‐alcoholic fatty liver disease (NAFLD) and type II diabetes.
2.1.2. Atherosclerosis
Atherosclerosis promotes cardiovascular disease, and lipid metabolism disorder is the pathological basis of atherosclerosis. Therefore, understanding abnormal lipid metabolism, such as high blood lipid, high cholesterol, and obesity, is vital. 36 , 37 Atherosclerosis is usually characterized by the deposition of lipids, cholesterol, and sugar complexes beginning from the intima and histiocytosis, leading to calcification. 38 Low‐density lipoprotein and apolipoprotein are closely related to blood lipid levels and have therefore been a focus of atherosclerosis research. In 2013, al‐Mashhadi et al. 39 generated proprotein convertase subtilisin/kexin type 9 (PCSK 9) mutation pigs, which exhibited reduced low‐density lipoprotein receptor (LDLR) levels and developed severe hypercholesterolemia and spontaneous atherosclerosis. Similarly, in 2014, Davis et al. 40 inserted a neomycin‐resistance cassette (NeoR) into the pig LDLR gene, disrupting its normal expression. In addition to spontaneous development of certain features of human atherosclerosis, atherosclerosis in LDLR mutant pig models could be accelerated by placing pigs on high‐fat and high‐cholesterol diets. The PCSK 9 transgenic pigs and the LDLR knockout pigs both focus on the regulation of low‐density lipoprotein to model human hypercholesterolemia. However, there is currently no evidence to prove that PCSK 9(D374Y) is functionally important in pigs. Compared to the PCSK 9 transgenic pigs, the LDLR−/− pigs have a shortened time to development of atherosclerosis. Focusing on apolipoprotein, a pig model of hypertriglyceridemia was developed in 2012 by Wei et al., 41 who targeted apolipoprotein (Apo) CIII, a key apolipoprotein in triglyceride metabolism. The pigs expressed human ApoCIII in the liver and intestinal tract. However, human ApoC III transgenic pigs are still the preferred tools for studying the mechanisms of hypertriglyceridemia‐associated diseases and for potential drug development, and it was unclear whether these pigs developed atherosclerosis. In 2018, Fang et al. 42 generated apolipoprotein E (ApoE) knockout pigs in which severe hypercholesterolemia and human‐like atherosclerotic lesions could be induced by a high‐fat, high‐cholesterol diet. The rate of cholesterol elevation under a high‐fat diet in ApoE −/− pigs is higher than in the PCSK9 transgenic pigs and the LDLR −/− pigs, and the hypertriglyceridemia phenotype was found in ApoE −/− pigs but not the PCSK 9 transgenic pigs or the LDLR −/− pigs, suggesting that ApoE −/− pigs may be a better model to simulate human atherosclerosis. Advances in gene‐editing technology led Huang et al. 43 to create ApoE and LDLR double gene knockout pigs in 2017. These pigs had significantly increased serum levels of low‐density lipoprotein cholesterol (LDL‐C) and total cholesterol (TC) and enriched the available models. Besides LDL, some cholesterol absorption relevant genes also influence the development of atherosclerosis. In 2015, Wang et al. 44 generated a pig model with InDels of NPC1L1, an important gene in cholesterol absorption.
In addition to abnormal lipid metabolism, atherosclerosis can be caused by abnormal glucose metabolism. 45 In 2017, Yang et al. 46 used zinc finger nuclease technology to create PPARγ mono‐allelic knockout pigs, which proved to be a good model for both atherosclerosis and type 2 diabetes. These pig models provide new research opportunities for early asymptomatic human atherosclerosis and other cardiovascular diseases that are difficult to study and treat.
2.1.3. Myocardial infarction
Myocardial infarction (MI) is a major cause of morbidity and mortality worldwide. Atherosclerosis is a risk factor for MI, as the rupture of atherosclerotic plaques leads to thrombus and sudden obstruction of the coronary artery, further resulting in myocardial ischemic necrosis. Various pig models of cardiovascular disease have been widely used in the development of treatments. In 2019, Hobby et al. 16 guided an angioplasty balloon through the femoral artery to the mid‐LAD past the first diagonal branch. The MI model generated by inflation of the balloon led to the discovery that cortical bone stem cells (CBSCs) influence cardiomyocyte and noncardiomyocyte cell death and immune cell recruitment in the heart following MI. 47 MicroRNAs have proven to be another rewarding avenue for MI research. MiR‐590‐3p was shown to suppress proliferation, migration, and differentiation of cardiac fibroblasts, whereas 13 MiR‐144‐3p and microRNA‐199a appear to induce these cardiac fibroblast programs. 12 , 48 At present, most research models of myocardial infarction are disposable models prepared by surgery, which have limitations for long‐term use. If a stable genetic model can be developed in the future, the research on myocardial infarction will be greatly accelerated (Table 3).
TABLE 3.
Pig models of metabolic diseases
| Human disease | Gene | Modification | References |
|---|---|---|---|
| Mody3 | HGF | Mutation | 32 |
| Type 2 diabetes | GIPR | Mutation | 29 |
| Diabetes, coronary heart disease | PPARγ | Knockout | 46 |
| Permanent neonatal diabetes mellitus | INS | Mutation | 27 |
| Type 2 diabetes | 11β‐HSD 1, HIAPP, CHOP | Knock‐in | 34 |
| Diabetes | INS | Knock‐in | 28 |
| Type 2 diabetes | hIAPP | Knockout | 31 |
| NAFLD | GIPR dn , hIAPP, PNPLA3 I148M | Knock‐in | 35 |
| Hypertriglyceridemia | ApoCIII | Knock‐in | 41 |
| Hypercholesterolemia, atherosclerosis | PCSK9 | Mutation | 39 |
| Hypercholesterolemia, atherosclerosis | LDLR | Knock‐in | 40 |
| Disorder of cholesterol absorption | NPC1L1 | Knockout | 44 |
| Serum LDL‐C and TC levels increase | ApoE and LDLR | Knockout | 43 |
| Hypercholesterolemia, atherosclerosis | ApoE | Knockout | 42 |
2.2. Neurodegenerative diseases
Neurodegenerative diseases are functional disorders caused by the loss of neurons and/or their myelin sheaths in the brain and spinal cord. The most common diseases include Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS).
