Abstract
Colocynth has a long history of use in traditional medicine for treatment of various inflammatory diseases where it is commonly roasted before being applied for medical purposes to reduce its toxicity. This study aims at tracking the effect of heat processing on the metabolic profile of the peels, pulps and seeds of colocynth fruit using UPLC-QqQ-MS-based metabolomics. The analysis resulted in tentative identification of 72 compounds belonging to different chemical classes. With roasting, a decline was observed in the relative amounts of chemical constituents where 42, 25 and 29 compounds were down-regulated in the peels, pulps and seeds, respectively. EC100 values resulting in 100% cell viability were all higher in roasted samples compared to their relevant raw ones. Correlation analysis indicated that the main cytotoxic chemical markers were cucurbitacin glycosides and their genins. Further, ex vivo anti-inflammatory activity testing multivariate models revealed that unprocessed samples correlated with inhibition of TNF-α, IL-1β and IFN-γ where quercetrin, calodendroside A, and hexanoic acid methyl ester were the most significant chemical markers, while processed samples showed correlation with IL-6 pro-inflammatory marker inhibition with protocatechuic and protocatechuic acid glycoside being the main correlated chemical markers.
Colocynth has a long history of use in traditional medicine for treatment of various inflammatory diseases where it is commonly roasted before being applied for medical purposes to reduce its toxicity.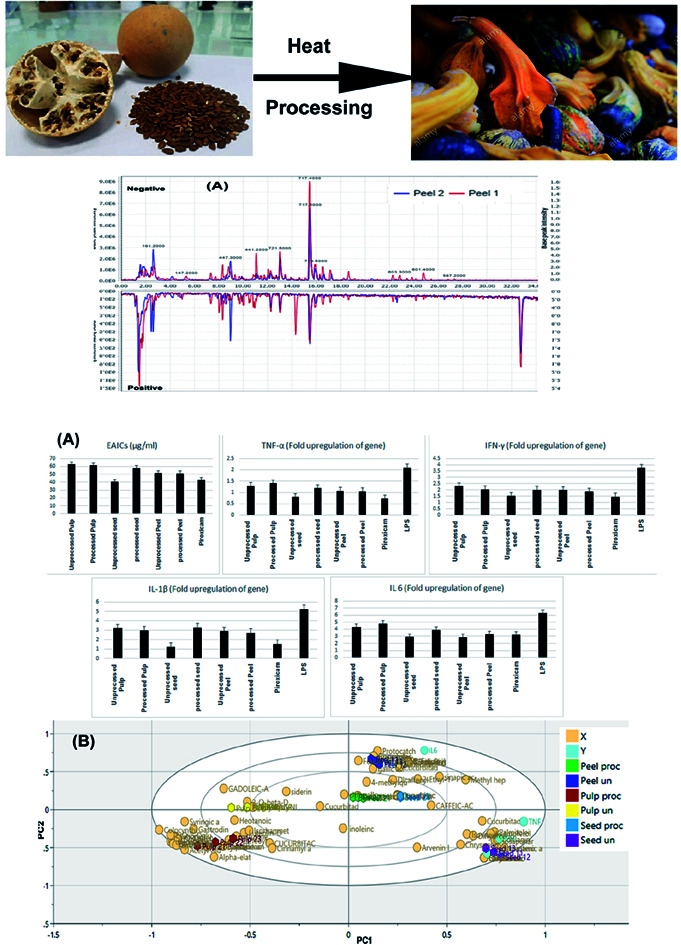
1. Introduction
The Cucurbitaceae family is considered as one of the families whose plant members such as colocynth (bitter apple), gourd, cucumber, watermelon, and pumpkin are used as food or fodder.1 Colocynth (Citrullus colocynthis L.) Schrad is an annual valuable plant widely distributed in the desert areas of the world since it possesses great resistance to drought and desert extreme conditions.2 It can be found in many Asian countries including India, Pakistan, Kuwait, Saudi Arabia, Jordan, Iran, Iraq, Turkey, Afghanistan, Yemen and Sri Lanka,2 as well as African tropical countries (e.g. Ethiopia, Somalia and Chad) and some countries of the Mediterranean region, such as Morocco, Egypt, Libya, Algeria and Tunisia.2
Colocynth has a long history of use as an anti-inflammatory drug where in traditional Iranian medicine, rubbing the colocynth pulp on a painful knee relieves its pain and in cases of sciatica, gout, backache, and paralysis. The oil of colocynth is used externally for ear pains, tinnitus, toothache, and hair loss.3 In Iraq, colocynth is used for the treatment of breast inflammation and joints pain.4 In Tunisia, it is widely used in folk medicine for the treatment of several inflammatory diseases, including rheumatism and rheumatoid arthritis.5
The medicinally used parts of colocynth include the peels pulp, seeds, leaves and roots.3 Previous report on the phytoconstituents of colocynth indicated that it is a rich source of as tannins, saponins, phenolic acids, flavonoids, terpenoids, alkaloids, steroids and cucurbitacins.6
Cucurbitacins are steroidal tetracyclic terpenes identified from plants in the Cucurbitaceae family and are considered part of the plants' natural defense mechanisms against animals and microorganisms. Owing to its high content of cucurbitacins, C. colocynthis is listed amongst the top 10 list of toxic plants.
In several Northern African and Arabian Peninsula, colocynth is usually roasted before being applied for medical purposes. This is claimed to reduce the toxicity of the fruits and seeds. Roasting is carried out on direct flames or in special ovens known as Clayton Ring the enigmatic early Egyptian portable ceramic ovens found in the Western Desert.7,8 Grilled or roasted colocynth is traditionally used in Iraq in order to treat heel spur, through putting the hot grilled colocynth on the painful area of the heel.9 Therefore, it is deemed necessary to track the changes imposed by processing on the chemical profile of the drug to reveal the changes that occur during the roasting process that is presumed to reduce colocynth toxicity.
In view of the above mentioned points, the study in hand aims at evaluating the impact of heat processing on the metabolic profile of different parts of colocynth fruit including the peel, pulp and seeds using UPLC/MS/MS as well as determination of the main chemical markers responsible for scrutiny of processed and unprocessed samples. Further, comparative evaluation of the samples ex vivo anti-inflammatory activity of different on LPS-stimulated white blood cells was attempted followed by determination of the main chemical markers correlated to bioactive discrimination of processed and unprocessed samples through implementation of different multivariate statistical analysis models.
2. Experimental
2.1. Plant collection
Colocynth samples (4500 g) (Fig. S1†) were purchased from the local market in Anbar Governorate, Iraq in November 2020, the sample was authenticated by Professor Sania Ahmad, Faculty of Science, Alexandria University via macroscopical and microscopical examination of the tested samples. A voucher specimen (CC 2020) was kept at the Department of Pharmacognosy-Faculty of Pharmacy-Alexandria University.
2.2. Samples preparation
The purchased colocynth sample was divided into two groups each of 2250 g, the first group was separated into five seeds, five pulp and five peel samples (a total of 15 unprocessed samples) each weighed 200 g. The influence of the roasting processes was evaluated by heat processing of 200 g of the pulps, seeds and peels samples in a hot air oven at 140 °C until reaching constant weight with occasional stirring according to previously reported method.10 The heat processing for each fruit part was repeated five times resulting in a total of 15 processed samples. 50 g of each sample (processed and unprocessed) were ground into powder by means of an electric grinding machine and then separately extracted using 100 ml of 70% ethanol by ultra-sonication for 30 minutes at 50 °C. The resulting extracts were then filtered and concentrated under reduced pressure to dryness. Maceration method of extraction was chosen due to its advantages, being a simple energy saving method that doesn't require a skilled operator. Further, it is a suitable method for best separation of metabolites as there are a prolonged contact with solvent.11
2.3. Chemical profiling of processed and unprocessed colocynth extracts using UPLC-MS/MS
2.3.1. Preparation of the ethanol extracts sample for UPLC-MS analysis
The dry extract was prepared at a concentration of 1 mg ml−1 using HPLC-grade methanol followed by its filtration using membrane disc filter (0.2 μm). Furthermore, sonication was used for sample degassing in order to prepare the sample before injection. The chromatographic column received a volume of 10 μl of the sample that injected in the full loop mode. Each sample was analyzed five times. Please refer to the ESI† for the details of standard solutions preparation.
2.3.2. Conditions of the UPLC experiment
Secondary metabolites present in C. colocynthis extracts were assigned using an UPLC XEVO TQD triple quadruple instrument Waters Corporation, Milford, MA01757, USA. The chromatographic system is composed of a Waters Acquity QSM pump, a LC-2040 autosampler, degasser in addition to Waters Acquity CM detector. The dimensions of Waters Acquity UPLC BEH C18 column that was used for the chromatographic separation was 50 mm (length), 2.1 mm (internal diameter) and 1.7 μm (particle size). The operation of the column was at a flow rate of 0.2 ml min−1 and the system was thermostated at 30 °C.
The mobile phase that used for analyses consisted of two phases; phase A and B, ultrapure water + 0.1% (v/v) formic acid were phase A, while methanol + 0.1% (v/v) formic acid were phase B. These mobile phase components were used in order to provide good separation and resolution for the compounds. Moreover, 0.1% formic acid is a suitable solvent regarding MS detectors, as it shows a distinct influence on the responses and ionization efficiency of analytes. Elution was gradient one and its program was as follows: 0.0–2.0 min, 10% eluent B; 2.0–5.0 min, 30% eluent B; 5.0–15.0 min, 70% eluent B; 15.0–22.0 min, 90% eluent B; 22.0–25.0 min, 90% eluent B; 26.0 min, 100% eluent B; 26.0–29.0 min, 100% eluent B; 30.0–40 min, 10% eluent B. 4 min were set at the initial conditions in order to re-equilibrate the column.
