Abstract
As neuroimagery datasets continue to grow in size, the complexity of data analyses can require a detailed understanding and implementation of systems computer science for storage, access, processing, and sharing. Currently, several general data standards (e.g., Zarr, HDF5, precomputed) and purpose-built ecosystems (e.g., BossDB, CloudVolume, DVID, and Knossos) exist. Each of these systems has advantages and limitations and is most appropriate for different use cases. Using datasets that don’t fit into RAM in this heterogeneous environment is challenging, and significant barriers exist to leverage underlying research investments. In this manuscript, we outline our perspective for how to approach this challenge through the use of community provided, standardized interfaces that unify various computational backends and abstract computer science challenges from the scientist. We introduce desirable design patterns and share our reference implementation called intern.
I. INTRODUCTION
In response to the growing number and size of large-scale volumetric neuroscience datasets, the community has developed a diverse set of tools and storage frameworks that balance ease of data manipulation and storage with efficiency and cost. These tools are often purpose-built, and feature team- or task-specific features that make them particularly well-suited for their host projects, such as version control, cloud-native implementations, efficient caching, multi-tier storage, targeted annotation or proofreading tasks and more [1], [2], [3], [4]. Historically, this has been advantageous, as it has enabled teams to develop tools quickly and effectively to address unique research challenges. This diverse ecosystem, however, has also led to community fragmentation and interoperability challenges because research organizations rely on standards for data storage and access that are often incompatible. As scientific questions continue to grow in ambition and scope, it is increasingly important that scientists are able to easily analyze, collaborate, and share their data using consistent formats and data-storage engines.
Though it is tempting to develop prescriptive data formats and standards, the fast-moving pace of the big-data neuroscience field — as well as the need for backward-compatibility with ongoing and past projects — will complicate the process of standardization. Instead, it is more feasible to standardize in abstraction: Rather than developing common data formats, it is more effective to build common data access strategies which can be applied to a variety of underlying datastores, file formats, and interfaces.
In response to collaborations that span data sizes from megabytes to petabytes, and that span institutional, international, and interdisciplinary boundaries from neuroscience to computer science to graph theory, interfacing tools are critical to reducing barriers for new and experienced scientists and enabling existing algorithms to scale to big data challenges. Data access toolkits and analysis tools (e.g., neuPrint [5], CloudVolume [6]) provide well-integrated solutions for their use cases.
We have developed intern, a Python client library for neuroscience data access. intern simplifies data transit between industry-standard data formats, and exposes a consistent and intuitive API for end-users so that code for an analysis performed on a dataset in a particular datastore format may be trivially ported to other datasets and datastores (i.e., ecosystems).
We explain our architecture and implementation details, and share several use cases common to scientific analysis which are simplified through the use of intern. We believe that this tool is helpful in providing seamless solutions when switching between cloud native, local, and file-based solutions, and offers an extensible software-design paradigm as new solutions are developed.
II. BACKGROUND
Most connectomics data management tools act as either a data-storage tool, which manages the (long-term) preservation of data, or a data-access tool – which enables an end user (whether human or automated) to access and interact with the data.
A. Data Storage Tools
Though many biological science disciplines rely on local, single-file data storage systems (e.g., HDF5, multipage TIFFs), the field of connectomics realized the need for reproducible, shareable, scalable datastores early in its evolution. These datastores are persistent, performant servers of volumetric data, and are often centralized into repositories holding information from multiple experiments and laboratories [2], [1], [3], [6], [4]. As the size of data increased, these datastores specialized in returning subvolumes of data based upon 3D user queries, rather than trying to transmit full datasets. Almost all of the most widely-used data storage tools now leverage chunked storage [7], an access-efficiency paradigm borrowed from domains such as astronomy and GIS [8]. This enabled databases to increase their bandwidth and serve more data-requests per second, because each subvolume could be accessed in parallel, reducing the file input/output and hard-drive read-speed bottlenecks.