2.2.1. Alzheimer’s disease
Alzheimer’s disease, accounting for approximately 50%~80% of human dementia cases, 49 is a neurodegenerative disease with hidden onset, characterized by general dementia such as memory impairment, aphasia, executive dysfunction, and personality behavior changes. Patients usually exhibit accumulation of extracellular amyloid‐beta (Aβ) to form senile plaques and intracellular neurofibrillary tangles of microtubule‐binding protein Tau in the gray matter of the brain. At present, amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) are considered to be pathogenic genes of familial AD. In 2009, Kragh et al. 50 generated an AD pig model of transgenic human APP695sw. Although high expression of the transgene was detected in different brain regions of this pig model, there was no elevated Aβ level in tissues or memory impairment in 1‐year‐old pigs. 51 In 2013, Jakobsen et al. 52 used recombinase‐mediated cassette exchange (RMCE) technology to generate a PSEN1 M146I mutant pig model. AD pigs carrying both APP 695sw and PSEN 1 M146I mutations were subsequently generated in 2016. These pigs were found to accumulate Aβ‐42 in their brains 53 at around 10–18 months. Several known pathogenic genes of familial AD have been modified in pig models. Also, AD pigs carrying triple mutations of hAPP (K670N/M671L, I716V, and V717I), hTau (P301L), and hPS1 (M146V and L286P) were generated using the polycistronic vector system. These pigs were similarly found to accumulate Aβ‐40 and Aβ‐42 in their brain, 54 a significant phenotype of AD patients.
2.2.2. Parkinson’s disease
Parkinson’s disease, also known as paralysis tremors, is a neurodegenerative disease caused by the degeneration of dopamine neurons in the substantia nigra and the presence of Lewy bodies in the neurons. 55 , 56 In 2014, Yao et al. 57 generated DJ‐1 gene knockout pigs using TALEN. Although the expression of DJ‐1 was inhibited at the protein level, defective cloning led to the early death of these animals. In 2014, Zhou et al. 58 generated a PARK2 and PINK1 double knockout pig with deficient protein levels of both gene products, and in 2016, Wang et al. 59 generated pigs with triple gene knockouts of DJ‐1, Parkin, and PINK1 using CRISPR/Cas9. In 2018, Zhu et al. 60 developed SCNA knock‐in pigs carrying three missense mutations (E46K, H50Q, and G51D) known to cause Parkinson’s disease. No typical symptoms of PD have been observed in any of these pig models, possibly because PD is a progressive disease that occurs mostly in the elderly.
2.2.3. Huntington’s disease
Huntington’s disease is a rare autosomal dominant genetic disorder. Due to variations in Huntington protein (HTT), patients typically develop motor symptoms, cognitive dysfunction, and mental disorders. In 2010, Yang et al. 61 generated HD pigs with HTT mutations that suffered significant involuntary movements. In 2018, Yan et al. 62 found that endogenous expression of full‐length HTT mutants in pigs elicited significant neuronal degeneration, which effectively mimics human Huntington’s disease. This single gene mutation has resulted in the current pig models that simulate Huntington’s disease well. Future use of these models to search for effective treatments will be an important application of these pig models (Table 4).
TABLE 4.
Pig models of neurodegenerative diseases
| Human disease | Gene | Modification | References |
|---|---|---|---|
| Alzheimer | APP695sw | Knock‐in | 50 |
| Alzheimer | PSEN1 MI46I | Mutation | 52 |
| Alzheimer | APP SW , PSEN1 MI46I | Mutation | 53 |
| Alzheimer | hAPP, hTau, hPS1 | Mutation | 54 |
| Huntington | HTT | Mutation | 61 |
| Huntington | HTT | Knock‐in | 62 |
| Parkinson | Parkin, DJ‐1 | Knockout | 57 |
| Parkinson | PARK2, DJ‐1, PINK1 | Knockout | 59 |
| Parkinson | SNCA | Knock‐in | 60 |
| Parkinson | PARK2, PINK1 | Knockout | 58 |
2.3. Genetic diseases
Genetic diseases generally refer to diseases caused by changes in genetic material or disease genes. In addition to the metabolic diseases and neurodegenerative diseases discussed above, pig models of cystic fibrosis, Duchenne muscular dystrophy, hemophilia, and various cancers have also been developed for medical research.
2.3.1. Cystic fibrosis
Cystic fibrosis (CF), a recessive genetic disease with a single gene mutation, is caused by dysfunction of the CF transmembrane conductance regulator (CFTR). The disease starts in early childhood and affects many tissues and organs, including the respiratory tract, lungs, gastrointestinal tract, pancreas, liver, reproductive tract, and sweat glands. Due to defective chloride ion channels in CF patients, respiratory mucus gland secretions become dehydrated and viscous, resulting in respiratory tract infection, airway obstruction, and meconium obstruction. Viscous secretions can additionally block the reproductive system, leading to male infertility. 63 , 64 The pig model of cystic fibrosis is an outstanding example of a genetically engineered pig as a model of human disease. In 2008, a pig model with the CFTR allele deletion and another with the most common mutation (ΔF508) were generated by Rogers et al., using a recombinant adeno‐associated virus (RAAV) delivery system. 65 While approximately 15% of CF patients are born with meconium blocking, meconium blocking rates were 100% in CFTR −/− pigs, and a little bit less in CFTR +/ΔF508 pigs. Subsequent studies have shown that CFTR ΔF508/ΔF508 pigs develop meconium blocking, abnormal pancreatic and bile secretion, 66 and lung diseases similar to those of CF patients, which develop spontaneously within a few weeks of birth. 67 Based on studies of the CFTR −/− pigs, Stoltz et al. 68 established a corrected model for intestinal expression in 2013, which successfully alleviated meconium obstruction. Thus, the CFTR −/− pig models replicate most of the features of human CF and have shown tremendous promise for translational therapies. 69
2.3.2. Duchenne muscular dystrophy
Muscular dystrophy is a genetic disorder characterized by progressive muscle weakness, wasting, and muscle degeneration. These diseases mainly include Duchenne muscular dystrophy (DMD), Becker muscular dystrophy (BMD), limb‐girdle muscular dystrophy (LGMD), congenital muscular dystrophy (CMD), and Emery‐Dreifuss Muscular dystrophy (EDMD). 70 , 71 DMD is an incurable X‐linked genetic disease caused by deletion, point mutation, or duplication of the DMD gene. 72 Patients tend to die in their 20s or 30s due to weaknesses in the muscles of the heart and lungs. In 2013, Klymiuk et al. 73 used gene targeting and SCNT to generate a pig model with a deletion of Exon 52 of DMD. This pig model developed symptoms similar to human DMD patients, for instance, elevated serum creatine kinase activity, myofibrosis, and loss of myotrophin. However, use of the DMD pig model has been greatly restricted by the considerable rates of pig neonatal death. Yu et al. 74 used CRISPR/Cas9 gene‐editing technology to accurately edit exon 27 of DMD, generating another DMD pig model in 2016. This model also displayed a phenotype similar to human DMD with loss of myotrophic protein and myocardial damage. However, similar to the previous model, these pigs are prone to premature death. Moretti et al. 75 found that a truncated DMD Δ51–52 pig model improved skeletal muscle function and heart rhythm as well as reducing neonatal death, and recent studies by Chiappalupi et al. 76 found that injection of porcine Sertoli cells can eliminate the inflammatory response and the expression of dystrophin. Overall, DMD is a disease with single gene mutations. Pig DMD models hold great promise in the development of drugs and treatments for DMD.
