Abstract
Introduction
There is an increased need for the development of novel blood‐based biomarkers for early detection, prevention, or intervention in Alzheimer's disease (AD). This study sought to determine whether serum glycopeptide analysis holds potential for identifying novel diagnostics and prognostics of AD.
Methods
The study involved 195 participants, including 96 patients with an AD diagnosis and 99 controls with no cognitive deficit. Utilizing a validated analytical mass spectrometry method, we monitored the site‐specific glycosylation of 52 serum glycoproteins.
Results
Partial least‐squares discriminant analysis revealed that changes in overall sialylation and fucosylation of serum glycoproteins may be indicators of an AD disease state. Loss of fucosylation of immunoglobulin G1 (IgG1) and IgG2 was indicative of AD diagnosis. Individual glycopeptide analysis found separation between the AD patients and controls on complement proteins and apolipoprotein B.
Discussion
The results of this study suggest that serum glycoprofiling may be a promising approach for biomarker discovery.
Keywords: Alzheimer's disease, blood‐based biomarker, mass spectrometry, site‐specific glycosylation
1. INTRODUCTION
Alzheimer's disease (AD) is a chronic neurodegenerative disease that begins decades before the onset of symptoms and discernible dementia. 1 Medications for the treatment for AD in the later stages of disease have been elusive, with even the newest and most promising approaches only being able to slow the progression of disease but not prevent or reverse it. 1 Given the hallmark characteristics of AD—namely amyloid beta (Aβ) deposition, neurofibrillary tangles involving hyperphosphorylated tau, and neurodegeneration—the current techniques for AD detection rely on cerebrospinal fluid (CSF) and plasma biomarkers (eg Aβ42/Aβ40 ratio) and images from positron emission tomography (PET) to evaluate the extent of neurodegeneration in the brain. 2 , 3 , 4 However, CSF‐based biomarkers require invasive sample collection, the PET imaging‐based detection is costly for screening purposes, and these detection approaches are generally uninformative about how to intervene to prevent or slow the progression of the disease. Thus, early, actionable detection approaches that could lead to the development of new disease prevention strategies are urgently needed.
Glycosylation is a common post‐translational modification that affects protein structure and function with diverse glycans. Multiple studies have revealed aberrant glycosylation in AD‐related proteins, including amyloid precursor protein (APP), tau, Beta‐secretase 1 (BACE1), and Nicastrin (NCSTN). 5 , 6 , 7 , 8 Furthermore, dysregulated glycosylation in AD brains affects multiple biological processes, such as neuroinflammation, cell adhesion, and cell signaling. 8 Plasma glycan‐based measurements have proven useful for the detection of other diseases, including ovarian cancer and breast cancer. 9 , 10
Multiple reaction monitoring (MRM) technology has enabled large‐scale glycoproteomic profiling of blood. Here we used an established MRM method 11 , 12 to evaluate the potential for plasma‐ or serum‐based glycoproteomic profiling as a viable biomarker approach for AD. We used serum samples obtained from 100 AD patients and 100 age‐ and gender‐matched cognitively normal controls whose samples are part of the UC Davis Alzheimer's Disease Research Center (ADRC) biorepository, to quantify serum glycopeptides of the most abundant serum proteins, and to determine the potential of serum glycoprofiling as a novel biomarker approach for AD.
2. EXPERIMENTAL SECTION
2.1. Study design
A power calculation was conducted on an exploratory set of samples involving 48 serum samples. Sample size calculation was performed prior to the study design as multiple two‐sample t‐tests with an FDR‐adjusted rate of 0.05, and a K factor of 22. A sample size of 100 per group was determined to be an adequate number of participants for a power of 80%. In this study, samples from 195 total participants were analyzed, which included 99 participants with no cognitive deficit as per clinical diagnosis, and 96 participants with an AD diagnosis. All AD patients had a clinical AD diagnosis made within 1 year of the blood draw based on the UC Davis ADRC clinical diagnosis criteria. The diagnosis criteria for AD involved autopsy confirmation with BRAAK state IV or higher with a likelihood of AD moderate to high, with those patients for whom autopsy confirmation was not available being diagnosed with “probable AD” based on the ADRC clinical diagnosis criteria. All human subjects provided informed consent as per the ADRC guidelines. The cohorts were selected to have equal percentages of males and females in both groups. In the control group, 50% of participants were male, whereas in the AD group 51% were male. Table 1 summarizes the demographic data of all participants including age, sex, body mass index (BMI), apolipoprotein E (APOE) genotype, and ethnicity, among other parameters. Among AD patients, 13.3% were African American, 10% were Hispanic, and 77% were White. Among controls, 20% were African American, 27% were Hispanic, and 53% were White. The median age was 79 (interquartile range [IQR]: 73‐83) for AD patients and 78 (IQR:73‐82) for the normal controls. The median BMI was 26.4 (IQR: 23.83‐29.64) for AD patients and 27.70 (IQR: 25.34‐31.01) for the normal controls.