Eventually, some datastores, including BossDB [1] and CloudVolume [6], moved to cloud storage systems such as Amazon AWS S3 [9] or Google Cloud Storage (GCS) [10]. These systems abstracted file-access even further and enabled high-speed network read- and write-operations, at the cost of renting — rather than owning — data storage space. While tools such as Knossos or DVID may be run on cloud resources as easily as on local compute infrastructure, other datastores such as BossDB are cloud-native, meaning that they fully leverage the scalability and parallelism of cloud-compute and serverless resources, and cannot be run on conventional compute hardware.
Data storage tools can be classified into two other large categories: Those with server-side compute resources, and those without. Tools like DVID, BossDB, and Knossos use devoted compute resources that perform functions such as mesh generation, cache management, and access-control authorization. Systems like CloudVolume or zarr-backed datastores require much simpler infrastructure to run, but cannot perform processes such as skeleton- or mesh-generation without client-side compute resources.
B. Data Access Tools
Some researchers may feel comfortable accessing data directly from one of the storage tools listed above (e.g., via RESTful services or object-level access), but most prefer to interact with the data through more familiar and intuitive interfaces, such as a Python library or a web interface. Almost every data storage tool mentioned above has its own devoted data access tool: DVID has Go and Python libraries; data stored in the precomputed format may be accessed with the cloud-volume Python library. BossDB may be accessed with either cloud-volume or intern Python libraries. A common frustration in the connectomics community is that with only a small handful of exceptions, though the underlying data may be the same in several data storage tools, most access tools are only capable of reading from their “partner” storage tool, and the interfaces vary in complexity and format. In order to integrate data and tools from our collaborators, we expanded our initial data access tool intern to support more data formats as well as more data storage systems, in a Resource-based system (Fig. 1). intern’s architecture was expanded to communicate with CloudVolume-accessible volumes, DVID-hosted datasets, and several other commonly-used data storage tools and formats.
Fig. 1.
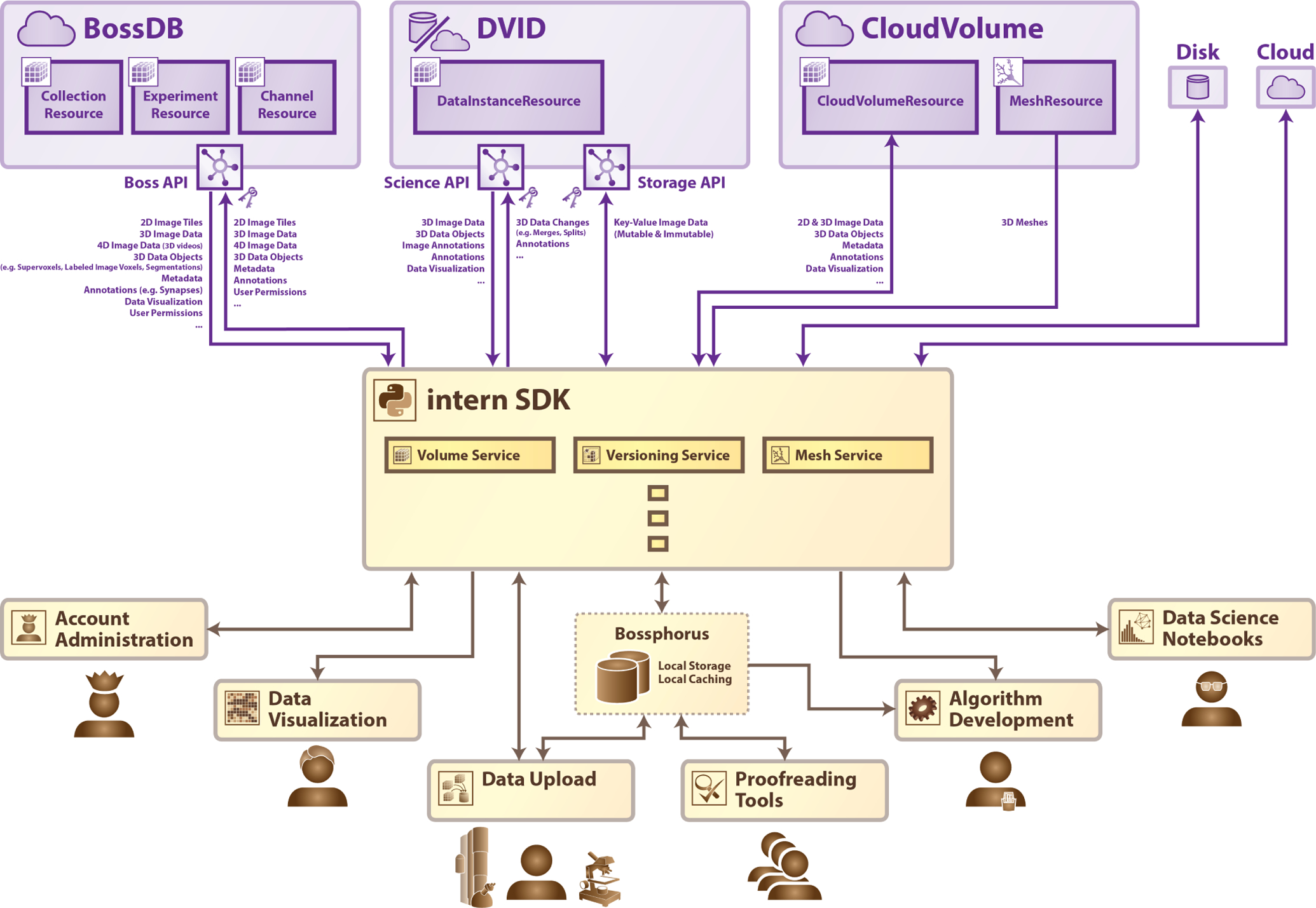
The intern Python library acts as a shock absorber to provide a consistent API to researchers, tool developers, and other users. A community-facing data-access tool should operate on all major data-storage systems (including CloudVolume, DVID, and BossDB), and remain flexible enough to enable common use cases (such as visualization, data upload/download, and data proofreading) without sacrificing performance.
