Abstract
Candida glabrata has been often isolated from AIDS patients with oropharyngeal candidiasis treated with azole antifungal agents, especially fluconazole. We recently showed that the ATP-binding-cassette (ABC) transporter gene CgCDR1 was upregulated in C. glabrata clinical isolates resistant to azole antifungal agents (D. Sanglard, F. Ischer, D. Calabrese, P. A. Majcherczyk, and J. Bille, Antimicrob. Agents Chemother. 43:2753–2765, 1999). Deletion of CgCDR1 in C. glabrata rendered the null mutant hypersusceptible to azole derivatives and showed the importance of this gene in mediating azole resistance. We observed that wild-type C. glabrata exposed to fluconazole in a medium containing the drug at 50 μg/ml developed resistance to this agent and other azoles at a surprisingly high frequency (2 × 10−4 to 4 × 10−4). We show here that this high-frequency azole resistance (HFAR) acquired in vitro was due, at least in part, to the upregulation of CgCDR1. The CgCDR1 deletion mutant DSY1041 could still develop HFAR but in a medium containing fluconazole at 5 μg/ml. In the HFAR strain derived from DSY1041, a distinct ABC transporter gene similar to CgCDR1, called CgCDR2, was upregulated. This gene was slightly expressed in clinical isolates but was upregulated in strains with the HFAR phenotype. Deletion of both CgCDR1 and CgCDR2 suppressed the development of HFAR in a medium containing fluconazole at 5 μg/ml, showing that both genes are important mediators of resistance to azole derivatives in C. glabrata. We also show here that the HFAR phenomenon was linked to the loss of mitochondria in C. glabrata. Mitochondrial loss could be obtained by treatment with ethidium bromide and resulted in acquisition of resistance to azole derivatives without previous exposure to these agents. Azole resistance obtained in vitro by HFAR or by agents stimulating mitochondrial loss was at least linked to the upregulation of both CgCDR1 and CgCDR2.
The number of life-threatening fungal infections observed worldwide has increased significantly (4). This situation is the consequence of the increasing use of immunosuppressive drugs and antibiotics given to cancer or organ transplant patients. The AIDS epidemic, which maintains human immunodeficiency virus-infected patients permanently immunocompromised, has also largely contributed to the higher incidence of fungal infections. The major agents of fungal infections are Candida species, and Candida albicans is the most frequent species, followed by C. glabrata and C. tropicalis (22). The need for antifungal agents has correspondingly increased but is restricted to a few agents. Among these, azole antifungal agents have been more extensively utilized, in particular, fluconazole since it is well tolerated.
Reports on resistance to azole antifungal agents were rare until the late 1980s. The first cases of resistance were reported in C. albicans after prolonged therapy with miconazole and ketoconazole (12–14). Fluconazole is used in a wide variety of clinical settings, and antifungal resistance to this agent has been more frequently reported (30). Resistance of yeast clinical isolates to azole antifungal agents can be mediated by the following different mechanisms: (i) the cellular content of the azole cellular target (Erg11p) can be elevated, (ii) the affinity of Erg11p to these agents can be decreased, (iii) cells can fail to accumulate these agents, and (iv) the ergosterol biosynthetic pathway can be altered by inactivation of the sterol Δ5,6-desaturase (25, 27, 29, 31). Since resistance to azole antifungal agents can develop as a stepwise process over time, one can expect these mechanisms to combine with each other (27).
C. glabrata is a yeast with intrinsic low susceptibility to azole derivatives and is often recovered from clinical samples originating from AIDS or cancer patients. This yeast is still able to acquire resistance to azole antifungal agents during treatment with these types of antifungals. We recently described the major mechanism of azole resistance in two separate C. glabrata isolates from AIDS patients with oropharyngeal candidiasis who were treated with fluconazole (26). The two azole-resistant isolates were accumulating less fluconazole than their azole-susceptible parents. This effect was due to the upregulation of CgCDR1, a gene encoding a multidrug transporter of the ATP-binding-cassette (ABC) transporter family. Deletion of CgCDR1 in an azole-resistant clinical isolate rendered the resulting mutant susceptible to azole derivatives, and it accumulated as much fluconazole as the azole-susceptible parent. We therefore considered CgCDR1 to be the major ABC transporter gene involved in the resistance of clinical isolates to azole derivatives. During our investigations on mechanisms of azole resistance in C. glabrata, we observed that azole-susceptible isolates exposed in vitro to fluconazole were generating azole-resistant clones at a surprisingly high frequency. In this study, we addressed the mechanisms operating in this process. We show here that the high frequency of azole resistance (HFAR) acquired in vitro is linked with mitochondrial loss and CgCDR1 upregulation. We also identified another ABC transporter gene, CgCDR2, which contributes to this in vitro-acquired resistance.
MATERIALS AND METHODS
Strains and media.
The yeast strains used in this study are listed in Table 1. They were grown at 30°C on complex medium (yeast extract-peptone-dextrose [YEPD]) with 2% (wt/vol) glucose, 1% (wt/vol) Bacto Peptone (Difco), and 0.5% (wt/vol) yeast extract (Difco Laboratories, Detroit, Mich.). YEPD agar plates contained 2% (wt/vol) agar (Difco) as a supplement. Yeast Nitrogen Base (YNB) (Difco) with 2% (wt/vol) glucose and 2% (wt/vol) agar (Difco), with amino acids and bases added as required, was used as a selective medium after transformation of Saccharomyces cerevisiae YKKB-13 and of C. glabrata. Agar plates containing 5-fluoroorotic acid (Toronto Research Chemicals, North York, Ontario, Canada) at 50 μg/ml were made for regeneration of the ura3 genetic marker in YNB selective medium with uridine at 50 μg/ml. Escherichia coli DH5α (11) was used as a host for plasmid constructions and propagation and was grown on Luria-Bertani (LB) medium with ampicillin.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype or phenotype | Parent strain | Reference |
|---|---|---|---|
| S. cerevisiae | |||
| YKKB-13 | MATa ura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 Δpdr5::TRP1 | YPH499 | 5 |
| DSY1693 | Contains pAAH5 | YKKB-13 | |
| DSY1694 | Contains pDS647 (CgCDR2 expression plasmid) | YKKB-13 | |
| DSY1696 | Contains pDS646 (CgCDR1 expression plasmid) | YKKB-13 | |
| C. glabrata | |||
| DSY562 | C. glabrata clinical isolate, susceptible to azoles | 26 | |
| DSY565 | C. glabrata clinical isolate, resistant to azoles | DSY562 | 26 |
| DSY1029 | ura3 | DSY565 | 26 |
| DSY1041 | Δcgcdr1::hisG-URA3-hisG | DSY1029 | 26 |
| DSY1612 | Δcgcdr2::hisG-URA3-hisG | DSY1029 | This study |
| DSY1613 | Δcgcdr2::hisG Δcgcdr1::hisG-URA3-hisG | DSY1056 | This study |
| DSY562-HFAR | HFAR strain | DSY562 | This study |
| DSY1041-HFAR | HFAR strain | DSY1041 | This study |
| DSY562-rho | Mitochondrion-deficient mutant isolated by EtBr treatment | DSY562 | This study |
| DSY1041-rho | Mitochondrion-deficient mutant isolated by EtBr treatment | DSY1041 | This study |
| DSY1613-rho | Mitochondrion-deficient mutant isolated by EtBr treatment | DSY1613 | This study |
Susceptibility testing.