TABLE 1.
Demographics data of participants with AD and controls
| Control | AD | |
|---|---|---|
| N | 99 | 96 |
| Age (years) (median [IQR]) | 78 [73–82] | 79 [78–83] |
| Gender = Male (%) | 49 (49.5) | 49 (51) |
| BMI (median [IQR]) | 27.7 [25.3–31] | 26.4 [27–29.5] |
| APOE genotype | ||
| e22 (%) | 1 (1.08) | 0 (0) |
| e23 (%) | 13 (14) | 1 (1.06) |
| e24 (%) | 1 (1.08) | 3 (3.19) |
| e33 (%) | 55 (59.1) | 35 (37.2) |
| e34 (%) | 23 (24.7) | 42 (44.7) |
| e44 (%) | 0 (0) | 13 (13.8) |
| APOE ε4 Positivity (%) | 30 (30.3) | 60 (62.5) |
| Ethnicity | ||
| African American (%) | 20 (20.4) | 12 (12.5) |
| Hispanic (%) | 26 (26.5) | 10 (10.4) |
| White (%) | 52 (53.1) | 74 (77.1) |
| Verbal Memory Score (median [IQR]) | 0.5 [−0.13–0.94] | −1.4 [−1.3–1] |
| Executive Function Score (median [IQR]) | 0.15 [−0.11–0.48] | −0.72 [−0.63–0.47] |
| Semantic Memory Score (median [IQR]) | 0.66 [−0.013–1.2] | −0.33 [−0.42–0.17] |
| Spatial Score (median [IQR]) | 0.45 [−0.11–0.82] | −0.51 [−0.47–0.14] |
| CDR (median [IQR]) | 0 [0–0.38] | 4.5 [5.3–7] |
| Total White Matter Hypertension (median [IQR]) | 5.7 [2.7–13] | 8.4 [15–18] |
| Intracranial Volume (median [IQR]) | 1300 [1200–1400] | 1300 [1300–1400] |
Note: Cognitive function scores and brain measurements had missing values in both groups. The number of data points for each measurement was listed in Table S3.
Abbreviation: AD, Alzheimer's disease.
2.2. Sample preparation
Serum samples were processed using a well‐established protocol coupled to the dynamic multiple‐reaction monitoring (dMRM) analytical method developed in our laboratory. 13 Briefly, serum samples were reduced with dithiothreitol, alkylated with iodoacetamide, and digested with trypsin in a water bath at 37˚C for 18 hours. For glycopeptide quantitative analysis, tryptic‐digested samples were analyzed directly with no further enrichment, as shown in Figure S1. Serum samples were then spiked with a synthetic peptide standard for inter‐batch reproducibility monitoring.
RESEARCH IN CONTEXT
Systematic Review: The authors reviewed the literature using traditional (eg, PubMed) sources and meeting abstracts and presentations. Although glycosylation alterations have been observed in the brains, cerebrospinal fluid, and serum of patients with Alzheimer's disease (AD), serum glycoprofiles are not widely studied. We sought to determine whether serum‐based glycoprofiling may be a useful approach for biomarker discovery in AD and related dementias.
Interpretation: Our results suggest that serum glycoprofiling may indeed be a powerful biomarker discovery platform that can be harnessed to identify new markers in diverse cohorts and new therapeutic targets, particularly those focused on immune‐related mechanisms.
Future Directions: Future studies should include the analysis of a larger sample size, particularly in ethnically diverse cohorts powered to account for age, sex, and ethnicity. Further work to refine the transitions monitored by the multiple reaction monitoring (MRM) mass spectrometry method that are specific to AD pathology would lead to higher precision and specificity.