Additionally, we believe that in order to enable cross-institutional collaboration in the community, it is important to bridge the gap between those data storage tools with server-side compute and those without. For this reason, we also introduced a Service-based system into intern that enables a user to run surrogates for the server-side processing tools of one data-storage system using the data from another. For example, we want a user to be able to request mesh representations of data from BossDB — a tool that supports server-side mesh generation — as well as from CloudVolume — a tool that supports client-side mesh generation — as well as from a dataset residing in a zarr archive in AWS S3 — a storage technique that does not support mesh generation at all. That these three tools differ in how their meshes may be generated should not matter to an end-user: The user should be able to use the same syntax to request mesh data from all of them with only minimal code changes.
Finally, we wrote intern to be easily extended to additional use cases and features as scientific needs grow. We believe the underlying design principles are common to many research questions and have value beyond the specific implementation described here.
C. The Connectomics Data-Access Ecosystem
When considering the data storage engine for a particular scientific question, several different factors should be considered, which we summarize as data size, versioning, authentication and user management, cloud services, performance, and accessibility/sharing. Each tool has a user community and powerful feature-sets: File-based solutions are simple and easily portable and understood, but are difficult to access and analyze by communities. CloudVolume excels in portability and simplicity, but does not provide user accounts, differential permissions, or data management services. DVID offers an excellent solution for terascale solutions and fast, efficient data-versioning, but does not leverage cloud-scale capabilities. BossDB is a managed cloud-native service with user permissions, access control, and a robust storage engine tested to hold and process petabytes of data, but cannot run locally and requires more infrastructural complexity than many research labs may have the expertise to maintain.
Although for the uninitiated these storage solutions may seem to introduce unnecessary complexity, managing and manipulating such large datasets and corresponding analysis derivatives (e.g., metadata) requires advanced technology. The intent of the paradigm introduced in this paper is to abstract from the user all of the challenges introduced by the scale of the data in order to allow methods to be easily run on these data while minimizing impedance mismatches.