Tests of susceptibility to azole antifungals were performed using the broth microdilution assay according to NCCLS protocol M27-A (21) using RPMI 1640 medium (Difco) and incubation at 35°C for 48 h. Endpoint readings were recorded with a microplate reader (Bio-Rad, Hercules, Calif.), and the azole concentration yielding at least 50% growth inhibition compared to growth in drug-free medium was defined as the MIC.
Susceptibility of the C. glabrata isolates and of S. cerevisiae strains containing CgCDR1 and CgCDR2 expression plasmids to different compounds was also tested qualitatively by spotting serial dilutions of yeast cultures onto complex YEPD medium agar plates with different drug concentrations. This provides easy visualization of growth differences between different yeast strains. Each plate contained 15 ml of agar. Preliminary tests were performed to optimize drug concentrations in YEPD plates so that growth differences between the different S. cerevisiae and C. glabrata strains used in this study could be observed. To perform the susceptibility tests, yeast strains were grown overnight at 30°C with constant shaking in YEPD liquid medium. The cultures were diluted to 2 × 107 cells per ml in 0.9% (wt/vol) NaCl. Five-microliter volumes of this suspension and serial dilutions of the yeast cultures were spotted onto each type of plate and incubated for 48 h at 30°C.
Isolation of C. glabrata RNA, genomic DNA, and mtDNA.
Small-scale isolation of RNA and DNA from C. glabrata was performed as previously described (26). Mitochondrial DNA (mtDNA) was extracted from growing C. glabrata cultures by spheroplasting of cells and then sedimentation of mitochondria as described in reference (8).
Northern and Southern blot assays.
Northern and Southern blot assays were carried out as described previously (26). 32P-labeled DNA probes were generated by random priming (9) and added to the hybridization solution overnight. DNA probes in Southern and Northern blots were for CgCDR1, a 1.8-kb XbaI-BamHI fragment from pNB126 described in reference (26). The CgCDR2 probe was a 1-kb EcoRI fragment isolated from pDS521 (see below). The CgURA3 probe was generated from genomic DNA by PCR as reported previously (26). Quantifications of Northern blots were performed by exposition of the hybridized membranes in an Instant Imager (Packard Instrument Company, Meriden, Conn.). Signals were integrated by the software supplied by the manufacturer and normalized to the CgURA3 probe (used as an internal standard).
Cloning of CgCDR2 and CgSNQ2.
Reverse-transcribed cDNA was prepared using the First Strand cDNA Synthesis Kit for reverse transcription (RT)-PCR from Roche Molecular Biochemicals (Rotkreuz, Switzerland) with total RNA prepared from C. glabrata strain DSY1041-HFAR (Table 1). This cDNA was used as the template for PCR amplification with primers PDR-GUESS R (5′-TTGTTCVACATTTARACCTTCACCWGSSAAC-3′) and PDR-GUESS 3 (5′-AAATTYCAATGYGGGATAAYGCHACVAGR-3′). The design of these primers was based on comparisons with DNA sequences of the S. cerevisiae PDR5, PDR10, and PDR15 genes and the C. glabrata CgCDR1 gene and corresponding to the Walker A and B ATP-binding motifs of ABC transporters. The products obtained by RT-PCR, with an expected size of approximately 2 kb, were subcloned either into vector pCR2.1 (Invitrogen Corporation, Carlsbad, Calif.) or into EcoRI-HindIII-digested pBluescript (Stratagene) after restriction digestion of the same PCR products by the same enzymes. Both strategies ensured the cloning of all of the possible PCR products obtained. Individual clones were sequenced and revealed the presence of two distinct nucleotide sequences. The first one, which was similar to portions of CgCDR1 and almost identical to a 2-kb portion of PDH1 (19), was cloned into pCR2.1 (Genbank accession number AF251023). The second one, which was most similar to ABC transporter gene SNQ2 from S. cerevisiae, was therefore named CgSNQ2 (Genbank accession number AF251022) and was cloned into the HindIII-EcoRI sites of pBluescript.
Disruption of CgCDR2.
pCR2.1, containing the CgCDR2 gene cloned by RT-PCR, was digested with EcoRI, and a fragment of approximately 1 kb was cloned into the EcoRI site of pBluescript. The resulting plasmid, pDS521, was digested with BglII, which cuts within CgCDR2, and the 3.7-kb BamHI-BglII fragment from pNKY51 (1) containing the hisG-URA3-hisG cassette was inserted at this site. The resulting plasmid, pDS547, was digested with EcoRI, and the 4.7-kb fragment containing the hisG-URA3-hisG cassette flanked by CgCDR2 sequences was used to transform DSY1029. One of the Ura+ clones (DSY1612) was selected, and the correct integration of the disruption cassette was verified by Southern blotting (data not shown). The ura3 genetic marker was regenerated by 5-fluoroorotic acid treatment of DSY1612. The ura3 derivative of DSY1612 was used for the disruption of CgCDR1 as described previously (26). The correct gene replacement of CgCDR1 by the hisG-URA3-hisG disruption cassette was also verified (data not shown). The resulting double deletion (CgCDR1 CgCDR2) strain was named DSY1613.
Expression of CgCDR1 and CgCDR2 in S. cerevisiae.
The CgCDR1 and CgCDR2 open reading frames (ORFs) were cloned into the HindIII site of pAAH5, which is a vector expressing ORFs under the control of the ADH1 promoter (2). The CgCDR1 and CgCDR2 ORFs were amplified by high-fidelity PCR (Expand PCR; Roche Molecular Biochemicals) with primers to which HindIII sites were added. The following pairs of primers were for CgCDR1 and CgCDR2, respectively (HindIII site underlined): 5′-GCGCAAGCTTACAATGTCTCTTGCAAGTGACAAGAAG-3′ and 5′-GCGCAAGCTTTTATTTCTTGGCAAGTTTACCAGATTT-3′; 5′-GCGCAAGCTTACAATGG CAATTGGTATATATACTGGAACG-3′ and 5′-GCGCAAGCTTCTAGAAGGGAATTAACCTTCTAATAAAATTAACC-3′. Genomic DNA from clinical isolate DSY562 served as the template in the PCR. The CgCDR1 and CgCDR2 expression plasmids obtained (pDS646 and pDS647, respectively) were transformed into S. cerevisiae YKKB-13. Verification that a functional protein was being produced from the genes contained in each plasmid was that each plasmid conferred resistance to azole antifungal agents when transformed into S. cerevisiae YKKB-13.
Isolation of mitochondrial mutants of C. glabrata.
C. glabrata strains were grown overnight in YEPD liquid medium and diluted 100-fold in 2 ml of fresh medium. Ethidium bromide (EtBr) was added from a sterile filtered stock solution to a final concentration of 20 μg/ml. The culture was grown overnight at 30°C, and an aliquot was plated on YEPD agar to isolate single colonies. After 2 days of incubation at 30°C, single colonies were selected and tested for their growth phenotypes on glycerol- and glucose-containing media. Colonies growing not on glycerol medium but on glucose medium were analyzed for the presence of mtDNA by staining with MitoTracker Green FM (see below) or by agarose gel electrophoresis. Mutants lacking mtDNA (considered rho0) were further selected.
Immunoblot assays.
