Abstract
Diabetes-related complications reflect longstanding damage to small and large vessels throughout the body. In addition to the duration of diabetes and poor glycemic control, genetic factors are important contributors to the variability in the development of vascular complications. Early heritability studies found strong familial clustering of both macrovascular and microvascular complications. However, they were limited by small sample sizes and large phenotypic heterogeneity, leading to less accurate estimates. We take advantage of two independent studies—UK Biobank and the Action to Control Cardiovascular Risk in Diabetes trial—to survey the single nucleotide polymorphism heritability for diabetes microvascular (diabetic kidney disease and diabetic retinopathy) and macrovascular (cardiovascular events) complications. Heritability for diabetic kidney disease was estimated at 29%. The heritability estimate for microalbuminuria ranged from 24 to 60% and was 41% for macroalbuminuria. Heritability estimates of diabetic retinopathy ranged from 6 to 33%, depending on the phenotype definition. More severe diabetes retinopathy possessed higher genetic contributions. We show, for the first time, that rare variants account for much of the heritability of diabetic retinopathy. This study suggests that a large portion of the genetic risk of diabetes complications is yet to be discovered and emphasizes the need for additional genetic studies of diabetes complications.
Introduction
Genome-wide association studies (GWAS) have identified >300 genetic loci associated with type 2 diabetes. Together these top GWAS signals explain >19% of the phenotypic variance in risk for the type 2 diabetes risk (1). Genetic exploration underlying type 1 diabetes has been heavily focused on the HLA region, although GWAS has identified >50 regions contributing to type 1 diabetes risk thus far (2–4). The area under the receiver operating characteristic curve of a genetic risk score for type 1 diabetes generated using top GWAS loci was estimated as high as 0.9 (5,6). Although there is strong inheritance of risk of developing diabetes, less is known about the heritability of diabetes complications. Poor glycemic control and duration of diabetes are two major risk factors for vascular injury (7,8). The progression of diabetes to the development of diabetes complications is heterogeneous, even in individuals with comparable glucose control and diabetes duration (9). This heterogeneity greatly complicates the prediction of risk and personalization of diabetes therapy.
Among other diabetes-associated diseases, diabetic kidney disease (DKD) has been extensively studied in family studies. Siblings with diabetes of probands with DKD had approximately two to four times the risk of developing DKD than siblings with diabetes of probands free of DKD (10–12). A heritability analysis of renal complications in type 1 diabetes estimated that 34–59% (adjusted for sex, diabetes duration, and age at diabetes onset; 24–42% unadjusted) of the variance was explained by common genetic variants, depending on the stages or phenotype definitions of DKD (13). A similar unadjusted analysis of DKD in individuals with type 2 diabetes estimated SNP heritability to be 8–12%, probably because of the phenotypic heterogeneity of kidney disease in type 2 diabetes (14).
Early family and twin studies suggested high concordance of diabetic retinopathy (DR) between family members (15,16). Of note, genetic components for the risk of DR appear more closely related to the severity of retinopathy rather than to the simple presence or absence of retinopathy (15,17). Heritability estimates from family studies range from 18 to 52% (18–20), while SNP heritability of severe DR due to common genetic variants is estimated at 7% (21).
Little is known about the genetic contributions to cardiovascular disease (CVD) heritability among individuals with diabetes. The heritability of coronary artery disease in the general population is estimated to be between 40 and 60% in family and twin studies (22–24) and ∼30% in studies of unrelated individuals using common genetic variants (25). However, the only heritability-based studies for CVD in diabetes populations come from small family studies of quantitative traits, including coronary artery calcification (26) and carotid intima-media thickness (27).
As the rising prevalence of diabetes has led to more people at risk for serious complications, elucidating the genetic contribution (i.e., heritability) to the development and progression of complications takes on greater urgency. A deeper understanding of these genetic connections with diabetes complications may identify those most in need of aggressive interventions, uncover new target pathways, and ultimately enhance our ability to use precision medicine for tailored disease prevention/treatment. In the current study, we conducted a comprehensive heritability analysis using two well-characterized cohorts—the UK Biobank (UKB) study and the Action to Control Cardiovascular Risk in Diabetes (ACCORD) trial—to investigate genetic components involved in the development and progression of diabetes complications. Our results highlight the importance of the genetic contribution, whether alone or in conjunction with environmental perturbations, to the development and progression of diabetes complications.
Research Design and Methods
Study Design and Participants
The UKB study recruited during the years 2006–2010 ∼500,000 individuals aged 40–69 from the general population across the U.K. Participants answered detailed demographic, socioeconomic, and health-related questions. Historical and follow-up information is provided by linking to health and medical records. Genome-wide genotype data have been collected on all participants, permitting study of the genetic basis of complex traits (28,29). This large-scale cohort study with linked health and longitudinal medical records enables use of a prospective study design to study incident diabetes complications.
From the UKB, we curated a diabetes cohort using the following sources: 1) baseline information (2006–2010) and subsequent assessments (2012–2013) at UKB assessment centers, including questionnaires, physical examination measurements, and biological samples, and 2) health-related records, which include hospital visits, a death registry, algorithmically defined event outcomes, first occurrences of medical conditions, and ongoing primary care data. Diabetes cases were ascertained according to the following criteria: 1) the first occurrence of any of the following: International Classification of Disease, Ninth (ICD-9) and ICD-10 codes for type 1, type 2, and unspecified diabetes; self-report of diabetes at a UKB Assessment center visit along with the interpolated date from the age of diagnosis; or a limited number of primary care codes mapped to the three-digit ICD10 code E10, E11, and E14; 2) the first occurrence of a more extensive list of diabetes-related primary care codes. Pregnancy or malnutrition-related diabetes was excluded. After excluding individuals with non-European ancestry, 26,387 non-Hispanic White (NHW) patients with diabetes were identified between the ages of 49 and 82 years as of 2020. We refer to this group of individuals as the UKB-NHW-Diabetes cohort (Supplementary Fig. 1). Among the UKB-NHW-Diabetes cohort, we defined the first recorded diagnosis of diabetes as the index date and any incident vascular complication “case” to be the first occurrence of the event after the index date.
