Abstract
Maturity-onset diabetes of the young (MODY) is an autosomal dominant form of monogenic diabetes, reported to be caused by variants in 16 genes. Concern has been raised about whether variants in BLK (MODY11), KLF11 (MODY7), and PAX4 (MODY9) cause MODY. We examined variant-level genetic evidence (cosegregation with diabetes and frequency in population) for published putative pathogenic variants in these genes and used burden testing to test gene-level evidence in a MODY cohort (n = 1,227) compared with a control population (UK Biobank [n = 185,898]). For comparison we analyzed well-established causes of MODY, HNF1A, and HNF4A. The published variants in BLK, KLF11, and PAX4 showed poor cosegregation with diabetes (combined logarithm of the odds [LOD] scores ≤1.2), compared with HNF1A and HNF4A (LOD scores >9), and are all too common to cause MODY (minor allele frequency >4.95 × 10−5). Ultra-rare missense and protein-truncating variants (PTV) were not enriched in a MODY cohort compared with the UK Biobank population (PTV P > 0.05, missense P > 0.1 for all three genes) while HNF1A and HNF4A were enriched (P < 10−6). Findings of sensitivity analyses with different population cohorts supported our results. Variant and gene-level genetic evidence does not support BLK, KLF11, or PAX4 as a cause of MODY. They should not be included in MODY diagnostic genetic testing.
Introduction
Maturity-onset diabetes of the young (MODY) is the most common subtype of monogenic diabetes. It is reported to be caused by heterozygous variants in 16 genes (1). MODY accounts for ∼3% of all diabetes cases diagnosed before 30 years of age (2,3). The prevalence of MODY is estimated to be 108 cases per million (4). An accurate genetic diagnosis is important for patients with MODY, as it can determine the correct treatment (1,5) and provides an accurate assessment of the risk of diabetes for future offspring. The advent of next-generation sequencing has enabled a paradigm shift in genetic testing from focusing on single gene testing to gene panel tests for diseases (6). While this can boost diagnostic yield, it also has the potential to increase the risk of reporting variants in genes that are not a cause of MODY if genes are not carefully selected. This is likely to occur with next-generation sequencing, as it enables large numbers of genes to be tested more easily. An incorrect genetic diagnosis could result in stopping insulin in a patient with type 1 diabetes. It could lead to inappropriate testing of family members, causing increased anxiety in unaffected relatives and inflicting the psychological burden of having a genetic disease. Therefore, it is crucial that the gene panel only includes genes with robust aetiological evidence to prevent misdiagnosis of MODY.
BLK, KLF11, and PAX4 are listed on Online Mendelian Inheritance in Man (OMIM) as MODY11, MODY7, and MODY9, but there is a need to reevaluate whether variants in these genes do cause MODY. Variants in BLK and PAX4 have been reported to cause MODY via haploinsufficiency (7,8), while variants in KLF11 were reported to cause the disease, potentially via a gain-of-function mechanism (9). These studies were conducted >10 years ago, before the availability of variant frequency in large population cohorts (7–9). KLF11 and PAX4 were identified based primarily on biological candidacy rather than the hypothesis-free genetic approach, which is now considered to be the most robust method for gene discovery studies. The only BLK coding variant (p.A71T) reported to cause MODY was later found to be very common in the population, raising doubt over the aetiological role of BLK (10). Rarity of a variant in a large control population as well as enrichment of variants in that gene in a disease cohort compared with a control population has become crucial evidence to support the gene-disease association alongside familial cosegregation (11,12).
Therefore, the aim of our study was to evaluate genetic evidence for variants in BLK, KLF11, and PAX4 as a cause of MODY. We evaluated the existing evidence for these genes and assessed the gene-disease association using a large MODY cohort and population cohorts. We demonstrate that there is a lack of robust genetic evidence to support the aetiological role of variants in BLK, KLF11, and PAX4 for MODY.
Research Design and Methods
Study Populations
MODY Cohort
We included 1,227 unrelated probands from the U.K. who were referred for genetic testing for MODY from routine clinical care to the Exeter Genomics Laboratory at the Royal Devon and Exeter Hospital. Cohort characteristics can be found in Supplementary Table 1. None of these individuals were reported to have islet autoantibodies by the referring clinicians. Of patients, 84% were of self-reported European ancestry, and the overall rate of monogenic diabetes was 22.5%. Informed consent was obtained from the probands or their parents/guardians, and the study was approved by the North Wales ethics committee (17/WA/03).
UK Biobank
UK Biobank is a population-based cohort from the U.K. with deep phenotyping data and genetic data for ∼500,000 individuals aged 40–70 years at recruitment (13,14). A subset of ∼200,000 DNA samples from UK Biobank participants underwent exome sequencing; this data set was recently made available for research (15). Of individuals included, 94% were of self-reported White ethnicity. The UK Biobank resource was approved by the UK Biobank Research Ethics Committee, and all participants provided written informed consent to participate.
gnomAD
We used Genome Aggregation Database (gnomAD) v2.1.1 (141,456 individuals) and v3 (76,156 individuals) for alternative control populations in supplementary analyses. A detailed description of the cohort has previously been published (16). gnomAD v2.1.1 includes individuals with exome (n = 125,748) and genome (n = 15,708) data, whereas v3 includes individuals with genome data. Of individuals in gnomAD v2.1.1, 46% are of non-Finnish European ancestry, while for v3 this is 45%.