2.3.3. ESI-MS conditions and metabolites identification
The sample was analyzed in negative and positive ionization modes, the triple quadrupole (TQD) mass spectrometer was the mass analyzer and with electrospray ionization (ESI) source.
The electrospray ionization source (ESI) was set to be in the negative and positive ion modes, in order to get a comprehensive picture of the metabolite profiles of the test extracts. The mass analyzer used was triple quadrupole (QqQ); it is used for tandem MS methods as its first and third quadrupoles act as filters of masses while its second quadrupole which is a radiofrequency-only quadrupole is considered as a collision cell where parent ions are fragmented as a result of the interaction with a collision gas. QqQ is considered as a powerful tool for providing highly discriminating and many important structural data for the ions of compounds of interest.12
ESI was operated at the following conditions: 3 kV (capillary voltage), 35 V (cone voltage). Regarding temperature, 150 °C was the ion source temperature, 35 psi was the pressure of the nitrogen gas (nebulizer), the temperature of drying and sheath gas (N2) was 440 °C and 350 °C, respectively. At 900 L h−1 and 50 L h−1, the drying and sheath gas flows were applied, respectively. The total run time of the analysis run time was 30 min. In order to achieve MS spectra, full range acquisition covering 50–1000 m/z was applied. Regarding automatic MS/MS fragmentation analyses of the parent ions, the first quadrupole (Q1) was used for mass-selection of parent ions, collision-induced dissociation (CID) technique was used for the fragmentation of parents ions in the second quadrupole (Q2) using energy ramp from 30 to 70 eV using collision gas (nitrogen gas). In the negative ion mode, the improvement of the fragmentations of flavonoids and diterpene acids was done through using collision energies ranges from 20 to 40 eV. Finally, monitoring of the daughter ions which yielded from the fragmentation was carried out in the third quadrupole, these ions are related to the molecular structure of the parent ions. Furthermore, MSn experiments included the same conditions of chromatography and mass spectrometry as described above. Metabolites were assigned based on the comparison of their retention times to that of external standards. Furthermore, quasi-molecular ions and characteristic MS/MS fragmentation patterns were used for metabolite assignment in comparison to our in-house database, data published in literature in addition to phytochemical dictionary of natural products database (CRC) in order to get a high confidence level of metabolite annotation.
2.4. Assessment of the cytotoxicity and anti-inflammatory activity of processed and raw samples
Assessment of cytotoxicity of the different extracts compared to piroxicam was carried out using MTT assay. The effective anti-inflammatory concentrations (EAICs) of each extract in lipopolysaccharides (LPS)-stimulated human WBC's culture were determined. Determination of IL-1β, IL 6, TNF-α and INF-γ expression level by real time polymerase chain reaction (PCR) was implemented. Results were expressed as means ± standard deviations of three individual replicates. Details of the procedures can be found out in ESI section.†
2.5. Statistical analysis
One-way analysis of the variance (ANOVA) was done using the software SPSS 26.0 (SPSS Inc., Chicago, IL. USA) for semi-quantitative analysis and biological activity testing. SIMCA 14 software (Umetrics, Malmo, Sweden) was utilized for metabolomics multivariate statistical data analysis. Metaboanalyst 4.0 (http://www.metaboanalyst.ca/),14 a web-based metabolomics data processing tool, was also employed for MS data analysis to construct hierarchical cluster analysis heat maps, volcano plots, coefficient plots and unsupervised self-organizing maps (SOM).
3. Results and discussion
3.1. Compounds identification in the different tested extracts
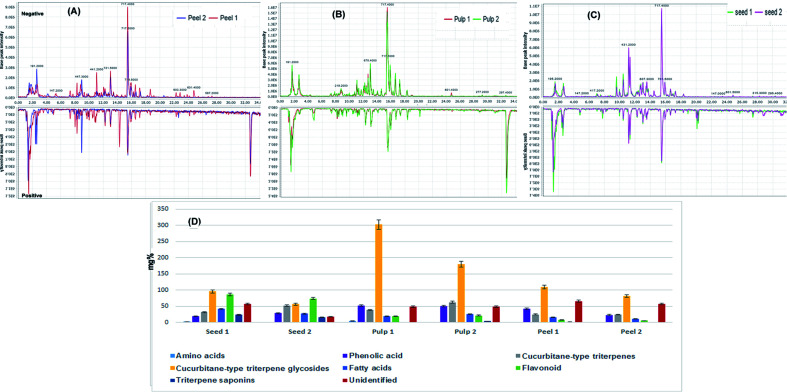
Base peak chromatograms (Fig. 1A–C) of the processed and unprocessed colocynth extracts in positive and negative ionization modes totally revealed the identification of 72 compounds that encountered several chemical classes such as: phenolic acids, amino acids, fatty acids, aliphatic acids, quinic acid derivatives, flavonoids, iridoids, coumarins, monoterpenes, stigmastanes, triterpenes mainly of cucurbitacin-type in addition to miscellaneous compounds.
Fig. 1. Base peak chromatograms collected in the negative and positive modes for (A) unprocessed peel (peel 1) and processed peels (peel 2), (B) unprocessed pulp (pulp 1) and processed pulp (pulp 2), (C) unprocessed seeds (seed 1) and processed seeds (seed 2). (D) Relative quantitation of the total content of different chemical classes identified in the processed and unprocessed samples of colocynth expressed as mg equivalents (eq.) per 100 g dry weight.
3.1.1. Phenolic acids
Nine peaks were identified as phenolic acids, among which peaks 5 and 21 were phenolic acid glycosides. Regarding the former it was identified as salicylic acid-O-glucoside, while the latter was tentatively identified protocatechuic acid-O-glucoside. Both compounds showed a characteristic peak (M − H-162) that result from glucose unit loss in addition to a peak that was corresponding to extra neutral loss of CO2 (M − H-44).15 Peak 6 showed characteristic MS2 fragments at 105 and 131 Da that result from the loss of CO2 and water, respectively. By referring to literature peak 6 was identified as cinnamic acid.16 Peaks 7 and 14 were found to be methoxylated phenolic acids as both peaks showed daughter peaks due to loss of CO2 (M − H-44) followed by additional loss of a methyl group (M–H–CO2–CH3), thus both compounds were tentatively identified as sinapic acid and ferulic acid, respectively.17,18 Moreover, peaks 34 was a methoxylated phenolic acid as it showed characteristic MS2 fragments at 182 Da (M–H–CH3), 167 (M–H–2CH3) and 153 Da (M–H–CO2) and this compound was identified as syringic acid.19 Peak 21 showed characteristic fragments at 163, 145 and 135 Da that are corresponding to loss of water and formic acid. Based on that information this compound was found to be caffeic acid.20 Peaks 8 and 16 showed pseudo-molecular ion peaks at 170 Da (M + H) and 154 (M − H), respectively. By referring to literature they were tentatively identified as gallic acid and protocatechuic acid.17,21
3.1.2. Quinic acid derivatives
This class included six peaks, peak 19, 20, 25, 44, 47 and 64. Quinic acid as a free acid was represented by peak 13 with a pseudo-molecular ion peak (M − H) at m/z value of 191 along with a daughter peak that result from water loss at 173 Da.22 Peak 25 was tentatively identified as feruloylquinic acid isomer which is considered as an ester of quinic acid and ferulic acid (hydroxy-methoxycinnamic acid). Such identification based on the presence of the characteristic daughter peaks at 191 and 173 Da which corresponds to quinic acid moiety and at 193 Da that corresponds to ferulic acid moiety.22 The ester of caffeic acid and quinic acid (chlorogenic acid) was represented by peak 44 that showed a quasi-molecular ion peak (M − H) at 353 Da along with MS2 fragments at 191 and 173 Da that belongs to quinic acid moiety and at 179 and 135 Da from caffeic acid moiety.22 Peak 64 was tentatively identified as digalloylquinic acid isomer based on its molecular ion peak (M − H) at 495 Da in addition to its MS2 fragments at 343 Da that indicated the loss of one gallic acid moiety in addition to quinic acid moiety fragment at 191 Da and the remaining gallic acid moiety fragment at 169 Da.23,24 Furthermore, peaks 20 and 47 were esters of shikimic acid (dehydrated quinic acid) with one caffeic acid moiety and two caffeic acid moieties, respectively. Both compounds showed characteristic daughter peaks at 179, 161 and 135 Da that belongs to caffeic acid moiety. Moreover, peak 47 showed a MS2 fragment at 335 Da that indicated the loss of one caffeoyl moiety. Base on this information, peaks 19 and 47 were identified as caffeoylshikimic acid and dicaffeoylshikimic acid isomers.25,26
3.1.3. Flavonoids
Nine peaks represented this class, among which 6 peaks (peaks 18, 32, 35, 36, 42 and 45) were identified as O-glycosides, 2 peaks (peaks 57 and 58) were C-glycosides, one peak (peak 30) represented flavonoid C,O-di-glycoside in addition to one flavan-3-ol (peak 70).