To allow detection of the proteins encoded by CgCDR1 and CgCDR2 (i.e., CgCdr1p and CgCdr2p) in yeast extracts, polyclonal antibodies against these proteins were raised in rabbits. The proteins for immunization were obtained in E. coli by fusion of the first 60 amino acids of each protein with glutathione S-transferase (GST). The GST fusion constructs were prepared by subcloning PCR fragments obtained using genomic DNA from C. glabrata DSY562 as the template. The following pairs of primers were used for CgCDR1 and CgCDR2, respectively (BamHI and EcoRI sites underlined): 5′-GCGCGGATCCATGTCTCTTGCAAGTGACAAGAAGGA-3′ and 5′-GCGCGGAATTCCAGGCAGAGTGTGTGTTCTTTCTTTTGATG-3′; 5′-GCGCGGATCCATGGAACACACCCGATGACTCTAGTTGT-3′ and 5′-GCGCGGAATTCCCAATGGCGCGCTGCCATCTGCGGGGGC-3′. The PCR fragments obtained were subcloned after restriction digestion with EcoRI and BamHI into the same sites of the vector pGEX-2T (Amersham Pharmacia Biotech, Dübendorf, Switzerland) to allow in-frame fusions with GST. The purification of GST fusion proteins for raising polyclonal rabbit antibody was achieved by affinity chromatography of E. coli cell extracts on glutathione-agarose using standard procedures described by the manufacturer (Amersham Pharmacia Biotech).
These antibodies were used to detect, by immunoblot assay, CgCdr1p and CgCdr2p from cellular proteins of C. glabrata or S. cerevisiae transformed with expression plasmids. Cell extracts were prepared, and immunoblot assays were performed essentially as described by Sanglard et al. (27).
Measurement of total cytochrome content.
Cytochromes aa3, b, and c were measured in whole cells by difference spectra of reduced versus oxidized cells. C. glabrata strains were first grown in liquid YEPD overnight at 30°C, and equivalent cell densities (8 × 1010 cells) were pelleted by centrifugation. The pelleted cells were resuspended in 6 ml of phosphate-buffered saline (PBS) and distributed in separate 5-ml quartz cuvettes. After baseline correction, sodium dithionite and H2O2 were added to each cuvette and the difference spectra were recorded from 500 to 650 nm with a Lambda 18 spectrophotometer (Perkin-Elmer International, Rotkreuz, Switzerland).
Staining of mitochondria.
C. glabrata isolates were grown overnight in liquid YEPD medium. Cells (2 × 107) were washed twice with PBS and resuspended in 0.5 ml of PBS containing 0.5% (wt/vol) glucose. MitoTracker Green FM (Molecular Probes Inc., Eugene, Oreg.) was added at a final concentration of 100 nM, and the mixture was incubated at room temperature for 30 min. Cells were directly observed for green fluorescence with a Zeiss Axioplan microscope equipped with fluorescence filter set 13.
Fluorescence microscopy.
Fluorescence microscopy and phase-contrast microscopy were performed with a Zeiss Axioplan microscope (Zeiss) equipped for epifluorescence microscopy with a 100-W high-pressure mercury bulb and Zeiss fluorescein-specific filter set 13. A Kappa DX30 digital camera with high resolution (Kappa Messtechnik GmbH, Gleichen, Germany) was used to record images, which were further processed using the computer program Adobe Photoshop 5.0 (Adobe Systems Incorporated, Mountain View, Calif.).
Other methods.
Rhodamine accumulation was measured by flow cytometry as previously described (26).
Chemicals.
Analytical-grade or equivalent chemicals were used in this study. Restriction enzymes and DNA-modifying enzymes were from Roche Molecular Biochemicals.
RESULTS
HFAR in C. glabrata is linked with upregulation of ABC transporter genes.
We recently showed that deletion of ABC transporter gene CgCDR1 in an azole-resistant C. glabrata clinical isolate (DSY1041) resulted in increased susceptibility of the mutant to azole derivatives (26). We observed, however, that when DSY1041 was incubated on agar medium containing several azole derivatives, including fluconazole and itraconazole, several colonies could still appear from individual cells. Plating a high density of the mutant DSY1041 onto rich agar medium containing fluconazole at 5 μg/ml gave rise to fluconazole-resistant cells at frequencies ranging from 2 × 10−4 to 4 × 10−4. These cells were referred to as HFAR cells (Fig. 1). HFAR cells could also be generated from azole-susceptible wild-type C. glabrata strain DSY562 at a frequency similar to that of DSY1041 but using agar medium containing as much as 50 μg of fluconazole per ml (Fig. 1). Azole MICs for HFAR derivatives and for their parents are shown in Table 2. Fluconazole MICs and itraconazole MICs for HFAR derivatives obtained from DSY562 and DSY1041 were higher than those of their respective parent strains. Furthermore, azole MICs for HFAR strains were found to be similar to those exhibited by azole-resistant clinical strain DSY565 (Table 2). To examine the mechanism of resistance operating in HFAR cells, accumulation of rhodamine 6G was first tested, since we showed previously that poor accumulation of this compound in azole-resistant cells was linked with upregulation of multidrug transporter genes, especially CgCDR1 (26). HFAR cells which arose from DSY1041 (DSY1041-HFAR) and DSY562 (DSY562-HFAR) accumulated 6.5- and 6-fold less rhodamine 6G than their azole-susceptible parents, respectively, suggesting that multidrug transporter genes could be upregulated in these strains. Expression of CgCDR1 could only be examined in DSY562-HFAR, since this gene was deleted in the DSY1041-HFAR strain. A 25-fold relative increase in CgCDR1 mRNA could be measured in DSY562-HFAR compared to DSY562. The increase in the CgCDR1 mRNA signal in DSY562-HFAR was approximately 2.5-fold greater than that obtained for the same gene in azole-resistant clinical strain DSY565 (Fig. 2). The increase in azole MICs measured for DSY562-HFAR could therefore be attributed, at least in part, to the upregulation of CgCDR1.
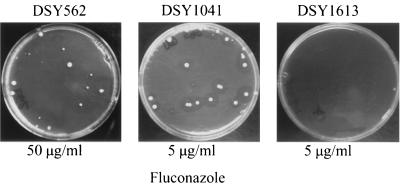
FIG. 1.
HFAR in C. glabrata strains DSY562, DSY1041, and DSY1613 following growth on YEPD containing fluconazole. The inoculum size of DSY562 on the YEPD plate containing fluconazole at 50 μg/ml was 3.3 × 104 cells. The frequency of HFAR cells in three independent experiments was 3.2 × 10−4 ± 0.71 × 10−4. Inoculum sizes for DSY1041 and DSY1613 were 4 × 104 and 5 × 104 cells, respectively. The frequency of HFAR cells from DSY1041 was similar to that obtained with DSY562. Incubation of the plates was for 4 days at 30°C. The fluconazole concentration used in YEPD is indicated for each plate.
TABLE 2.
Azole MICs for C. glabrata isolates used in this study
| C. glabrata strain | MIC (μg/ml) of:
|
|
|---|---|---|
| Fluconazole | Itraconazole | |
| DSY562 | 4 | 0.25 |
| DSY565 | >128 | 4 |
| DSY562-HFARa | >128 | 2 |
| DSY562-rhob | >128 | 2 |
| DSY1041 | 8 | 0.5 |
| DSY1041-HFARa | 16 | 1 |
| DSY1041-rhob | 16 | 1 |
| DSY1613 | 4 | 0.25 |
| DSY1613-rhob | 8 | 0.5 |
Strain obtained by HFAR.
Strain obtained by EtBr treatment.