Genetic Testing
MODY Cohort
We undertook targeted next-generation sequencing of BLK, PAX4, and KLF11 as well as HNF1A and HNF4A for probands suspected to have MODY, as previously described (6). Targets were covered at a mean read depth of 460X per base, and all bases had a mean coverage depth of at least 30 reads across the cohort. Variants were annotated against Genome Reference Consortium Human Build 37 (GRCh37) with Alamut Batch (Interactive Biosoftware, Rouen, France) using a RefSeq transcript: BLK NM_001715.3, KLF11 NM_003597.4, PAX4 NM_001366110.1, HNF1A NM_000545.6, and HNF4A NM_175914.4.
UK Biobank
We included 185,898 unrelated individuals from all ethnicities with exome sequencing data (17). Detailed sequencing methodology for UK Biobank samples is provided by Szustakowski et al. (15) and is available from https://biobank.ctsu.ox.ac.uk/showcase/label.cgi?id=170. Variants were called against Genome Reference Consortium Human Build 38 (GRCh38). We lifted the variants over to GRCh37 (18) to ensure compatibility with the variants from our MODY cohort and then annotated variants using Alamut Batch with a RefSeq transcript: BLK NM_001715.3, KLF11 NM_003597.4, PAX4 NM_001366110.1, HNF1A NM_000545.6, and HNF4A NM_175914.4.
gnomAD
The gnomAD consortium performed joint variant calling of the samples using a standardized BWA-Picard-GATK pipeline (16). GnomAD was quality controlled and analyzed with use of the Hail open-source framework for scalable genetic analysis (https://gnomad.broadinstitute.org/about). Variants in v2.1.1 were called against GRCh37 and in v3 against GRCh38. We lifted over v3 to GRCh37 and then annotated all gnomAD variants using Alamut Batch with use of a RefSeq transcript: BLK NM_001715.3, KLF11 NM_003597.4, PAX4 NM_001366110.1, HNF1A NM_000545.6, and HNF4A NM_175914.4. As gnomAD is an agglomeration of different sequencing projects, some genomic regions have low coverage in some samples; therefore, to control for this, we removed the variants from both the MODY cohort and gnomAD cohorts if they were in a region of low coverage (≤10× coverage in ≤80% of samples) in either cohort or flagged as low quality in gnomAD.
Cosegregation Analysis of Putative Pathogenic Variants
We used author-provided LOD (logarithm of the odds) scores where available for the first published variants in BLK, PAX4, KLF11, HNF1A, and HNF4A, which suggested the causal role of those variants in MODY. This was only available for BLK p.A71T (7). If the LOD score was not provided, we calculated it based on the Gene Clinical Validity Curation Process Standard Operating Procedure (19). We summed the LOD scores for multiple pedigrees where possible based on this guidance to calculate a combined LOD score. Using a binomial test we compared the observed proportion of family members with diabetes and a putative variant with the expected proportion of 0.5 if the variant was not associated with diabetes.
Statistical Analysis
For each analysis, variant frequency was defined in the MODY cohort plus the control cohort combined. We compared the frequency of ultra-rare (allele count = 1) protein-truncating variants (PTV) (essential splice site, stop-gain, and frameshift variants, excluding those in the last exon) and missense variants in each gene in the MODY cohort with that in the UK Biobank population cohort. We also provided the evidence of an association in terms of Bayesian false discovery probabilities (BFDP) as previously described (20). We replicated our analysis using two alternative control populations: gnomAD v2.1.1 (141,456 individuals) and gnomAD v3 (76,156 individuals) (16).
We used synonymous variants as a control to assess the difference in sequencing technologies and analysis pipeline. We also compared the frequency of rare variants (minor allele frequency [MAF] <0.0001) and the frequency of all PTV (no frequency filter) to test whether there was an undue influence of ultra-rare variants due to differences in capture platforms.
The most common HNF1A pathogenic variant is a frameshift variant (p.G292Rfs*25) in exon 4 due to a duplication of a C nucleotide. This variant is difficult to detect robustly in exome/genome sequencing data due to its location in a repetitive poly-C tract and the presence of a common variant that adds an additional 5′ C nucleotide to the tract (rs56348580 G>C, MAF = 0.26). Since we were unable to perform confirmatory Sanger sequencing in the UK Biobank or gnomAD cohorts, we excluded this variant from our analysis from all study cohorts.
We used Fisher exact test to assess variant enrichment in our MODY cohort and compute odds ratios (ORs) with 95% CIs. We used a threshold P value of 0.01 (0.05/5), as we tested five genes. We used Stata 16 (StataCorp, College Station, TX) for this analysis. BFDP was computed using a “gap” R package. We used a prior probability of association of 0.2. We calculated the variance of the prior log(OR), as described by Wakefield (20). We also explored different plausible priors as a sensitivity analysis.
Data and Resource Availability
UK Biobank data are accessible via application: https://www.ukbiobank.ac.uk/enable-your-research. GnomAD data are publically available: https://gnomad.broadinstitute.org/. The MODY cohort data are not publicly available due the limitations of the current ethics and to protect patient confidentiality but are available from the corresponding authors on reasonable request. No applicable resources were generated or analyzed during the current study.