2.3. UPLC−ESI‐QqQ−MS analysis of serum glycoproteomic
The mixed samples were analyzed using ultrahigh‐performance liquid chromatography (UPLC) coupled with triple quadrupole mass spectrometry as described previously. 12 Briefly, 2 μL samples were injected and separated by an Agilent Eclipse plus C18 column (rapid resolution high definition [RRHD] 1.8 μm, 2.1×100 mm) coupled with a C18 guard column (RRHD 1.8 μm, 2.1×5 mm). An Agilent 1290 infinity liquid chromatography (LC) system (Agilent Technologies, Santa Clara, CA) was used, and the separation was performed with a 70 minute binary gradient consisting of solvent A of 3% acetonitrile, 0.1% formic acid, solvent B of 90% acetonitrile, and 0.1% formic acid in nano pure water (v/v) at a flow rate of 0.5 mL per minute. The UPLC system was coupled to an Agilent 6490 triple quadrupole (QQQ) mass spectrometer (Agilent Technologies, Santa Clara, CA) with the mass spectrometry (MS) conditions as described previously. 11 The dMRM method used predetermined collision‐induced dissociation from a previously determined study; however, LC retention times were adjusted. The list of the serum glycoproteins monitored are shown in Table S1, which involved over 400 transitions. A deep learning method PB‐Net (Peak Boundary Neural Network) was used for fully automatic chromatographic peak integration. 14 Relative glycopeptide abundances were calculated using the area under the curve of the glycopeptide and normalized to its reference non‐glycosylated peptide as described previously. 13
2.4. Data processing and statistical analysis
Instrument reproducibility was monitored with commercially available digested serum samples (Sigma‐Aldrich) as quality controls. The digested serum pool was used as a quality control for monitoring MRM transitions every 10 samples for a total of 20 technical replicates. A hierarchical clustering was then performed using the relative abundance of all glycopeptides with average linkage to detect any outlier samples. Samples were determined to be outliers at a branch cutoff at height 15.5 using hierarchical clustering and were removed from analysis. The coefficient of variation (CV) was calculated for all glycopeptides based on these pooled serum samples. Glycopeptides that were below the limit of quantitation for the instrument or with CV >30% were excluded from analysis. The naming convention of the glycopeptides used throughout the text follows the convention, Protein_Glycosite_Glycan. For example, IgG1_297_5510 reads immunoglobulin G1 at glycosite N297 with N‐glycan composition of Hex(5)HexNAc(5)Fuc(1)Neu5Ac(0).
The fractions of sialylated and fucosylated peptides were calculated as the sum of relative glycopeptide abundances of non‐, mono‐, di‐, or poly‐glycosylated peptides relative to that of the total peptides. Relative glycopeptide abundances of individual glycopeptides and the fractions of sialylated and fucosylated peptides were used in subsequent analyses. All statistical analyses were performed using the R statistical package (R version 4.1.0). Linear models were constructed for univariate and multivariate analysis that included each potential covariate separately. Each simple multivariate model had one covariate at a time: glycopeptide abundance ∼ Diagnosis + covariate. These simple multivariate models were compared to the univariate model, which included diagnosis group as the only variable to evaluate the impact of these covariates on glycopeptide concentrations. Potential covariates included age, sex, ethnicity, presence of the APOE ε4 allele, and BMI. Linear models with interaction terms were used to analyze the sex‐by‐AD diagnosis interaction and ethnicity‐by‐AD diagnosis interaction: glycopeptide abundance ∼ diagnosis + sex + diagnosis * sex or glycopeptide abundance ∼ diagnosis + ethnicity + Diagnosis * ethnicity. For sex‐by‐AD diagnosis interaction, a post hoc analysis was done to estimate the sex‐specific diagnosis effects on serum glycopeptide abundance.
Differential expression analysis was performed as linear models using the limma package in R. Coefficients related to diagnosis were tested according to the null hypothesis (being zero) using t‐tests moderated in a Bayesian fashion. Raw P‐values were then adjusted for multiple hypothesis testing using the Benjamini‐Hochberg (BH) correction. The differences in glycopeptide abundance between normal control and AD are presented as natural log fold‐change.
A partial least‐squares discriminant analysis (PLS‐DA) was performed to assess the discriminatory potential of serum glycopeptides. We scaled glycopeptide abundances to the variance of 1 and conducted leave‐one‐out cross‐validation (LOOCV) to identify the best number of latent components using 1 through 20 with the train() function from the caret package. The importance of independent variables in the PLS‐DA model was measured using variable importance in projection (VIP) scores. Spearman's correlation analysis was used to evaluate the association between glycopeptides, cognitive scores, Spanish and English Neuropsychological Assessment Scales (SENAS) scores and clinical dementia rating (CDR).
3. RESULTS
A hierarchical clustering was first performed to target outlier samples that can result in false positives. The hierarchical clustering results show three outlier samples that were excluded from all further analysis (B10, C13, and A67) as shown in Figure S2. CVs were calculated for all 372 glycopeptides monitored in this study and their distribution is listed in Figure S3. Of the 372 glycopeptides monitored, 14 glycopeptides had CV values greater than 30% as listed in Table S2.
3.1. Fucosylated and sialylated glycopeptides in AD
We grouped glycopeptides by their fucosylation and sialylation status separately and assessed whether the fucosylation and sialylation status of glycopeptides discriminates serum samples from AD patients and normal controls by looking at patterns of fucosylated and sialylated glycopeptides generated from PLS‐DA analysis. The fucosylation and sialylation status of glycopeptides did not explicitly separate AD samples from normal controls with PLS‐DA models (Figure 1A,C). However, the models show that several changes in overall sialylation and fucosylation of serum glycoproteins may be indicators of AD disease state. The PLS‐DA model for fucosylation status had 12 glycopeptides with VIP scores >1.5. A VIP score >1.5 is considered to enable discrimination between groups. Notably, non‐fucosylated and mono‐fucosylated immunoglobulin G1 (IgG1) and IgG2 had high contributions to the model with VIP score >2, indicating the fucosylation status of IgG1 and IgG2 may be potential biomarkers for AD (Figure 1B). The PLS‐DA analysis for sialylation identified 19 glycopeptides with VIP scores >1.5, which includes inflammation response glycoproteins such as Kininogen‐1 (KNG1) and immune response glycoproteins such as Complement factor I (CFAI) as well as lipid metabolism glycoproteins Apolipoprotein C3 (APOC3) and Clusterin (CLUS) (Figure 1D).
