Abstract
Therapeutic vascular endothelial growth factor (VEGF) replenishment has met with limited success for the management of critical limb-threatening ischemia. To improve outcomes of VEGF therapy, we applied single-cell RNA sequencing (scRNA-seq) technology to study the endothelial cells of the human diabetic skin. Single-cell suspensions were generated from the human skin followed by cDNA preparation using the Chromium Next GEM Single-cell 3′ Kit v3.1. Using appropriate quality control measures, 36,487 cells were chosen for downstream analysis. scRNA-seq studies identified that although VEGF signaling was not significantly altered in diabetic versus nondiabetic skin, phospholipase Cγ2 (PLCγ2) was downregulated. The significance of PLCγ2 in VEGF-mediated increase in endothelial cell metabolism and function was assessed in cultured human microvascular endothelial cells. In these cells, VEGF enhanced mitochondrial function, as indicated by elevation in oxygen consumption rate and extracellular acidification rate. The VEGF-dependent increase in cell metabolism was blunted in response to PLCγ2 inhibition. Follow-up rescue studies therefore focused on understanding the significance of VEGF therapy in presence or absence of endothelial PLCγ2 in type 1 (streptozotocin-injected) and type 2 (db/db) diabetic ischemic tissue. Nonviral topical tissue nanotransfection technology (TNT) delivery of CDH5 promoter–driven PLCγ2 open reading frame promoted the rescue of hindlimb ischemia in diabetic mice. Improvement of blood flow was also associated with higher abundance of VWF+/CD31+ and VWF+/SMA+ immunohistochemical staining. TNT-based gene delivery was not associated with tissue edema, a commonly noted complication associated with proangiogenic gene therapies. Taken together, our study demonstrates that TNT-mediated delivery of endothelial PLCγ2, as part of combination gene therapy, is effective in diabetic ischemic limb rescue.
Introduction
Critical limb-threating ischemia is a severe form of peripheral arterial disease (PAD) with markedly reduced blood flow in lower extremities (1,2). Type 2 diabetes mellitus (T2DM) increases the incidence and severity of limb ischemia by an estimated two- to fourfold (3,4). Distal limb vessels, often affected by diabetic PAD, suffer from limited potential for collateral vessel development, which poses a barrier to revascularization (5). Hyperglycemia-induced endothelial cell dysfunction (ECD) is recognized as a contributor to such limitation (6). ECD encompasses impairment in endothelial cell barrier function as well as defective proliferative, angiogenic, and tube-formation capacities (7,8). ECD accounts for the increased risk of pathological outcomes, such as skin lesions and ulcerations in the diabetic foot (9). In the context of diabetic PAD, the cutaneous microcirculation is of critical interest (10). Diabetic ECD, caused by hyperglycemia, has a direct impact on microcirculatory function as well as vascular inflammation (11,12).
In patients with diabetes, ECD is characterized by downregulation of hypoxia inducible factor (HIF-1α) and attenuated release of vascular factors, such as vascular endothelial growth factor (VEGF). Thus, ischemia causes angiogenic incompetence (6,13). It has been shown in two different models of mice with T2DM (high-fat diet–induced and db/db) that the increase in VEGF expression is attenuated after ischemic injury (14,15). Additionally, VEGF levels remain low in NOD mice, a polygenic model for autoimmune T1DM, in ischemic muscle compared with wild-type mice (16). This low VEGF expression is often associated with impairment of perfusion recovery and neocapillary formation in ischemic tissue (14–16). Therapeutic angiogenesis aiming at VEGF replenishment has been tested for the management of critical limb-threating ischemia (17). While preclinical animal studies and the initial VEGF gene therapy studies produced encouraging results, follow-up clinical trials were disappointing (17–24). To improve outcomes of VEGF therapy, we sought to apply single-cell RNA-sequencing (scRNA-seq) technology to study the endothelial cells of the human diabetic skin. Phospholipase Cγ2 (PLCγ2) was identified as the top candidate that was downregulated in diabetic versus nondiabetic cells. In this work, we tested the significance of endothelial PLCγ2 deficiency in VEGF therapy.