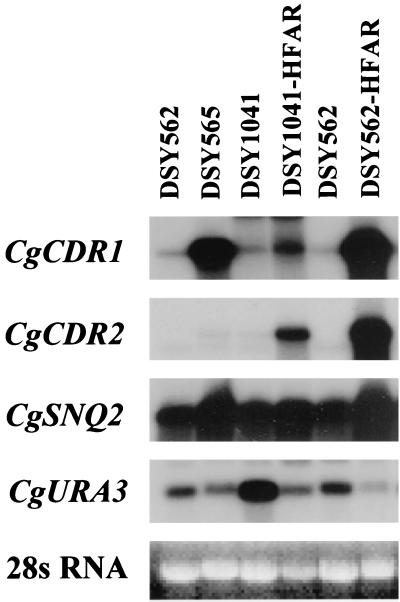
FIG. 2.
Expression of ABC transporter genes in fluconazole-resistant isolates of C. glabrata. RNA was extracted from C. glabrata clinical isolates DSY562 and DSY565, from CgCDR1 deletion mutant DSY1041, and from HFAR cells obtained with DSY562 and DSY1041. The Northern blot was probed sequentially with 32P-labeled probes specific for all of the genes (CgCDR1, CgCDR2, CgSNQ2, and CgURA3) as indicated. The mRNA-hybridizing band detected in RNA of DSY1041-HFAR is probably due to cross-hybridization of the CgCDR1 probe with CgCDR2 mRNA, as discussed by Sanglard et al. (26). Due to variations in CgURA3-specific signals, EtBr-stained 28S RNA is shown to indicate that similar RNA quantities were loaded on the agarose gel. Using CgURA3 signals for normalization, CgCDR1 signals were increased 25- and 9-fold in DSY562-HFAR and DSY565, respectively, compared to those detected in parent strain DSY562.
Since CgCDR1 was deleted in DSY1041 and since it was possible to obtain HFAR cells from this strain, we considered that another or several other multidrug transporter genes were upregulated. The reduced accumulation of rhodamine 6G in DSY1041-HFAR was consistent with this hypothesis. To address this question, degenerate primers corresponding to a region conserved among yeast ABC transporter genes were used in a PCR carried out with reverse-transcribed mRNA from DSY1041-HFAR. From the expected PCR products of approximately 2 kb, two different sequences were obtained. One was similar to a portion of ABC transporter gene SNQ2 from S. cerevisiae, whereas the other was similar to CgCDR1 but identical to the C. glabrata ABC transporter gene PDH1 reported recently (19) (see Materials and Methods for further details). These genes were named CgSNQ2 and CgCDR2, respectively, in this work. The nucleotide sequence of CgCDR2 cloned in this work was 98% similar to the comparable portion of PDH1. Labeled probes made from CgSNQ2 and CgCDR2 were hybridized to a Northern blot containing RNAs extracted from several C. glabrata strains. Whereas the expression of CgSNQ2 was little affected by azole resistance, the intensity of the mRNA signal obtained with CgCDR2 was increased in HFAR strains (Fig. 2). We calculated an up-to-100-fold increase in the CgCDR2 mRNA signal in strain DSY562-HFAR compared to the signals obtained in strain DSY562. Therefore, of the two genes isolated by RT-PCR, only CgCDR2 seemed important for the development of azole resistance. This view was confirmed by examination of CgCDR2 expression in strain DSY1041-HFAR, in which CgCDR1 was deleted (Fig. 2). In this strain, the mRNA signal corresponding to CgCDR2 was increased 16-fold compared to that in azole-susceptible strain DSY1041. Therefore, it is likely that the azole resistance restored in DSY1041-HFAR was due, at least in part, to CgCDR2 upregulation. However, the role of other, still uncharacterized, multidrug transporter genes in the HFAR phenomenon cannot be ruled out yet.
Deletion of CgCDR2 in C. glabrata.
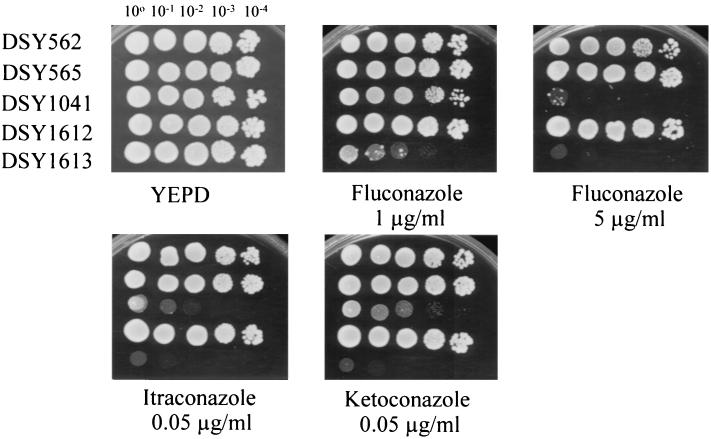
To address the importance of CgCDR2 in the acquisition of azole resistance by C. glabrata, this gene was deleted by targeted deletion in DSY1041. Southern blot hybridization confirmed that the deletion of CgCDR2 had taken place at the correct genomic locus (data not shown). The resulting double deletion (CgCDR1 CgCDR2) mutant was named DSY1613. The deletion of both CgCDR1 and CgCDR2 in DSY1613 resulted in the absence of full-length CgCDR-specific mRNAs (Fig. 3A) and, consistently, in the absence of detectable CgCdr1p and CgCdr2p (Fig. 3B). DSY1613 was more susceptible to azole derivatives than its parent DSY1041 when tested in media containing low concentrations of fluconazole, ketoconazole, and itraconazole (Fig. 4). More importantly, this strain was not capable of forming HFAR cells on a medium containing fluconazole at 5 μg/ml under conditions in which DSY1041 formed HFAR cells (Fig. 1). Thus, it appears that both CgCDR1 and CgCDR2 are important for the in vitro acquisition of azole resistance in C. glabrata. Furthermore, since HFAR cells were obtained from DSY1041, CgCDR2 upregulation can compensate for the absence of CgCDR1 and can contribute to decreased azole susceptibility.
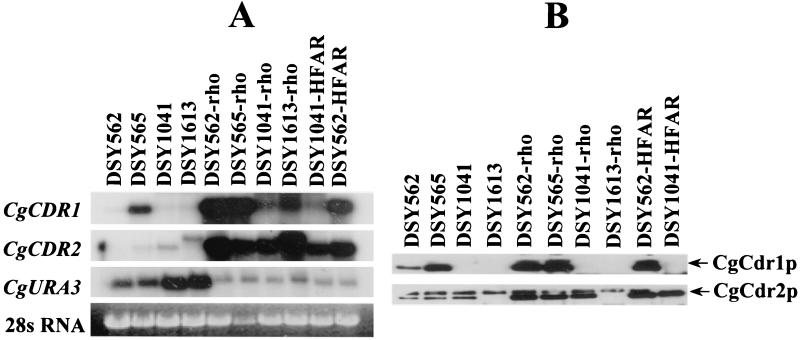
FIG. 3.