FIGURE 1.
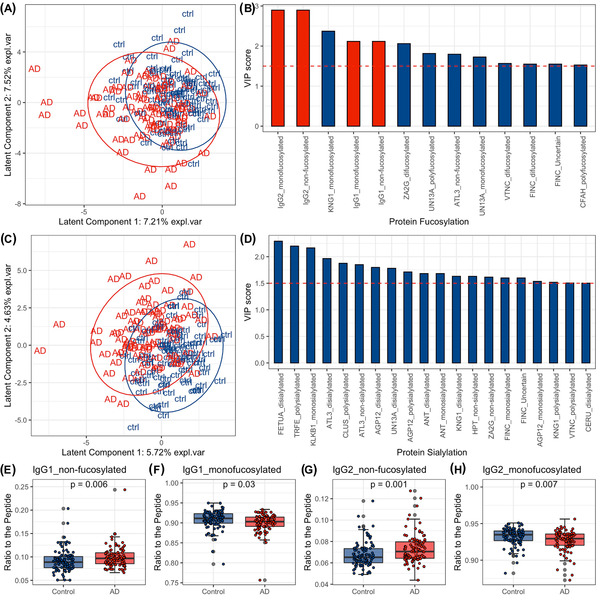
Changes in fucosylation and sialylation status in serum samples from patients with clinically diagnosed Alzheimer's disease (AD) compared to controls (ctrl) using partial least‐squares discriminant analysis (PLS‐DA). (A) The scores plot shows the distribution of subjects across latent components 1 and 2 given the PLS‐DA model for fucosylation status. (B) Variables with variable importance in projection (VIP) scores ≥1.5 and their VIP scores in the PLS‐DA model for fucosylation status. Variables colored in red are those selected for differential analysis and are visualized in E‐H. (C) The scores plot shows the distribution of participants across latent components 1 and 2 given the PLS‐DA model for sialylation status. (D) Variables with VIP scores ≥1.5 and their VIP scores in the PLS‐DA model for sialylation status. (E‐H) Boxplots showing the differences in abundance of non‐fucosylated and mono‐fucosylated immunoglobulin G1 (IgG1) and IgG2 proteins from control and AD samples
3.2. Identification of aberrantly fucosylated or sialylated glycopeptides in AD
The PLS‐DA analysis for fucosylation and sialylation showed that AD and control samples were located in two clusters in the scores plot with some overlap. We then identified fucosylated and sialylated glycopeptides that drive the discrimination for AD versus normal controls using differential analysis. For fucosylation, six statistically significant glycoproteins, which correspond to 10 glycopeptides, differed (P‐values < .05) between AD and normal controls in the univariate model, including glycopeptides with a high contribution to the fucosylation PLS‐DA model (non‐fucosylated IgG1, monofucosylated IgG1, non‐fucosylated IgG2, mono‐fucosylated IgG2, and so on) (Figure 1E‐H and Figure S4A). For sialylation, eight proteins with 10 glycopeptides differed between AD and normal controls (Figure S4B).
3.3. Glycopeptide signatures of AD
We then used PLS‐DA analysis to model the individual glycopeptide data and found a separation between the AD patients and normal controls, although as with the fucosylation and sialylation data there was overlap between the groups (Figure 2A). The model had 51 glycopeptides that had a VIP score >1.5, including proteins involved in lipid metabolism (APOB, APOC3, APOH), immunity (IgG1, IgG2, IgM), and inflammation response (CO3, CFAI, VTNC, ANT) (Figure 2B). We then determined whether there were significant differences between patients and controls in the abundance of the glycopeptides that had a high contribution to the PLS‐DA model using differential analysis. CFAI_494_5402 and CO3_85_5200 were significantly increased and APOB_983_5401 was significantly decreased in the AD patients compared to controls (P‐values < .05). The three glycopeptides are all non‐fucosylated (Figure 2C‐E).
FIGURE 2.