The ACCORD study was a double-blind, two-by-two factorial, randomized controlled, parallel treatment trial with 10,251 participants (30,31). The glucose-lowering component of the ACCORD study was to evaluate the effectiveness of a more aggressive treatment target to reduce the rate of macrovascular and microvascular complications (32). The ACCORD study included participants with type 2 diabetes with HbA1c concentrations of ≥7.5% (58.5 mmol/mol) and who were aged 40–79 years with a history of CVD or 55–79 years with evidence of significant atherosclerosis, albuminuria, left ventricular hypertrophy, or at least two risk factors for CVD (dyslipidemia, hypertension, smoking, or obesity). As with the UKB analysis, we excluded individuals with non-European ancestry from the ACCORD cohort because having people of different ancestral groups can inflate heritability estimates. Analyses for other ethnic groups were not conducted because of their small sample sizes. For example, the next biggest ethnic group in the ACCORD was African Americans, with a sample size of 935 after data preprocessing steps. Details of the design and principal results of the ACCORD trial were reported previously (30,31).
Outcome Definitions
In the UKB-NHW-Diabetes cohort, we defined incident cases as the first occurrence of the event after the first diabetes diagnosis. For each type of incident case, we excluded individuals with documented occurrences of the event before the diagnosis of diabetes. For the control cases of DKD, we additionally required them to have no event of interest during the entire observation period with at least 5 years of follow-up. Key phenotypes are detailed in Table 1. A complete list of codes and data fields used in the definition of diabetes, diabetes complications, and their date of the first occurrence are presented in the Supplemental Material.
Table 1.
Outcome definitions for UKB and ACCORD
| Outcome | Definition | |
|---|---|---|
| UKB | DR | Composite for diabetic eye disease in self-reported, primary care, or hospital admission records |
| DKD | Chronic/DKD in self-reported, primary care, hospital, or death records | |
| Macroalbuminuria | UACR >33.9 mg/mmol at either UKB visit | |
| Microalbuminuria | UACR >3.4 mg/mmol at either UKB visit | |
| CVD | Composite for CVD. MI, ischemic stroke, unstable angina, or percutaneous coronary intervention | |
| MI | MI from self-report, primary care, hospital admissions, or death records. Control subjects were required to have no evidence of certain CVDs | |
| Stroke any | Ischemic, hemorrhagic, or unspecified stroke | |
| Stroke infarct | Ischemic stroke | |
| ACCORD | Retin1 | Retinal photocoagulation or vitrectomy to treat retinopathy |
| Retin2 | Eye surgery for cataract extraction | |
| Retin3 | Three-line change in visual acuity | |
| Retin4 | Severe vision loss (Snellen fraction <20/200) | |
| Neph1 | Doubling of baseline serum creatinine or >20 mL/min per 1.73 m2 decrease in estimated GFR | |
| Neph2 | Development of macroalbuminuria. UACR ≥33.9 mg/mmol | |
| Neph3 | ESRD (i.e., initiation of dialysis or a rise of serum creatinine to 3.3 mg/dL [292 μmol/L]) | |
| Neph4 | Development of Neph1, Neph2, or Neph3 | |
| Neph5 | Development of microalbuminuria. UACR ≥3.4 mg/mmol | |
| Primary | Composite for CVD. Nonfatal MI, nonfatal/total stroke, death from cardiovascular causes, new/worsening congestive heart failure, or major CHD | |
| Nonfatal MI | Nonfatal myocardial infarction | |
| Major CHD | Major coronary heart disease | |
| Total mortality | All-cause mortality | |
| CVD mortality | Mortality from cardiovascular causes | |
| Nonfatal stroke | Nonfatal stroke | |
| Total stroke | Total stroke |
UACR, urinary albumin-to-creatinine ratio.
In the ACCORD trial, all outcomes were prespecified and adjudicated by the outcome committee. The prespecified ACCORD primary cardiovascular (CVD) outcome was the first occurrence of nonfatal myocardial infarction (MI) or nonfatal stroke or death from cardiovascular causes. We expanded this primary CVD outcome by including individual outcomes of new or worsening congestive heart failure, total stroke, and major coronary heart disease (CHD). For microvascular complications, taking advantage of the adjudicated broad combination of microvascular outcomes (illustrated in Table 1), we analyzed a spectrum of outcomes ranging from severe microvascular complications (e.g., Neph3 and Retin1) to less advanced conditions. A detailed description of the prespecification of the ACCORD outcomes was documented previously (30,31).
Genotyping and Imputation in UKB and ACCORD
We analyzed the genotyping and imputation (version 3) data released by the UKB in 2017. Details on genotyping and imputation have been extensively described elsewhere (29). In summary, genome-wide genotyping was performed on all UKB participants using the UKB Axiom array. Approximately 850,000 variants were directly genotyped, and >90 million variants were imputed using the merged UK10K and 1000 Genomes Phase 3 (33) reference panels. Only autosomal SNPs were included for all analyses. In the analyses involving imputation data, we discarded SNPs with imputation info score >0.3, missing genotype rate >0.05, Hardy-Weinberg equilibrium test P < 1 × 10−6, and minor allele frequency (MAF) <0.0001, yielding a total of 33,932,888 autosomal SNPs.
Detailed accounts on DNA extraction, genotyping, and quality control procedures in ACCORD were reported previously (34). After retrieving the ACCORD genetic study data from dbGap, we used genetic variants genotyped on Affymetrix Axiom Biobank chips from the University of North Carolina and merged data under two different Institutional Review Board (IRB) protocols—HMB-IRB (73941) and DS-CDKD-IRB (73944). There were 6,291 individuals (2,335 women and 3,956 men) with 546,800 SNPs in the merged data set. Based on self-reported ethnicity, there were 4,369 NHW, 935 African Americans, 381 Hispanics, and 606 others. After preimputation quality control steps, imputation was performed on the genotype data using a two-step approach: prephasing genotype calls (35) and imputation (36). After imputed SNPs with R2 < 0.3 and MAF < 0.0003 were discarded, the total number of SNPs was 25,667,109. Additional details are provided in the Supplementary Material.
Statistical Analysis
Overview of Methods
We use three different methods to compute heritability: single-component genomic restricted maximum likelihood estimation (GREML-SC), GREML-LDMS-I, and Stratified LD Score Regression (S-LDSC). GREML-SC is a single-component variance component approach that is typically applied to common SNPs (MAF ≥ 0.01) (37). GREML-LDMS-I is a multiple variance components approach that bins imputed SNPs by their MAF and individual levels of linkage disequilibrium (LD) (37,38). Compared with GREML-SC, GREML-LDMS-I can attenuate the bias arising from a mismatch between the MAF distribution of the causal variants and that of the SNPs used to generate the genetic relationship matrix (GRM) (39). We selected the GREML-LDMS-I approach over other multicomponent approaches such as GREML-LDMS-R, which allocates SNPs by the MAF and regional LD, since GREML-LDMS-I was shown to be the least biased method (40). While both GREML-SC and GREML-LDMS-I require individual-level genotypes and phenotypes data, S-LDSC relies only on GWAS summary statistics. S-LDSC partitions SNP heritability by functional genomic annotations, as opposed to SNP properties such as MAF or LD in GREML-LDMS-I. For a survey of heritability estimation methods, see Evans et al. (40).