In flavonoid-O-glycosides, the sugar part size and its structure can be identified in MS2 experiments as in the second compartment of the triple quadrupole analyzer collision induced dissociation result in the cleavage of the O–C glycosidic linkage resulting in formation of characteristic MS2 fragments; (M − H-162), (M − H-146) and (M − H-132) that are corresponding to the loss of O-hexose, O-deoxyhexose or O-pentose sugar units, respectively.27 Peaks 42 was claimed to be a rhamnoside, this was deduced from the presence of a characteristic daughter ion peak at (M − H-146) that results from loss of rhamnose. Moreover, its characteristic MS2 fragments at 147 Da (B1.3)− and 151 Da (A1.3)− that result from retro-Diels-Alder rearrangement (RDA) revealed the aglycone part to be quercetin and peak 45 was tentatively identified as quercetin-O-rhamnoside (quercitrin).28 Peaks 36 and 42 were hexosides as both of them showed (M − H-162) ion peak that indicated the loss of one hexose unit. Based on MS2 data and by referring to literature they were tentatively identified as chrysoeriol-O-hexoside29 and luteolin-O-hexoside,30 respectively. Peaks 18, 30 and 32 were identified as diglycosides. The sugar part of peaks 32 and 35 was rhamnose and hexose as they showed a characteristic daughter fragment at (M − H-308) that indicated the loss of rutinose unit and proved that the rhamnose and glucose are united together in the same position of substitution in the aglycone part.13 Based on this information, they were identified as isorhamnetin rutinoside and kaempferol rutinoside, respectively.29,31 Peaks 18 was identified as flavanone di-glucosides. Based on the fragmentation data and by searching literature both compounds were tentatively identified as calodendroside A.32
Peak 30 was tentatively identified as isosaponarin (isovitexin 4′-O-glucoside), a flavonoid C,O-di-glycosides which represented this class. It showed (M − H) ion peak at 593 Da and characteristic daughter peaks at 503 Da (M − H-90) and 473 Da (M − H-120) that result from the cross cleavage of glucose unit. Moreover, daughter ions at 341 Da (M − H-90-162), and 311 Da (M − H-120-162) due to loss of glucose moiety. These findings indicated the presence of C-glucosyl and O-glucosyl units (Li et al.,33 2017).
The fragmentation of C-glycosides mainly results from the retro-Diels–Alder rearrangement in addition to cross cleavage of the sugar units. Peaks 57 and 58 showed (M − H) ion peaks at 432 Da and 464 Da, respectively. Peak 57 was tentatively identified as isovitexin (apigenin-6-C-glucoside), as it showed characteristic ion fragments that result from the crossing cleavage of glucose unit at 341 Da and 311 Da m/z. Moreover, the loss of CO from the m/z fragment ion (311 Da) and the loss of H2O from the m/z fragment ion (341 Da) result in the formation of two daughter fragments at 283 Da and 323 Da, respectively.34 Based on the fragmentation data and by referring to literature, peak 58 was identified as isoorientin 3′-O-methyl ether.34,35
Peak 70 was tentatively identified as gallocatechin36 that belongs to flavan-3-ol class, its major daughter ion peaks were due to loss of water, CO2 and gallic acid. Furthermore, fragment ions at 169 Da result from the intact gallic acid anion, while that at 125 Da indicated the intact A-ring of the catechin structure.
3.1.4. Cucurbitacin and fried-oleanane type triterpenes
Twenty-one peaks represented this class, among which 13 peaks (38–40, 43, 46, 49–51, 54, 59, 60, 63, and 65) were glycosylated triterpenes as they showed the characteristic a fragment ion (M − H-162) that indicated from loss of the one glucose unit.37
The triterpene nucleus of cucurbitacins has a general fragmentation pattern in which the side-chain plays a key role in their mass spectrum fragmentation. The most prevalent ion peak at 96 Da, that corresponds to the composition C6H8O, which caused by side-chain cleavage, as validated by high resolution mass measurements.38 The production of this ion is most likely due to a side-chain rupture and concomitant hydrogen migration from the hydroxyl at C-20 as shown in peaks 39, 43, 50, 51, 52, 54, 59, 68 and 69. In this series of compounds, the occurrence of this peak in the mass spectrum is effective in detecting this side-chain.
Another distinctive fragment was at 403 Da that results from a straightforward cleavage of the cucurbitacins' C-20–C-22 bond without hydrogen transfer and with the positive charge on C-20 retained, allowing us to attribute hydroxyl, ketonic, and olefinic functional groups to C-20, C-22, and C-23, respectively as an exemplified in peaks numbered as 48, 49, 51, 54, 59 and 66. The presence of a hydroxyl group at C-20 was confirmed by removing one water molecule from the 403 Da ion peak, resulting in a strong signal at 385 Da (M − H-18)39 as in case of compounds 31, 49, 51, 54, 55, 59 and 66.
The breakage of the C-17–C-20 link produced another peak at 356 Da (C22H28O4), demonstrating the presence of two oxygen functions in rings C/D38,40 as shown in compounds 40, 43, 48, 54, 56 and 59.
Another aspect to be considered is the existence or absence of a double bond between C-1 and C-2, where in the lack of one in compounds 43, 51, 52, 54 and 56, a peak corresponds to the breakage of the C-7–C-8 bond, which is preceded by a McLafferty type41 of rearrangement involving the 11-keto group and a hydrogen at C-1 has been appeared.
On the other hand, cleavage of the McLafferty type involving the 11-keto group is not conceivable when a double bond exists between C-1 and C-2 in ring A as in case of compounds 39, 48, 49, 50, 59, 66 and 69. In this case, a retro-Diels–Alder rupture of ring B is truly noteworthy, yielding a particularly conjugated ion fragment that emerges as an intense peak at 164 Da.37,38,40
Furthermore, cucurbitacins A, B, and C nuclei also had an extra peak that formed at (M − H-60) when the acetic acid components were removed from the molecule. In addition, in cucurbitacins A and C, the presence of a –CH2OH group at C-9 results in the ion fragment (M − H-60-30). The transfer of the hydroxyl hydrogen on the carbonyl oxygen at C −11 causes CH2O to be ejected from the primary alcohol function, resulting in a 30 Da loss.39
Based on MS, MS/MS data in addition to literature comparison,40,42–44 the peaks 38–40, 43, 46, 49–51, 54, 59, 60, 63, and 65 were tentatively identified as glycosylated triterpenes and defined as colocynthosides A, cucurbitacin E-2-O-glucopyranoside, cucurbitacin L-2-O-glucoside, cucurbitacin A-2-O-glucopyranoside, colocynthosides B, colocynthin B, cucurbitacin I-glucoside, datiscoside, arvenin I,2-O-glucopyranosyl-16-20-dihydroxycucurbita-1,5,23,25(26)-tetraen-3,11,22-trione, 6′-acetyl-2-O-glucopyranosyl cucrbitacin E, 22-deoxocucurbitoside B and colocynthin A, respectively.
Moreover, the peaks numbered as 31, 48, 52, 55, 56, 61, 68 and 69 were tentatively annotated as cucurbitacin F, cucurbitacin L, cucurbitacin D, cucurbitacin S, dihydrocucurbitacin C, dihydrocucurbitacin E, cucurbitacin B and cucurbitacin E, respectively.
On the other hand, fried-oleanane triterpenes class was represented by only one peak (peak 37), it showed (M − H) ion peak at 455 Da along with its characteristic mass fragments that results from at 437 Da, 411 Da, 441 Da, 409 Da that result from loss of water, CO2, CH3 and formic acid, respectively.45
3.2. Chemical profiling of the peels, pulps and seeds of Citrullus colocynthis fruit using UPLC/MS/MS analysis
A total of 124 metabolites were detected in the extracts of peels, pulps and seeds of Citrullus colocynthis fruit with the tentative identification of 72 metabolites belonging to different chemical classes including amino acids, triterpene saponins, cucurbitane-type triterpene saponins, cucurbitane-type triterpene glycosides, fatty acids, phenolic acids and flavonoids (Table 1). Compounds were identified through comparison of their retention time, quasi-molecular ions as well as their MS/MS fragment ions with those reported in literature besides databases including Dictionary of natural products and MassBank.
Metabolites identified in processed and processed pulps, peels and seeds samples of colocynth fruit using UPLC-MS in positive and negative ionization modes.
