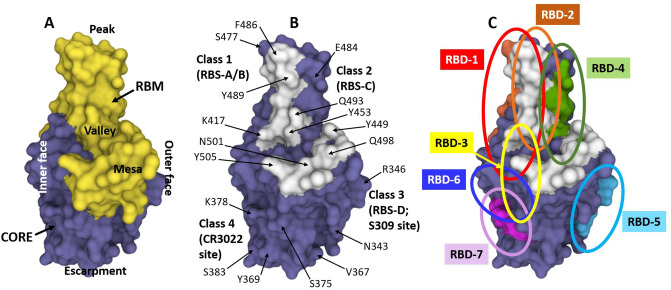
Fig. 4.
A, B Structure of the SARS-CoV-2 RBD (from PDB ID 7CH5) showing the receptor binding motif (RBM; in yellow, residues 437–507 as per OSM Fig. S2) and the core (non-RBM sequences of RBD in blue). The landscape of the RBM, including the peak (top as shown here; characterized by residues S477, E484 and F486), valley (the “indentation motif”; characterized by residues K417 and Y453), and the mesa (large hump; characterized by residues Q498 and N501), are shown as described by Hastie et al. [290]. The escarpment is the area occupying the base under the steep slope, as depicted by residues N343, V367, Y369, and S375. The inner face, which is buried when the RBD is in the closed (or down) position, is on the left. The outer face, which is exposed with the RBD in both the closed (down) and open (up) positions, is on the right of the RBD as depicted here. B The 17 residues that specifically contact human ACE2 receptor (see OSM Fig. S2) are shown in white, and several residues throughout the RBD are labeled for reference. As depicted here, the Class I antibodies described by Barnes et al. [276], as well as the RBS-A and B antibodies of Yuan et al. [277] tend to bind in the area of the peak toward the inner face; Class 2 antibodies [276] and RBS-C epitope antibodies [277] tend to bind on the outer face of peak area, Class 3 [276]/RBS-D [277] antibodies bind at the lower outer face, also known as the S309 site or the proteoglycan epitope site [277] and typically interact with the glycan attached to residue N343 [277], and Class 4 antibodies [276] such as CR3022 bind on the inner side of the lower part in the area of residues Y369 and S383. C The RBD with the overlapping epitope groups described by Hastie et al. [290], RBD-4, RBD-5, and RBD-7 are shown in red, orange, green, light blue and pink, respectively. These seven epitope groups described by Hastie et al. [290] and adopted herein are shown in more detail in Table 5. The PDB program [201, 202] was used to generate and annotate the structures. ACE2 angiotensin-converting enzyme-2, PDB Protein Data Bank, RBD receptor binding domain, RBM receptor binding motif, RBS receptor binding site