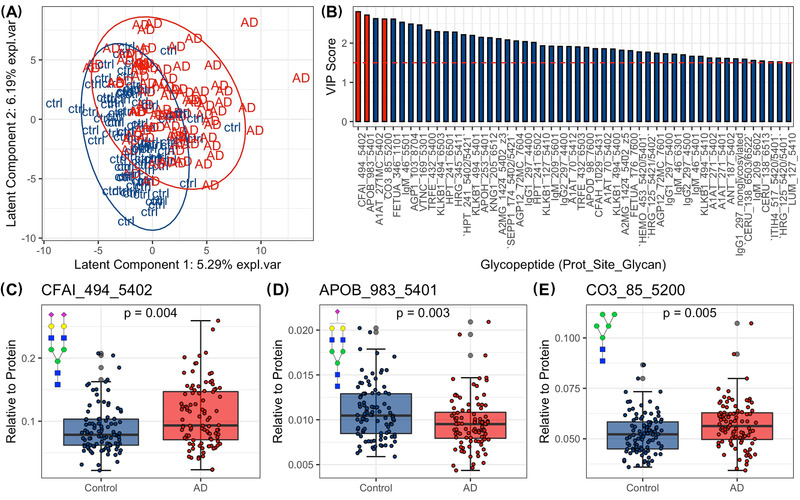
Changes in serum glycopeptide abundance in serum samples from Alzheimer's disease (AD) patients compared to controls using PLS‐DA. (A) The scores plot shows the distribution of participants across latent components 1 and 2 given the PLS‐DA model for glycopeptides. (B) Variables with VIP scores ≥1.5 and their VIP scores. Variable colored in red are those selected for differential analysis and are visualized in C‐E. (C‐E) Boxplots showing the differences in abundances of glycopeptides from control and AD samples. Protein complement factor I (CFAI), apolipoprotein B (apolipoprotein B) and Complement C3 (CO3). N‐glycan symbol key: yellow circles, galactose (Gal); green circles, mannose (Man); blue squares, N‐acetylglucosamine (GlcNAc); red triangles, fucose (Fuc); purple diamonds, N‐acetylneuraminic acid (Neu5Ac)
3.4. Identification of aberrant glycopeptides in AD
We next performed differential analysis to identify which glycopeptide abundances differed in the serum of AD patients versus normal controls. With the univariate model, among the 372 glycopeptide transitions monitored accounting for 53 of the 57 glycoproteins, 35 glycopeptide abundances were altered in individuals with AD, including 19 upregulated glycopeptides and 16 downregulated glycopeptides (P‐values < .05, Figure 3A). Among the 35 glycopeptides that were differentially expressed in AD patients, most belong to proteins involved in immune function, including immunoglobulins and acute‐phase proteins such as alpha‐1‐antitrypsin (A1AT), alpha‐2‐macroglobulin (A2MG), alpha‐1‐acid‐glycoprotein (AGP1), alpha‐2‐HS‐glycoprotein (FETUA), and complement C3 (CO3). After adjusting for sex and age separately, these differences remain statistically significant. With the models adjusting for ethnicity, APOE ε4, and BMI separately, the majority of the differentially expressed glycopeptides remained statistically significant; however, new differences were also detected in these multivariate models as shown in the x‐axis of Figure 3A. In the final multivariate model, all covariates including sex, age, ethnicity, APOE ε4 status, and BMI were included. This analysis yielded 24 significant AD‐associated glycopeptides (Figure 3A). The glycopeptides with the highest VIP scores in the PLS‐DA model, such as CFAI_494_5402, APOB_983_5401, A1AT_271MC_5402, FETUA_346_1101, and so on, were also significantly different by differential expression analysis (P‐values < .05), indicating that they are important in distinguishing AD patients from controls.
FIGURE 3.
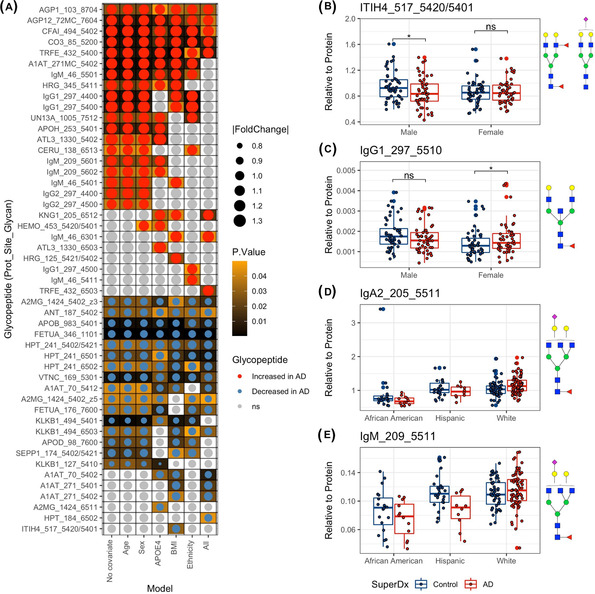
Differential analysis of glycopeptides and the impact of confounders on glycopeptide abundance. (A) The heatmap shows glycopeptides significantly different in Alzheimer's disease (AD) patients compared to controls using univariate and multivariate linear regressions. The dot size shows the effect size (absolute fold change). Darker background color refers to smaller P‐value (before multiple testing correction). (B‐C) The boxplot shows the sex specific diagnosis effects on serum glycopeptide abundance. Differences with P‐value < .05 are denoted by asterisk, whereas differences with P > .05 are denoted as “ns” (not significant). (D‐E) Boxplot showing the ethnicity specific diagnosis effects on serum glycopeptide abundance. Protein abbreviations: Inter‐alpha‐trypsin inhibitor heavy chain H1 (ITIH), immunoglobulin G1 (IgG1), immunoglobulin A2 (IgA2), and immunoglobulin M (IgM). N‐glycan symbol key: yellow circles, galactose (Gal); green circles, mannose (Man); blue squares, N‐acetylglucosamine (GlcNAc); red triangles, fucose (Fuc); purple diamonds, N‐acetylneuraminic acid (Neu5Ac)
3.5. Potential confounders
Factors including sex, ethnicity, age, BMI, and APOE ε4 genotype had impacts on glycopeptide abundances in serum. Sex and ethnicity were the two major confounders that influenced glycopeptide abundance in controls and AD. We analyzed the contributions of sex and ethnicity on serum glycopeptide abundances as covariates in the linear regression models. Among 372 glycopeptides, 135 glycopeptide abundances differed by sex, including 50 that were significantly higher in females and 85 that were significantly higher in males (adjusted P‐values < .05, Figure S5 A). Thirty‐eight glycopeptides significantly differed across ethnicity groups, where KLKB1_127_5410 had the lowest P‐value (adjusted P‐values < .05, Figure S5 B‐D). The impacts of other confounders are shown in Figure S5 E‐G.