| # | Compound name | Retention time | Ion type | Class | m/z value | Molecular weight | Molecular formula | MSn fragments |
|---|---|---|---|---|---|---|---|---|
| 1 | Asparagine | 1.20 | M + H | Amino acid | 132.1 | 132.12 | C4H8N2O3 | 116, 98, 87.07 |
| 2 | Heptanoic-acid | 1.38 | M − H | Saturated fatty acid | 129.1 | 130.1 | C7H14O2 | 111, 85, 59 |
| 3 | Gastrodin | 1.48 | M − H | Alcohol | 285.3 | 286.2 | C13H18O7 | 123, 105 |
| 4 | Citral | 1.54 | M − H | Monoterpenoid | 151.1 | 152.24 | C10H16O | 83, 93 |
| 5 | Salicylic acid-O-glucoside | 8.16 | M − H | Phenolic acid glycosides | 299.2 | 300.2 | C13H16O8 | 137, 93 |
| 6 | Cinnamic acid | 1.62 | M + H | Phenolic acid | 149.3 | 148.1 | C9H8O2 | 105, 131 |
| 7 | Sinapic acid | 1.64 | M − H | Phenolic acid | 223.5 | 224.2 | C11H12O5 | 208, 179, 164.2 |
| 8 | Gallic acid | 8.21 | M + H | Phenolic acid | 171.2 | 170.12 | C7H6O5 | 125, 107 |
| 9 | Cinnamyl alcohol | 1.81 | M − H | Alcohol | 133.1 | 134.17 | C9H10O | 115 |
| 10 | Histidine | 2.37 | M + H | Amino acid | 156.2 | 155.15 | C6H9N3O2 | 110, 93 |
| 11 | Citronellol | 2.61 | M + H | Monoterpenoid | 157.1 | 156.3 | C10H20N3O | 137, 122, 68 |
| 12 | Methyl-heptanone | 2.69 | M + H | Aliphatic ketone | 129.3 | 128.21 | C8H16O | 86, 73 |
| 13 | Quinic acid | 2.72 | M − H | Quinic acid derivatives | 191.4 | 192.17 | C7H12O6 | 173 |
| 14 | Ferulic acid | 2.81 | M − H | Phenolic acid | 193.2 | 194.1 | C10H10O4 | 149, 178 |
| 15 | Protocatechuic acid-O-glucoside | 8.32 | M − H | Phenolic acid glycoside | 315.4 | 316.2 | C13H16O9 | 153, 109 |
| 16 | Protocatechuic acid | 9.69 | M − H | Phenolic acid | 153.1 | 154.12 | C7H6O4 | 109 |
| 17 | Methylquinoline | 4.85 | M + H | Quinolines | 144.3 | 143.18 | C10H9N | 116, 129, 143 |
| 18 | Calodendroside A | 7.51 | M − H | Flavanone-O-glycoside | 627.3 | 628.5 | C27H32O17 | 465, 303, 537, 507, 287, 197 |
| 19 | Caffeoyl shikimic acid isomer | 7.74 | M − H | Quinic acid derivatives | 335.2 | 336.3 | C16H16O8 | 179, 161, 135 |
| 20 | Caprylic acid | 1.59 | M − H | Saturated fatty acids | 143.2 | 144.21 | C8H16O2 | 125, 99, 59 |
| 21 | Caffeic-acid | 1.67 | M + H | Phenolic acid | 181.3 | 180.16 | C9H8O4 | 163, 145, 135 |
| 22 | Gadoleic-acid | 4.31 | M − H | Unsaturated fatty acids | 309.1 | 310.5 | C20H38O2 | 291, 265, 54 |
| 23 | Siderin | 8.79 | M − H | Coumarin | 219.1 | 220.22 | C12H12O4 | 204, 191, 189 |
| 24 | Feruloylquinic acid | 9.01 | M − H | Quinic acid derivatives | 267.2 | 368.3 | C17H20O9 | 193, 191, 173, 149, |
| 25 | 2-Ethyl-1-hexanol | 9.33 | M –H | Alcohol | 129.1 | 130.2 | C8H18O | 111, 114, 99 |
| 26 | Dimethyl azelate | 9.55 | M − H | Aliphatic acid ester | 215.2 | 216.27 | C11H20O4 | 183, 151 |
| 27 | Caproic acid | 3.06 | M − H | Saturated fatty acids | 115.2 | 116.15 | C6H12O2 | 97, 71, 59 |
| 28 | Alpha-spinasterol | 9.81 | M + H | Stigmastane-type sterol | 413.4 | 412.7 | C29H48O | 105, 310, 395 |
| 29 | Heptadecanoic acid (margaric acid) | 9.91 | M − H | Saturated fatty acid | 269.3 | 270.45 | C17H34O2 | 251, 225, 59 |
| 30 | Isosaponarin | 9.92 | M − H | Flavone glycoside (C,O-diglycoside) | 593.2 | 594.5 | C27H30O15 | 503, 473, 341, 311, |
| 31 | Cucurbitacin F | 10.03 | M + HCOOH − H | Cucurbitacin-type triterpenes | 563.1 | 518.7 | C30H46O7 | 517.4, 499.2, 385.3 |
| 32 | Isorhamnetin 3-O-rhamnoside hexoside | 10.46 | M − H | Flavanone-O-glycoside | 623.2 | 624 | C28H32O16 | 315, 300 |
| 33 | Octacosanol | 10.47 | M + H | Fatty alcohol | 411.5 | 410.7 | C28H58O | 393, 365, 337 |
| 34 | Syringic acid | 11.09 | M − H | Phenolic acid | 197.2 | 198.17 | C9H10O5 | 18, 167, 153 |
| 35 | Kaempferol rhamnoside–hexoside | 11.19 | M − H | Flavanone-O-glycoside | 593.2 | 594 | C27H30O15 | 285, 257, 151 |
| 36 | Chrysoeriol-O-glucoside | 11.36 | M − H | Flavanone-O-glycoside | 461.4 | 462 | C22H22O11 | 299, 341, 323 |
| 37 | Bryonolic acid | 11.79 | M − H | Friedooleanane-type triterpenoid | 455.5 | 456.7 | C30H48O3 | 437, 411, 441, 409 |
| 38 | Colocynthoside A | 11.95 | M + Na | Cucurbitacin-type triterpenes glycosides | 757.4 | 734.8 | C38H54O14 | 697.3188, 365.1202, 572.8 |
| 39 | Cucurbitacin E-2-O-glucopyranoside | 12.11 | M + Na | Cucurbitacin-type triterpenes glycosides | 741.3 | 718.3 | C38H54O13 | 681.2, 597.8, 556.2, 349.1164.1, 96.1 |
| 40 | 2-O-β-d-Glucopyranosyl cucurbitacin L | 12.41 | M + H | Cucurbitacin-type triterpenes glycosides | 79.5 | 678.84 | C36H54O12 | 679, 661, 517, 499, 481, 356 |
| 41 | Lauric acid | 12.42 | M + H | Saturated fatty acid | 201.3 | 200.31 | C12H24O2 | 181, 155, 59 |
| 42 | Luteolin-O-glucoside | 12.68 | M − H | Flavanone-O- glycoside | 447.3 | 448.4 | C21H20O11 | 285, 151, 133 |
| 43 | Cucurbitacin A 2-O-β-d glucopyranoside | 12.72 | M + Na | Cucurbitacin-type triterpenes glycosides | 759.2 | 736.3 | C38H56O14 | 715.3, 634.3, 574.2, 556.2460, 365.1, 96.1 |
| 44 | Caffeoyl quinic acid (chlorogenic acid) | 12.74 | M − H | Quinic acid derivatives | 353.3 | 354.1 | C16H18O9 | 191, 179, 173, 135 |
| 45 | Quercitrin | 12.77 | M − H | Flavanone-O-glycoside | 447.4 | 448 | C21H20O11 | 301, 179, 151 |
| 46 | Colocynthoside B | 12.79 | M − H | Cucurbitacin-type triterpenes glycosides | 806.9 | 807 | C42H62O15 | 645, 498 |
| 47 | Di-caffeoyl shikimic acid | 13.01 | M + H | Quinic acid derivatives | 499.1 | 498 | C22H25O13 | 335, 179, 161, 135 |
| 48 | Cucurbitacin L | 13.02 | M − H | Cucurbitacin-type triterpenes | 515.3 | 516.7 | C30H44O7 | 142, 164, 219, 341, 403, 480, 498, 356 |
| 49 | Colocynthin B | 13.03 | M + H | Cucurbitacin-type triterpene | 691.6 | 690.8 | C37H54O12 | 528, 510, 496, 478, 403, 385, 164 |
| 50 | Cucurbitacin I 2-O-β-d-glucopyranoside | 13.07 | M + Na | Cucurbitacin-type triterpenes | 699.5 | 676.8 | C36H52O12 | 671, 598, 514, 349, 164, 96 |
| 51 | Cucurbitacin D dehydroepirhamnoside (datiscoside) | 13.08 | M − H | Cucurbitacin-type triterpenes glycosides | 515.6 | 516.7 | C30H44O7 | 624, 498, 481, 458, 455, 403, 385, 369, 219, 144, 127, 126, 112, 111, 105, 100, 96 |
| 52 | Cucurbitacin D | 13.15 | M + Na | Cucurbitacin-type triterpenes | 539.5 | 516.7 | C30H44O7 | 342.9, 181.1, 96.1 |
| 53 | Corilagin | 13.29 | M − H | Hydrosable tannin (ellagitannin) | 633.2 | 634.4 | C27H22O18 | 463, 301 |
| 54 | Arvenin I (cucurbitacin B 2-O-beta-d-glucoside) | 13.42 | M − H | Cucurbitacin-type triterpenes glycosides | 719.6 | 720.8 | C38H56O13 | 556, 403, 385, 96, 180, 162, 120, 90 |
| 55 | Cucurbitacin S | 13.51 | M + H | Cucurbitacin type triterpenes | 499.2 | 498.2 | C30H42O6 | 481.3, 317.2, 385.2, 463.3 |
| 56 | Dihydrocucurbitacin C | 13.54 | M + HCOOH − H | Cucurbitacin-type triterpenes | 607.5 | 562.7 | C32H50O8 | 561, 483, 501, 543, 356 |
| 57 | Isovitexin | 13.64 | M − H | Flavone-C-glycosides | 431.3 | 432.4 | C21H20O10 | 341, 323, 311, 283 |
| 58 | Isoorientin 3′-O-methyl ether (retention time) Dr Eman | 15.46 | M − H | Flavone-C-glycoside | 621.1 | 462.4 | C22H22O11 | 371, 341, 311, 299 |
| 59 | 2-O-β -d-Glucopyranosyl-16-20R-dihydroxycucurbita-1,5,23E,25(26)-tetraen-3,11,22-trione | 15.74 | M + H | Cucurbitacin-type triterpenes glycosides | 659.2 | 658.34 | C36H50O11 | 496.2861, 495.2784, 478.2730, 454.2392, 400.2240, 382.2126, 356.1699, 203.1063, 164.0775, 163.0697, 96.0575 |
| 60 | Acetyl glucocucurbitacin E** | 15.91 | M + Na | Cucurbitacin-type triterpenes glycosides | 783.1 | 760.86 | C40H56O14 | 722, 579, 518, 348 |
| 61 | Dihydrocucurbitacin E | 16.21 | M − 2H + Na | Cucurbitacin-type triterpenes | 579.5 | 558.7 | C32H46O8 | 551.68, 593.68 |
| 62 | Catalposide | 16.65 | M − H | Iridoid glycoside | 481.1 | 482.4 | C20H26O12 | 319, 275, 244.7 |
| 63 | 22-Deoxocucurbitoside B | 17.25 | M + HCOOH − H | Cucurbitacin-type triterpenes glycosides | 835.2 | 790.8 | C42H62O14 | 789.4, 643.4, 628.6, 505.3 |
| 64 | Digalloylquinic acid | 18.34 | M − H | Quinic acid derivatives | 495.2 | 496.4 | C21H20O14 | 343, 191, 169 |
| 65 | Colocynthin A | 18.35 | M + H | Cucurbitacin-type triterpene | 659.5 | 658.8 | C36H50O11 | 496, 478, 403, 385, 366, 340, 164 |
| 66 | Palmitoleic acid | 20.20 | M − H | Unsaturated fatty acids | 253.3 | 254.41 | C16H30O2 | 235, 209, 59 |
| 67 | Hexanoic acid methyl ester (caproic acid methyl ester) | 22.60 | M − H | Saturated fatty acid methyl ester | 129.1 | 130.18 | C7H14O2 | 114, 73 |
| 68 | Cucurbitacin B | 22.84 | M + Na | Cucurbitacin-type triterpenes | 581.4 | 558.7 | C32H46O8 | 581.5, 521.3, 429.6, 96.1 |
| 69 | Alpha-elaterin (cucurbitacin E) | 24.80 | M − H | Cucurbitacin type triterpenes | 555.5 | 556.7 | C32H44O8 | 325.3, 495.4, 555.5, 591.4, 601.2, 618.5, 164.2, 96.1 |
| 70 | Gallocatechin | 24.80 | M − H | Flavan-3-ol | 305.3 | 306.2 | C15H14O7 | 287, 261, 263, 153, 169, 125 |
| 71 | Docosanyl-acetate | 27.40 | M − H | Saturated fatty acid ester | 267.3 | 368.6 | C24H48O2 | 308, 73 |
| 72 | Linolenic acid | 32.72 | M − H | Unsaturated fatty acid | 277.1 | 278.43 | C18H30O2 | 260, 234, 54 |
The precision and reproducibility of the data were evaluated using the standard mixing solution and QC samples to ensure data quality. The standard mixed solution was continuously injected 10 times (5 times each in positive and negative ion modes) over the course of 5 days (5 times each in positive and negative ion modes).