We first computed a GRM from all autosomal SNPs in genotype data using the Relatedness Estimation in Admixed Populations (REAP) approach (41). We then selectively excluded one of any pair of individuals with an estimated kinship greater than the separation between full and half-siblings (estimated kinship > (1/2)5/2 = 0.1768) to maximize the remaining sample size (42). This step was done to avoid inflation caused by cryptic relatedness. After the pruning step, we estimated heritability in the NHW cohort. Based on the GRM constructed from the REAP, heritability was computed using the GREML-SC approach via the software package Genome-wide Complex Trait Analysis (GCTA) (43). For the UKB data, the following covariates were accounted for: sex, age in 2010, and the top 10 principal components. For the ACCORD data, we adjusted for sex, age at baseline, history of CVD at baseline, and the top five principal components. Within the UKB-NHW-Diabetes cohort, sensitivity analysis was also conducted by additionally adjusting for systolic blood pressure for the DKD outcome. In ACCORD, analysis was also conducted by excluding subjects with CVD history at baseline.
To calculate the narrow-sense heritability of diabetes complications from imputed data sets, we first applied GREML-LDMS-I. Following Evans et al. (40), we first calculated segment-based LD scores using the default settings in the GCTA software and stratified SNPs into high and low LD score groups using the median as a threshold. In each LD group, SNPs were further partitioned into four MAF bins. Then GRMs were computed for each of the eight groups. Finally, we estimated the heritability of each binary phenotype with fixed covariates.
Next, we applied S-LDSC (44,45). After acquiring statistics from logistic regression, we performed an analysis with 53 overlapping functional categories used in Finucane et al. (44) and a tissue-specific heritability enrichment analysis. In the tissue-specific analysis, we used the specifically expressed gene annotations generated by Finucane et al. (45) with the Genotype-Tissue Expression (GTEx) project (46). For all S-LDSC analyses, we used 1000 Genomes Project Phase 3 (33) European population SNPs as an LD reference panel. For more details on methods, see Supplemental Material.
Note that our heritability estimates do not take population prevalence/incidence into account. We display estimates without population ascertainment correction because the UKB and ACCORD reflect longitudinal and prospective intervention designs, respectively, rather than ascertained case-control studies. In the latter design, the proportion of case subjects is often overrepresented. In fact, our sample proportion of cases agrees with the prevalence/incidence reported in the literature. For example, the proportion of DKD cases in the UKB-NHW-Diabetes cohort is 0.256, which is similar to the prevalence of any DKD among U.S. adults with diabetes (0.262; 95% CI, 0.226–0.299) reported in Afkarian et al. (47). The proportions of incident cases for the primary CVD outcome and total stroke in the ACCORD group are 0.106 and 0.018, respectively, while the hospital discharge record in 2016 reported the proportion of cases to be 0.0753 and 0.0136 (48).
Data and Resource Availability
The data that support the findings of this study are available from open access repositories. ACCORD study data are available in the biologic Specimen and Data Repository Information Coordinating Center (BioLincc), https://biolincc.nhlbi.nih.gov/, with the permission of BioLincc. The UKB data are retrieved under Project ID 48152. Data are available at https://www.ukbiobank.ac.uk, with the permission of UKB.
Results
Characteristics of the NHW samples used in the UKB and ACCORD analyses are presented in Tables 2 and 3, respectively. Supplementary Figs. 1 and 2 show participant flow in the UKB and ACCORD analyses, respectively.
Table 2.
Characteristics of the non-Hispanic White participants used in the UKB analyses
| Characteristics | N = 26,387 |
|---|---|
| Age in 2010, years | 60.9 ± 7.0 |
| Age at first diabetes diagnosis, years | 56.4 ± 12.4 |
| BMI, kg/m2 | 31.5 ± 5.7 |
| Lipids at baseline, mmol/L | |
| HDL | 1.2 ± 0.3 |
| LDL | 3.0 ± 0.9 |
| Triglycerides | 2.3 ± 1.3 |
| Blood pressure at baseline, mmHg | |
| Systolic | 143.5 ± 18.8 |
| Diastolic | 83.3 ± 10.9 |
| HbA1c at initial visit, mmol/mol | 48.8 ± 13.3 |
| HbA1c at repeat visit, mmol/mol | 48.6 ± 11.1 |
| Sex | |
| Male | 61.4 |
| Female | 38.6 |
| Current/former smoking | |
| Yes | 56.1 |
| No | 43.4 |
| Missing | 0.5 |
| Diabetes type | |
| Type 1 | 2.9 |
| Type 2 | 69.0 |
| Unspecified | 28.1 |
The initial visit occurred between 2006 and 2010, depending on the individual. Data are presented as mean ± SD or percentage of patients.
Table 3.
Characteristics of the NHW participants used in the ACCORD analyses
| Characteristics | N = 4,318 |
|---|---|
| Age at baseline, years | 63.2 ± 6.4 |
| Years since diabetes diagnosis | 10.7 ± 7.4 |
| Lipids at baseline, mg/dL | |
| HDL | 40.2 ± 10.6 |
| LDL | 102.8 ± 33.1 |
| Triglycerides | 208.7 ± 158.4 |
| Blood pressure at baseline, mmHg | |
| Systolic | 135.2 ± 17.3 |
| Diastolic | 74.2 ± 10.8 |
| HbA1c at baseline, % | 8.2 ± 1.0 |
| Sex | |
| Female | 34.4 |
| Male | 65.6 |
| Smoked cigarettes in last 30 days | |
| Yes | 12.4 |
| No | 87.6 |
| Smoked >100 cigarettes during lifetime | |
| Yes | 50.4 |
| No | 37.9 |
| No answer | 11.7 |
| CVD history at baseline | |
| Yes | 36.1 |
| No | 63.9 |
| Glycemic treatment arm | |
| Intensive | 49.8 |
| Standard | 50.2 |
Data are presented as mean ± SD or as percentage of patients.
Heritability
We computed the heritability of phenotypes from the SNPs on the genotyping array using the GREML-SC approach (37). After pruning related individuals and extracting NHW samples, there remained 26,387 samples for the UKB and 4,318 samples for the ACCORD.