Results
BLK, KLF11, and PAX4 Variants Had Poor Cosegregation in the Published Pedigrees
Variants that are highly penetrant causes of MODY would be expected to show strong cosegregation with the disease. To evaluate the genetic evidence of cosegregation with disease, we reviewed published pedigrees for putative variants in BLK, KLF11, and PAX4 causing MODY (Supplementary Table 2). We identified one BLK, three KLF11, and one PAX4 pedigrees with more than three individuals with variants to calculate LOD scores (7–9). KLF11 and PAX4 variants showed poor cosegregation with diabetes in the families, with LOD scores of 1.2 and 0.6, respectively (Table 1). In line with low LOD scores, these variants were not associated with diabetes in family members in these pedigrees (P > 0.5) (Table 1). The BLK variant p.A71T also had a low LOD score of 1.16 and was modestly associated with diabetes in family members (P = 0.02). In contrast, the variants reported in the first articles for HNF1A (21) and HNF4A (22), which are well-established causes of MODY, showed strong cosegregation with diabetes, with combined LOD scores for the first reported variants of 9.63 and 15.05, respectively (Table 1).
Table 1.
Cosegregation of BLK, KLF11, and PAX4 published variants with diabetes
| Gene | No. pedigree used in analysis | Variants (reference no.) | Combined LOD score | No. of family members with diabetes and variant/total no. of family members with diabetes and genotype information | Proportion of family members with diabetes and variant (95% CI) | Binomial test P value (against expected proportion of 0.5) |
|---|---|---|---|---|---|---|
| BLK | 1 | p.A71T (7) | 1.16 | 9/10 | 0.9 (0.55–1) | 0.02 |
| KLF11 | 3 | p.A347S (9), p.T220M (9)† | 1.2 | 2/4 | 0.5 (0.068–0.93) | 1 |
| PAX4 | 1 | p.R164W (8) | — | 2/2 | 1 (0.16–1) | 0.5 |
| HNF1A | 7 | p.G292Rfs*25 (21), p.P447L (21), p.V380Sfs*4 (21), p.E548Rfs*112 (21), p.R131Q (21), c.1768 + 1G>A (21), c.1108-2A>G (21) | 9.63 | 38/39 | 0.97 (0.87–1) | 1.4 × 10−10 |
| HNF4A | 2 | p.Q255* (43), p.R141* (43) | 15.05 | 49/50 | 0.98 (0.89–1) | 9 × 10−14 |
Table shows the LOD scores and association of variants with diabetes in family members for variants where there were families with three or more people with the variant. We used author-provided LOD scores where available for the first published variants, which suggested the causal role of those variants in MODY. If the LOD score was not provided, we calculated it based on the Gene Clinical Validity Curation Process Standard Operating Procedure (19). We summed the LOD score for each pedigree to calculate the combined LOD score.
Two pedigrees with p.T220M were included in the combined LOD score calculation.
Putative Pathogenic Variants in BLK, KLF11, and PAX4 Are Common in the Population
The frequency of a putative pathogenic variant should not exceed the expected prevalence of the commonest variant in the commonest genetic subtype of the disease. MODY is estimated to have a population frequency of 1.08 per 10,000 (4). We used the framework developed by Whiffin et al. (23,24) to calculate the maximum tolerated allele count in the population (gnomAD v2.1.1 [n = 141,456]) for a putative pathogenic variant causing MODY. We used HNF1A, the most common cause of MODY, as a model to calculate the maximum tolerated allele count in the population. HNF1A accounts for 52% of MODY cases (4), and the most common mutation (p.G292Rfs*25) accounts for 19% of HNF1A cases (25). At 50% penetrance, the framework suggests that a pathogenic variant causing MODY should be present three or fewer times (frequency <2.1 × 10−5) in gnomAD v2.1.1 for HNF1A. As other genes will account for far fewer MODY cases, the putative pathogenic variants in BLK, KLF11, and PAX4 should be even rarer.
We looked at the frequency of variants in BLK, KLF11, and PAX4 that were reported to cause MODY before large-scale population data were made available publicly in 2016 (26) (Table 2). Publications on the variants since 2016 should have included the frequency of the variant in these databases as part of their screening process and thus would be expected to have only included rare variants (see Supplementary Table 2 for full list of Human Gene Mutation Database [HGMD] variants in these genes).
Table 2.
Population frequency of variants in BLK, KLF11, and PAX4 published as MODY causing
| Gene | Variant | Allele count/total alleles in gnomAD v2.1.1 | Allele frequency in gnomAD v2.1.1 | Allele count in ancestry with maximum frequency/ total alleles in the ancestry | Maximum allele frequency in a single ancestry in gnomAD v2.1.1 (ancestry) | Reference no. for variants |
|---|---|---|---|---|---|---|
| BLK | p.A71T | 3,281/282,812 | 0.012 | 420/10,368 | 0.041 (Ashkenazi Jewish) | 7 |
| KLF11 | p.Q62R | 25,823/282,778 | 0.091 | 1,497/10,370 | 0.144 (Ashkenazi Jewish) | 9 |
| p.T220M | 1,207/282,762 | 4.27 × 10−3 | 1,098/24,958 | 0.044 (African/African American) | 9 | |
| p.A347S | 36/282,304 | 1.28 × 10−4 | 17/35,410 | 4.80 × 10−4 (Latino/Admixed American) | 9 | |
| PAX4 | p.R31L | 105/250,972 | 4.18 × 10−4 | 102/30,616 | 0.003 (South Asian) | 44 |
| p.R164W | 14/282,800 | 4.95 × 10−5 | 3/24,948 | 1.2 × 10−4 (African/African American) | 8 | |
| p.R192H | 2,214/282,856 | 7.83 × 10−3 | 2,182/19,946 | 0.109 (East Asian) | 8 | |
| HNF1A | p.P447L | 3/249,186 | 1.20 × 10−5 | 1/20,812 | 4.81 × 10−5 (Finnish European) | 21 |
| p.V380Sfs*4 | 0 | 0 | 0 | 0 | 21 | |
| p.E548Rfs*112 | 0 | 0 | 0 | 0 | 21 | |
| p.R131Q | 1/251,390 | 3.98 × 10−6 | 1/113,698 | 8.80 × 10−6 (non-Finnish European) | 21 | |
| c.1768 + 1G>A | 0 | 0 | 0 | 0 | 21 | |
| c.1108-2A>G | 0 | 0 | 0 | 0 | 21 | |
| HNF4A | p.Q255* | 0 | 0 | 0 | 0 | 42 |
| p.R141* | 0 | 0 | 0 | 0 | 43 |
Allele frequency taken from gnomAD v2.1.1. The table provides coding variants reported before 2016 that are reported to cause MODY, as the release of ExAC (26) that year meant for variants published since then investigators have had access to a large control population as part of their screening process. The HNF1A and HNF4A variants included here for comparison are those from the original articles used in the LOD score calculations in Table 1.