III. METHODS
Our intern library implements the philosophy of abstracting computer science requirements by offering consistent data access trait interfaces, which are categorized into Services, Resources, and Remotes. This system of abstraction acts as a shock absorber to differing data formats, data processing, and tool functionality, and serves to enable reproducible and extensible connectomics analysis. We describe our intern reference implementation, and explore how other tool-developers may choose to expand intern or develop their own community-ready software using the same paradigm.
A. Architecture
As the field of connectomics evolves rapidly, a library must strike a balance between accessibility and adaptability. We designed our toolkit such that even minimal coding skills and copy-pasting of simple design patterns can be leveraged to reduce user burden. As the community continues to formalize use cases and data storage paradigms, programmatic workflows like SABER [11], LONI [12], Luigi [13], or other workflow managers [14], [15], [16] may allow for additional simplification and can directly leverage these functions. Point and click graphical interfaces may also follow.
In order to facilitate extension of the intern Python library by the community, we have published extensive online documentation for both software engineering beginners as well as professionals. The library is split into three types of trait-based interfaces; Remotes, Resources, and Services.
1). Remotes:
Remotes represent data storage tools, such as databases, on-disk chunked or non-chunked files, and other providers of volumetric-data access APIs. A Remote must at least allow the retrieval of volumetric data, and may allow upload, manipulation, user permissions, or project management as well.
2). Resources:
Resources are pointers to atomic units or groupings of data from a Remote. For example, in the hierarchical BossDB data paradigm, the BossResource implementation interfaces with a CollectionResource, an ExperimentResource and a ChannelResource [1]. In the DVIDRemote implementation, a DataInstanceResource points to a specific dataset at a specific version in its history.
3). Services:
Services are features or manipulations that act upon data retrieved from a Remote. Services either call upon the server-side compute of a Remote, or instead a Service may implement a standalone local algorithm that can act as a surrogate for a Remote that does not have a service available. For example, a CloudVolumeRemote has an associated CloudVolumeMeshService that invokes the built-in cloud-volume meshing functionality, but a ZarrRemote may use a locally-executed MeshService with the same API.
Provided the underlying data are the same, the output from different Services will be consistent (give-or-take obvious differences in performance/timing or scalability, as well as differences in parameters). In this way, raw image data from any database (i.e. Remote) can be treated the same; segmentation from any database can be treated the same; and annotation byproducts can be treated the same.
B. Performance and Design Considerations
Due to the chunked-storage approach used by large-scale volumetric datastores, intern was developed with an intention to maximize the performance of the underlying datastores by optimizing Remote-specific parameters such as chunk size, parallelism, and data compression. Chunk size, or the amount of data downloaded in a single request, has a significant effect on end-to-end performance for downloading and uploading data. Default chunk size for BossDB downloads were empirically derived to reduce latency for the user. Ideally, data chunk sizes should be optimized per-remote, and ideal cutout sizes may vary based upon client resources such as network throughput or compute (Fig. 2). Furthermore, intern utilizes parallel downloading in order to saturate client bandwidth without causing bottlenecks at the data decompression or reconstitution stage. Convenience features consolidates the extract, load, and transform data pipeline into a few straightforward functions.
Fig. 2.