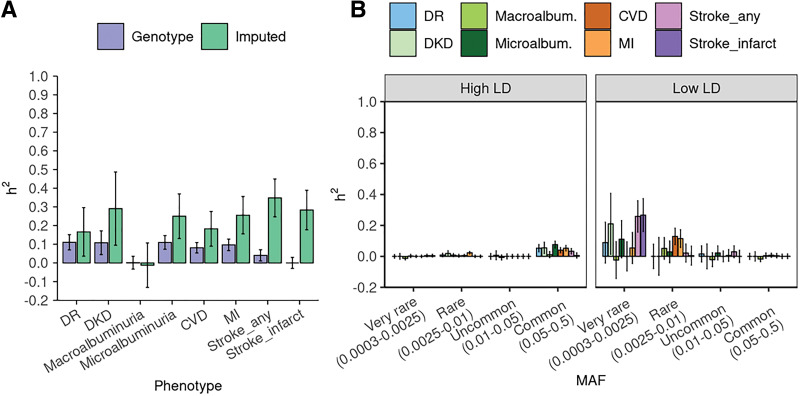
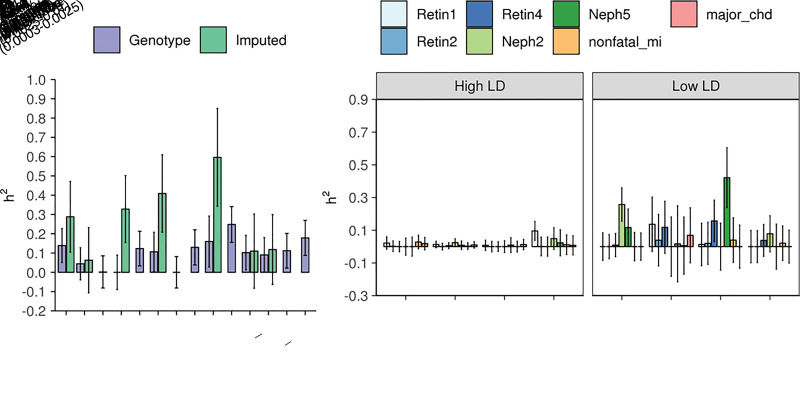
Heritability estimates from the UKB genotype data are illustrated in Table 4 and as purple bars in Fig. 1A. Interestingly, estimates of the UKB phenotypes are smaller in magnitude than those of the ACCORD. While the composite CVD phenotype from the ACCORD (primary) is 0.248 (SE 0.093), the composite CVD outcome from UKB is 0.081 (SE 0.028). Heritability estimates for the ACCORD data are displayed in Table 5 and as purple bars in Fig. 2A. The estimate for the composite nephropathy outcome among type 2 diabetes (Neph4) is 0.129 (SE 0.091), which is comparable with estimates from a similar analysis (0.12 for chronic kidney disease and 0.08 for DKD among subjects with type 2 diabetes) (14). We also ran an additional GREML-SC analysis that includes interaction with the intensive glycemic treatment arm (Supplementary Fig. 3). Interestingly, variance component for the gene-treatment interaction appears to explain a large part of phenotypic variance in microalbuminuria (Neph5). Heritability estimates from the UKB genotype data tend to have smaller error bars than those from the ACCORD genotype data due to the larger sample size in the UKB-NHW-Diabetes cohort.
Table 4.
GREML-SC and GREML-LDMS estimates for individuals with diabetes using the UKB genotype data
| Phenotype | Diabetes (N = 26,387) | Diabetes: type 2 only (N = 18,198) | No diabetes (N = 26,387) | Diabetes and no diabetes (N = 52,774) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Proportion of cases | n | V(G)/V(p) (SE) | Proportion of cases | n | V(G)/V(p) (SE) | V(G)/V(p) (SE) | V(G)/V(p) (SE) | ||
| GREML-SC | GREML-LDMS | GREML-LDMS | GREML-LDMS | GREML-LDMS | |||||
| DR | 0.541 | 11,739 | 0.110 (0.041) | 0.166 (0.130) | 0.514 | 7,516 | 0.029 (0.199) | NA | NA |
| DKD | 0.256 | 7,707 | 0.108 (0.064) | 0.291 (0.196) | 0.259 | 4,969 | 0.237 (0.300) | 0.453 (0.15) | 0.217 (0.089) |
| Macroalbuminuria | 0.029 | 13,246 | 0.001 (0.034) | 0.000 (0.119) | 0.026 | 9,332 | NA | NA | 0.026 (0.085) |
| Microalbuminuria | 0.238 | 13,246 | 0.110 (0.037) | 0.250 (0.119) | 0.230 | 9,332 | 0.214 (0.163) | 0.155 (0.201) | 0.226 (0.079) |
| CVD | 0.159 | 17,540 | 0.081 (0.028) | 0.183 (0.093) | 0.170 | 11,506 | 0.208 (0.139) | 0.176 (0.069) | 0.085 (0.041) |
| MI | 0.094 | 16,310 | 0.097 (0.031) | 0.256 (0.100) | 0.099 | 10,610 | 0.192 (0.149) | 0.278 (0.072) | 0.141 (0.043) |
| Stroke any | 0.087 | 16,002 | 0.041 (0.030) | 0.348 (0.101) | 0.095 | 10,434 | 0.590 (0.151) | NA | 0.199 (0.044) |
| Stroke infarct | 0.042 | 15,429 | 0.000 (0.029) | 0.283 (0.106) | 0.047 | 10,030 | 0.330 (0.158) | 0.141 (0.073) | 0.167 (0.045) |
GREML-LDMS estimates were for individuals with 1) diabetes (both type 1 and type 2), 2) type 2 diabetes only, 3) without diabetes, and 4) groups with diabetes and no diabetes combined using the UKB imputed data. Individuals with no diabetes (N = 26,387) in this table were randomly sampled from a pool of 296,315 individuals with no diabetes in the UKB data. Estimates for the last column were obtained from combining groups with diabetes and no diabetes (N = 52,774). N, number of total samples in the group; n, number of samples without missing phenotype; NA, under GREML-LDMS, the GREML analysis failed to run due to the small sample size; V(G)/V(p), proportion of phenotypic variance explained by genotypes (i.e., heritability, as observed in the study population).
Figure 1.
Heritability estimates and SEs of diabetes complication outcomes using the UKB data. A: Estimates from genotype and imputed data are obtained using the GREML-SC and GREML-LDMS-I approaches, respectively. We adjusted for sex, age in 2010, and the top 10 genetic principal components. B: GREML-LDMS estimates with eight bins (two LD bins for each of the four MAF bins). For each phenotype, the sum of estimates from the eight bins of MAF shown in panel B is equal to the estimate represented as the green bar on panel A.
Table 5.