Expression of multidrug efflux transporters in C. glabrata isolates with in vitro-acquired azole resistance. (A) Northern blot analysis. Approximately 5 μg of total RNA from each indicated yeast strain was loaded on the agarose gel. The Northern blot was hybridized sequentially with each 32P-labeled probe. The CgCDR2-specific signal from DSY1613 has a larger size than that from DSY1041 and probably corresponds to an aberrant RNA product. The Northern blots were revealed by exposure of Fuji XAR film at −80°C. Normalized signals for CgCDR1, compared to those detected in DSY562, were increased 7-, 62-, 67-, and 90-fold in DSY565, DSY562-rho, DSY565-rho, and DSY562-HFAR, respectively. Normalized signals for CgCDR2, compared to those detected in DSY562, were increased 1.7-, 1-, 68-, 40-, 22-, 26-, and 80-fold in DSY565, DSY1041, DSY562-rho, DSY565-rho, DSY1041-rho, DSY1041-HFAR, and DSY562-HFAR, respectively. (B) Immunodetection of CgCdr1p and CgCdr2p in cellular extracts. A 10-μg sample of total protein was loaded on a sodium dodecyl sulfate–10% (wt/vol) polyacrylamide gel and separated by electrophoresis. Western blots were incubated separately with CgCdr1p and CgCdr2p antisera, and signals were revealed by chemiluminescence on Fuji XAR film. Absence of signals in protein extracts from DSY1041 and DSY1613 is consistent with the deletion of CgCDR1 and CgCDR2 in these strains.
FIG. 4.
Susceptibility of C. glabrata multidrug efflux transporter mutants to azole derivatives. Each yeast strain is indicated at the left. Azole derivatives were added to YEPD medium at the indicated concentrations. Plates were incubated for 48 h at 30°C.
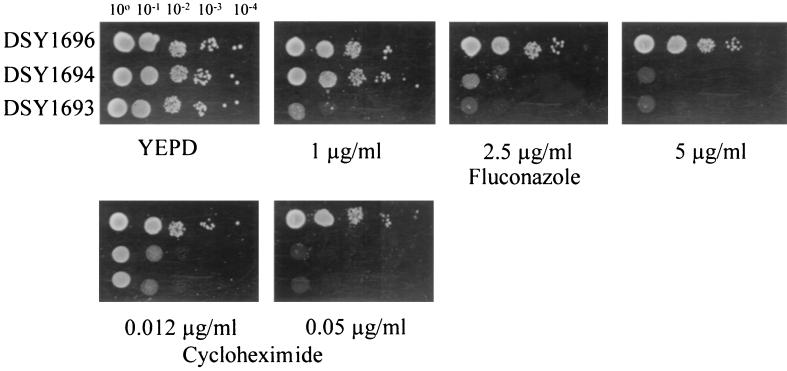
The level of azole resistance obtained by CgCDR2 upregulation, however, seemed lower than that obtained by CgCDR1, since no HFAR cells could be isolated in a medium containing fluconazole at a concentration of 50 μg/ml (data not shown). To test this hypothesis, CgCDR1 and CgCDR2 were expressed under the control of the same ADH1 promoter in azole-hypersusceptible S. cerevisiae strain YKKB-13, in which the ABC transporter gene PDR5 has been deleted. Figure 5 shows that DSY1696, in which CgCDR1 was expressed, was still able to grow in a medium containing fluconazole at 5 μg/ml, whereas DSY1694 expressing CgCDR2 could only grow at lower drug concentrations. The same relationship was observed in a medium containing cycloheximide, which is a known substrate for CgCDR1 (26). These results confirmed our above-mentioned hypothesis that CgCDR1 has a higher capacity than CgCDR2 for the development of fluconazole resistance.
FIG. 5.
CgCDR1 and CgCDR2 expression in S. cerevisiae. Susceptibility tests of yeast transformants were performed with fluconazole and cycloheximide at the indicated concentrations. Plates were incubated for 48 h at 30°C.
The growth defect of HFAR cells is paralleled by loss of mitochondria.
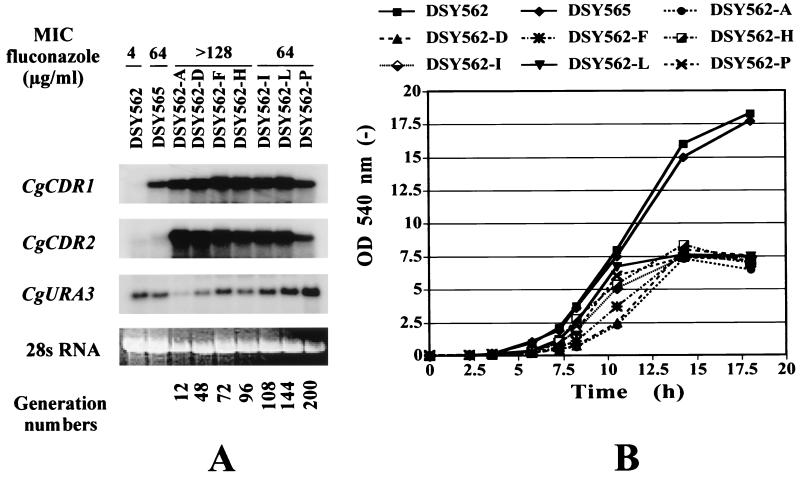
Upon repeated subculture of DSY562-HFAR in fluconazole-free medium, the fluconazole MIC for this strain dropped from >128 μg/ml (subculture A) to 64 μg/ml (subculture P), which corresponds approximately to a span of 200 generations after the start of the experiment. Figure 6A shows that upregulation of both CgCDR1 and CgCDR2 was still detected for up to 200 generations in drug-free medium. Quantification of CgCDR1 and CgCDR2 mRNA signals normalized with the CgURA3 probe, however, revealed a tendency to decrease with increasing subculture. In DSY562-A, the signals for CgCDR1 and CgCDR2 mRNAs were increased 60- and 90-fold, respectively, compared to those detected in DSY562, and the increases declined to 10- and 5-fold in DSY562-P. Increases in normalized mRNA signals for CgCDR1 and CgCDR2 in DSY562-P compared to DSY562 were similar to those obtained in azole-resistant clinical strain DSY565. The correlation between multidrug transporter gene expression and fluconazole MICs could not be well established because the fluconazole concentration in the MIC test was limited to 128 μg/ml. However, although multidrug transporter gene expression declined with repeated subculture but was still comparable to the expression observed in azole-resistant clinical isolates, we considered that azole resistance could be maintained in HFAR cells under drug-free culture conditions. When growth of HFAR cells was recorded, optical densities did not reach the values measured in cultures of parent strains DSY562 and DSY565 in late lag phase (Fig. 6B). Subtle differences were, however, noticed in the initial growth rate of HFAR cells, depending on the degree of subculture. While early subcultures showed initial slow growth (for example, subculture DSY562-A or DSY562-D), subsequent subcultures (DSY562-L or DSY562-P) grew faster in their log phase. This trend closely followed the relative increases in CgCDR1 and CgCDR2 expression, where both genes, being highly upregulated in first subculture, were gradually less upregulated in subsequent subculture. The cell densities of DSY562-HFAR strains obtained in late log phase, however, remained almost identical. The restoration of the measured initial growth rate seemed to be related to the decrease in CgCDR1 and CgCDR2 expression, as shown in Fig. 6A. These results suggest that high expression levels of these multidrug transporter genes could have adverse effects, the consequence of which results in reduced growth rate.
FIG. 6.
Maintenance of azole resistance of HFAR cells in drug-free medium. (A) Expression of CgCDR1 and CgCDR2 in DSY562-HFAR in drug-free YEPD medium over subcultures A (no. 1) to P (no. 16). Intensities of CgCDR1 and CgCDR2 signals were normalized to those obtained with the CgURA3 probe. Normalized signals for CgCDR1, compared to those detected in DSY562, were increased 60-, 40-, 31-, 35-, 26-, 23-, and 8-fold in DSY562-A to -P, respectively, and 12-fold in DSY565. Normalized signals for CgCDR2, compared to those detected in DSY562, were increased 90-, 33-, 22-, 22-, 18-, 15-, and 5-fold fold in DSY562-A to -P, respectively, and 2.5-fold in DSY565. (B) Growth of clinical isolate DSY562, DSY565 (azole-resistant), and DSY562-HFAR subcultures (A to P) in YEPD.