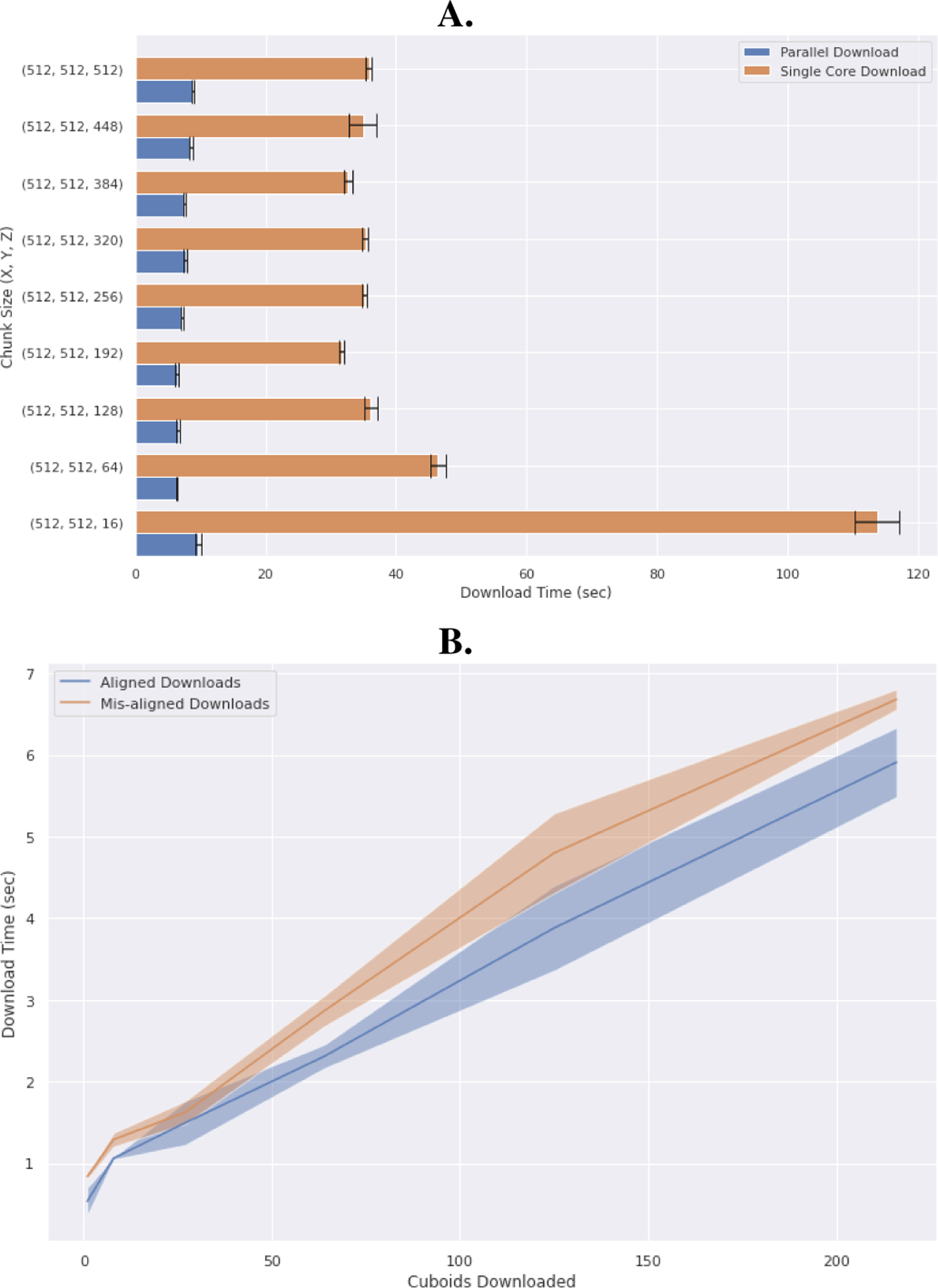
Many factors impact data download rate. As an illustration, we tuned the chunk-size parameters for parallel- and non-parallel downloads from the BossDB remote. A. Performance was impacted by client-side compute speed (for data decompression) as well as network throughput, illustrating possible avenues for further abstraction of other remotes. B. Chunked data stores benefit from data requests that are aligned to the cuboid subdivisions in the server backend. This effect is more pronounced in filesystem-based data-stores such as CloudVolume or Zarr, as the cuboid periphery must be downloaded and cropped on local compute resources. In contrast, data stores with cloud-side compute (such as DVID or BossDB) can perform this cropping operation prior to data download, and so the additional egress burden is not incurred.
C. Use Cases
1). Transferring data between Remotes:
Since Remotes provide unique task-specific capabilities that are exclusive to a particular data store or data type, a common use-case of intern is to transfer data between remotes to leverage their unique capabilities.