GREML-SC and GREML-LDMS estimates using the ACCORD genotype and imputed data, respectively
| Phenotype | Proportion of cases in the sample | n | V(G)/V(p) (SE) | |
|---|---|---|---|---|
| GREML-SC | GREML-LDMS | |||
| Retin1 | 0.084 | 4,318 | 0.139 (0.088) | 0.288 (0.183) |
| Retin2 | 0.158 | 4,318 | 0.044 (0.083) | 0.063 (0.169) |
| Retin3 | 0.360 | 4,318 | 0.002 (0.084) | NA |
| Retin4 | 0.068 | 4,318 | 0.000 (0.089) | 0.328 (0.174) |
| Neph1 | 0.591 | 4,318 | 0.123 (0.090) | NA |
| Neph2 | 0.070 | 3,866 | 0.106 (0.101) | 0.409 (0.201) |
| Neph3 | 0.028 | 4,318 | 0.000 (0.082) | NA |
| Neph4 | 0.616 | 4,318 | 0.129 (0.091) | NA |
| Neph5 | 0.241 | 2,912 | 0.160 (0.132) | 0.596 (0.254) |
| Primary | 0.106 | 4,318 | 0.248 (0.093) | NA |
| Nonfatal MI | 0.071 | 4,318 | 0.102 (0.090) | 0.110 (0.192) |
| Major CHD | 0.129 | 4,318 | 0.090 (0.089) | 0.118 (0.181) |
| Total mortality | 0.066 | 4,318 | 0.013 (0.088) | NA |
| CVD mortality | 0.028 | 4,318 | 0.094 (0.089) | NA |
| Nonfatal stroke | 0.015 | 4,318 | 0.112 (0.090) | NA |
| Total stroke | 0.018 | 4,318 | 0.179 (0.091) | NA |
NA under GREML-LDMS, the GREML analysis failed to run due to the small sample size; n, number of samples without missing phenotype; V(G)/V(p), proportion of phenotypic variance explained by genotypes (i.e., heritability, as observed in the study population).
Figure 2.
Heritability estimates and SEs of diabetes complications using the ACCORD data. A: Estimates from genotype and imputed data are obtained using the GREML-SC and GREML-LDMS-I approaches, respectively. We adjusted for sex, age at baseline, CVD history at baseline, and the top five genetic principal components. B: GREML-LDMS estimates with eight bins (two LD bins for each of the four MAF bins). For each phenotype, the sum of estimates from the eight bins of MAF shown in panel B is equal to the estimate represented as the green bar on panel A.
Heritability estimates using imputed data sets and the GREML-LDMS-I method are provided as green bars in Figs. 1A and 2A for the UKB and ACCORD, respectively (also see Tables 4 and 5). In UKB, the heritability of DKD is estimated to be 0.29 (SE 0.20). Microalbuminuria estimates range from 0.25 (SE 0.12) to 0.60 (SE 0.25), while macroalbuminuria estimates are up to 0.41 (SE 0.20). In ACCORD, heritability estimates of DR range from 0.06 (SE 0.17) to 0.33 (SE 0.17), depending on the definition of phenotype. Although still less than family study estimates for broad-sense heritability—0.27 for DR (20) and as high as 0.52 (SE 0.31) for proliferative retinopathy among adults with type 1 diabetes (19)—our estimates are close to pedigree heritability estimates.
Of note, we observed higher estimates with more advanced retinopathy: 0.29 and 0.33 for Retin1 (retinal photocoagulation or vitrectomy) and Retin4 (severe vision loss), respectively, as opposed to 0.06 and ∼0 for Retin2 (cataract extraction) and Retin3 (three-line change in visual acuity), respectively. On the other hand, diabetic nephropathy phenotypes do not exhibit such a pattern. While the heritability of macroalbuminuria phenotype within ACCORD is estimated at 0.41, that of microalbuminuria from ACCORD is at 0.60. Estimates for either Neph1 or Neph3 are unavailable despite larger sample sizes (4,318 for both Neph1 and Neph3 and 3,866 and 2,912 for Neph2 and Neph5, respectively). This pattern or lack thereof is consistent with earlier heritability studies that implicated genetic components in the severity of DR and the presence/absence of diabetic nephropathy (15,17). Although we cannot confirm the trend of DR in the UKB data (due to the absence of more granular outcome definitions for DR), when restricting the analysis to participants with type 2 diabetes in UKB only, we found the heritability of DR reduces to 0.029 (SE 0.199) from 0.166 (SE 0.130) (Table 4). As DR recorded in primary care data tends to be less severe, it is consistent with the observations from ACCORD.
Heritability analyses using imputed data reveal a substantial contribution of low-frequency/rare variants to the predisposition for complications. While the heritability of severe DR from common genetic variants among individuals with type 2 diabetes was estimated to be 0.07 in a previous study (21) and up to 0.14 in our analysis (see GREML-SC results of Retin1 and Retin4 in Table 5), heritability estimates of advanced DR among individuals with type 2 diabetes were higher (0.29 and 0.33 for Retin1 and Retin4 in ACCORD) when calculated from directly typed plus imputed genetic markers. The distribution of heritability across the MAF spectrum for other complication phenotypes, including retinopathy, is found in Figs. 1B and 2B. Notably, the UKB results (Fig. 1B) show a more pronounced contribution pattern with small error bars and heritability heavily concentrated in very rare variants (0.0003 ≤ MAF < 0.0025).
To corroborate our results, we applied the same set of phenotyping rules to a sample of 26,387 individuals with no diabetes in the UKB to estimate heritability of chronic kidney disease and CVD using the definition outlined above. This group was randomly sampled from 296,315 individuals with no diabetes in the UKB to match the sample size of the group with diabetes and ease the computational burden. We also computed estimates after combining groups with and without diabetes and adjusting for diabetes status as a covariate. These results can be found in Table 4. Overall, heritability shows higher estimates for kidney disease in the group without diabetes (i.e., 0.291 [SE 0.196] in diabetes vs. 0.453 [SE 0.15] without diabetes), while the heritability estimate for microalbuminuria is higher than that in the group without diabetes (i.e., 0.250 [SE 0.119] in diabetes vs. 0.155 [SE 0.201] without diabetes).
GWAS
Association results identified multiple significant peaks (P < 5 × 10−8) in the UKB-NHW-Diabetes cohort. For macrovascular complications in the UKB-NHW-Diabetes cohort (CVD and MI), variants on chromosome 9p21 reached genome-wide significance. Association of the regions on chromosome 9p21 with type 2 diabetes and progression of CVD was reported previously (49). For DR in the UKB-NHW-Diabetes cohort, 22 variants on 6p21 reached genome-wide significance (P < 5 × 10−8) with rs9273367 (P = 1.23 × 10−9, odds ratio 1.18) being the most significant SNP. These variants were in or near HLA regions, whose previous associations with type 1 diabetes have been well documented (50). For DKD, 17 variants had P < 5 × 10−8, and 11 of these SNPs were on chromosome 3q26.31, and 6 were in UMOD and PDILT genes (lead SNP rs77924615 with P = 7.82 × 10−9, odds ratio 0.75) on chromosome 16p12.3. UMOD was previously reported to be associated with estimated glomerular filtration rate in the meta-analysis combining patients with type 1 and type 2 diabetes of European and Asian ancestry (14). Although some variants were below the genome-wide significance threshold in the ACCORD cohort, they were not as prominent as in the UKB-NHW-Diabetes cohort.