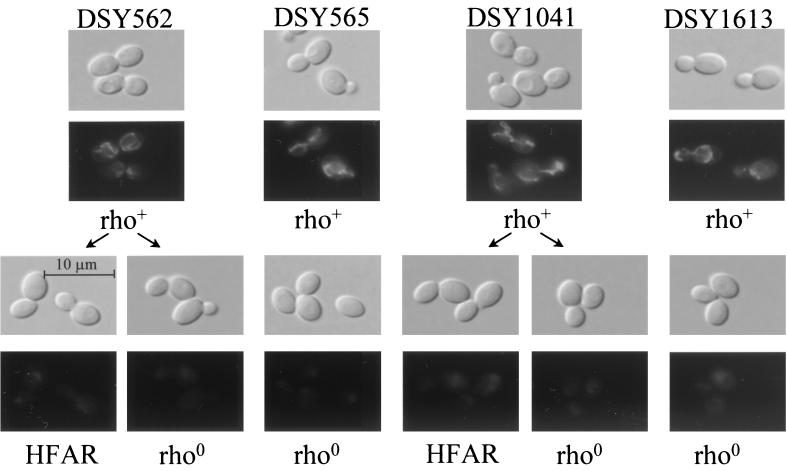
When tested for the ability to grow on different carbon sources, HFAR cells did not grow on ethanol and arabinitol, which are nonfermentable carbon sources (data not shown). The lack of growth on these carbon sources suggested alterations in the mitochondrial respiratory capacity of these yeasts. A similar phenomenon is known in S. cerevisiae, and cells with a respiratory deficiency are called petite mutants. These cells often lack mtDNA. Since the reduced respiratory capacity detected in C. glabrata could probably be attributed to a loss of mitochondria, staining with the mitochondrion-specific fluorescent dye MitoTracker Green FM was applied to C. glabrata in growing cultures. Figure 7 shows positive staining in respiratorily competent (i.e., rho+) cells, whereas weak or absent staining was seen in HFAR cells. The absence of mtDNA was verified in HFAR cells by subjecting their mitochondrial cell fractions to agarose gel electrophoresis, and no mtDNA bands could be stained with EtBr (data not shown). The loss of mitochondria in these cells can therefore account for their above-mentioned growth defect. Petite mutants can be easily obtained in S. cerevisiae by treatment with EtBr. We applied this technique to C. glabrata and obtained cells with the petite characteristics. These cells could not be stained with MitoTracker Green FM (Fig. 7) and failed to reveal mtDNA banding on gel electrophoresis analysis (data not shown), as in the case of HFAR cells. Petite mutants obtained by EtBr treatment were therefore considered rho0 according to the nomenclature accepted for S. cerevisiae. These cells were referred as to DSY-rho strains in this work. Although generated by different chemical treatments, HFAR and rho0 cells possess similar characteristics. First, when total cytochrome content was measured in whole cells of HFAR and rho0 isolates by difference spectra, no absorption peak could be measured corresponding to those of cytochromes aa3 and b, which are gene products encoded by mtDNA (data not shown). Second, the azole MICs obtained for HFAR and rho0 strains were similar. As summarized in Table 2, the fluconazole and itraconazole MICs for DSY562-HFAR and DSY562-rho were >128 and 2 μg/ml, respectively, whereas those for DSY1041-HFAR and DSY1041-rho were 16 and 1 μg/ml. It is remarkable that DSY-rho derivatives obtained by EtBr treatment acquired azole resistance without previous exposure to these antifungals.
FIG. 7.
Staining of mitochondria in C. glabrata. Strains DSY562, DSY565, DSY1041, and DSY1613 are respiratorily competent and therefore were designated rho+. From each of these strains, HFAR cells obtained by fluconazole exposure or rho0 cells obtained by EtBr treatment were incubated with MitoTracker Green FM and examined by either phase-contrast (top rows) or epifluorescence (bottom rows) microscopy.
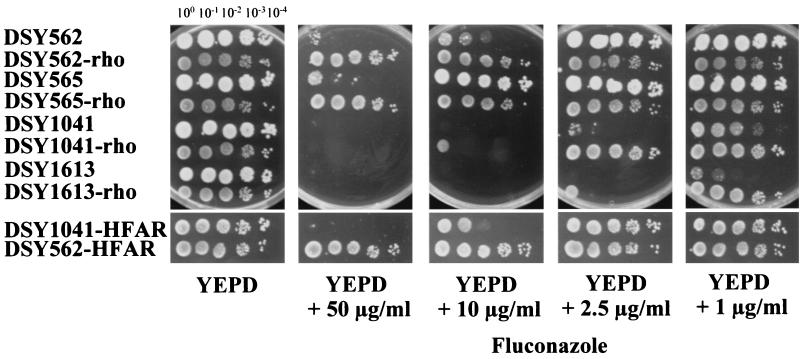
Depending on the genetic background used to generate rho0 derivatives, variable degrees of azole resistance were obtained. Figure 8 shows, for example, that DSY562-rho could grow on a medium containing fluconazole at up to 50 μg/ml, whereas DSY1041-rho could only grow distinctly in a medium containing the same drug at only 2.5 μg/ml. Interestingly, the double mutant (CgCDR1 CgCDR2) strain DSY1613 rendered rho0 by EtBr treatment (DSY1613-rho) acquired resistance to fluconazole. Azole resistance obtained in DSY1613-rho was only visible when a low fluconazole concentration (i.e., 1 μg/ml) was present in YEPD plates. As shown in Fig. 8, DSY1613-rho grew well at this drug concentration whereas parent strain DSY1613 grew weakly in the same medium.
FIG. 8.
Mitochondrial loss is linked to the acquisition of azole resistance in C. glabrata. Wild-type, HFAR, and rho0 cells were spotted in serial dilutions onto YEPD agar and fluconazole-containing YEPD medium. Plates were incubated for 48 h at 30°C.
The rho0 cells accumulated less rhodamine 6G than their wild-type parents, and the acquisition of this azole resistance was as stable as in the case of HFAR cells (data not shown). The expression of ABC transporter genes was tested in different rho0 cells with specific genetic backgrounds. CgCDR1 and CgCDR2 were upregulated in these cells compared to rho+ cells, to levels similar to those measured in HFAR cells (Fig. 3A). For example, CgCDR1-normalized mRNA signals were elevated 90- and 62-fold in DSY652-rho and DSY562-HFAR cells, respectively, while CgCDR2-normalized mRNA signals were increased 80- and 68-fold in the same strains. Accordingly, CgCdr1p and CgCdr2p levels were higher in rho0 and HFAR derivatives than in rho+ cells, except in cells in which the genes encoding these proteins were deleted (Fig. 3B).