Our study also provided the evidence of sex‐by‐AD diagnosis and ethnicity‐by‐AD diagnosis interactions. For example, ITIH4_517_5420/5401 was decreased in male AD patients versus controls (P‐value < .05) but was not different in female AD patients versus controls (Figure 3B). In contrast, IgG1_297_5510 was increased in AD female patients versus controls (P‐value < .05) but was not different in male patients versus controls (Figure 3C). For the ethnicity‐specific effects, diagnosis had a different effect in glycopeptides such as IgA2 _201_5511, IgM _209_5511 depending on the ethnicity of subjects (Figure 3D,E).
3.6. Associations between glycopeptides and clinical cognitive scores
Correlation analysis revealed associations between cognitive scores and glycopeptides that were important in discriminating AD patients and controls. For example, non‐fucosylated and mono‐fucosylated IgG2, the former of which was increased and the latter of which was decreased in AD, were positively and negatively correlated with the CDR (Figure S6, A&B), respectively. CO3_85_5200, which highly contributed to the PLS‐DA model and which was increased in AD patients versus controls, was negatively associated with semantic score, and positively associated with CDR (Figure S6, C&D).
4. DISCUSSION
In this study we set out to determine whether serum‐based glycopeptide analysis may be a useful approach for identifying novel actionable diagnostics for AD. Our results suggest that serum glycopeptide profiling is a promising approach for the development of new biomarkers. Even with this platform, which is not yet tailored for AD‐specific glycan markers, and instead is an application of an existing platform developed originally for cancer diagnostics, our results indicate that glycopeptide profiling can uncover important underlying disease mechanisms and point to future biomarker development. The importance of the fucosylation status of IgG in discriminating between controls and AD patients stood out as an important finding (Figure 1B). As the most abundant antibody in human blood, the importance of IgG in inflammation, infections, metabolic health, and autoimmunity is well established. 15 , 16 Specifically, an increased abundance of non‐fucosylated IgG1 and IgG2 was observed in AD patients compared with controls (Figure 1, E&G). Previous investigations have associated dementia with IgG N‐glycans; however, this previous work lacked site‐specific information and did not include a diverse cohort in the population selection. 17 The extent of IgG fucosylation and sialylation has been found previously to be associated with the pro‐ versus anti‐inflammatory signaling of IgG, with non‐fucosylated and non‐sialylated IgG preferentially binding to pro‐inflammatory Fcγ receptors. 18 In addition, PLS‐DA analysis of glycopeptides was able to separate AD patients from controls (Figure 2A) and proteins with VIP score >1.5 also involved immunoglobulins IgG1, IgG2, IgA, and IgM, among others (Figure 2B). Together our results highlight the dysregulation of glycosylation of immune response proteins, particularly immunoglobulins, in AD pathology. Other glycopeptides with VIP score >1.5 included proteins involved in lipid metabolism (APOB, APOC3, APOH) and inflammation response (CO3, CFAI, VTNC, ANT) (Figure 2B). For example, the mono‐sialylated glycopeptide APOB_N983_5401 was decreased in AD patients compared to controls (Figure 2D). Previous studies have shown a relationship between the glycosylation state of lipoprotein‐associated proteins and the pro‐inflammatory capacity and functionality of their lipoprotein carrier. 19 , 20
The univariate model showed 35 altered glycopeptide abundances in AD patients from the 372 glycopeptide transitions monitored (Figure 3A). All altered proteins are involved in immune function and immune response, including immunoglobulins and acute phase proteins. Of interest, all immunoglobulin glycopeptides were observed to be increased in AD patients. The study cohort involved equal numbers of males and females, and equal average age in both groups; thus when adjusting for sex and age, all glycopeptide remained statistically significant as expected. Age and sex are the covariates that are typically accounted for in biomarker discovery; however, our study suggests that ethnicity is also an important variable that can contribute to variability (Figure 3D&9). Research on racial disparities in biomarkers for AD has shown that analyses of molecular biomarkers of AD should adjust for race. Given the low sample size for participants of different ethnic groups in this preliminary study, we are not able to fully explore the contribution of ethnicity to the variability in serum glycopeptide profiles. However, our results indicate that serum glycoprofiling may be an exceptionally useful tool for the development of biomarkers in a disease such as AD, in which ethnic‐specific differences in disease pathophysiology and biomarker profiles are marked. For instance, a differential effect on total tau and phosphorylated tau181 has been shown to differ in African American individuals compared to White individuals. 21 Our data revealed that certain glycopeptides such as KLKB1_127_5410 strongly distinguished between AD patients and controls only in African American participants but not in White or Hispanic participants, highlighting the potential for the development of glycan‐based diagnostics that have the ability to identify individuals at risk of developing AD across a broad diversity of individuals (Figure S5, B‐D).