Supplementary Data 1 reports all genome-wide significant GWAS loci for diabetes complications. Supplementary Figs. 4–7 show Manhattan and QQ plots for GWAS.
Heritability Enrichment by Functional Annotations
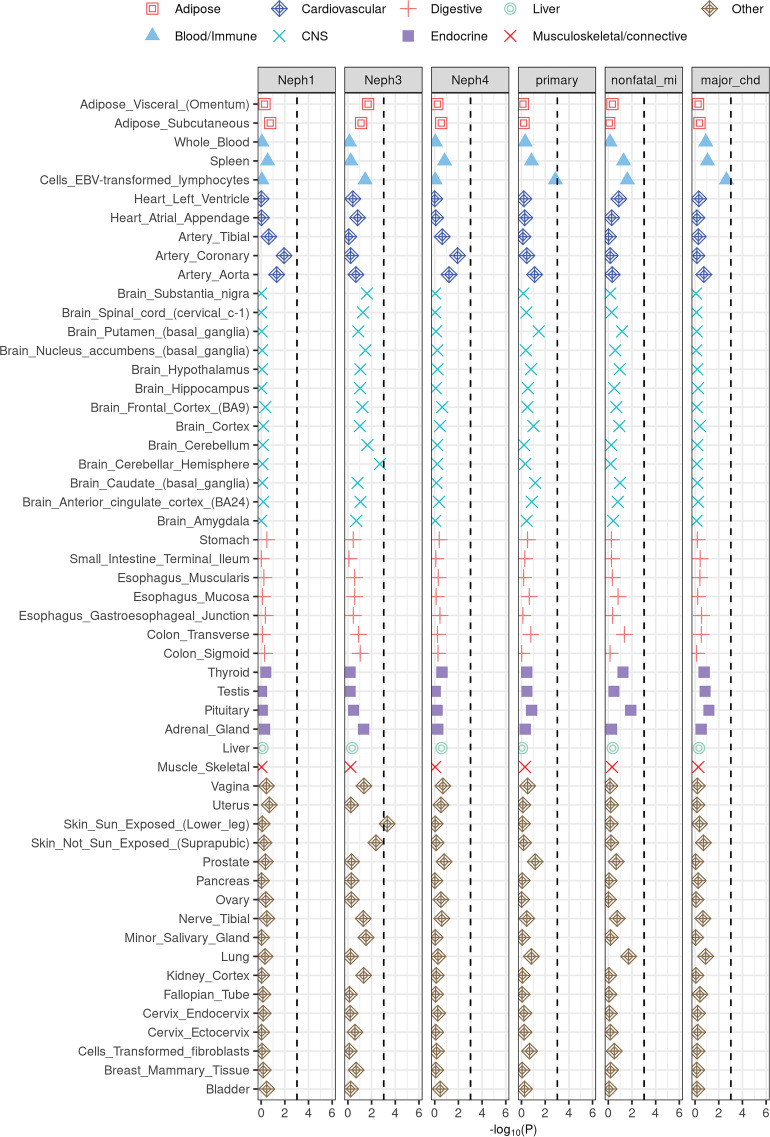
We applied S-LDSC to identify disease-relevant tissues and cell types. Results for the selected ACCORD phenotypes are illustrated in Fig. 3 (also see Supplementary Figs. 8 and 9). Renal failure or end-stage renal disease (ESRD) phenotype (Neph3) exhibit skin-specific (sun-exposed skin P = 4.82 × 10−4; non–sun-exposed skin P = 4.29 × 10−3) and brain-specific enrichments (brain cerebellar hemisphere P = 1.99 × 10−3). The skin-specific enrichment captures dermatologic manifestations of ESRD (51). Macrovascular complications (primary and major CHD) show enrichments in Epstein-Barr virus–transformed lymphocytes (P = 1.38 × 10−3 and P = 2.25 × 10−3, respectively). This finding reflects the mechanism of macrovascular complications involving inflammatory cells (e.g., monotypes and T lymphocytes) (52). Despite the larger sample size, no tissues were enriched for the heritability of diabetic complications from the UKB (Supplementary Figs. 10 and 11).
Figure 3.
Enrichment of the selected ACCORD phenotypes in tissue-specific gene expression annotations used in Finucane et al. (45). The black dashed lines indicate the Bonferroni significance threshold (P < 0.05/53). CNS, central nervous system; EBV, Epstein-Barr virus.
Results from the S-LDSC analysis partitioning heritability into 53 (overlapping) categories used in Finucane et al. (44) are illustrated in Supplementary Fig. 12. In the UKB data, only the coding region shows Bonferroni-corrected (0.05/53 = 9.43 × 10−4) significant enrichment in DKD (P = 6.55 × 10−4). Although only nominally significant, H3K9ac is enriched in the microalbuminuria phenotype (P = 0.04). H3K9ac enrichment agrees with the findings from Salem et al. (53) that the top signal (TAMM41) for microalbuminuria is close to the histone marks—H3K27ac, H3K9ac, and H3k4me1. In the ACCORD data, none of the categories for any phenotype passed the Bonferroni significance threshold (0.05/53 = 9.43 × 10−4), given the small sample size. Some categories are still noteworthy, however. Promoter region showed enrichment in the retinopathy phenotype (Retin1; P = 2.82 × 10−2), and H3K27ac showed enrichment in the composite nephropathy phenotype (Neph4; P = 4.64 × 10−2).
Discussion
In this report, we have provided a comprehensive assessment of SNP heritability for diabetes microvascular and macrovascular complications. Estimates from the imputed data revealed a substantial contribution of low-frequency/rare variants in low LD with neighboring variants for variation of diabetes complications. Our estimates are higher than those obtained from common SNPs in GWAS but approach pedigree heritability. Our findings imply that a large portion of the genetic risk of diabetes complications is yet to be discovered. Additional sensitivity analyses adjusting for the common risk factor (blood pressure measures at baseline) and excluding participants with CVD history in ACCORD cohort did not change the heritability estimates in our studies.