2.3.3. Cancer
Carcinoma is the most common type of malignant tumor originating from epithelial tissue. In 2010, Luo et al. 77 reported a pig model with a knockout of the breast cancer‐associated gene (BRCA1) mediated by adenovirus. Although the BRCA1 +/∆11 pigs were able to develop to term, they had high perinatal mortality. No one pig survived more than 18 days, leading the model to be adjusted further. In 2012, Flisikowska et al. 78 produced abnormal lesions and adenomas in large intestines of pigs by mutating adenomatous polyposis coli (APC) at sites 1311 and 1016. In these pigs, a single allele mutation of APC was sufficient to initiate the well‐characterized precancer sequence leading to growths similar to those in patients with familial adenomatous polyposis in human colorectal lesions, which has not been possible in the mouse models. RUNX 3 is considered to be a tumor suppressor gene associated with gastric adenocarcinoma. In 2016, Kang et al. 79 established a pig model with a RUNX 3 knockout, providing opportunities for gastric cancer research. In 2016, Saalfrank et al. 80 generated a targeted TP53 knockout pig, which developed osteosarcoma in the long bone, skull, and mandible. Some genes tend to cause more than one type of cancer. In 2014, Sieren et al. 81 generated pigs with a mutant TP53 gene that developed multiple tissue lesions such as lymphoma, Wilm’s neuroblastoma, and bone‐derived tumor. In 2015, Schook et al. 82 constructed a pig model that could be conditionally induced to express various tumor types via mutation of KRAS G12D and TP53 R167H via Cre recombinase expression. In 2017, Wang et al. 83 used TALEN and SCNT techniques to produce pigs simulating human non‐small cell lung cancer (NSCLC). These pigs achieved time–space and site‐specific expression of the mutant proteins by Cre induction of rearrangement of echinoderm microtubule‐associated protein 4 (EML4) and anaplastic lymphoma kinase (ALK) genes. This inducible system may be used to study many other cancers.
2.3.4. Other genetic diseases
Additional pig models have been developed to recapitulate various other genetic diseases over the years. Von Willebrand disease is an inherited hemorrhagic disorder generally caused by an autosomal dominant plasma vWF deficiency. In 2014, Hai et al. 84 generated a vWF knockout pig model of von Willebrand disease, which showed significant prolonged bleeding and defective coagulation.
Hemophilia comprises a group of recessive X‐linked inherited clotting disorders in patients lacking various clotting factors. Hemophilia B is caused by lack of factor IX (F9) gene. In 2020, Chen et al. 85 reported that targeted pig knockouts lacking a functional F9 gene showed obvious symptoms of hemophilia B, such as cruor disorder, synovitis, and cartilage destruction. Moreover, the symptoms were significantly rescued by knocking the human F9 gene into the knockout pigs. This research suggests new ways to correct hemophilia B in the future by genome editing.
Hutchinson‐Gilford progeria syndrome (HGPS) is a rare genetic disorder that often causes premature aging and cardiovascular complications. Introducing heterozygous mutations of the LMNA gene into pigs induced growth retardation, lipodystrophy, skin and bone changes, cardiovascular disease, and death in adolescence. 86 The mean lifespan of these pigs is just about 6 months, making them good models for longevity studies in clinics.
Loss‐of‐function mutations in the COL2A1 gene are the etiology of type II collagenopathy. COL2A1 mutant pigs exhibit bone dysplasia and tracheal collapse, modeling aspects of human spondyloepiphyseal dysplasia and stickler syndrome type I. 87
Waardenburg’s disease is a syndrome of deafness, white hair, and eye disease. Wang et al. 88 generated MITF mutant pigs using CRISPR/Cas9, which also developed white fur and hearing impairments. Then in 2021, Yao et al. 89 successfully rescued anophthalmia and hearing loss in the cloned pigs using single‐stranded oligodeoxynucleotide (ssODN) and long donor plasmid DNA as the repair template.
Another epidermal disorder, oculocutaneous albinism type I was modeled in pigs by either TYR gene fragment knockout or point mutation. 58 , 90 The pigs completely lost dark pigment in skin, hair, and eyes, showing visible signs of the disease, but this model is still worth further analysis.
Unlike mice, pigs have a high cone density and dense photoreceptor retinal area, similar to humans. Cloned pigs with a rhodopsin (Rho) mutation showed reduced light sensitivity, similar to patients with inherited retinal degeneration 91 , 92 , 93 ; ELOVL4 mutant pigs, which simulate Stargardt disease type 3, showed photoreceptor loss and reduced retinal response. 94
Hereditary tyrosinemia type I (HT1) is caused by a deficiency of fumaryl acetoacetic acid hydrolase (FAH), which leads to liver failure. Hickey et al. 95 generated FAH +/− cloned pigs with an adeno‐associated virus‐mediated gene targeting strategy. The FAH −/− offspring showed severe liver damage, but unlike humans, FAH‐deficiency in pigs causes a lethal defect in utero, and interestingly the defect of FAH could be cured by 2‐(2‐nitro‐4‐trifluoromethylbenzoyl)‐1,3 cyclohexanedione (NTBC).
Phenylketonuria, caused by a deficiency of phenylalanine hydroxylase (PAH), can lead to neurocognitive impairment, behavioral problems, eczema, and hypopigmentation. Koppes et al. 96 generated a pig model of phenylketonuria with symptoms including hyperphenylalaninemia, growth retardation, hypoplasia, ventricular dilation, and decreased gray matter volume. But they did not show devastating neurocognitive and neurological clinical characteristics.
In summary, pig models have been widely used to simulate human diseases, and most genetic diseases can be studied by preparing pig models. Especially when the causal gene in humans is known (Table 5).
TABLE 5.
Pig models of genetic diseases
| Human disease | Gene | Modification | References |
|---|---|---|---|
| Cystic fibrosis | CFTR | Knockout, mutation | 64 |
| Cystic fibrosis | CFTR | Knockout | 67 |
| Cystic fibrosis | CFTR | Knockout | 68 |
| Duchenne muscular dystrophy | DMD | Knockout | 73 |
| Duchenne muscular dystrophy | DMD | Knockout | 74 |
| Breast cancer | BRCA1 | Knockout | 77 |
| Colorectal cancer | APC 1311 , APC 1016 | Mutation | 78 |
| Lymphoma, wilm‐blastoma, and bone tumors | TP53 R167H | Mutation | 81 |
| Cancer | KRAS G12D , TP53 R167H | Mutation | 82 |
| Gastric cancer | RUNX3 | Knockout | 79 |
| Osteosarcoma | TP53 | Knockout | 80 |
| Lung cancer | EML4, ALK | Knock‐in | 83 |
| Von Willebrand disease | vWF | Knockout | 84 |
| Hemophilia B | hF9 | Knock‐in | 85 |
| Hutchinson‐Gilford progeria syndrome | LMNA | Mutation | 86 |
| Waardenburg’s | MITF | Knockout | 88 |
| Ocular skin albinism type 1 | TYR | Knockout | 58 |
| Retinitis pigmentosa | Rho | Mutation | 91 |
| Retinitis pigmentosa | Rho | Mutation | 92 |
| Retinitis pigmentosa | Rho | Mutation | 93 |
| Stargardt disease type 3 (STGD 3) | ELOVL4 | Knockout, mutation | 94 |
| Tyrosinemia type I | FAH | Knockout | 95 |
| Phenylketonuria | PAH | Knockout | 96 |
2.4. Xenotransplantation
One of the most important roles of pigs in the biomedical field is as tissue and organ donors. There is currently a serious shortage of life‐saving tissues and organs for human clinical transplantation. The structure and function of organs are similar between pigs and humans. Because of this, pigs have attracted great interest in the field of xenotransplantation. Corneas, hearts, kidneys, livers, lungs, nerve cells, and islets of pigs have been studied as candidates for xenotransplantation.