Of note, our data revealed that non‐fucosylated IgG2 was positively correlated with CDR while mono‐fucosylated IgG2 was negatively correlated with CDR (Figure S6 A and B). This result suggests a direct relationship between immunoglobulin fucosylation and clinical measures of cognitive function loss. Likewise, a non‐sialylated, non‐fucosylated glycopeptide of CO3 was also positively correlated with CDR (Figure S6 C), highlighting the loss of fucosylation and sialylation at specific sites of immune‐related proteins as a potential mediator and/or biomarker of AD. Major challenges in the development of blood‐based biomarkers for AD include a high degree of patient heterogeneity and the potential existence of multiple clinical phenotypes. 22 , 23 , 24 It is important to note that new biomarkers are needed not just for diagnostic purposes but also for determining prognosis and monitoring therapeutic efficacy. In this pilot work, we sought to determine whether serum‐based glycoprofiling may be a useful approach to address these critical questions in the field. Our work for the first time utilized a previously developed MRM analytical method to monitor glycan alterations in serum glycoproteins in a site‐specific manner to determine whether serum glycoprofiling can discriminate between AD patients and controls, but also simultaneously point to potential therapeutic approaches to prevent, treat, or reverse AD. Our results suggest that serum glycoprofiling may indeed be a powerful biomarker discovery platform that can be harnessed to identify new markers in diverse cohorts and new therapeutic targets, particularly those focused on immune‐related mechanisms. Future studies including a larger sample size, particularly in ethnically diverse cohorts powered to account for age, sex, and ethnicity, are needed to develop glycan‐based biomarkers for AD. Further work to refine the transitions monitored by the MRM method that are specific to AD would improve the sensitivity and specificity of the biomarkers.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Designed the study: Lee‐Way Jin, Angela M. Zivkovic, Carlito B. Lebrilla, Jennyfer Tena, Danielle Harvey. Supervised the study: Angela M. Zivkovic, Carlito B. Lebrilla. Performed Experiments: Jennyfer Tena, Qingwen Zhou. Performed statistical analysis: Xinyu Tang, Jennyfer Tena, Danielle Harvey. Analyzed data: Jennyfer Tena, Qingwen Zhou, Maria Barajas‐Mendoza, Xinyu Tang. Interpreted data: Angela M. Zivkovic, Jennyfer Tena, Xinyu Tang, Carlito B. Lebrilla. Wrote the manuscript: Angela M. Zivkovic, Xinyu Tang, Carlito B. Lebrilla, Jennyfer Tena
Supporting information
SUPPORTING INFORMATION
SUPPORTING INFORMATION
SUPPORTING INFORMATION
SUPPORTING INFORMATION
SUPPORTING INFORMATION
SUPPORTING INFORMATION
SUPPORTING INFORMATION
ACKNOWLEDGMENT
Research was financially supported by the National Institute on Aging of the National Institutes of Health under Award Number R01AG062240. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Tena J, Tang X, Zhou Q, et al. Glycosylation alterations in serum of Alzheimer's disease patients show widespread changes in N‐glycosylation of proteins related to immune function, inflammation, and lipoprotein metabolism. Alzheimer's Dement. 2022;14:12309. 10.1002/dad2.12309
Jennyfer Tena and Xinyu Tang contributed equally.