For example, DVID provides best-in-class data-versioning of large scale image segmentation, and it may be preferable to use DVID for this sort of data-versioning rather than try to replicate this feature in other datastores. Volumetric data that is stored in, e.g., BossDB can be downloaded from the cloud for local processing and uploaded into a DVID repository using intern. Once the proofreading is completed, the final annotated data can be re-uploaded to BossDB in order to be cached internationally and served publicly. We note that certain data storage tools, such as BossDB, have high-throughput ingest systems available [1], which may be faster than an intern-based data transfer, but which are not available in all remotes. Additionally, users may wish to only work with a portion of a dataset, in which case ingestion services may be overly complex.
2). Shock-Absorption:
Though such software abstractions place an additional engineering burden on developers, we assert that developing flexible, ecosystem-agnostic tools is a fundamental need of the dynamic connectomics community in lieu of more formal data-standards. To meet this requirement, we developed intern with such flexibility in mind: intern acts as a “shock-absorber” for common connectomics use-cases by implementing database-agnostic Services (e.g. mesh generation, skeleton generation, segmentation proofreading), which can run regardless of data source. An intern Service definition includes a list of its required Resources, and any Remote or other data-source that meets this interface can run the Service.
As a concrete example, a local marching-cubes MeshService converts 3D segmentation to OBJ- or precomputed-formatted meshes. This Service requires only a VolumeResource provider, and so it can run on, for example, a BossRemote, a CloudVolumeRemote, or even, e.g., on a raw ZarrVolumeResource.
This approach enables the end-user to reproducibly run the same analysis code, changing only one line to specify from where the data should be pulled. In other words, a user may confidently change a line of code from
BossRemote#mesh(id)
to
DVIDRemote#mesh(id)
, regardless of whether the data-sources themselves support the meshing operation. The provenance of these operations may be stored alongside the data products, in order to aid in future reproducibility.
3). Local Data Caching:
Like many projects in the big-data neuroscience community, one of the most painful bottlenecks in much of our work is the speed with which data can be uploaded and downloaded from user-facing machines for visualization and analysis. In order to mitigate this challenge, we developed Bossphorus, a data relay that uses intern to fetch data from “upstream” data storage tools in their respective dialects and which serves data “forward” in the BossDB-flavored REST API dialect [1]. As a result, Bossphorus instances can be daisy-chained as a multi-tier cache. This enables an end-user to quickly browse data from a variety of sources with low latency, even if the datastore in question does not support caching. With a Bossphorus instance running locally using our publicly available Docker image, or a Bossphorus instance running on on-premise hardware at an academic institute (or indeed with both running in series), a user can interactively browse large volumes of data from multiple data sources with sub-second latency. This enables realtime data manipulation and visualization. intern’s interfaces are designed to be highly compatible with common data-science tools like numpy[17] and pandas[18]; popular data standards like DataJoint [16]; as well as visualization tools such as neuroglancer[19], substrate[20], matplotlib[21], and plotly[22].
4). Processing:
Tool and algorithm developers commonly target specific data storage ecosystems in order to reduce the burden of supporting several disparate ecosystems and data-standards [23], [11]. By leveraging shock-absorber tools like intern, algorithm developers can write code once and deploy it to a variety of datastores. As a proof of concept, we adopted a synapse-detection algorithm based upon the U-net architecture [11], [24]. This algorithm Service targets data downloaded from an intern VolumeResource, which means that it is trivially portable to data downloaded from any supported volumetric data storage service. One advantage of tightly coupling volumetric data access with such machine learning algorithms is addressing the challenge of stitching subvolumes of data together. Using intern for data management helps address this problem by storing data products in the cloud, rather than in task-specific cache files or on users’ drives.
Just as tool designers can use intern to develop and test their software, the intern Python library is production-ready, and is verified to work at petabyte scale. We believe that reproducible and repeatable algorithm design extends past tool-design, and continues to be a fundamental aspect of responsible computational science in public-facing research. Flexible tools like intern equip peer institutes and collaborating researchers with the ability to quickly and accurately reproduce, verify, and build upon scientific claims.