One of the key problems in xenotransplantation is immune rejection. The presence of α‐1,3‐galactose (α‐Gal) epitopes on pig cells is a major obstacle to successful xenotransplantation. α‐galactosyl transferase 1 (GGTA 1) is an important gene involved in the biosynthesis of α‐1,3‐galactose. Researchers have established GGTA 1 knockout or mutant pig models. 97 , 98 , 99 , 100 , 101 Similarly, N‐glycolylneuraminic acid (NeuGc) is a non‐Gal xenoantigen in pigs which can compromise successful transplantation to human hosts. This challenge was met by the establishment of a CMP‐Neu5Ac hydroxylase (CMAH) knockout pig model. 102 , 103 Since immune rejection is often not controlled by a single gene, researchers have also generated a combined knockout of GGTA 1 and CMAH, as well as some other xenoantigen genes such as iGb3S and β4 GalNT2. 104 , 105 , 106 , 107 In addition to xenoantigens, major histocompatibility complex class I (MHC I) 108 , 109 , 110 and NK cells 111 are important factors in host immunity, for which pig models have been established to address potential problems. Furthermore, the establishment of several pig models with severe combined immunodeficiency and inactivation of porcine endogenous retroviruses has reduced concerns about the spread of zoonotic diseases and has provided important materials for the advancement of xenotransplantation. 112 , 113 , 114 , 115 , 116 , 117
Solid organ xenotransplantation between pig and non‐human primates is also a key research priority before human clinical trials. In recent years, with the development of xenotransplantation, several types of solid organ xenotransplantation have been tested in non‐human primates with some success, including heart, 118 , 119 kidney, 120 lung, 121 and liver. 122 Even more exciting, the world’s first gene‐edited pig heart transplant into a human was carried out in January 2022. Although the patient died unfortunately after two months, this is still a milestone in the search for a solution to the shortage of human organs. Almost at the same time, the world’s first pig kidney transplant into a human was reported. 123 We expect that in the future, gene‐modified pigs will certainly provide new opportunities for the shortage of human organs (Table 6).
TABLE 6.
Pig models of xenotransplantation
| Human disease | Gene | Modification | References |
|---|---|---|---|
| Immunological rejection (αGal) | GGTA1 | Knockout, mutation | 97, 98, 99, 100, 101 |
| Immunological rejection (non‐Gal) | CMAH | Knockout | 102, 103 |
| Immunological rejection | GGTA1, CMAH | Knockout | 105, 106 |
| Immunological rejection | GGTA1, CMAH, iGb3S | Knockout | 104 |
| Immunological rejection | GGTA1, β4GalNT22, CMAH | Knockout | 107 |
| Immunological rejection (MHC I) | SLA | Knockout | 110 |
| Immunological rejection (MHC I) | B2M | Knockout | 108, 109 |
| Immunological rejection (NK cell) | ULBP1 | Knockout | 111 |
| Severe combined immunodeficiency | RAG2 | Knockout | 117 |
| Severe combined immunodeficiency | RAG1/2 | Knockout | 116 |
| Severe combined immunodeficiency | RAG2, IL2RG | Knockout | 113 |
| Severe combined immunodeficiency | IL2RG | Knockout | 112, 114 |
| Inactivation of porcine endogenous retroviruses | PERV | Knockout | 115 |
3. CONCLUSION
Currently, there are pig models for a variety of human diseases including cardiovascular, metabolic, neurodegenerative, and other genetic diseases, which have provided considerable support for the analysis and treatment of human diseases. Recently, the COVID‐19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has led to a serious global public health crisis. The analysis of the pathogenesis of infection, the development of diagnostic and therapeutic methods, and the validation of vaccine and drug products all require large animal models similar to human clinical pathogenesis. Du et al. 124 replaced pig angiotensin‐converting enzyme 2 (ACE2) by site‐specific knock‐in of human hACE2 and found that primary epithelial cells isolated from the lungs and kidneys of this humanized pig model were highly sensitive to SARS‐CoV‐2 infection. In conclusion, pig models have great potential to advance the study of human diseases, from the study of pathogenesis to the development and utilization of drugs, and even as tissue and organ donors.
In addition, there is much that needs improving in pig gene editing, in vitro embryo culture, and assisted reproduction. In recent years, research on pig pluripotent stem cells has also provided new opportunities for the production of cloned pigs. Although the emergence of gene‐editing technology has greatly accelerated progress in pig models for studying genetic background and for testing drugs, therapeutics, and methods of delivery, safety, and ethical issues cannot be ignored. On the one hand, humans and pigs are different in many ways, and drugs and treatments developed in pig models must be determined to be safe before clinical tests. On the other hand, because of the existence of zoonosis, care must be taken at every stage of the experiment to avoid cross‐contamination and the spread of disease. Apart from safety and ethical issues, animal welfare also affects society’s willingness to condone animal research. The health of the animal used as a model is not only critical to obtaining reliable results but is also a responsibility for every researcher. Improving the nutrition, physical environment, health, behavioral interactions, and mental state of pigs will promote the development and social acceptance of pig models. 125 By addressing the importance of these issues, pig models will continue to be an important source of support for the advancement of human medicine in the future.
CONFLICT OF INTEREST
The authors declared no conflicts of interest.
AUTHOR CONTRIBUTIONS
Naipeng Hou conceived and wrote the original draft of the manuscript. Xuguang Du and Sen Wu revised the manuscript. All authors critically read and contributed to the manuscript, and approved its final version.
ACKNOWLEDGMENT
We thank Dr. Lara Carroll (University of Utah) for the careful reading of the manuscript. This work was supported by the National Key Research and Development Program of China (Grant No. 2021YFA0805900), the 2020 Research Program of Sanya Yazhou Bay Science and Technology City (Grant No. 202002011), the National Natural Science Foundation of China (Grant No. 32002180) and the Key Research and Development Program of Hainan Province, China (Grant No. ZDYF2021SHFZ230).