REFERENCES
- 1. Association A . 2021 Alzheimer ’ s disease facts and figures Race, ethnicity and Alzheimer's in America. Alzheimers Dement. 2021;13(3):1‐104. [Google Scholar]
- 2. Jack CR, Bennett DA, Blennow K, et al. NIA‐AA research framework: toward a biological definition of Alzheimer's disease. Alzheimer's Dement. 2018;14(4):535‐562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging‐Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dement. 2011;7(3):263‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Molinuevo JL, Ayton S, Batrla R, et al. Current state of Alzheimer's fluid biomarkers. Acta Neuropathol. 2018. 10.1007/s00401-018-1932-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Akasaka‐Manya K, Manya H. The role of APP o‐glycosylation in Alzheimer's disease. Biomolecules. 2020;10(11):1‐14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schedin‐Weiss S, Winblad B, Tjernberg LO. The role of protein glycosylation in Alzheimer disease. FEBS J. 2014;281(1):46‐62 [DOI] [PubMed] [Google Scholar]
- 7. Schedin‐Weiss S, Gaunitz S, Sui P, et al. Glycan biomarkers for Alzheimer disease correlate with T‐tau and P‐tau in cerebrospinal fluid in subjective cognitive impairment. FEBS J. 2020;287(15):3221‐3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Q, Ma C, Chin LS, Li L. Integrative glycoproteomics reveals protein n‐glycosylation aberrations and glycoproteomic network alterations in Alzheimer's disease. Sci Adv. 2020;6(40):1‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kirmiz C, Li B, An HJ, et al. A serum glycomics approach to breast cancer biomarkers. Mol Cell Proteomics. 2007;6(1):43‐55 [DOI] [PubMed] [Google Scholar]
- 10. Ruhaak LR, Kim K, Stroble C, et al. Protein‐specific differential glycosylation of immunoglobulins in serum of Ovarian cancer patients. J Proteome Res. 2016;15(3):1002‐1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hong Q, Lebrilla CB, Miyamoto S, Ruhaak LR. Absolute quantitation of immunoglobulin G and its glycoforms using multiple reaction monitoring. Anal Chem. 2013;85(18):8585‐8593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong Q, Ruhaak LR, Stroble C, et al. A method for comprehensive glycosite‐mapping and direct quantitation of Serum glycoproteins. J Proteome Res. 2015;14(12):5179‐5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Q, Kailemia MJ, Merleev AA, et al. Site‐specific glycosylation quantitation of 50 serum glycoproteins enhanced by predictive glycopeptidomics for improved disease biomarker discovery. Anal Chem. 2019;91(8):5433‐5445 [DOI] [PubMed] [Google Scholar]
- 14. Wu Z, Serie D, Xu G, PB‐Net Zou J.: Automatic peak integration by sequential deep learning for multiple reaction monitoring. J Proteomics. 2020;223(October 2019):103820 [DOI] [PubMed] [Google Scholar]
- 15. Biermann MHC, Griffante G, Podolska MJ, et al. Sweet but dangerous—the role of immunoglobulin G glycosylation in autoimmunity and inflammation. Lupus. 2016;25(8):934‐942 [DOI] [PubMed] [Google Scholar]
- 16. Plomp R, Ruhaak LR, Uh HW, et al. Subclass‐specific IgG glycosylation is associated with markers of inflammation and metabolic health. Sci Rep. 2017;7(1):1‐10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang X, Yuan H, Lyu J, et al. Association of dementia with immunoglobulin G N‐glycans in a Chinese Han Population. NPJ Aging Mech Dis. 2021;7(1):1‐12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maverakis E, Kim K, Shimoda M, et al. Glycans in the immune system and The Altered Glycan Theory of Autoimmunity: a critical review. J Autoimmun. 2015;57:1‐13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krishnan S, Shimoda M, Sacchi R, et al. HDL glycoprotein composition and site‐specific glycosylation differentiates between clinical groups and affects IL‐6 secretion in lipopolysaccharide‐stimulated monocytes. Sci Rep. 2017;7(March):1‐15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu C, Wong M, Li Q, et al. Site‐specific glycoprofiles of HDL‐associated ApoE are correlated with HDL functional capacity and unaffected by short‐term diet. J Proteome Res. 2019;18(11):3977‐3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morris JC, Schindler SE, McCue LM, et al. Assessment of racial disparities in biomarkers for Alzheimer disease. JAMA Neurol. 2019;76(3):264‐273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hampel H, O'Bryant SE, Molinuevo JL, et al. Blood‐based biomarkers for Alzheimer disease: mapping the road to the clinic. Nat Rev Neurol. 2018;14(11):639‐652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tarawneh R. Biomarkers: our path towards a cure for Alzheimer disease. Biomark Insights. 2020;15. 10.1177/1177271920976367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Henriksen K, Bryant SEO, Hampel H, et al. The future of blood‐based biomarkers for Alzheimer ’ s disease. Alzheimer's Dement. 2014;10:115‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION
SUPPORTING INFORMATION
SUPPORTING INFORMATION
SUPPORTING INFORMATION
SUPPORTING INFORMATION
SUPPORTING INFORMATION
SUPPORTING INFORMATION