All the identified metabolites were relatively quantified using standard compounds that were successfully utilized to calculate the relative concentrations of fatty acids, flavonoids, phenolic acids, organic acids, amino acids, cucurbitacin and cucurbitacin glycosides. The quantified compounds were expressed as mg standard equivalents per g dry extract of each tested extract (Table S2†).
Fig. 1D displays the relative concentrations of each chemical class in the different fruit parts before and after processing. As indicated, the highest relative amount of cucurbitane-type triterpene glycosides was confined within the fruit pulp followed by the seeds and then the peels.
Cucurbitane-type triterpene glycosides dominated the active constituents identified in the pulps of the fruits followed by cucurbitane-type triterpene saponins, phenolic acids and flavonoids. The same pattern was detected in the peels while the seeds exhibited the presence of significant amounts of flavonoids comparable to that of cucurbitane-type triterpene glycosides. In contrary to other parts, the seeds accumulated considerable amounts of fatty acids and amino acids.
As a general trend, heat processing leads to a decrease in the total amount of chemical constituents. Seeds were the least affected organ by heat processing while the peels were the most affected. A decline was observed in the relative amounts of chemical constituents except for cucurbitane-type triterpene genins which showed a surge in their amounts, particularly in the pulps, suggesting the conversion of cucurbitane-type triterpene glycosides to their genins under the effect of heat processing.
The above findings are consistent with the only available previous research on the effect of roasting on colocynth seeds which reported that the extractable content of cucurbitacin glucosides is significantly reduced by roasting.46
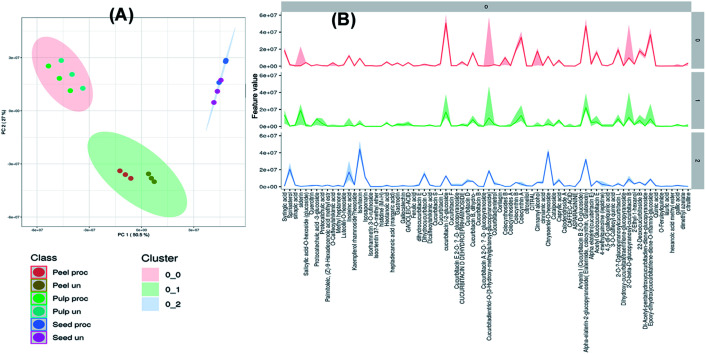
3.3. Unsupervised pattern recognition of variation in chemical composition of heat processed and raw samples
The collected data of the different raw and processed peels, pulps and seeds samples was processed by MetaboAnalyst 5.0 and subjected to unsupervised self-organizing map (SOM) analysis, a neural network-based dimensionality reduction algorithm. PC1 and PC2 explained 50.5% and 27.1% of variation within the samples. As depicted in Fig. 2A, samples were segregated into three main clusters each comprising samples of a specific part of the fruit indicating the variation in their chemical composition. Fig. 2B displays the most significant features of each cluster where the dark lines represent the mean value of each feature in the cluster members. As depicted, cucurbitacin E glucoside was the most abundant identified metabolite in all clusters while linoleic acid, spinasterol, isovitexin, dihydrocucurbitacin C and chryseriol-O-hexoside were the most significant features of the seeds cluster. Meanwhile, sinapic acid, protocatechuic acid glucoside, cucurbitacin I glucoside, cucurbitacin I glucoside, colocynthin B and cucurbitadienetriol-hydroxy-methylglutaroylglucoside were the most significant features of the peels cluster. On the other hand, cucurbitacin I glucoside, dihydro-cucurbitacin B, sinapic acid, cucurbitadienetriol-hydroxy-methylglutaroylglucoside, colocynthin B, colocynthoside B, cucurbitacin E, cucurbitacin L glucoside and cucurbitacin B glucoside were the most significant features of the pulps cluster (Fig. 2B).
Fig. 2. Unsupervised self-organizing map (SOM) of the processed and unprocessed peel, pulp and seed samples of colocynth fruit (A). The most significant features of each cluster graph (B) (dark lines represent the mean value of each feature in the cluster members).
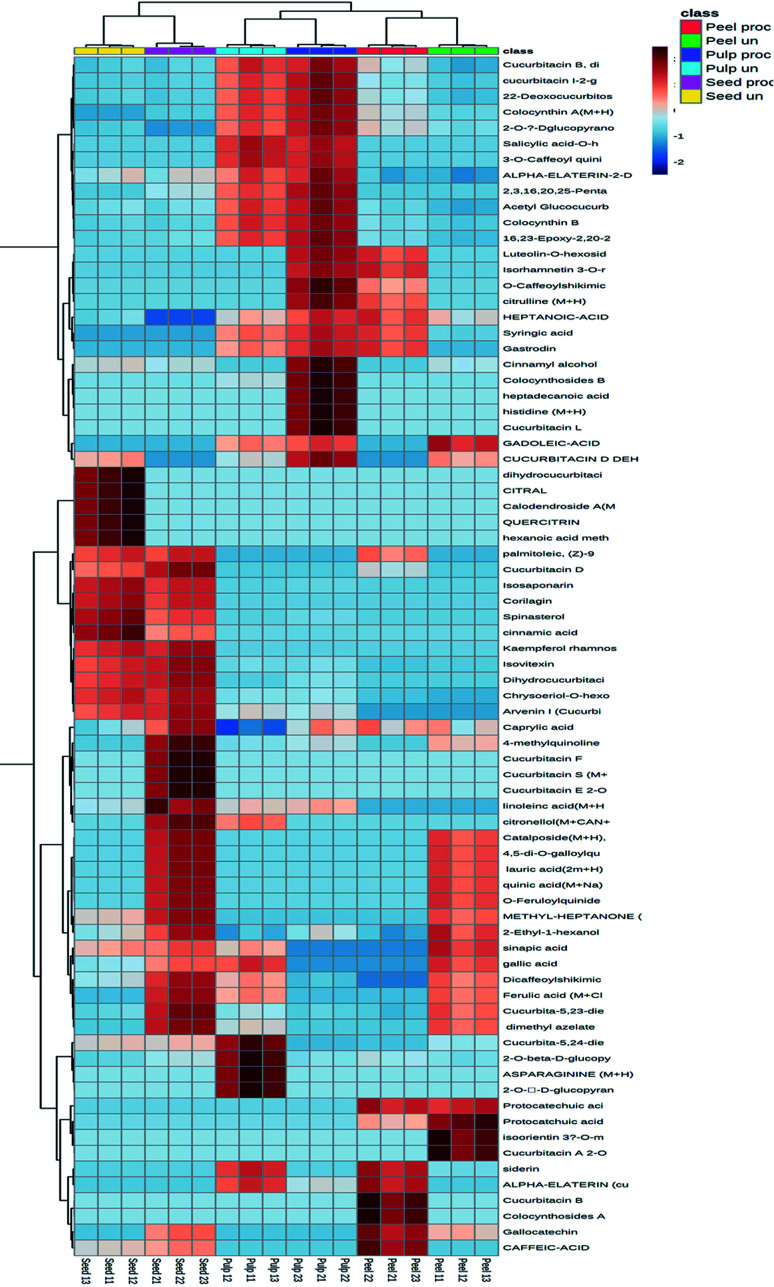
The hierarchical clustering analysis heat-map to track the changes in the compounds in each fruit part upon heat processing was constructed. As indicated by Fig. 3, processed and unprocessed seeds formed a separate cluster, while processed and unprocessed peels and pulps were clustered together with each part forming a separate cluster. Processed and unprocessed samples of the different fruit parts were separately sub-clustered indicating that there were significant differences in the composition of each fruit part upon heat processing.