Hou N, Du X, Wu S. Advances in pig models of human diseases. Anim Models Exp Med. 2022;5:141–152. doi: 10.1002/ame2.12223
Funding information
The National Key Research and Development Program of China (Grant No. 2021YFA0805900), the 2020 Research Program of Sanya Yazhou Bay Science and Technology City (Grant No. 202002011), the National Natural Science Foundation of China (Grant No. 32002180) and the Key Research and Development Program of Hainan Province, China (Grant No. ZDYF2021SHFZ230)
REFERENCES
- 1. Rydell‐Törmänen K, Johnson JR. The applicability of mouse models to the study of human disease. Methods Mol Biol. 2019;1940:3‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yue F, Cheng Y, Breschi A, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355‐364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Palliyaguru DL, Vieira Ligo Teixeira C, Duregon E, et al. Study of longitudinal aging in mice: presentation of experimental techniques. J Gerontol A Biol Sci Med Sci. 2021;76:552‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perrin S. Preclinical research: make mouse studies work. Nature. 2014;507:423‐425. [DOI] [PubMed] [Google Scholar]
- 5. Wernersson R, Schierup MH, Jørgensen FG, et al. Pigs in sequence space: a 0.66X coverage pig genome survey based on shotgun sequencing. BMC Genomics. 2005;6:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bähr A, Wolf E. Domestic animal models for biomedical research. Reprod Domest Anim. 2012;47:59‐71. [DOI] [PubMed] [Google Scholar]
- 7. Ballarin C, Povinelli M, Granato A, et al. The brain of the domestic bos taurus: weight, encephalization and cerebellar quotients, and comparison with other domestic and wild cetartiodactyla. PLoS ONE. 2016;11:e0154580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Herndon JG, Tigges J, Klumpp SA, Anderson DC. Brain weight does not decrease with age in adult rhesus monkeys. Neurobiol Aging. 1998;19:267‐272. [DOI] [PubMed] [Google Scholar]
- 9. Lossi L, D’Angelo L, De Girolamo P, Merighi A. Anatomical features for an adequate choice of experimental animal model in biomedicine: II. Small laboratory rodents, rabbit, and pig. Ann Anat. 2016;204:11‐28. [DOI] [PubMed] [Google Scholar]
- 10. Louey S, Cock ML, Harding R. Long term consequences of low birthweight on postnatal growth, adiposity and brain weight at maturity in sheep. J Reprod Dev. 2005;51:59‐68. [DOI] [PubMed] [Google Scholar]
- 11. Lunney JK, Van Goor A, Walker KE, Hailstock T, Franklin J, Dai C. Importance of the pig as a human biomedical model. Sci Transl Med. 2021;13:eabd5758. [DOI] [PubMed] [Google Scholar]
- 12. Yuan X, Pan J, Wen L, et al. MiR‐144‐3p enhances cardiac fibrosis after myocardial infarction by targeting PTEN. Front Cell Dev Biol. 2019;7:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yuan X, Pan J, Wen L, et al. MiR‐590‐3p regulates proliferation, migration and collagen synthesis of cardiac fibroblast by targeting ZEB1. J Cell Mol Med. 2020;24:227‐237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. López E, Sánchez‐Margallo FM, Álvarez V, et al. Identification of very early inflammatory markers in a porcine myocardial infarction model. BMC Vet Res. 2019;15:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Valina C, Pinkernell K, Song YH, et al. Intracoronary administration of autologous adipose tissue‐derived stem cells improves left ventricular function, perfusion, and remodelling after acute myocardial infarction. Eur Heart J. 2007;28:2667‐2677. [DOI] [PubMed] [Google Scholar]
- 16. Hobby ARH, Sharp TE 3rd, Berretta RM, et al. Cortical bone‐derived stem cell therapy reduces apoptosis after myocardial infarction. Am J Physiol Heart Circ Physiol. 2019;317:H820‐H829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moriguchi Y, Tateishi K, Ando W, et al. Repair of meniscal lesions using a scaffold‐free tissue‐engineered construct derived from allogenic synovial MSCs in a miniature swine model. Biomaterials. 2013;34:2185‐2193. [DOI] [PubMed] [Google Scholar]
- 18. Peck Y, He P, Chilla GS, Poh CL, Wang DA. A preclinical evaluation of an autologous living hyaline‐like cartilaginous graft for articular cartilage repair: a pilot study. Sci Rep. 2015;5:16225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang P, Lu Z, He M, Shi B, Lei X, Shan A. The effects of endoplasmic‐reticulum‐resident selenoproteins in a nonalcoholic fatty liver disease pig model induced by a high‐fat diet. Nutrients. 2020;12(3):692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rodríguez RR, González‐Bulnes A, Garcia‐Contreras C, et al. The Iberian pig fed with high‐fat diet: a model of renal disease in obesity and metabolic syndrome. Int J Obes (Lond). 2020;44:457‐465. [DOI] [PubMed] [Google Scholar]
- 21. Oliver PL, Davies KE. New insights into behaviour using mouse ENU mutagenesis. Hum Mol Genet. 2012;21:R72‐R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hai T, Guo W, Yao J, et al. Creation of miniature pig model of human Waardenburg syndrome type 2A by ENU mutagenesis. Hum Genet. 2017;136:1463‐1475. [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y, Xue Y, Cao C, et al. Thyroid hormone regulates hematopoiesis via the TR‐KLF9 axis. Blood. 2017;130:2161‐2170. [DOI] [PubMed] [Google Scholar]
- 24. Hai T, Cao C, Shang H, et al. Pilot study of large‐scale production of mutant pigs by ENU mutagenesis. eLife. 2017;6:e26248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jia Q, Cao C, Tang H, et al. A 2‐bp insertion (c.67_68insCC) in MC1R causes recessive white coat color in Bama miniature pigs. J Genet Genomics. 2017;44:215‐217. [DOI] [PubMed] [Google Scholar]
- 26. American DA. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2014;37:S81‐S90. [DOI] [PubMed] [Google Scholar]
- 27. Renner S, Braun‐Reichhart C, Blutke A, et al. Permanent neonatal diabetes in INS(C94Y) transgenic pigs. Diabetes. 2013;62:1505‐1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cho B, Kim SJ, Lee E‐J, et al. Generation of insulin‐deficient piglets by disrupting INS gene using CRISPR/Cas9 system. Transgenic Res. 2018;27:289‐300. [DOI] [PubMed] [Google Scholar]
- 29. Renner S, Fehlings C, Herbach N, et al. Glucose intolerance and reduced proliferation of pancreatic beta‐cells in transgenic pigs with impaired glucose‐dependent insulinotropic polypeptide function. Diabetes. 2010;59:1228‐1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nauck MA, Baller B, Meier JJ. Gastric inhibitory polypeptide and glucagon‐like peptide‐1 in the pathogenesis of type 2 diabetes. Diabetes. 2004;53:S190‐S196. [DOI] [PubMed] [Google Scholar]