DISCUSSION
We recently showed how CgCDR1 could contribute to azole resistance in C. glabrata clinical isolates (26). In this study, we further detailed the molecular basis of azole resistance in this yeast species by exploring the role of another ABC transporter gene, CgCDR2, in this phenomenon. We isolated CgCDR2, which is identical to PDH1 (19), through observations which revealed the possible expression of other multidrug transporter genes in a CgCDR1 deletion strain exposed in vitro to fluconazole. The results shown in this study indicate that the regulatory system of these transporters might be quite complex. On the one hand, while the upregulation of CgCDR1 is significant in azole-resistant clinical isolate DSY565, CgCDR2 is only moderately expressed in the same strain. On the other hand, when azole resistance is established by HFAR or EtBr treatment from an azole-susceptible strain, both CgCDR1 and CgCDR2 are upregulated to high levels. This suggests that one or several regulatory systems should exist in C. glabrata enabling the differential regulation of these genes. Both genes possess in their promoter regions matching the consensus for Pdr1p-Pdr3p binding sites (15); therefore, one can expect that they can be controlled coordinately by Pdr1p-Pdr3p-like transcription factors. In the light of the results obtained here, it is likely that other regulatory elements assist the differential expression of CgCDR1 and CgCDR2.
C. glabrata ABC transporter gene PDH1, reported by Miyazaki et al. (19), has a sequence which is almost identical to that of CgCDR2 in the region corresponding to the PCR-amplified fragment. Miyazaki et al. (19) reported that PDH1 was upregulated up to fourfold in C. glabrata clinical strains resistant to fluconazole. In this study, slight upregulation of the same gene could be detected in azole-resistant clinical isolate DSY565. We examined the expression of CgCDR2 in additional azole-susceptible or resistant clinical strains of C. glabrata and observed that this gene was also expressed at moderate levels. Upregulation of CgCDR2 was noticed in a few azole-resistant strains, whereas CgCDR1 upregulation was always clearly manifested (D. Sanglard and D. Calabrese, unpublished data). Further comparisons of the two transporters are under way. The data presented here favor the hypothesis that CgCDR1 is more potent in its ability to confer azole resistance than is CgCDR2. First, HFAR cells derived from wild-type C. glabrata strains could be obtained on medium containing a high fluconazole concentrations (50 μg/ml; Fig. 1), whereas they could be obtained with DSY1041 (the CgCDR1 deletion strain) only on medium with a lower fluconazole concentration (5 μg/ml; Fig. 1). Second, the heterologous expression of CgCDR1 and CgCDR2 in S. cerevisiae showed that CgCDR1 expression resulted in higher fluconazole resistance levels than in the case of CgCDR2 expression (Fig. 5). It is already known that ABC transporter genes with high similarity (for example, the C. albicans CDR1 and CDR2 genes) differ in the ability to confer resistance to the same drugs (28). Whether this effect is due to altered substrate specificity or reduced drug efflux capability has still not been precisely determined and deserves further investigation.
Isolation of azole-resistant C. glabrata strains using miconazole has been reported, but no growth defects were observed in the azole-resistant strains obtained (23). In this work, we established a link in C. glabrata between loss of mitochondria and acquisition of azole resistance (HFAR) mediated at least by the ABC transporter genes CgCDR1 and CgCDR2. There are discrepancies between the HFAR in C. glabrata described here and the phenomenon of heteroresistance reported in C. neoformans (20) and more recently in C. albicans (18). Heteroresistance implies that, from a single progenitor, different reversible drug resistance phenotypes can be obtained which are reflected by different drug MICs. In the present work, HFAR cells appeared from a single progenitor with similar resistance phenotypes and with uniform and irreversible mitochondrial loss. Thus, it is reasonable to distinguish HFAR from heteroresistance. It is likely that loss of mitochondria in C. glabrata will affect the expression of other genes, some of which are able to play a role in azole resistance. A good candidate could be the C. glabrata ERG11 gene, the upregulation of which was coupled with azole resistance in some C. glabrata strains (16). In this work, it is striking that CgURA3 expression was decreased in HFAR and rho0 mutant strains, although identical RNA quantities were loaded in the Northern blots of Fig. 2, 3, and 6. Expression studies on a genome-wide scale would be very helpful in determining the number of genes with altered expression upon loss of mitochondria. Recently, the acquisition of azole resistance by mitochondrial loss in C. glabrata was reported by Defontaine et al. (8); however, the authors of that study did not work out the molecular mechanisms behind this observation. One important question resulting from both the work of Defontaine et al. (8) and ours is the relevance of this phenomenon to clinical situations. One can expect that by being exposed to azole antifungals, azole-resistant C. glabrata strains would appear by HFAR and therefore be devoid of mitochondria. Under azole-selective pressure, these HFAR strains would have a clear advantage over still susceptible strains. However, due to their defective respiratory capacity, HFAR strains may be not as competitive as wild-type cells under the environmental conditions of the host. If the antifungal selection is removed in a patient, HFAR cells may be very rapidly overgrown by other, more growth-competent strains. It would be interesting to test in animal models or in patients if HFAR cells could be detected when azole treatment is initiated and if these cells remain present after drug removal. Data recently reported by Bouchara et al. (6) reveal that C. glabrata mitochondrial mutants were obtained from stool samples taken from a bone marrow transplant patient undergoing fluconazole therapy, thus showing the potential relevance of this phenomenon to clinical situations.
Loss of mitochondria is a phenomenon well described in S. cerevisiae which can arise spontaneously or in response to DNA-targeting drugs (7). It is coupled with known phenotypes, one of them being growth deficiency in nonfermentable carbon sources. S. cerevisiae belongs to the group of petite-positive yeasts, for which loss of mitochondrial functions is not lethal. The mtDNA encodes essential functions for some other yeasts, and these yeasts are called petite negative. C. glabrata undoubtedly belongs to the group of petite-positive organisms. The close evolutionary relatedness between C. glabrata and S. cerevisiae helps to explain this characteristic. The fact that rho0 mutants of this yeast could be obtained by using an antifungal agent (i.e., fluconazole) other than a DNA-targeting drug has not been often reported. The capacity to form HFAR cells upon fluconazole exposure is not too surprising, since the upregulation of ABC transporter genes, the products of which use fluconazole as substrates, is one of the consequences of mitochondrial loss. With convenient EtBr treatment inducing mitochondrial loss in C. glabrata, it has been possible to isolate rho0 mutants of strain DSY1613. Although this yeast does not possess functional CgCDR1 and CgCDR2 genes, it was possible to observe acquisition of a slight degree of azole resistance in the DSY1613-rho derivative (Fig. 8). DSY1613-rho cells reduced their accumulation of rhodamine 6G to 60% compared to DSY1613 cells (data not shown) and therefore we considered this result a consequence of upregulation of one or several putative additional multidrug transporter genes which could also belong to the ABC transporter family. This is a likely explanation given that a multiplicity of ABC transporter genes has been documented in other yeast species. We are currently attempting the cloning of a putative transporter expressed in DSY1613-rho cells by the RT-PCR strategy outlined above. The results obtained in this study can also help to define other conditions for the generation of azole-resistant derivatives in C. glabrata. Exposing C. glabrata to fluconazole-containing media with nonfermentable carbon sources will allow us to obtain azole-resistant strains with mitochondria, since these organelles are required for the assimilation of these carbon sources. Such strains might be more comparable to respiratorily competent clinical isolates that are azole resistant. The analysis of such mutants is in progress in our laboratory.