Fig. 3. Hierarchical analysis heat maps of all identified constituents in the processed and unprocessed peel, pulp and seed samples of colocynth fruit. Brick red and blue indicate higher and lower abundances, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
3.4. Determination of potential discriminatory metabolites between processed and unprocessed raw samples
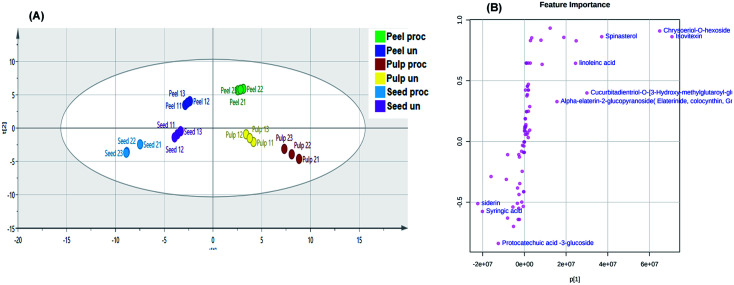
A predictive OPLS-DA model was then constructed to discriminate the different studied samples. The model comprised 1 orthogonal and 5 predictive components with R2X, R2Y, and Q2 values of 0.854, 0.993, and 0.964, respectively. The score scatter plot of the OPLS-DA model (Fig. 4A) inferred in-between class discrimination between samples of the different parts of the fruits. A clear within-class discrimination was observed between processed and unprocessed peels samples indicating that they were the most affected by heat processing. The S-plot (Fig. 4B) was generated to determine the potential discriminatory metabolites between processed and unprocessed colocynth fruits samples. Data points at the two ends of the “S” plot represent the characteristic chemical markers with the most confidence to each group. It can be observed that chrysoeriol-O-hexoside, isovitexin, alpha elaterin glucopyranoside, cucurbitadienetriol-hydroxy-methylglutaroylglucoside and linoleic acid were the most characteristic markers for unprocessed samples, while, protocatechuic acid glucoside, syringic acid and siderin were the most characteristic markers for processed samples.
Fig. 4. Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) score scatter plot (A). S-plot of the constructed OPLS-DA model (B).
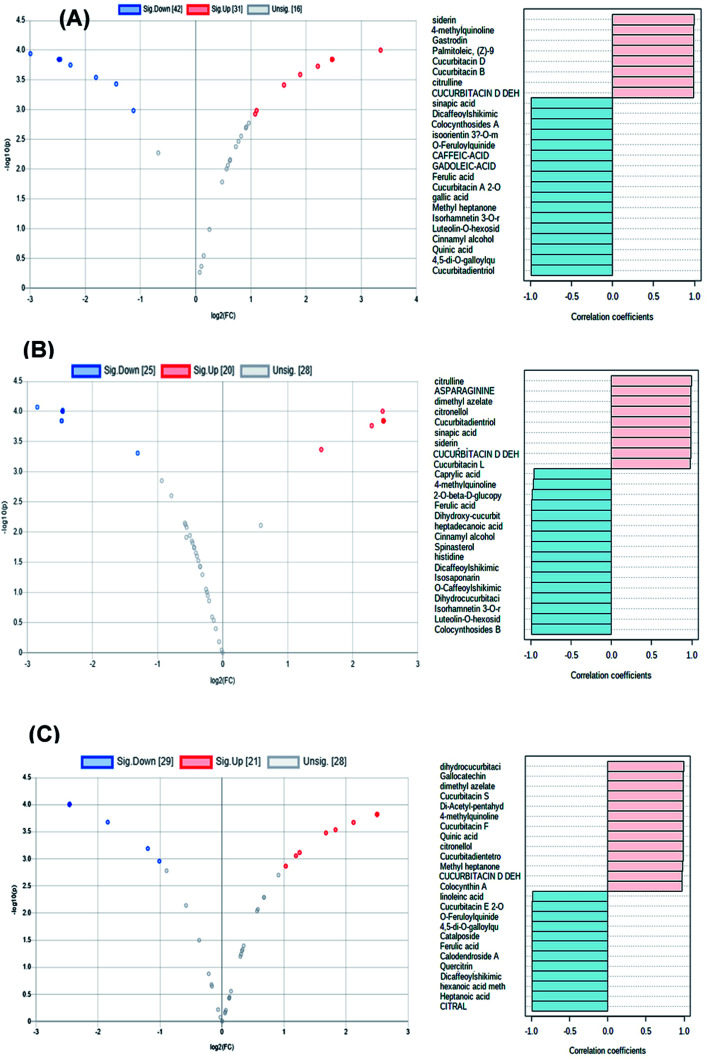
Volcano and coefficient plots were utilized to visualize the up-regulated and down-regulated metabolites within the peels, pulps and seeds of the fruit upon heat processing. As shown in Fig. 5A, 31 metabolites were up-regulated (red scatter points), 42 were down-regulated (blue scatter points) and 16 metabolites showed insignificant change upon heat processing of the peels samples. Siderin, 4-methyl quinolone, gastrodin, palmitoleic, cucurbitacin D, cucurbitacin B and citrulline were the metabolites with the highest correlation coefficient to processed peels samples, while cucurbitadienetriol-hydroxy-methylglutaroylglucoside, 4,5-di-O-galloylquinic acid, cinnamyl alcohol, luteolin-O-hexoside, isorhamnetin 3-O-rutinoside and cucurbitacin A 2-O-glucoside were among the metabolites with the highest correlation coefficient to unprocessed peels samples (Fig. 5A). Meanwhile, 20 metabolites were up-regulated (red scatter points), 25 were down-regulated (blue scatter points) and 28 metabolites showed insignificant change upon heat processing of the pulp samples (Fig. 5B). Citrulline, asparaginine, dimethyl azaleate, cucurbitadienetriol, sinapic acid, siderin and cucurbitacin L possessed the highest correlation coefficients to heat processed pulp samples, while, colocynthoside B, luteolin-O-hexoside, isorhamnetin 3-O-rutinoside, isosaponarin, 2-O-glucopyranosylcucurbitacin L and dihydroxy-cucurbitatetraentrione-glucopyranoside were among the metabolites with the highest correlation coefficient to unprocessed pulp samples. On the other hand, 21 metabolites were up-regulated (red scatter points), 29 were down-regulated (blue scatter points) and 28 metabolites showed insignificant change upon heat processing of the seed samples (Fig. 5C). Dihydrocucurbitacin E, gallocatechin, dimethyl azaleate, cucurbitacin S, cucurbitacin F, quinic acid, methyl heptanone, cucurbitadientetrol, and dehydrocucurbitacin D displayed the highest correlation coefficients to processed seeds samples while citral, heptanoic acid, hexanoic acid methyl ester, dicaffeoyl shikimic acid, quercetrin, calodendroside A, ferulic acid, catalposide and cucurbitacin E glucoside were among the metabolites with the highest correlation coefficient to unprocessed seeds samples.
Fig. 5. Volcano and coefficient plots of processed and unprocessed peels (A), pulps (B) and seeds (C) samples of colocynth fruit.
3.5. In vitro cytotoxicity and anti-inflammatory activity of processed and unprocessed samples
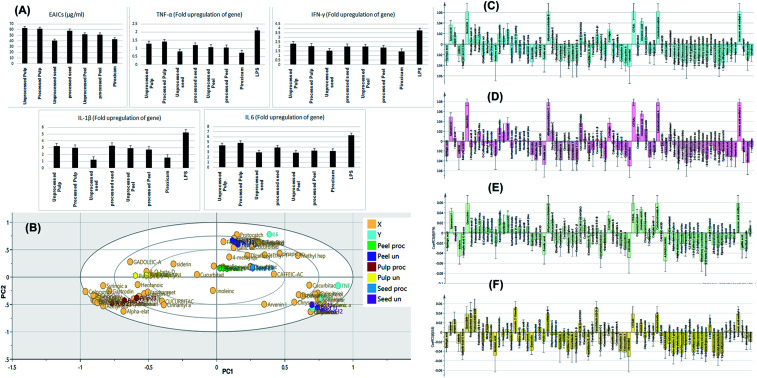
Safety of the tested extracts was assessed using MTT test and EC100 values resulting in 100% cell viability were calculated for all the tested extracts and the standard piroxicam. Piroxicam exhibited EC100 value of 1000 μg ml−1. The processed peel extract showed the highest EC100 value (1500 μg ml−1) followed by processed pulp extract, processed seed extract, unprocessed seeds extract, unprocessed peel extract and unprocessed pulp extract as they showed EC100 values of 1239 μg ml−1, 1008 μg ml−1, 887.2 μg ml−1, 732.1 μg ml−1 and 409.2 μg ml−1, respectively. These results indicated that roasted samples possessed higher safety profile than the relevant unprocessed ones, with the exception of the seeds. After that, the effective anti-inflammatory concentrations (EAICs) of tested extracts and piroxicam was determined (Fig. 6A). Unprocessed seed extract showed EAIC value of 40.1 μg ml−1 which is lower than that of piroxicam (42.5 μg ml−1) indicating its efficacy, while other extracts showed EAIC values higher than that of piroxicam but still comparable to it indicating their efficacy. The mechanism of anti-inflammatory activity of the tested extracts was determined through investigating of the gene expression of four pro-inflammatory markers (TNF-α, IL-1β, IFN-γ, IL-6) by real time polymerase chain reaction (PCR) in normal WBCs and the treated WBCs with lipopolysaccharide (LPS). Gram-negative bacteria's cell wall contains proteins that called lipopolysaccharides (LPS) which degraded into O-antigen, core protein and lipid-A. Among lipid-A possess high immunogenic and pro-inflammatory effects.47 TNF-α, IL-1β, INF-γ, IL-6 are pro-inflammatory cytokines that are directly related to immune system and inflammation process such as, vasodilatation and edema addition to their critical roles in chronic inflammation and autoimmune diseases.48–51
Fig. 6. Bar charts showing the effective anti-inflammatory concentrations (EAICs) of tested extracts and piroxicam as well as the levels of TNF-α, IL-1β, IFN-γ, IL-6 (expressed as fold change) of the tested samples (A). OPLS-DA biplot of the tested samples in correlation to the pro-inflammatory markers inhibition levels (B). Correlation analysis of differential metabolites identified and TNF-α levels (C), IL-1β (D), IFN-γ (E) and IL-6 (F) in the tested samples.