We have used two independent studies to estimate the heritability for diabetes complications. Although a meta-analysis from the two studies would have increased the sample size, we conducted two separate analyses to reduce the risk of phenotypic heterogeneity. Our analyses show some discordance in findings between the two data sets. Heritability estimates obtained using imputed data sets tend to be larger in the ACCORD study than in the UKB study despite a larger sample size in the UKB-NHW-Diabetes cohort. Additionally, no tissue enrichment is observed in the UKB-NHW-Diabetes cohort. Differences in study designs and potential biases may provide a basis for such discordant findings. First, the ACCORD is a clinical trial that offers adjudicated outcomes in a well-controlled clinical trial setting. In contrast, the UKB reflects a cohort in “real-world” scenarios and is based on electronic medical records, which typically have a high noise-to-signal ratio and many possible sources of bias.
Second, there is an underlying risk of sampling or selection bias of the two research studies. While the ACCORD cohort consisted of adults at increased risk for CVD with a longer duration of diabetes and higher glycated hemoglobin level (Table 2), the UKB participants were younger and relatively healthy (Table 1).
We have also shown that genetic contributions to chronic kidney disease are larger in the group without diabetes than in the group with diabetes, while heritability for macrovascular complications stays similar between the two groups. Several reasons may explain the differences: 1) outcome misclassifications due to electronic health record-based phenotyping, 2) unaccounted confounders, such as medications, and 3) higher heritability of kidney diseases among the general population than that of DKD among diabetes (54). The heritability of macrovascular complications was similar between groups with and without diabetes. It may be because GWAS hits for CVD among the population with diabetes tend to coincide with those in the general population (55).
This heritability analysis represents the first systematic investigation of SNP heritability for diabetes complications in the White subset of UKB and ACCORD cohorts. It adds to the existing heritability information derived primarily from family or small cohort studies and supports the need for further genetic investigation of diabetes complications, both for general disease outcomes and for specific phenotypes. Replication studies will be instrumental in strengthening conclusions in this area.
Article Information
Funding. This research was partially funded by grants from the National Institute of General Medical Sciences (R35GM141798 to H.Z.), the National Human Genome Research Institute (R01HG006139 to H.Z. and J.J.Z.), the National Science Foundation (DMS-2054253 to H.Z. and J.J.Z.), the National Institute of Diabetes and Digestive and Kidney Diseases (P30DK116073 to S.R. and K01DK106116 to J.J.Z.), the National Heart, Lung, and Blood Institute (R21HL150374 to J.J.Z.), the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (2020R1A6A3A03037675 to S.K.), and the U.S. Department of Veterans Affairs (IK2-CX001907 to S.R.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. J.K., A.J., and S.K., performed the analysis. J.K., S.R., L.S.P., A.H., Y.S., H.Z., P.R., and J.J.Z. wrote the manuscript and approved the final manuscript. J.K., H.Z., and J.J.Z. conceived the contents of the manuscript. J.J.Z. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.19122485.
References
- 1. Vujkovic M, Keaton JM, Lynch JA, et al.; HPAP Consortium; Regeneron Genetics Center; VA Million Veteran Program . Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet 2020;52:680–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barrett JC, Clayton DG, Concannon P, et al.; Type 1 Diabetes Genetics Consortium . Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nyaga DM, Vickers MH, Jefferies C, Perry JK, O’Sullivan JM. The genetic architecture of type 1 diabetes mellitus. Mol Cell Endocrinol 2018;477:70–80 [DOI] [PubMed] [Google Scholar]
- 4. Onengut-Gumuscu S, Chen WM, Burren O, et al.; Type 1 Diabetes Genetics Consortium . Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet 2015;47:381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Noble JA, Valdes AM. Genetics of the HLA region in the prediction of type 1 diabetes. Curr Diab Rep 2011;11:533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ferrat LA, Vehik K, Sharp SA, et al.; TEDDY Study Group & Committees . A combined risk score enhances prediction of type 1 diabetes among susceptible children. Nat Med 2020;26:1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. American Diabetes Association . Implications of the United Kingdom Prospective Diabetes Study. Diabetes Care 1998;21:2180–2184 [DOI] [PubMed] [Google Scholar]
- 8. Nathan DM, Genuth S, Lachin J, et al.; Diabetes Control and Complications Trial Research Group . The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med 1993;329:977–986 [DOI] [PubMed] [Google Scholar]
- 9. Bowden DW. Genetics of diabetes complications. Curr Diab Rep 2002;2:191–200 [DOI] [PubMed] [Google Scholar]
- 10. Borch-Johnsen K, Nørgaard K, Hommel E, et al. Is diabetic nephropathy an inherited complication? Kidney Int 1992;41:719–722 [DOI] [PubMed] [Google Scholar]
- 11. Quinn M, Angelico MC, Warram JH, Krolewski AS. Familial factors determine the development of diabetic nephropathy in patients with IDDM. Diabetologia 1996;39:940–945 [DOI] [PubMed] [Google Scholar]
- 12. Harjutsalo V, Katoh S, Sarti C, Tajima N, Tuomilehto J. Population-based assessment of familial clustering of diabetic nephropathy in type 1 diabetes. Diabetes 2004;53:2449–2454 [DOI] [PubMed] [Google Scholar]
- 13. Sandholm N, Van Zuydam N, Ahlqvist E, et al. The FinnDiane Study Group; The DCCT/EDIC Study Group; GENIE Consortium; SUMMIT Consortium . The genetic landscape of renal complications in type 1 diabetes. J Am Soc Nephrol 2017;28:557–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Zuydam NR, Ahlqvist E, Sandholm N, et al.; Finnish Diabetic Nephropathy Study (FinnDiane); Hong Kong Diabetes Registry Theme-based Research Scheme Project Group; Warren 3 and Genetics of Kidneys in Diabetes (GoKinD) Study Group; GENIE (GEnetics of Nephropathy an International Effort) Consortium; Diabetes Control and Complications Trial (DCCT)/Epidemiology of Diabetes Interventions and Complications (EDIC) Research Group; SUrrogate markers for Micro- and Macrovascular hard endpoints for Innovative diabetes Tools (SUMMIT) Consortium . A genome-wide association study of diabetic kidney disease in subjects with type 2 diabetes. Diabetes 2018;67:1414–142729703844 [Google Scholar]
- 15. The Diabetes Control and Complications Trial Research Group . Clustering of long-term complications in families with diabetes in the diabetes control and complications trial. Diabetes 1997;46:1829–1839 [PubMed] [Google Scholar]
- 16. Leslie RDG, Pyke DA. Diabetic retinopathy in identical twins. Diabetes 1982;31:19–21 [DOI] [PubMed] [Google Scholar]
- 17. Hallman DM, Huber JC Jr, Gonzalez VH, Klein BEK, Klein R, Hanis CL. Familial aggregation of severity of diabetic retinopathy in Mexican Americans from Starr County, Texas. Diabetes Care 2005;28:1163–1168 [DOI] [PubMed] [Google Scholar]