Research Design and Methods
Human Subjects
Under Institutional Review Board–approved protocol of the Indiana University, surgically discarded and deidentified human skin was obtained from individuals undergoing surgeries who were clinically diagnosed with T2DM or who did not have diabetes. The demographics of subjects included in this study have been provided in Supplementary Table 7. A total of nine nondiabetic and eight T2DM skin tissue samples were used for this study. Out of these, three T2DM and five nondiabetic skin samples were used for scRNA-seq analysis, and others were used for validation studies (Supplementary Table 7). The patients with T2DM (n = 8; 7 male, 1 female, 7 Caucasian, and 1 African American) had a mean age of 62.6 ± 9.7 years (mean ± SD). Among subjects with T2DM studied, six were clinically diagnosed with diabetic neuropathy, five with hypertension, and five with hyperlipidemia. The group without diabetes (n = 9; 3 male, 6 female, 5 Caucasian, and 4 African American) had a mean age of 46.6 ± 14.7 years (mean ± SD). The subjects with T2DM (n = 8) had an HbA1c value of 8.5 ± 2.9% (mean ± SD), and five subjects were on insulin.
scRNA-seq
Single-cell suspensions were generated from the unwounded (UW) human skin. Briefly, the tissue was chopped into small pieces and subjected to enzymatic dissociation using a whole-skin dissociation kit (cat. no. 130-101-540) in MACS C Tubes (cat. no. 130-093-237; Miltenyi Biotec, San Diego, CA). The samples were incubated in a water bath at 37°C for ∼3 h. After incubation, the samples were diluted by adding 0.5 mL of cold complete cell culture medium to stop the enzymatic reaction. The C Tubes were then tightly closed and attached upside down onto the sleeve of the gentleMACS Octo Dissociator, and a UW human skin dissociation program was performed. The cell suspension was subjected to a 70-µm preseparation filter and placed on a 15-mL tube to remove the debris. RBCs were lysed using 10X RBC lysis buffer (cat. no. 420301; BioLegend, San Diego, CA). The resulting cell suspension was used for scRNA-seq using the 10x Genomics platform using Chromium Next GEM Single Cell 3′ GEM, Library & Gel Bead Kit v3.1 (10x Genomics) and sequenced on an Illumina NovaSeq 6000 (Illumina).
Raw Data Processing and Quality Control for Cell Exclusion
Seurat package (v.3.1.1) in R (v.3.5.1) (25,26) was used for preprocessing and visualization. The initial data set, containing 48,769 cells from 8 samples (3 diabetic skin and 5 nondiabetic skin) and 27,393 genes, was integrated using Canonical Correlational Analysis as described previously (27). The values of the genes were log normalized and scaled to 10,000 transcripts/cell. The top 2,000 variable genes were identified. Then, principal component analysis was performed. For quality control, cells with >15% mitochondrial RNA, <200, or >6,000 detected transcripts and cells with <1,000 or >40,000 of the total number of counts were excluded. Finally, genes that were detected in fewer than three cells were excluded. After quality control, 36,487 cells and 27,393 genes were maintained for downstream analysis. Data for all of the cells are presented in Supplementary Fig. 1C, with black representing the excluded cells and green representing the included cells. Preprocessing was performed again after excluding low-quality cells, and the top 20 principal components were used for clustering the cells, resulting in 12 clusters. The t-distributed stochastic neighbor embedding (tSNE) algorithm was used for dimensionality reduction (28).