All putative MODY-causing variants in BLK, KLF11, and PAX4 with publication prior to 2016 were too common in the population to cause MODY. The allele count in gnomAD v2.1.1 was 4–8,608 times higher than the maximum tolerable allele count for the commonest cause of MODY (Table 2). The least common was PAX4 p.R164W, which is seen 14 times in the whole of gnomAD v2.1.1 at a frequency of 4.95 × 10−5 but seen at higher frequency, of 1.2 × 10−4 (3 of 24,948), in the African/African American population. In contrast, the first reported variants in HNF1A and HNF4A, which were reported in the 1990s, are rare in the population, with the most common (p.P447L) present three times in gnomAD v2.1.1 (1.20 × 10−5) (Table 2).
Rare Variants in BLK, KLF11, and PAX4 Are Not Enriched in a MODY Cohort
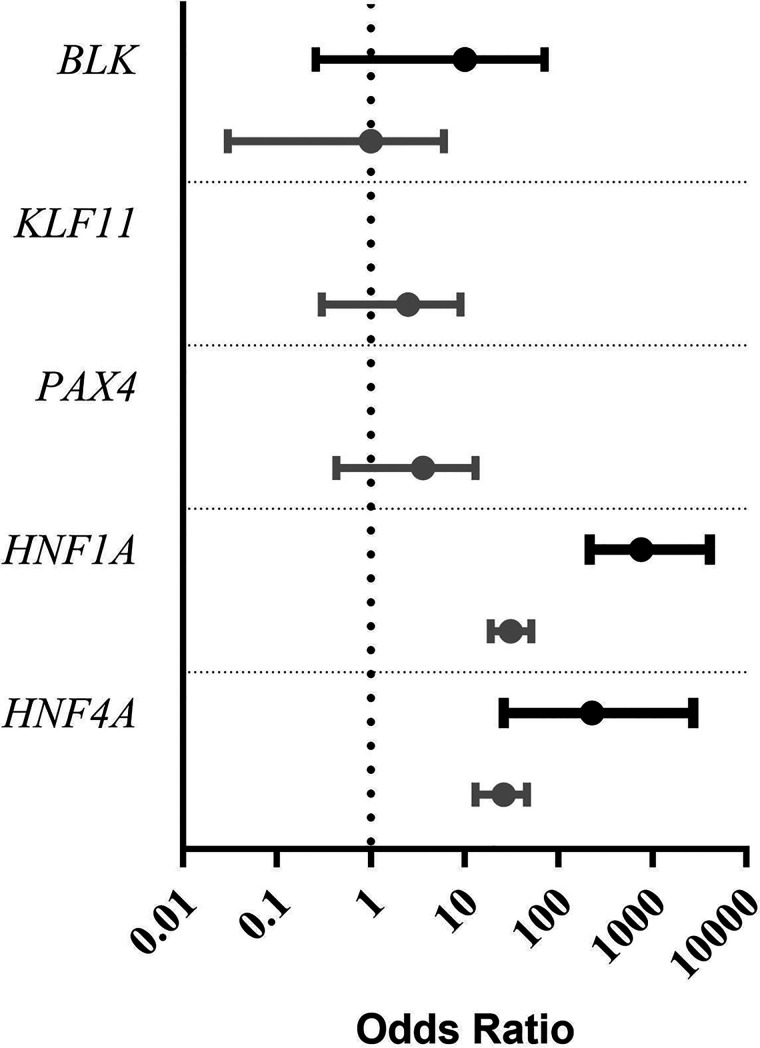
Having conducted variant-level analyses on published variants in these genes, we then carried out a gene-level analysis to establish whether other rare variants in these genes are likely to be pathogenic for MODY. To assess this, we carried out a gene burden test comparing the frequency of ultra-rare coding variants in a cohort of 1,227 patients referred for MODY genetic testing with the frequency in the unrelated 185,898 exome-sequenced individuals from the UK Biobank population cohort (Table 3 and Fig. 1).
Table 3.