- 31. Zou X, Ouyang H, Yu T, et al. Preparation of a new type 2 diabetic miniature pig model via the CRISPR/Cas9 system. Cell Death Dis. 2019;10:823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Umeyama K, Watanabe M, Saito H, et al. Dominant‐negative mutant hepatocyte nuclear factor 1α induces diabetes in transgenic‐cloned pigs. Transgenic Res. 2009;18:697‐706. [DOI] [PubMed] [Google Scholar]
- 33. Yamagata K. Regulation of pancreatic beta‐cell function by the HNF transcription network: lessons from maturity‐onset diabetes of the young (MODY). Endocr J. 2003;50:491‐499. [DOI] [PubMed] [Google Scholar]
- 34. Kong S, Ruan J, Xin L, et al. Multi‐transgenic minipig models exhibiting potential for hepatic insulin resistance and pancreatic apoptosis. Mol Med Rep. 2016;13:669‐680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang K, Tao C, Xu J, et al. CD8+ T cells involved in metabolic inflammation in visceral adipose tissue and liver of transgenic pigs. Front Immunol. 2021;12:690069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Linton MF, Yancey PG, Davies SS, et al. The role of lipids and lipoproteins in atherosclerosis. In: Feingold KR, Anawalt B, Boyce A, et al., eds. Endotext. MDText.com, Inc.; 2019. [Google Scholar]
- 37. Poirier P, Giles TD, Bray GA, et al. Obesity and cardiovascular disease: pathophysiology, evaluation, and effect of weight loss. Arterioscl Throm Vas. 2006;26:968‐976. [DOI] [PubMed] [Google Scholar]
- 38. Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317‐325. [DOI] [PubMed] [Google Scholar]
- 39. Al‐Mashhadi RH, Sørensen CB, Kragh PM, et al. Familial hypercholesterolemia and atherosclerosis in cloned minipigs created by DNA transposition of a human PCSK9 gain‐of‐function mutant. Sci Transl Med. 2013;5:166ra1. [DOI] [PubMed] [Google Scholar]
- 40. Davis BT, Wang X‐J, Rohret JA, et al. Targeted disruption of LDLR causes hypercholesterolemia and atherosclerosis in Yucatan miniature pigs. PLoS ONE. 2014;9:e93457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wei J, Ouyang H, Wang Y, et al. Characterization of a hypertriglyceridemic transgenic miniature pig model expressing human apolipoprotein CIII. FEBS J. 2012;279(1):91‐99. [DOI] [PubMed] [Google Scholar]
- 42. Fang B, Ren X, Wang Y, et al. Apolipoprotein E deficiency accelerates atherosclerosis development in miniature pigs. Dis Model Mech. 2018;11:dmm036632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang L, Hua Z, Xiao H, et al. CRISPR/Cas9‐mediated ApoE−/− and LDLR−/− double gene knockout in pigs elevates serum LDL‐C and TC levels. Oncotarget. 2017;8:37751‐37760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Y, Du Y, Shen B, et al. Efficient generation of gene‐modified pigs via injection of zygote with Cas9/sgRNA. Sci Rep. 2015;5:8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nicholls SJ, Uno K. Peroxisome proliferator‐activated receptor (PPAR α/γ) agonists as a potential target to reduce cardiovascular risk in diabetes. Diab Vasc Dis Res. 2012;9:89‐94. [DOI] [PubMed] [Google Scholar]
- 46. Yang D, Yang H, Li W, et al. Generation of PPARγ mono‐allelic knockout pigs via zinc‐finger nucleases and nuclear transfer cloning. Cell Res. 2011;21:979‐982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sharp TE 3rd, Schena GJ, Hobby AR, et al. Cortical bone stem cell therapy preserves cardiac structure and function after myocardial infarction. Circ Res. 2017;121:1263‐1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gabisonia K, Prosdocimo G, Aquaro GD, et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature. 2019;569:418‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Winblad B, Amouyel P, Andrieu S, et al. Defeating Alzheimer’s disease and other dementias: a priority for European science and society. Lancet Neurol. 2016;15:455‐532. [DOI] [PubMed] [Google Scholar]
- 50. Kragh PM, Nielsen AL, Li J, et al. Hemizygous minipigs produced by random gene insertion and handmade cloning express the Alzheimer’s disease‐causing dominant mutation APPsw. Transgenic Res. 2009;18:545‐558. [DOI] [PubMed] [Google Scholar]
- 51. Søndergaard LV, Ladewig J, Dagnæs‐Hansen F, Herskin MS, Holm IE. Object recognition as a measure of memory in 1‐2 years old transgenic minipigs carrying the APPsw mutation for Alzheimer’s disease. Transgenic Res. 2012;21:1341‐1348. [DOI] [PubMed] [Google Scholar]
- 52. Jakobsen JE, Johansen MG, Schmidt M, et al. Generation of minipigs with targeted transgene insertion by recombinase‐mediated cassette exchange (RMCE) and somatic cell nuclear transfer (SCNT). Transgenic Res. 2013;22:709‐723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jakobsen JE, Johansen MG, Schmidt M, et al. Expression of the Alzheimer’s disease mutations aβpp695sw and psen1m146i in double‐transgenic göttingen minipigs. J Alzheimers Dis. 2016;53:1617‐1630. [DOI] [PubMed] [Google Scholar]
- 54. Lee S‐E, Hyun H, Park M‐R, et al. Production of transgenic pig as an Alzheimer’s disease model using a multi‐cistronic vector system. PLoS ONE. 2017;12:e0177933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kalia LV, Lang AE. Parkinson’s disease. Lancet. 2015;386(9996):896‐912. [DOI] [PubMed] [Google Scholar]
- 56. Shulman JM, De Jager PL, Feany MB. Parkinson’s disease: genetics and pathogenesis. Annu Rev Pathol. 2011;6:193‐222. [DOI] [PubMed] [Google Scholar]
- 57. Yao J, Huang J, Hai T, et al. Efficient bi‐allelic gene knockout and site‐specific knock‐in mediated by TALENs in pigs. Sci Rep. 2014;4:6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhou X, Xin J, Fan N, et al. Generation of CRISPR/Cas9‐mediated gene‐targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci. 2015;72:1175‐1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang X, Cao C, Huang J, et al. One‐step generation of triple gene‐targeted pigs using CRISPR/Cas9 system. Sci Rep. 2016;6:20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhu X‐X, Zhong Y‐Z, Ge Y‐W, Lu K‐H, Lu S‐S. CRISPR/Cas9‐mediated generation of guangxi bama minipigs harboring three mutations in α‐synuclein causing Parkinson’s disease. Sci Rep. 2018;8:12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang D, Wang C‐E, Zhao B, et al. Expression of Huntington’s disease protein results in apoptotic neurons in the brains of cloned transgenic pigs. Hum Mol Genet. 2010;19:3983‐3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yan S, Tu Z, Liu Z, et al. A Huntingtin knockin pig model recapitulates features of selective neurodegeneration in Huntington’s disease. Cell. 2018;173:989‐1002.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dinwiddie R. Pathogenesis of lung disease in cystic fibrosis. Respiration. 2000;67:3‐8. [DOI] [PubMed] [Google Scholar]