Treatment of LPS-treated WBCs with the tested extracts decreased the gene upregulation of both cytokines to levels lower than that exerted by LPS. Among the tested extracts, unprocessed seed extract showed the highest anti-inflammatory activity regarding inhibiting the upregulation of IL-1β gene INF-γ as the extract decreased such upregulation to be 1.21-fold and 1.53-fold, respectively. Finally, IL-6 gene expression was upregulated by 6.3-fold upon addition of LPS to WBCs and decreased to be 3.2-fold after treatment of cells with piroxicam. Among the tested extracts, processed peel extracts extract showed the highest anti-inflammatory activity regarding this pro-inflammatory marker.
These results come in accordance to previous studies that assessed the anti-inflammatory effect of colocynth fruit. For instance, a study showed that colocynth cream exhibited anti-inflammatory activity upon topical application on animals and this effect was deduced to be due to inhibition of the pro-inflammatory markers, TNF-α and IL-6.52 Moreover, a previous study showed that colocynth extract significantly reduced the expression of TNF-α, PGE2, IL-1β, NO, iNOS, COX-2.53
3.6. Determination of the ant-inflammatory discriminatory markers of raw and roasted colocynth samples
Discrimination of samples in relation to their tested in vitro anti-inflammatory activity as well as determination of the chemical markers responsible for such discrimination was attempted through construction of an orthogonal projection to latent structures discriminant analysis model (OPLS-DA). The model comprised 1 orthogonal and 4 predictive components with R2X, R2Y, and Q2 values of 0.993, 0.991, and 0.984, respectively. For validation, cross-validation and random permutation test (200 times) were performed on the corresponding OPLS-DA model. Obtained results indicated the predictability and goodness of fit of the constructed OPLS-DA model.
Fig. 6B depicts the OPLS-DA biplot which shown in-between class discrimination of un-processed and processed pulp samples which formed as separate cluster along the negative side of PC1 from other samples, while within class discrimination was observed for the un-processed seed samples which were clustered along the positive side of PC1 and negative side of PC2 from processed seeds, as well as unprocessed and processed peel samples. Un-processed seed samples showed a strong spatial correlation with inhibiting TNF-α, IL-1β and IFN-γ while processed seeds, as well as unprocessed and processed peel samples showed spatial correlation with inhibiting IL-6 indicating a considerable difference in the effect of processed and un-processed seeds on the measured pro-inflammatory markers. Studying the biplot revealed that the discrimination of unprocessed seeds samples is attributed to their content of dihydrocucurbitacin C, dihydrocucurbitacin E, hexanoic acid methyl ester, spinatsterol, corilagin, cinnamic acid, isovitexin, kaempferol rhamnoside and chrysoeriol hexoside.
In an attempt to reveal the main anti-inflammatory discriminatory chemical markers of the tested extracts, correlation analysis was implemented. As indicated from Fig. 6C and D, quercetrin, dihydrocucurbitacin E, citral, calodendroside A, hexanoic acid methyl ester, spinatsterol, corilagin, cinnamic acid and sinapic acids were the main chemical markers positively correlated to the inhibition of TNF-α and IL-1β levels. Meanwhile, quercetrin, dihydrocucurbitacin E, citral, calodendroside A, hexanoic acid methyl ester, spinatsterol, corilagin, dihydrocucurbitacin C, dehydrocucurbitacin D, kaempferol rhamnoside and chrysoeriol hexoside and isovitexin were positively correlated to lowered levels of the pro-inflammatory marker IFN-γ (Fig. 6E). On the other hand, protocatechuic acid, prtocatechuic acid glycoside, isoorientin methyl ether, dihydrocucurbitacin E, cucurbitacin A glycoside and sinapic acid were positively correlated to the downregulation of IL-6 (Fig. 6F).
3.7. Determination of the cytotoxic discriminatory markers of raw and roasted colocynth samples
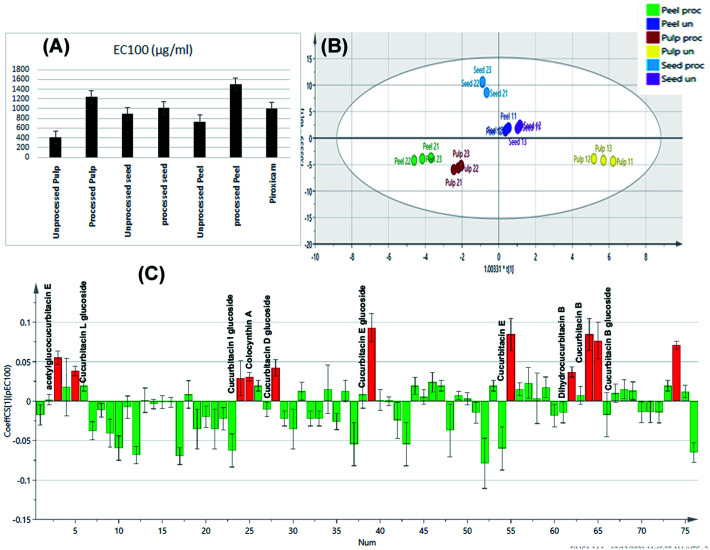
Construction of an orthogonal projection to latent structures discriminant analysis model was implemented for sample discrimination in relation to their tested cytotoxic activity on normal cells as indicated by their EC100 values (Fig. 7A). With R2X, R2Y, and Q2 values of 0.994, 0.992, and 0.985, the constructed OPLS-DA model has one orthogonal and three predictive components. The model was validated using cross-validation and a random permutation test (200 times). The obtained findings demonstrated the constructed OPLS-DA model's predictability and quality of fit.
Fig. 7. Bar charts showing the EC100% resulting in 100% cell viability of tested extracts and piroxicam (A). OPLS-DA score scatter plot of the tested samples in correlation to the cytotoxic activity (B). Correlation analysis of differential metabolites identified and cytotoxic activity of the tested samples (C).
The score scatter plot of the OPLS-DA model (Fig. 7B) revealed in-between class discrimination of raw un-processed samples which were segregated along the positive side of PC1 and roasted processed samples which were clustered along the negative side of PC1. Meanwhile, within class discrimination was observed for the un-processed pulp samples which possessed the lowest EC100 value and thus highest cytotoxicity on normal cells. In addition, within-class discrimination was observed for roasted seed samples from roasted peel and pulp ones which could be attributed to the fact that the former samples showed the least change in their EC100 values with roasting.
Correlation analysis was then applied (Fig. 7C) to disclose the main cytotoxic discriminatory chemical markers of the tested extracts, As expected, the main cytotoxic chemical markers were cucurbitacin glycosides and their genins including cucurbitacin B and its glucoside, dihydrocucurbitacin B, cucurbitacin E and its glucoside, acetyl glucocucurbitacin E, colocynthin A, cucurbitacin D glucoside, cucurbitacin I glucoside and cucurbitacin L glucoside. The obtained results come in accordance to previous reports which attributed the cytotoxicity of colocynth to its content of cucurbitacins where they have been reported as highly toxic compounds.54
4. Conclusion
The study in hand offers the first comprehensive study on the changes imposed by heat processing on the metabolic profile of the seeds, pulps and peels of Citrullus colocynthis fruit. UPLC/MS/MS in combination to multivariate data analysis tools were successfully employed to unravel the discriminatory chemical markers amid the unprocessed and heat processed samples. It was evident that the roasting process had a significant effect on the chemical profile of the fruit particularly the peels. Correlation analysis was then applied to disclose the main cytotoxic discriminatory chemical markers which were identified as cucurbitacin glycosides and their genins. Determination of the anti-inflammatory discriminatory markers of the processed and unprocessed samples was achieved through combination of ex vivo anti-inflammatory activity testing and multivariate statistical models which indicated that unprocessed seeds acted through inhibition of the pro-inflammatory markers TNF-α, IL-1β and IFN-γ while most processed samples showed correlation with IL-6 pro-inflammatory marker inhibition. Overall, the study provided insights into the rationality of processing from the perspective of chemical composition as well biological activity of the different parts of colocynth fruit and sets bases for studying the beneficial effects of the different parts of both raw and roasted colocynth fruits. The obtained results shed light on the changes imposed by processing on the chemical profile of the different parts of colocynth fruit to revealing the changes that occur during the roasting process in an attempt for safer application of the fruit as an anti-inflammatory agent through reduction of its toxicity. Further in vivo studies are needed to get more insights about the safety and toxicity of colocynth as an anti-inflammatory drug.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra07751a