Results of gene burden tests comparing the frequency of variants in BLK, KLF11, and PAX4 in a disease cohort with that in a population cohort
| Variant type | Gene | Allele count in MODY cohort | Allele frequency in MODY cohort | Allele count in population cohort (UK Biobank) | Allele frequency in population cohort (UK Biobank) | OR (95% CI) | P | Prior probability | BFDP |
|---|---|---|---|---|---|---|---|---|---|
| Ultra-rare PTV | BLK | 1 | 4.10 × 10−4 | 14 | 3.77 × 10−5 | 10 (0.26–71) | 0.09 | 0.2 | 0.71 |
| KLF11 | 0 | 0 | 15 | 4.03 × 10−5 | 0 (0–39) | 1 | 0.2 | 0.8 | |
| PAX4 | 0 | 0 | 6 | 1.61 × 10−5 | 0 (0–97) | 1 | 0.2 | 0.8 | |
| HNF1A | 15 | 6.11 × 10−3 | 3 | 8.07 × 10−6 | 762 (215–4,108) | 1.37 × 10−30 | 0.2 | 1.20 × 10−9 | |
| HNF4A | 3 | 1.22 × 10−3 | 2 | 5.38 × 10−6 | 228 (26–2,724) | 2.79 × 10−6 | 0.2 | 0.03 | |
| Ultra-rare missense variants | BLK | 1 | 4.10 × 10−4 | 141 | 3.79 × 10−4 | 1 (0.03–6.00) | 0.6 | 0.2 | 0.82 |
| KLF11 | 2 | 8.10 × 10−4 | 122 | 3.28 × 10−4 | 2.5 (0.3–9.0) | 0.2 | 0.2 | 0.78 | |
| PAX4 | 2 | 8.10 × 10−4 | 84 | 2.26 × 10−4 | 3.6 (0.43–13.00) | 0.1 | 0.2 | 0.70 | |
| HNF1A | 21 | 8.56 × 10−3 | 102 | 2.74 × 10−4 | 31 (19–51) | 1.70 × 10−23 | 0.2 | 5.80 × 10−38 | |
| HNF4A | 14 | 5.70 × 10−3 | 83 | 2.23 × 10−4 | 26 (13–46) | 4.41 × 10−15 | 0.2 | 1.20 × 10−25 |
The frequency of ultra-rare (allele count = 1) PTV and missense variants in a MODY cohort (n = 1,227) was compared with the frequency in the UK Biobank population cohort (n = 185,898).
Figure 1.
Graph showing the ORs for ultra-rare (allele count = 1) PTV (black lines) and missense variants (gray lines) in a MODY cohort (n = 1,227) compared with the population cohort UK Biobank (n = 185,898). Dotted line shows an OR of 1. Bar not shown for KLF11 and PAX4 PTV, as OR is 0, which cannot be plotted on a log axis.
Ultra-rare (allele count = 1) PTV and missense variants in BLK, KLF11, and PAX4 are not enriched in our MODY cohort compared with the UK Biobank (all P values ≥0.09) (Table 3). The BFDP for ultra-rare PTV and missense variants was ≥0.70 for BLK, KLF11, and PAX4 (Table 3). The results of BFDP remained ≥0.37 with use of other plausible priors (Supplementary Table 3). In contrast, variants in HNF1A and HNF4A, which are well-established causative genes for MODY, were greatly enriched in our MODY cohort (all P values ≤2.79 × 10−6) with a very low BFDP (all ≤6.74 × 10−5).
Lack of Enrichment of Rare Variants in BLK, KLF11, and PAX4 Is Not Due to Technical Artifacts
To ensure that our results are not due to differences in sequencing technologies or analysis pipelines between case and control subjects, we performed a series of sensitivity analyses. Firstly, we analyzed synonymous variant frequency in our MODY cohort and control population and showed that the frequency of synonymous variants in all five genes was similar in our MODY cohort and the UK Biobank population (all P > 0.05) (Supplementary Table 4).
Secondly, we replicated our gene burden analysis using gnomAD v2.1.1 and v3 as two alternative population cohorts with sequencing on different platforms (exome vs. genome, respectively) and with a different analysis pipeline versus the UK Biobank. Despite these differences, we found similar results, with no enrichment in PTV or missense variants in BLK, KLF11, or PAX4 (Supplementary Tables 5 and 6).
Finally, to remove any undue influence of ultra-rare variants caused by differences in capture platforms, we performed a gene burden analysis for rare PTV and missense variants (MAF <0.0001). We also compared the frequency of all PTV in our MODY cohort and control population, as all PTV in these genes are considered to be pathogenic. These analyses showed results similar to those of our main analysis: rare PTV and missense variants, and all PTV, in BLK, KLF11, and PAX4 were not enriched in our MODY cohort, whereas all of these variant subsets in HNF1A and HNF4A showed great enrichment in our MODY cohort. (Supplementary Tables 7 and 8).
Discussion
Variant- and gene-level genetic evidence presented in this study suggests that variants in BLK, KLF11, and PAX4 do not cause MODY. The lack of cosegregation of published MODY-causing variants, presence in the population at high frequency, and lack of enrichment of rare variants in a MODY cohort are consistent with these genes not causing MODY. The robustness of our approach is demonstrated by the results supporting the well-established causality of HNF1A and HNF4A variants.
Variants in BLK, KLF11, and PAX4 were reported to cause MODY >10 years ago, before large-scale variant population frequency became available (7–9). Only small numbers of control subjects were available for ruling out variants being present in the population.
BLK was first described in 2009 (7) through follow-up of linkage to the 8p23 region (27) in six MODY families and identification of variants in BLK in three of the families. The frequency of the BLK variants was tested in 336 White control individuals and, for one variant, an additional 577 African American control individuals. BLK was identified via a linkage approach; it is possible that another candidate gene within the region of linkage is responsible for the disease in those families. Bonnefond et al. (10) found that the only nonsynonymous variant in BLK reported to cause MODY was common in normoglycemic individuals. This is the variant (p.A71T) that has a positive LOD score in the published pedigree; however, as BLK was identified by linkage, the LOD score would necessarily be positive regardless of the pathogenicity of the variant and, as also demonstrated by its frequency in gnomAD, the variant is clearly too common to cause MODY. No large MODY pedigrees with cosegregation have been described for BLK since the initial report. Noncoding variants in BLK were also reported to cause MODY (7); however, as our main cohorts consisted of targeted and exome sequencing data, we were unable to investigate noncoding variants. It is unlikely that noncoding variants would be pathogenic given the lack of evidence for coding variants in BLK as a cause of MODY and that both coding and noncoding variants were proposed to cause the disease via loss of function.