- 64. Rogers CS, Abraham WM, Brogden KA, et al. The porcine lung as a potential model for cystic fibrosis. Am J Physiol Lung Cell Mol Physiol. 2008;295:L240‐L263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rogers CS, Hao Y, Rokhlina T, et al. Production of CFTR‐null and CFTR‐ΔF508 heterozygous pigs by adeno‐associated virus–mediated gene targeting and somatic cell nuclear transfer. J Clin Invest. 2008;118:1571‐1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Uc A, Giriyappa R, Meyerholz DK, et al. Pancreatic and biliary secretion are both altered in cystic fibrosis pigs. Am J Physiol Gastrointest Liver Physiol. 2012;303:G961‐G968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ostedgaard LS, Meyerholz DK, Chen J‐H, et al. The ΔF508 mutation causes CFTR misprocessing and cystic fibrosis‐like disease in pigs. Sci Transl Med. 2011;3:74ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stoltz DA, Rokhlina T, Ernst SE, et al. Intestinal CFTR expression alleviates meconium ileus in cystic fibrosis pigs. J Clin Invest. 2013;123:2685‐2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li X, Tang XX, Vargas Buonfiglio LG, et al. Electrolyte transport properties in distal small airways from cystic fibrosis pigs with implications for host defense. Am J Physiol Lung Cell Mol Physiol. 2016;310(7):L670‐L679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dalkilic I, Kunkel LM. Muscular dystrophies: genes to pathogenesis. Curr Opin Genet Dev. 2003;13:231‐238. [DOI] [PubMed] [Google Scholar]
- 71. Bushby K, Norwood F, Straub V. The limb‐girdle muscular dystrophies—diagnostic strategies. Biochim Biophys Acta. 2007;1772:238‐242. [DOI] [PubMed] [Google Scholar]
- 72. Aartsma‐Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading‐frame rule. Muscle Nerve. 2006;34:135‐144. [DOI] [PubMed] [Google Scholar]
- 73. Klymiuk N, Blutke A, Graf A, et al. Dystrophin‐deficient pigs provide new insights into the hierarchy of physiological derangements of dystrophic muscle. Hum Mol Genet. 2013;22:4368‐4382. [DOI] [PubMed] [Google Scholar]
- 74. Yu H‐H, Zhao H, Qing Y‐B, et al. Porcine zygote injection with cas9/sgrna results in DMD‐modified pig with muscle dystrophy. Int J Mol Sci. 2016;17:1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Moretti A, Fonteyne L, Giesert F, et al. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat Med. 2020;26:207‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chiappalupi S, Salvadori L, Luca G, et al. Do porcine Sertoli cells represent an opportunity for Duchenne muscular dystrophy? Cell Prolif. 2019;52:e12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Luo Y, Li J, Liu Y, et al. High efficiency of BRCA1 knockout using rAAV‐mediated gene targeting: developing a pig model for breast cancer. Transgenic Res. 2011;20:975‐988. [DOI] [PubMed] [Google Scholar]
- 78. Flisikowska T, Merkl C, Landmann M, et al. A porcine model of familial adenomatous polyposis. Gastroenterology. 2012;143:1173‐1175.e7. [DOI] [PubMed] [Google Scholar]
- 79. Kang JT, Ryu J, Cho B, et al. Generation of RUNX3 knockout pigs using CRISPR/Cas9‐mediated gene targeting. Reprod Domest Anim. 2016;51:970‐978. [DOI] [PubMed] [Google Scholar]
- 80. Saalfrank A, Janssen KP, Ravon M, et al. A porcine model of osteosarcoma. Oncogenesis. 2016;5:e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sieren JC, Meyerholz DK, Wang X‐J, et al. Development and translational imaging of a TP53 porcine tumorigenesis model. J Clin Invest. 2014;124:4052‐4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Schook LB, Collares TV, Hu W, et al. A genetic porcine model of cancer. PLoS ONE. 2015;10:e0128864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang K, Jin Q, Ruan D, et al. Cre‐dependent Cas9‐expressing pigs enable efficient in vivo genome editing. Genome Res. 2017;27:2061‐2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hai T, Teng F, Guo R, Li W, Zhou Q. One‐step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24:372‐375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chen J, An B, Yu B, et al. CRISPR/Cas9‐mediated knockin of human factor IX into swine factor IX locus effectively alleviates bleeding in hemophilia B pigs. Haematologica. 2021;106:829‐837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dorado B, Pløen GG, Barettino A, et al. Generation and characterization of a novel knockin minipig model of Hutchinson‐Gilford progeria syndrome. Cell Discov. 2019;5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang B, Wang C, Zhang Y, et al. A CRISPR‐engineered swine model of COL2A1 deficiency recapitulates altered early skeletal developmental defects in humans. Bone. 2020;137:115450. [DOI] [PubMed] [Google Scholar]
- 88. Wang X, Zhou J, Cao C, et al. Efficient CRISPR/Cas9‐mediated biallelic gene disruption and site‐specific knockin after rapid selection of highly active sgRNAs in pigs. Sci Rep. 2015;5:13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yao J, Wang Y, Cao C, et al. CRISPR/Cas9‐mediated correction of MITF homozygous point mutation in a Waardenburg syndrome 2A pig model. Mol Ther Nucleic Acids. 2021;24:986‐999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Li Z, Duan X, An X, et al. Efficient RNA‐guided base editing for disease modeling in pigs. Cell Discov. 2018;4:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Petters RM, Alexander CA, Wells KD, et al. Genetically engineered large animal model for studying cone photoreceptor survival and degeneration in retinitis pigmentosa. Nat Biotechnol. 1997;15:965‐970. [DOI] [PubMed] [Google Scholar]
- 92. Kraft TW, Allen D, Petters RM, Hao Y, Peng YW, Wong F. Altered light responses of single rod photoreceptors in transgenic pigs expressing P347L or P347S rhodopsin. Mol Vis. 2005;11:1246‐1256. [PubMed] [Google Scholar]
- 93. Ross JW, Fernandez de Castro JP, Zhao J, et al. Generation of an inbred miniature pig model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2012;53:501‐507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sommer JR, Estrada JL, Collins EB, et al. Production of ELOVL4 transgenic pigs: a large animal model for Stargardt‐like macular degeneration. Br J Ophthalmol. 2011;95:1749‐1754. [DOI] [PubMed] [Google Scholar]