5). Visualization and Meshing:
The Remote, Resource and Service based architecture allows all Remote data-stores to benefit from all implementations of Services. An example of this is intern’s MeshService, which allows users to generate meshes using local compute resources. Any Remote that implements volumetric data retrieval as a VolumeResource (namely, all currently implemented Remotes) will automatically have this meshing capability. Due to this trait-based architecture, any future Remote implementation for new databases or data standards will likewise have this meshing capability without any further development required.
Any Service can also be used independent of the rest of the intern library. The MeshService described above, for example, will produce a Mesh object when passed a volume of 3D data either as an ndarray or as a VolumeResource (Fig. 3). This mesh object can then be converted into the common obj format or into the Neuroglancer precompute format [19].
Fig. 3.

Using the intern library, we downloaded nanometer-resolution 3D imagery and pixel segmentation from the public Witvliet 2020 et al. dataset on BossDB [25]. We then used the meshing service to produce 3D mesh files for visualization in 3D software such as Blender or Neuroglancer. Pictured here is Dataset 2, a C. elegans nematode brain imaged during the L1 larval stage.
While we have provided a simple, Python-native, CPU-based marching-cubes implementation, the user community is encouraged to package other specialized or distributed meshing or post-processing algorithms in the same Service-based class interface. Tools written to meet this specification will likewise be applicable to data from any data storage tool supported by the tool.
IV. DISCUSSION
In this work we highlight data accessibility, a common challenge in contemporary computational neuroscience, which has become particularly acute as data volumes grow in size and data ecosystems proliferate. New and experienced users will benefit greatly by adopting the concept of a computer science shock-absorber, which we illustrate in our solution (intern). Such tools are particularly valuable in domains such as connectomics, where cross-institutional collaborations and data reuse are not only common but increasingly necessary. Other complementary APIs and software libraries also exist to support approaches in the field and are well-suited for particular ecosystems and workflows. Many of these tools offer solutions that abstract many of the most challenging and repetitive aspects of large scale neuroscience discovery and also avoid common errors of interpretation. This is especially important to broaden accessibility of large, publicly-funded datasets for secondary analysis, including by new members of the community or those with complementary expertise (e.g., machine learning, statistical modeling). This work directly addresses the retrieval of volumetric data products but not object-level metadata such as synapse or neuron attributes, or the algorithms used to create derivative data products; these aspects are also important to consider when building standardized analysis workflows. Furthermore, as the user demand for such tools increases, we will continue to mature implementations of intern in other commonly used data science languages, such as Julia, R, and Node.
By developing user-facing tools such as intern that are flexible and provide an integrated interface to key community data storage systems, the connectomics community will be able to greatly benefit from shared, collaborative science, as well as large-scale, public, easily-accessible data.
ACKNOWLEDGMENT
We thank Joshua Vogelstein and Randal Burns for discussions that informed the design and vision for this project. We would also like to acknowledge Dean Kleissas for his support in the initial design and code of the intern library and the BossDB system. We would like to thank Cora Mentor Roy and Kevin Romero for their help with usability testing and community resource development. This manuscript has been released as a pre-print at https://doi.org/10.1101/2020.05.15.098707 [26].
This material is based upon work supported by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA), via IARPA Contract No. 2017-17032700004-005 under the MICrONS program. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the ODNI, IARPA, or the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright annotation therein. Research reported in this publication was also supported by the National Institute of Mental Health of the National Institutes of Health under Award Numbers R24MH114799 and R24MH114785. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was completed with the support of the CIRCUIT Office www.circuitinstitute.org, and JHU/APL Internal Research Funding.
Footnotes
Supplemental Data
Architecture details and a user guide for the intern library can be found at https://bossdb.org/tools/intern.
Data Availability Statement
The datasets analyzed for this study can be found at bossdb.org. The code used to demonstrate intern can be found at bossdb.org/tools/intern.