KLF11 was proposed as a cause of MODY, with a candidate gene approach, in 2005 (9). The frequency of the reported KLF11 variants was judged in only 313 normoglycemic individuals and 313 patients with type 2 diabetes. In functional studies with use of Gal4 reporter assays, a possible mechanism of action was suggested for the variants via gain of function causing increased KLF11 repression activity. If pathogenic variants in KLF11 act via gain of function then we would not expect to see enrichment of PTV in a MODY cohort but we might expect to see enrichment of missense variants. We did not see enrichment of either type of variant, and the previously identified KLF11 variants are too common in the population to be disease causing.
PAX4 was associated with MODY in patients from Thailand (8). The variants were screened in a maximum of 344 individuals without diabetes. While the control subjects of this study were from the same population as the case subjects, in using data from gnomAD we now know that p.R192H is common in East Asians and both this variant and p.R164W are too common to cause MODY (p.R192H seen 2,214 times in gnomAD v2.1.1 and p.R164W seen 14 times). Plengvidhya et al. (8) used luciferase reporter assays to show that p.R164W impairs the repressor activity of PAX4 on the insulin and glucagon promoters. However, they stated that the impairment was relatively small; thus, it is possible that the reduction may be insufficient to result in a clinical phenotype. No large MODY pedigrees with cosegregation for a variant in PAX4 have been described since the initial report.
In our study we had a large cohort of MODY cases and took advantage of the availability of large population cohorts. The lack of enrichment for BLK, KLF11, and PAX4 PTV and missense variants in a MODY cohort compared with a population cohort is consistent with these genes not causing MODY. However, alternative explanations may be that the mechanism of action for these genes is not loss of function (as has been suggested for KLF11 [9]) or they are an extremely rare cause of MODY. However, we did not see enrichment in missense variants (at either allele count = 1 or MAF <0.0001), suggesting that this is unlikely. In line with our results, gnomAD pLI (probability of being loss of function intolerant) and missense constraint scores for these genes are low, suggesting that these genes are not under strong negative selection—in contrast to HNF1A and HNF4A, which have high constraint scores. These data suggest that variants in these genes do not cause a rare monogenic disorder.
Variants in these genes could still be acting as polygenic risk factors for diabetes. PAX4 has been reported in the literature as a type 2 diabetes risk factor in East Asian populations (28,29). Fuchsberger et al. (30), in a study of 6,504 type 2 diabetes case and 6,436 control subjects, found a lack of exome-wide enrichment of PTV and deleterious missense variants for BLK (P ≈ 0.001), KLF11 (P > 0.05), and PAX4 (P > 0.05). However, the PAX4 p.R192H variant (a proposed MODY variant) showed association with type 2 diabetes in East Asian case subjects (OR 1.79) but not with age of diabetes diagnosis (P = 0.64), suggesting that this variant influences risk of type 2 diabetes rather than early-onset MODY. Similar to this, in a recent large type 2 diabetes case-control study of 20,791 case and 24,440 control subjects investigators did not find an exome wide-significant association in these genes, except for the PAX4 p.R192H variant, which was associated with type 2 diabetes in East Asians (31). However, the lack of exome-wide significance may reflect the relatively small size of these studies.
A limitation of our study is that by virtue of using publicly available control populations there were cross-platform differences between case and control individuals. This issue was mitigated by removal of genomic positions with low coverage in one cohort from the other and in our sensitivity analyses with use of synonymous variants as a negative control and testing alternative population control cohorts. Despite using a large MODY cohort, we still had a relatively limited sample size of cases, which could trend our gene burden tests of ultra-rare variants toward negative results. To ensure a lack of power was not determining the results we also used sensitivity analyses with MAF <0.001, and these did not suggest there was an association between BLK, KLF11, or PAX4 and MODY. One other caveat to our burden testing results is the fact that both our MODY cohort and the UK Biobank are predominantly European ancestry. We cannot rule out that an enrichment might be seen in MODY cohorts from other ancestries, particularly for PAX4, which was originally reported in East Asian ancestry. It must be acknowledged that the power of cosegregation analysis was limited, particularly for BLK and PAX4, as they are only based on one family each. However, in detailed review of all the published articles on putative pathogenic variants we did not identify additional large published pedigrees for cosegregation analysis.