- 95. Hickey RD, Mao SA, Glorioso J, et al. Fumarylacetoacetate hydrolase deficient pigs are a novel large animal model of metabolic liver disease. Stem Cell Res. 2014;13:144‐153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Koppes EA, Redel BK, Johnson MA, et al. A porcine model of phenylketonuria generated by CRISPR/Cas9 genome editing. JCI Insight. 2020;5:e141523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lai L, Kolber‐Simonds D, Park KW, et al. Production of alpha‐1,3‐galactosyltransferase knockout pigs by nuclear transfer cloning. Science. 2002;295:1089‐1092. [DOI] [PubMed] [Google Scholar]
- 98. Phelps CJ, Koike C, Vaught TD, et al. Production of alpha 1,3‐galactosyltransferase‐deficient pigs. Science. 2003;299:411‐414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Petersen B, Frenzel A, Lucas‐Hahn A, et al. Efficient production of biallelic GGTA1 knockout pigs by cytoplasmic microinjection of CRISPR/Cas9 into zygotes. Xenotransplantation. 2016;23:338‐346. [DOI] [PubMed] [Google Scholar]
- 100. Chuang CK, Chen CH, Huang CL, et al. Generation of GGTA1 mutant pigs by direct pronuclear microinjection of CRISPR/Cas9 plasmid vectors. Anim Biotechnol. 2017;28:174‐181. [DOI] [PubMed] [Google Scholar]
- 101. Tanihara F, Hirata M, Nguyen NT, et al. Efficient generation of GGTA1‐deficient pigs by electroporation of the CRISPR/Cas9 system into in vitro‐fertilized zygotes. BMC Biotechnol. 2020;20:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kwon D‐N, Lee K, Kang M‐J, et al. Production of biallelic CMP‐Neu5Ac hydroxylase knock‐out pigs. Sci Rep. 2013;3:1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tu C‐F, Chuang C‐K, Hsiao K‐H, et al. Lessening of porcine epidemic diarrhoea virus susceptibility in piglets after editing of the CMP‐N‐glycolylneuraminic acid hydroxylase gene with CRISPR/Cas9 to nullify N‐glycolylneuraminic acid expression. PLoS ONE. 2019;14:e0217236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Li P, Estrada JL, Burlak C, et al. Efficient generation of genetically distinct pigs in a single pregnancy using multiplexed single‐guide RNA and carbohydrate selection. Xenotransplantation. 2015;22:20‐31. [DOI] [PubMed] [Google Scholar]
- 105. Fischer K, Kraner‐Scheiber S, Petersen B, et al. Efficient production of multi‐modified pigs for xenotransplantation by 'combineering', gene stacking and gene editing. Sci Rep. 2016;6:29081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gao H, Zhao C, Xiang X, et al. Production of α1,3‐galactosyltransferase and cytidine monophosphate‐N‐acetylneuraminic acid hydroxylase gene double‐deficient pigs by CRISPR/Cas9 and handmade cloning. J Reprod Dev. 2017;63:17‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhang R, Wang Y, Chen L, et al. Reducing immunoreactivity of porcine bioprosthetic heart valves by genetically‐deleting three major glycan antigens, GGTA1/β4GalNT2/CMAH. Acta Biomater. 2018;72:196‐205. [DOI] [PubMed] [Google Scholar]
- 108. Wang Y, Du Y, Zhou X, et al. Efficient generation of B2m‐null pigs via injection of zygote with TALENs. Sci Rep. 2016;6:38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sake HJ, Frenzel A, Lucas‐Hahn A, et al. Possible detrimental effects of beta‐2‐microglobulin knockout in pigs. Xenotransplantation. 2019;26:e12525. [DOI] [PubMed] [Google Scholar]
- 110. Reyes LM, Estrada JL, Wang ZY, et al. Creating class I MHC‐null pigs using guide RNA and the Cas9 endonuclease. J Immunol. 2014;193:5751‐5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Joanna Z, Magdalena H, Agnieszka N‐T, et al. The production of UL16‐binding protein 1 targeted pigs using CRISPR technology. 3 Biotech. 2018;8:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kang J‐T, Cho B, Ryu J, et al. Biallelic modification of IL2RG leads to severe combined immunodeficiency in pigs. Reprod Biol Endocrinol. 2016;14:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lei S, Ryu J, Wen K, et al. Increased and prolonged human norovirus infection in RAG2/IL2RG deficient gnotobiotic pigs with severe combined immunodeficiency. Sci Rep. 2016;6:25222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ren J, Yu D, Fu R, et al. IL2RG‐deficient minipigs generated via CRISPR/Cas9 technology support the growth of human melanoma‐derived tumours. Cell Proliferat. 2020;53:e12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Niu D, Wei H‐J, Lin L, et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR‐Cas9. Science. 2017;357:1303‐1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Huang J, Guo X, Fan N, et al. RAG1/2 knockout pigs with severe combined immunodeficiency. J Immunol. 2014;193:1496‐1503. [DOI] [PubMed] [Google Scholar]
- 117. Lee K, Kwon D‐N, Ezashi T, et al. Engraftment of human iPS cells and allogeneic porcine cells into pigs with inactivated RAG2 and accompanying severe combined immunodeficiency. Proc Natl Acad Sci U S A. 2014;111:7260‐7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Mohiuddin MM, Singh AK, Corcoran PC, et al. Chimeric 2C10R4 anti‐CD40 antibody therapy is critical for long‐term survival of GTKO.hCD46.hTBM pig‐to‐primate cardiac xenograft. Nat Commun. 2016;7:11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Längin M, Mayr T, Reichart B, et al. Consistent success in life‐supporting porcine cardiac xenotransplantation. Nature. 2018;564:430‐433. [DOI] [PubMed] [Google Scholar]
- 120. Kim SC, Mathews DV, Breeden CP, et al. Long‐term survival of pig‐to‐rhesus macaque renal xenografts is dependent on CD4 T cell depletion. Am J Transplant. 2019;19:2174‐2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Watanabe H, Ariyoshi Y, Pomposelli T, et al. Intra‐bone bone marrow transplantation from hCD47 transgenic pigs to baboons prolongs chimerism to >60 days and promotes increased porcine lung transplant survival. Xenotransplantation. 2020;27:e12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Shah JA, Patel MS, Elias N, et al. Prolonged survival following pig‐to‐primate liver xenotransplantation utilizing exogenous coagulation factors and costimulation blockade. Am J Transplant. 2017;17:2178‐2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Porrett PM, Orandi BJ, Kumar V, et al. First clinical‐grade porcine kidney xenotransplant using a human decedent model. Am J Transplant. 2022. doi: 10.1111/ajt.16930 [DOI] [PubMed] [Google Scholar]
- 124. Du X, Guo Z, Fan W, et al. Establishment of a humanized swine model for COVID‐19. Cell Discov. 2021;7:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Kells NJ. Review: The Five Domains model and promoting positive welfare in pigs. Animal. 2021;100378. [DOI] [PubMed] [Google Scholar]