References
- Hussain A. I. Rathore H. A. Sattar M. Z. A. Chatha S. A. S. Sarker S. D. Gilani A. H. Ethnopharmacology. 2014;155:54–66. doi: 10.1016/j.jep.2014.06.011. [DOI] [PubMed] [Google Scholar]
- Mazher M. Ishtiaq M. Mushtaq W. Maqbool M. Zahid N. Husain T. Mazher M. J. Pharm. Res. 2020;4:1–13. [Google Scholar]
- Rahimi R. Amin G. Ardekani M. R. S. J. Altern. Complementary Med. 2012;18:551–554. doi: 10.1089/acm.2011.0297. [DOI] [PubMed] [Google Scholar]
- Al-Snafi A. E. IOSR J. Pharm. 2018;8:32–95. [Google Scholar]
- Marzouk B. Marzouk Z. Fenina N. Bouraoui A. Aouni M. Eur. Rev. Med. Pharmacol. Sci. 2011;15:665–672. [PubMed] [Google Scholar]
- Al-Snafi A. E. IOSR J. Pharm. 2016;6:57–67. [Google Scholar]
- Pashmforosh M. Vardanjani H. R. Vardanjani H. R. Pashmforosh M. Khodayar M. J. Medicina. 2018;54(4):51. doi: 10.3390/medicina54040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnold R. A. Sandford K. S. Shaw W. B. K. Moreau R. E. Riley N. D. Hutchinson T. Craig V. F. Geogr. J. 1933;82:211. doi: 10.2307/1786558. [DOI] [Google Scholar]
- Al-douri N. A. Pharm. Biol. 2000;38:74–79. doi: 10.1076/1388-0209(200001)3811-BFT074. [DOI] [PubMed] [Google Scholar]
- François G. Nathalie B. Jean-Pierre V. Daniel P. Didier M. Grasas Aceites. 2006;57:409–414. [Google Scholar]
- Mohammed G. R. Int. J. Basic Appl. Sci. 2018:10–14. [Google Scholar]
- Ghallab D. S. Mohyeldin M. M. Shawky E. Metwally A. M. Said Ibrahim R. LWT--Food Sci. Technol. 2021;136(part 1):110298. doi: 10.1016/j.lwt.2020.110298. [DOI] [Google Scholar]
- Xia J. Sinelnikov I. V. Han B. Wishart D. S. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J. Soufan O. Li C. Caraus I. Li S. Bourque G. Wishart D. S. Xia J. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramberger K. Barlič-Maganja D. Bandelj D. Baruca Arbeiter A. Peeters K. Miklavčič Višnjevec A. Pražnikar Z. J. Metabolites. 2020;10:1–25. doi: 10.3390/metabo10100403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. Y. Fan M. X. Wu X. Wang H. J. Yang J. Si N. Bian B. L. J. Chromatogr. Sci. 2013;51:273–285. doi: 10.1093/chromsci/bms138. [DOI] [PubMed] [Google Scholar]
- Sinosaki N. B. M. Tonin A. P. P. Ribeiro M. A. S. Poliseli C. B. Roberto S. B. da Silveira R. Visentainer J. V. Santos O. O. Meurer E. C. J. Braz. Chem. Soc. 2020;31:402–408. [Google Scholar]
- Oszmiański J. Kolniak-Ostek J. Wojdyło A. Eur. Food Res. Technol. 2013;236:699–706. doi: 10.1007/s00217-013-1925-x. [DOI] [Google Scholar]
- El-Sayed M. A. Abbas F. A. Refaat S. El-Shafae A. M. Fikry E. Egypt. J. Chem. 2021;64:793–806. [Google Scholar]
- Wu Z. J. Ma X. L. Fang D. M. Qi H. Y. Ren W. J. Zhang G. L. Eur. J. Mass Spectrom. 2009;15:415–429. doi: 10.1255/ejms.992. [DOI] [PubMed] [Google Scholar]
- Kumar S. Singh A. Kumar B. J. Pharm. Anal. 2017;7:214–222. doi: 10.1016/j.jpha.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang N. Yu S. Prior R. L. J. Agric. Food Chem. 2002;50:3579–3585. doi: 10.1021/jf0201327. [DOI] [PubMed] [Google Scholar]
- Clifford M. N. Stoupi S. Kuhnert N. J. Agric. Food Chem. 2007;55:2797–2807. doi: 10.1021/jf063533l. [DOI] [PubMed] [Google Scholar]
- Abu-Reidah I. M. Ali-Shtayeh M. S. Jamous R. M. Arráez-Román D. Segura-Carretero A. Food Chem. 2015;166:179–191. doi: 10.1016/j.foodchem.2014.06.011. [DOI] [PubMed] [Google Scholar]
- Gouveia S. Castilho P. C. Food Chem. 2011;129:333–344. doi: 10.1016/j.foodchem.2011.04.078. [DOI] [PubMed] [Google Scholar]
- Ben Said R. Hamed A. I. Mahalel U. A. Al-Ayed A. S. Kowalczyk M. Moldoch J. Oleszek W. Stochmal A. Int. J. Mol. Sci. 2017;18:1–18. doi: 10.3390/ijms18030512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachlicki P. Piasecka A. Stobiecki M. Marczak L. Molecules. 2016;21:1–21. doi: 10.3390/molecules21111494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.-H. Guo H. Xu W.-B. Ge J. Li X. Alimu M. He D.-J. J. Chromatogr. Sci. 2016;54:805–810. doi: 10.1093/chromsci/bmw016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. Price W. E. Ashton J. Tapsell L. C. Johnson S. Food Chem. 2016;211:215–226. doi: 10.1016/j.foodchem.2016.05.052. [DOI] [PubMed] [Google Scholar]
- March R. E. Lewars E. G. Stadey C. J. Miao X. S. Zhao X. Metcalfe C. D. Int. J. Mass Spectrom. 2006;248:61–85. doi: 10.1016/j.ijms.2005.09.011. [DOI] [Google Scholar]
- March R. E. Miao X.-S. Int. J. Mass Spectrom. 2004;231:157–167. doi: 10.1016/j.ijms.2003.10.008. [DOI] [Google Scholar]
- Abu-Reidah I. M. Ali-Shtayeh M. S. Jamous R. M. Arráez-Román D. Segura-Carretero A. Int. Food Res. 2015;70:74–86. doi: 10.1016/j.foodres.2015.01.023. [DOI] [PubMed] [Google Scholar]
- Li S. Wan C. He L. Yan Z. Wang K. Yuan M. Zhang Z. Rev. Bras. Farmacogn. 2017;27:188–194. doi: 10.1016/j.bjp.2016.10.003. [DOI] [Google Scholar]
- Shao S. Y. Ting Y. Wang J. Sun J. Guo X. F. Acta Chromatogr. 2020;32:228–237. [Google Scholar]
- Olennikov D. N. Chirikova N. K. Kashchenko N. I. Nikolaev V. M. Kim S. W. Vennos C. Front. Pharmacol. 2018;12(9):756. doi: 10.3389/fphar.2018.00756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miketova P. Schram K. H. Whitney J. Li M. Huang R. Kerns E. Valcic S. Timmermann B. N. Rourick R. Klohr S. J. Mass Spectrom. 2000;35:860–869. doi: 10.1002/1096-9888(200007)35:7<860::AID-JMS10>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Nayab D. Perveen S. Ahmed Z. Malik A. Helv. Chim. Acta. 2010;93:1012–1018. doi: 10.1002/hlca.200900341. [DOI] [Google Scholar]
- Nayab D. Ali D. Arshad N. Malik A. Choudhary M. I. Ahmed Z. Nat. Prod. Res. 2006;20:409–413. doi: 10.1080/14786410500044997. [DOI] [PubMed] [Google Scholar]
- Audier H. E. Das B. C. Tetrahedron Lett. 1966;7:2205–2210. doi: 10.1016/S0040-4039(00)72402-6. [DOI] [Google Scholar]
- Isaev M. I. Chem. Nat. Compd. 2000;36(3):236–238. doi: 10.1007/BF02238339. [DOI] [Google Scholar]
- McLafferty F. W. Anal. Chem. 1959;31:82–87. doi: 10.1021/ac60145a015. [DOI] [Google Scholar]
- Hatam N. A. R. Whiting D. A. Yousif N. J. Phytochemistry. 1989;28:1268–1271. doi: 10.1016/0031-9422(89)80230-4. [DOI] [Google Scholar]
- Ul Haq F. Ali A. Khan M. N. Shah S. M. Z. Kandel R. C. Aziz N. Adhikari A. Choudhary M. I. Ur-Rahman A. El-Seedi H. R. Musharraf S. G. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-52404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. L., Liu Y.-M. and V Wood K., in Modern Phytochemical Methods, ed. N. H. Fischer, M. B. Isman and H. A. Stafford, Springer US, Boston, MA, 1991, pp. 251–269 [Google Scholar]
- Honda C. Suwa K. Takeyama S. Kamisako W. Chem. Pharm. Bull. 2002;50:467–474. doi: 10.1248/cpb.50.467. [DOI] [PubMed] [Google Scholar]
- Ojiako O. A. Ogbuji C. A. Agha N. C. Onwuliri V. A. J. Med. Food. 2010;13:1203–1209. doi: 10.1089/jmf.2009.0221. [DOI] [PubMed] [Google Scholar]
- Darwish R. S. Hammoda H. M. Ghareeb D. A. Abdelhamid A. S. A. El Naggar E. B. Harraz F. M. Shawky E. Ethnopharmacology. 2020;259:1–12. doi: 10.1016/j.jep.2020.112971. [DOI] [PubMed] [Google Scholar]
- Zelova H. Hosek J. Inflammation Res. 2013;62:641–651. doi: 10.1007/s00011-013-0633-0. [DOI] [PubMed] [Google Scholar]
- Mühl H. Pfeilschifter J. Int. Immunopharmacol. 2003;3:1247–1255. doi: 10.1016/S1567-5769(03)00131-0. [DOI] [PubMed] [Google Scholar]
- Rena K. Torres R. Brain Res. Rev. 2009;60:57–64. doi: 10.1016/j.brainresrev.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K. Liu C. Tu Z. Xu Q. Chen S. Zhang Y. Wang X. Zhang J. Hu C.-A. A. Liu Y. Oxid. Med. Cell. Longevity. 2020;2020:1–14. doi: 10.1155/2020/5803639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashmforosh M. Vardanjani H. R. Vardanjani H. R. Pashmforosh M. Khodayar M. J. Medicina. 2018;54:1–11. doi: 10.3390/medicina54040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhzari M. Mirghiasi S M. Mol. Biol. 2015;4:147–151. doi: 10.4172/2168-9547.1000147. [DOI] [Google Scholar]
- Kaushik U. Aeri V. Mir S. R. Pharmacogn. Rev. 2015;9:12–18. doi: 10.4103/0973-7847.156314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.