Our study results have important implications for genetic diagnostic laboratories worldwide who offer testing for MODY. Based on our results, we recommend that BLK, KLF11, and PAX4 should not be included in the gene panels for genetic testing for MODY and should not be reported as a cause of MODY. Studies are still reporting variants in these genes as a cause of MODY, and they are routinely tested in clinical practice (32–36). In our systematic review of the National Center for Biotechnology Information Genetic Testing Registry we found that 19 of 25 panels offered by diagnostic genetic laboratories still have at least one of these genes on their panel. The results of our study remove the ambiguity of the etiological role of these genes for MODY and provide the clearest results to date that refute their role as causative genes for MODY. Excluding these genes from diagnostic panels will prevent misdiagnosis of MODY and reduce workload for laboratories. The results from our study provide much needed evidence to gene curation efforts such as Clinical Genome Resource (ClinGen) and the Gene Curation Coalition (GenCC) to support the removal of these three genes from MODY genetic panels (37,38). The ClinGen curation panel also came to a similar conclusion using their own scoring system (12) independently of our study. They classified BLK and PAX4 as “Refuted” genes (https://search.clinicalgenome.org/kb/genes/HGNC:1057; https://search.clinicalgenome.org/kb/genes/HGNC:8618) and KLF11 as a “Disputed” gene (https://search.clinicalgenome.org/kb/genes/HGNC:11811). However, it is important to note that in addition to their own approach, they used our current work as previously published as a conference abstract to reach their conclusion. We also strongly recommend that variants in BLK, KLF11, and PAX4 should be removed as a cause of MODY on databases such as HGMD (39), OMIM (40), ClinVar, and PanelApp (41) that are widely used by diagnostic laboratories and geneticists worldwide.
In conclusion, we present evidence from reanalysis of published variants in BLK, KLF11, and PAX4 that they are too common to cause MODY and have poor cosegregation with diabetes in those families and, since their initial description, no large MODY families with cosegregation of a variant have been published. We have shown a lack of enrichment of rare variants in these genes in a MODY cohort compared with a population cohort, providing evidence that rare variants in these genes do not cause MODY. Overall, the evidence does not support BLK, KLF11, or PAX4 as causes of MODY, and they should not be included in diagnostic genetic testing.
Article Information
Acknowledgments. The authors acknowledge use of the University of Exeter High-Performance Computing (HPC) facility in carrying out this work.
Funding. This research was conducted with use of the UK Biobank resource under application no. 49847. The current work was funded by Diabetes UK (19/0005994) and the Medical Research Council (grant no. MR/T00200X/1). T.W.L. is supported by a lectureship funded by the Research England Expanding Excellence in England (E3) fund. K.A.P. has a Career Development fellowship funded by the Wellcome Trust (219606/Z/19/Z). A.T.H. was a recipient of a Wellcome Trust Senior Investigator award (grant WT098395/Z/12/Z), is employed as a core member of staff within the National Institute for Health Research (NIHR)-funded Exeter Clinical Research Facility, and is an NIHR senior investigator. For the purpose of open access, the authors have applied a CC BY public copyright license to any author accepted manuscript version arising from this submission.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. C.F.W., S.E., A.T.H., M.N.We., and K.A.P. designed the study. K.C., S.E., A.T.H., and K.A.P. recruited patients and performed clinical phenotyping. T.W.L., M.N.Wa., and O.K. analyzed the MODY cohort targeted sequencing data. M.N.We. and K.A.P. analyzed the UK Biobank exome sequencing data. T.W.L., M.N.Wa., O.K., M.N.We., and K.A.P. carried out statistical analysis. T.W.L. and K.A.P. wrote the manuscript. All authors reviewed and approved the manuscript. K.A.P. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.19099838.
References
- 1. Owen KR. Monogenic diabetes in adults: what are the new developments? Curr Opin Genet Dev 2018;50:103–110 [DOI] [PubMed] [Google Scholar]
- 2. Pihoker C, Gilliam LK, Ellard S, et al.; SEARCH for Diabetes in Youth Study Group . Prevalence, characteristics and clinical diagnosis of maturity onset diabetes of the young due to mutations in HNF1A, HNF4A, and glucokinase: results from the SEARCH for Diabetes in Youth. J Clin Endocrinol Metab 2013;98:4055–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shepherd M, Shields B, Hammersley S, et al.; UNITED Team . Systematic population screening, using biomarkers and genetic testing, identifies 2.5% of the U.K. pediatric diabetes population with monogenic diabetes. Diabetes Care 2016;39:1879–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shields BM, Hicks S, Shepherd MH, Colclough K, Hattersley AT, Ellard S. Maturity-onset diabetes of the young (MODY): how many cases are we missing? Diabetologia 2010;53:2504–2508 [DOI] [PubMed] [Google Scholar]
- 5. Hattersley AT, Patel KA. Precision diabetes: learning from monogenic diabetes. Diabetologia 2017;60:769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ellard S, Lango Allen H, De Franco E, et al. Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia 2013;56:1958–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borowiec M, Liew CW, Thompson R, et al. Mutations at the BLK locus linked to maturity onset diabetes of the young and β-cell dysfunction. Proc Natl Acad Sci U S A 2009;106:14460–14465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Plengvidhya N, Kooptiwut S, Songtawee N, et al. PAX4 mutations in Thais with maturity onset diabetes of the young. J Clin Endocrinol Metab 2007;92:2821–2826 [DOI] [PubMed] [Google Scholar]
- 9. Neve B, Fernandez-Zapico ME, Ashkenazi-Katalan V, et al. Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic beta cell function. Proc Natl Acad Sci U S A 2005;102:4807–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bonnefond A, Yengo L, Philippe J, et al. Reassessment of the putative role of BLK-p.A71T loss-of-function mutation in MODY and type 2 diabetes. Diabetologia 2013;56:492–496 [DOI] [PubMed] [Google Scholar]