References
- [1].Kleissas D, Hider R et al. , “The block object storage service (bossDB): A cloud-native approach for petascale neuroscience discovery,” bioRxiv, p. 217745, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Katz WT and Plaza SM, “DVID: Distributed Versioned Image-Oriented Dataservice,” 2019. [DOI] [PMC free article] [PubMed]
- [3].Helmstaedter M, Briggman KL, and Denk W, “High-accuracy neurite reconstruction for high-throughput neuroanatomy,” Nature Neuroscience, vol. 14, no. 8, p. 1081–1088, Jul 2011. [DOI] [PubMed] [Google Scholar]
- [4].Saalfeld S, Cardona A, Hartenstein V, and Tomancak P, “Catmaid: collaborative annotation toolkit for massive amounts of image data,” Bioinformatics, vol. 25, no. 15, p. 1984–1986, Apr 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Clements J, Dolafi T et al. , “neuprint: Analysis tools for em connectomics,” bioRxiv, Jan 2020. [Google Scholar]
- [6].Silversmith W, “Cloudvolume: client for reading and writing to neuroglancer precomputed volumes on cloud services.”
- [7].Vogelstein JT, Perlman E et al. , “A community-developed open-source computational ecosystem for big neuro data,” Nature Methods, Oct 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Soroush E, Balazinska M, and Wang D, “Arraystore,” International Conference on Management of Data - SIGMOD, 2011. [Google Scholar]
- [9].Amazon Web Services, Inc., “Amazon simple storage service (s3),” Retrieved May 2021, https://aws.amazon.com/s3/.
- [10].“Cloud Storage: Object Storage,” Retrieved May 2020, https://cloud.google.com/storage.
- [11].Johnson EC, Wilt M et al. , “Toward a reproducible, scalable framework for processing large neuroimaging datasets,” Gigascience, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rex DE, Ma JQ, and Toga AW, “The loni pipeline processing environment,” NeuroImage, vol. 19, no. 3, p. 1033–1048, Jul 2003. [DOI] [PubMed] [Google Scholar]
- [13].Spotify, “Luigi,” Retrieved Feb 2020, https://github.com/spotify/luigi.
- [14].Apache, “Airflow,” Retrieved June 2018, https://airflow.apache.org.
- [15].Amazon Web Services, Inc., “AWS Batch,” Retrieved June 2018, https://aws.amazon.com/batch/.
- [16].Yatsenko D, Reimer J et al. , “Datajoint: managing big scientific data using matlab or python,” bioRxiv, Nov 2015. [Google Scholar]
- [17].Oliphant TE, A guide to NumPy. Trelgol Publishing; USA, 2006. [Google Scholar]
- [18].McKinney W et al. , “Data structures for statistical computing in python,” in Proceedings of the 9th Python in Science Conference, vol. 445. Austin, TX, 2010, pp. 51–56. [Google Scholar]
- [19].Maitin-Shepard J, “Neuroglancer,” Retrieved Sept 2018, https://github.com/google/neuroglancer.
- [20].Matelsky JK, Downs J et al. , “A substrate for modular, extensible data-visualization,” Big Data Analytics, vol. 5, no. 1, Feb 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hunter JD, “Matplotlib: A 2d graphics environment,” Computing in science & engineering, vol. 9, no. 3, pp. 90–95, 2007. [Google Scholar]
- [22].Plotly Technologies Inc., “Collaborative data science,” Montreal, QC, 2015, https://plot.ly. [Google Scholar]
- [23].Wu J, Silversmith WM, Lee K, and Seung HS, “Chunkflow: hybrid cloud processing of large 3D images by convolutional nets,” Nature Methods, pp. 1–2, 2021. [DOI] [PubMed] [Google Scholar]
- [24].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” MICCAI, 2015. [Google Scholar]
- [25].Witvliet D, Mulcahy B et al. , “Connectomes across development reveal principles of brain maturation in c. elegans,” bioXriv, Apr 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Matelsky JK, Rodriguez L et al. , “intern: Integrated Toolkit for Extensible and Reproducible Neuroscience,” BiorXiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed for this study can be found at bossdb.org. The code used to demonstrate intern can be found at bossdb.org/tools/intern.