In summary, we have shown here that C. glabrata exposed to fluconazole in vitro has the ability to acquire ABC transporter-mediated resistance through the loss of mitochondrial functions. Preliminary data obtained with S. cerevisiae mitochondrial mutants revealed a similar relationship (D. Sanglard, unpublished data). Azole resistance as a consequence of mitochondrial loss has not been reported in other yeasts pathogenic for humans until now. It might not be possible to address this question for some yeast species that are a priori petite negative. The respiratory capacity of these yeasts might, however, be transiently inhibited by specific inhibitors and their effects on azole resistance and expression of multidrug resistance genes could be investigated. Among yeast pathogens, mitochondrial mutants have been described only in C. albicans so far (3, 10, 24), despite the fact that C. albicans was considered a petite-negative yeast (17). Interestingly, C. albicans mitochondrial mutants are resistant to the antimicrobial peptide histatin 5 (10). It might be interesting to test the azole susceptibility of mitochondrial mutants of C. albicans.
ACKNOWLEDGMENTS
This work was supported by a grant from the Swiss National Research Foundation (3100-055901.98/1) to D.S.
We are thankful to P. A. Majcherczyk for technical assistance with FACS analysis.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ammerer G. Expression of genes in yeast using the ADC1 promoter. Methods Enzymol. 1983;101:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- 3.Aoki S, Ito-Kuwa S. Induction of petite mutation with acriflavine and elevated temperature in Candida albicans. J Med Vet Mycol. 1987;25:269–277. doi: 10.1080/02681218780000611. [DOI] [PubMed] [Google Scholar]
- 4.Beck-Sague C M, Jarwis W R. Secular trends in the epidiemology of nosocomial fungal infections in the United States, 1980–1990. National Nosocomial Infections Surveillance System. J Infect Dis. 1993;167:1247–1251. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 5.Bissinger P H, Kuchler K. Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. A yeast ABC transporter conferring mycotoxin resistance. J Biol Chem. 1994;269:4180–4186. [PubMed] [Google Scholar]
- 6.Bouchara J P, Zouchair R, Le Boudouil S, Renier G, Filmon R, Chabasse D, Hallet J N, Defontaine A. In vivo selection of an azole-resistant petite mutant of Candida glabrata. J Med Microbiol. 2000;49:977–984. doi: 10.1099/0022-1317-49-11-977. [DOI] [PubMed] [Google Scholar]
- 7.Chen X J, Clark-Walker G D. The petite mutation in yeasts: 50 years on. Int Rev Cytol. 2000;194:197–238. doi: 10.1016/s0074-7696(08)62397-9. [DOI] [PubMed] [Google Scholar]
- 8.Defontaine A, Bouchara J P, Declerk P, Planchenault C, Chabasse D, Hallet J N. In-vitro resistance to azoles associated with mitochondrial DNA deficiency in Candida glabrata. J Med Microbiol. 1999;48:663–670. doi: 10.1099/00222615-48-7-663. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg A, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 10.Gyurko C, Lendenmann U, Troxler R F, Oppenheim F G. Candida albicans mutants deficient in respiration are resistant to the small cationic salivary antimicrobial peptide histatin 5. Antimicrob Agents Chemother. 2000;44:348–354. doi: 10.1128/aac.44.2.348-354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning. A practical approach. Oxford, England: IRL Press; 1985. pp. 109–135. [Google Scholar]
- 12.Holt R J, Azmi A. Miconazole-resistant Candida. Lancet. 1978;i:50–51. doi: 10.1016/s0140-6736(78)90403-8. [DOI] [PubMed] [Google Scholar]
- 13.Horsburgh C R, Kirkpatrick C H. Long-term therapy of chronic mucocutaneous candidiasis with ketoconazole: experience with twenty-one patients. Am J Med. 1983;74(Suppl. 1B):23–29. doi: 10.1016/0002-9343(83)90511-9. [DOI] [PubMed] [Google Scholar]
- 14.Johnson E M, Richardson M D, Warnock D W. In vitro resistance with imidazole antifungals in Candida albicans. J Antimicrob Chemother. 1984;13:541–558. doi: 10.1093/jac/13.6.547. [DOI] [PubMed] [Google Scholar]
- 15.Katzmann D J, Hallstrom T C, Mahe Y, Moye-Rowley W S. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J Biol Chem. 1996;271:23049–23054. doi: 10.1074/jbc.271.38.23049. [DOI] [PubMed] [Google Scholar]
- 16.Marichal P, Vanden Bossche H, Odds F C, Nobels G, Warnock D W, Timmerman V, Vanbroeckhoven C, Fay S, Moselarsen P. Molecular-biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1997;41:2229–2237. doi: 10.1128/aac.41.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marmiroli N, Tedeschi F, Truzzi G, Ferrari C, Puglisi P P. Relationship between growth inhibition and mitochondrial function in petite-negative yeasts. I. Effects of antibiotics and dyes upon pathogenic and non-pathogenic Candida species. Biol Cell. 1985;53:67–74. doi: 10.1111/j.1768-322x.1985.tb00353.x. [DOI] [PubMed] [Google Scholar]
- 18.Marr K A, Lyons C N, Ha K, Rustad T R, White T C. Inducible azole resistance associated with a heterogeneous phenotype in Candida albicans. Antimicrob Agents Chemother. 2001;45:52–59. doi: 10.1128/AAC.45.1.52-59.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyazaki H, Miyazaki Y, Geber A, Parkinson T, Hitchcock C, Falconer D J, Ward D J, Marsden K, Bennett J E. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob Agents Chemother. 1998;42:1695–1701. doi: 10.1128/aac.42.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mondon P, Petter R, Amalfitano G, Luzzati R, Concia E, Polacheck I, Kwon-Chung K J. Heteroresistance to fluconazole and voriconazole in Cryptococcus neoformans. Antimicrob Agents Chemother. 1999;43:1856–1861. doi: 10.1128/aac.43.8.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Commitee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeast. Tentative standard M27-A. Villanova, Pa: National Commitee for Clinical Laboratory Standards; 1995. [Google Scholar]
- 22.Nguyen M H, Peacock J E, Morris A J, Tanner D C, Nguyen M L, Snydman D R, Wagener M M, Rinaldi M G, Yu V L. The changing face of candidemia—emergence of non-Candida albicans species and antifungal resistance. Am J Med. 1996;100:617–623. doi: 10.1016/s0002-9343(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 23.Nicholas R O, Burton J A, Kerridge D, Wayman F J. Isolation of mutants of Candida glabrata resistant to miconazole. Crit Rev Microbiol. 1987;15:103–110. doi: 10.3109/10408418709104453. [DOI] [PubMed] [Google Scholar]
- 24.Roth-Ben Arie Z, Altboum Z, Berdicevsky I, Segal E. Isolation of a petite mutant from a histidine auxotroph of Candida albicans and its characterization. Mycopathologia. 1998;141:127–135. doi: 10.1023/a:1006988119891. [DOI] [PubMed] [Google Scholar]
- 25.Sanglard D, Ischer F, Calabrese D, De Micheli M, Bille J. Multiple resistance mechanisms to azole antifungals in yeast clinical isolates. Drug Resist Updates. 1998;1:255–265. doi: 10.1016/s1368-7646(98)80006-x. [DOI] [PubMed] [Google Scholar]
- 26.Sanglard D, Ischer F, Calabrese D, Majcherczyk P A, Bille J. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob Agents Chemother. 1999;43:2753–2765. doi: 10.1128/aac.43.11.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents—characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 29.Sanglard D, Kuchler K, Ischer F, Pagani J L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanden Bossche H, Marichal P, Odds F C. Molecular mechanisms of drug resistance in fungi. Trends Microbiol. 1994;2:393–400. doi: 10.1016/0966-842x(94)90618-1. [DOI] [PubMed] [Google Scholar]
- 31.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]