- 11. MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature 2014;508:469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Strande NT, Riggs ER, Buchanan AH, et al. Evaluating the clinical validity of gene-disease associations: an evidence-based framework developed by the clinical genome resource. Am J Hum Genet 2017;100:895–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Szustakowski JD, et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. 4 November 2020 [preprint]. medRxiv: 2020.11.02.20222232 [DOI] [PubMed] [Google Scholar]
- 16. Karczewski KJ, Francioli LC, Tiao G, et al.; Genome Aggregation Database Consortium . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Exome sequences - genomics. Accessed 28 July 2021. Available from https://biobank.ctsu.ox.ac.uk/showcase/label.cgi?id=170
- 18. Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res 2002;12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clinical Genome Resource Gene Curation Working Group . Gene clinical validity curation process: standard operating procedure, 2019. Accessed 17 September 2021. Available from https://www.clinicalgenome.org/site/assets/files/3975/gene-disease_validity_standard_operating_procedures_version_ 7-1.pdf
- 20. Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet 2007;81:208–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamagata K, Oda N, Kaisaki PJ, et al. Mutations in the hepatocyte nuclear factor-1α gene in maturity-onset diabetes of the young (MODY3). Nature 1996;384:455–458 [DOI] [PubMed] [Google Scholar]
- 22. Yamagata K, Furuta H, Oda N, et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1). Nature 1996;384:458–460 [DOI] [PubMed] [Google Scholar]
- 23. Whiffin N, Minikel E, Walsh R, et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet Med 2017;19:1151–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ware J. alleleFrequencyApp, 2016. Accessed 17 September 2021. Available from https://www.cardiodb.org/allelefrequencyapp/
- 25. Colclough K, Bellanne-Chantelot C, Saint-Martin C, Flanagan SE, Ellard S. Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha and 4 alpha in maturity-onset diabetes of the young and hyperinsulinemic hypoglycemia. Hum Mutat 2013;34:669–685 [DOI] [PubMed] [Google Scholar]
- 26. Lek M, Karczewski KJ, Minikel EV, et al.; Exome Aggregation Consortium . Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim S-H, Ma X, Weremowicz S, et al. Identification of a locus for maturity-onset diabetes of the young on chromosome 8p23. Diabetes 2004;53:1375–1384 [DOI] [PubMed] [Google Scholar]
- 28. Shimajiri Y, Sanke T, Furuta H, et al. A missense mutation of Pax4 gene (R121W) is associated with type 2 diabetes in Japanese. Diabetes 2001;50:2864–2869 [DOI] [PubMed] [Google Scholar]
- 29. Kwak SH, Chae J, Lee S, et al. Nonsynonymous variants in PAX4 and GLP1R are associated with type 2 diabetes in an East Asian population. Diabetes 2018;67:1892–1902 [DOI] [PubMed] [Google Scholar]
- 30. Fuchsberger C, Flannick J, Teslovich TM, et al. The genetic architecture of type 2 diabetes. Nature 2016;536:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Flannick J, Mercader JM, Fuchsberger C, et al.; Broad Genomics Platform; DiscovEHR Collaboration; CHARGE; LuCamp; ProDiGY; GoT2D; ESP; SIGMA-T2D; T2D-GENES; AMP-T2D-GENES . Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature 2019;570:71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kleinberger JW, Copeland KC, Gandica RG, et al. Monogenic diabetes in overweight and obese youth diagnosed with type 2 diabetes: the TODAY clinical trial. Genet Med 2018;20:583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chapla A, Mruthyunjaya MD, Asha HS, et al. Maturity onset diabetes of the young in India - a distinctive mutation pattern identified through targeted next-generation sequencing. Clin Endocrinol (Oxf) 2015;82:533–542 [DOI] [PubMed] [Google Scholar]
- 34. Doddabelavangala Mruthyunjaya M, Chapla A, Hesarghatta Shyamasunder A, et al. Comprehensive maturity onset diabetes of the young (MODY) gene screening in pregnant women with diabetes in India. PLoS One 2017;12:e0168656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anık A, Çatlı, Ayhan Abacı, et al. Molecular diagnosis of maturity-onset diabetes of the young (MODY) in Turkish children by using targeted next-generation sequencing. 2015;28:1265–1271 [DOI] [PubMed] [Google Scholar]
- 36. Ming-Qiang Z, Yang-Li D, Ke H, et al. Maturity onset diabetes of the young (MODY) in Chinese children: genes and clinical phenotypes. J Pediatr Endocrinol Metab 2019;32:759–765 [DOI] [PubMed] [Google Scholar]
- 37. Rehm HL, Berg JS, Brooks LD, et al.; ClinGen . ClinGen--the Clinical Genome Resource. N Engl J Med 2015;372:2235–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. GenCC: The Gene Curation Coalition . Available from https://thegencc.org/
- 39. Stenson PD, Ball EV, Mort M, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat 2003;21:577–581 [DOI] [PubMed] [Google Scholar]
- 40. Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015;43:D789–D798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martin AR, Williams E, Foulger RE, et al. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat Genet 2019;51:1560–1565 [DOI] [PubMed] [Google Scholar]
- 42. Furuta H, Iwasaki N, Oda N, et al. Organization and partial sequence of the hepatocyte nuclear factor-4 alpha/MODY1 gene and identification of a missense mutation, R127W, in a Japanese family with MODY. Diabetes 1997;46:1652–1657 [DOI] [PubMed] [Google Scholar]
- 43. Lindner T, Gragnoli C, Furuta H, et al. Hepatic function in a family with a nonsense mutation (R154X) in the hepatocyte nuclear factor-4alpha/MODY1 gene. J Clin Invest 1997;100:1400–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chapla A, Mruthyunjaya MD, Asha HS, et al. Maturity onset diabetes of the young in India - a distinctive mutation pattern identified through targeted next-generation sequencing. Clin Endocrinol (Oxf) 2015;82:533–542 [DOI] [PubMed] [Google Scholar]