- 18. Looker HC, Nelson RG, Chew E, et al. Genome-wide linkage analyses to identify Loci for diabetic retinopathy. Diabetes 2007;56:1160–1166 [DOI] [PubMed] [Google Scholar]
- 19. Hietala K, Forsblom C, Summanen P; FinnDiane Study Group . Heritability of proliferative diabetic retinopathy. Diabetes 2008;57:2176–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arar NH, Freedman BI, Adler SG, et al.; Family Investigation of Nephropathy and Diabetes Research Group . Heritability of the severity of diabetic retinopathy: the FIND-Eye study. Invest Ophthalmol Vis Sci 2008;49:3839–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meng W, Shah KP, Pollack S, et al.; Wellcome Trust Case Control Consortium 2 (WTCCC2), Surrogate markers for Micro- and Macro-vascular hard endpoints for Innovative diabetes Tools (SUMMIT) study group . A genome-wide association study suggests new evidence for an association of the NADPH oxidase 4 (NOX4) gene with severe diabetic retinopathy in type 2 diabetes. Acta Ophthalmol 2018;96:e811–e819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zdravkovic S, Wienke A, Pedersen NL, Marenberg ME, Yashin AI, De Faire U. Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. J Intern Med 2002;252:247–254 [DOI] [PubMed] [Google Scholar]
- 23. Zdravkovic S, Wienke A, Pedersen NL, de Faire U. Genetic influences on angina pectoris and its impact on coronary heart disease. Eur J Hum Genet 2007;15:872–877 [DOI] [PubMed] [Google Scholar]
- 24. McPherson R, Tybjaerg-Hansen A. Genetics of coronary artery disease. Circ Res 2016;118:564–578 [DOI] [PubMed] [Google Scholar]
- 25. Simonson MA, Wills AG, Keller MC, McQueen MB. Recent methods for polygenic analysis of genome-wide data implicate an important effect of common variants on cardiovascular disease risk. BMC Med Genet 2011;12:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wagenknecht LE, Langefeld CD, Bowden DW, Carr JJ, Freedman BI, Rich SS. Familial aggregation of coronary artery calcium in families with type 2 diabetes. Diabetes 2001;50:861–866 [DOI] [PubMed] [Google Scholar]
- 27. Lange LA, Bowden DW, Langefeld CD, et al. Heritability of carotid artery intima-medial thickness in type 2 diabetes. Stroke 2002;33:1876–1881 [DOI] [PubMed] [Google Scholar]
- 28. Fry A, Littlejohns TJ, Sudlow C, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol 2017;186:1026–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ismail-Beigi F, Craven T, Banerji MA, et al.; ACCORD trial group . Effect of intensive treatment of hyperglycaemia on microvascular outcomes in type 2 diabetes: an analysis of the ACCORD randomised trial. Lancet 2010;376:419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gerstein HC, Miller ME, Byington RP, et al.; Action to Control Cardiovascular Risk in Diabetes Study Group . Effects of intensive glucose lowering in type 2 diabetes. N Engl J Med 2008;358:2545–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zoungas S, Arima H, Gerstein HC, et al.; Collaborators on Trials of Lowering Glucose (CONTROL) group . Effects of intensive glucose control on microvascular outcomes in patients with type 2 diabetes: a meta-analysis of individual participant data from randomised controlled trials. Lancet Diabetes Endocrinol 2017;5:431–8437 [DOI] [PubMed] [Google Scholar]
- 33. The 1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature 2015;526:68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shah HS, Gao H, Morieri ML, et al. Genetic predictors of cardiovascular mortality during intensive glycemic control in type 2 diabetes: findings from the ACCORD clinical trial. Diabetes Care 2016;39:1915–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Loh P-R, Danecek P, Palamara PF, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet 2016;48:1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet 2016;48:1284–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet 2010;42:565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang J, Bakshi A, Zhu Z, et al.; LifeLines Cohort Study . Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet 2015;47:1114–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang J, Manolio TA, Pasquale LR, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet 2011;43:519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Evans LM, Tahmasbi R, Vrieze SI, et al.; Haplotype Reference Consortium . Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat Genet 2018;50:737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thornton T, Tang H, Hoffmann TJ, Ochs-Balcom HM, Caan BJ, Risch N. Estimating kinship in admixed populations. Am J Hum Genet 2012;91:122–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W-M. Robust relationship inference in genome-wide association studies. Bioinformatics 2010;26:2867–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011;88:76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Finucane HK, Bulik-Sullivan B, Gusev A, et al.; ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium . Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 2015;47:1228–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Finucane HK, Reshef YA, Anttila V, et al.; Brainstorm Consortium . Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet 2018;50:621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. GTEx Consortium . Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Afkarian M, Zelnick LR, Hall YN, et al. Clinical manifestations of kidney disease among US adults with diabetes, 1988-2014. JAMA 2016;316:602–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Centers for Disease Control and Prevention . National Diabetes Statistics Report, 2020. Accessed 21 February 2022. Available from https://www.cdc.gov/diabetes/pdfs/data/statistics/national-diabetes-statistics-report.pdf
- 49. Helgeland Ø, Hertel JK, Molven A, et al. The chromosome 9p21 CVD-and T2D-associated regions in a Norwegian population (the HUNT2 survey). Int J Endocrinol 2015;2015:164652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Noble JA, Erlich HA. Genetics of type 1 diabetes. Cold Spring Harb Perspect Med 2012;2:a007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Galperin TA, Cronin AJ, Leslie KS. Cutaneous manifestations of ESRD. Clin J Am Soc Nephrol 2014;9:201–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cade WT. Diabetes-related microvascular and macrovascular diseases in the physical therapy setting. Phys Ther 2008;88:1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Salem RM, Todd JN, Sandholm N, et al.; SUMMIT Consortium, DCCT/EDIC Research Group, GENIE Consortium . Genome-wide association study of diabetic kidney disease highlights biology involved in glomerular basement membrane collagen. J Am Soc Nephrol 2019;30:2000–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang J, Thio CHL, Gansevoort RT, Snieder H. Familial aggregation of CKD and heritability of kidney biomarkers in the general population: the Lifelines Cohort Study. Am J Kidney Dis 2021;77:869–878 [DOI] [PubMed] [Google Scholar]
- 55. Cole JB, Florez JC. Genetics of diabetes mellitus and diabetes complications. Nat Rev Nephrol 2020;16:377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]