Determination of Cell-Type Identity
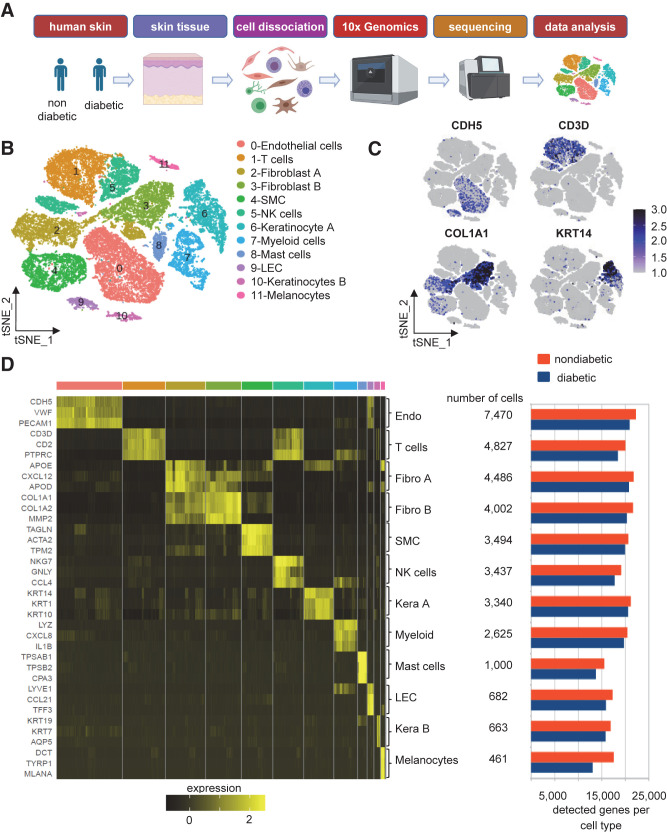
SingleR package in R was used to identify cell types (29). The Human Primary Cell Atlas data set was chosen to be a reference data set, which contains data from 713 microarrays classified into 38 main cell types and 169 subtypes. The similarity between our data set and the Human Primary Cell Atlas data set was computed using SingleR algorithm for unbiased recognition of cell types. Then, the final cell types assignment was determined using cell type–specific marker genes that were previously described in the literature. These include CDH5, VWF, and PECAM1 for endothelial cells; CD3D, CD2, and PTPRC for T cells; APOE, CXCL12, and APOD for fibro 1; COL1A1, COL2A2, and MMP2 for fibro 2; TAGLN, ACTA2, and TPM2 for smooth muscle cells (SMCs); NKG7, GNLY, and CCL4 for natural killer (NK) cells; KRT14, KRT1, and KRT10 for kera A; LYZ, CXCL8, and IL1B for myeloid cells; TPSAB1, TPSB2, and CPA3 for mast cells; LYVE1, CCL21, and TFF3 for lymphatic endothelial cells (LECs); KRT19, KRT7, and AQP5 for kera B; and DCT, TYRP1, and MLANA for melanocytes (Fig. 1C and D).
Figure 1.
Identification of 12 distinct clusters within the skin tissue in individuals with diabetes and without diabetes. A: The overall design of the study. Schematic diagram was created with BioRender.com. B: tSNE projection of the filtered data (36,487 cells). Each cell is represented as a dot. C: Expression level of selected markers: endothelial cells, CDH5 (further elaborated in Supplementary Fig. 4A); T cells, CD3D; fibroblasts, COL1A1; and keratinocytes, KRT14. D: Heat map showing relative expression of the top three markers for each cluster (left) and the total number of detected cells and genes in each cluster (right).
Differentially Expressed Gene Analysis
After preprocessing and determination of cellular identities, Wilcoxon rank sum test was used to perform differentially expressed gene (DEG) analysis. Cluster markers were identified by comparing each cluster with the rest of the cells. In addition, each cell type was compared between the diabetic cells and nondiabetic cells (12 comparisons) to identify the DEG at cluster level with an adjusted P value <0.05 and log-fold change (FC) cutoff ±0.25. Gene set enrichment analysis (GSEA) was performed using GSEA software for identified genes in the endothelial cell cluster (cluster 0) with an adjusted P value of <0.05 (30). A total of 1,796 genes were sorted based on their logFC values, and GSEA was performed using the GSEA preranked module with default parameters and a maximum size of gene sets to include 1,000 genes. Enrichment was performed against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (31,32). Heat maps were created using Genesis software (33).
Cell–Cell Interaction Analysis
Cellchat package in R was used to infer the cell–cell interactions (CCIs) between different skin cell types in diabetic and normal skin (34). Cellchat database is a collection of KEGG pathways in addition to other recent signaling molecules from the literature. The database contains 2,021 validated molecular interactions (as of March 2021) classified into 60% paracrine/autocrine signaling interactions, 21% receptor–extracellular matrix interactions, and 19% cell–cell contact interactions. All interactions were classified based on literature into 1 of 229 functionally related signaling pathways. Cellchat considers known composition of ligand–receptor complexes and cofactor types—agonists, antagonists, or costimulatory or coinhibitory membrane-bound receptors. After identification of differentially expressed ligands and receptors, Cellchat associates each interaction with a probability value to quantify outgoing and incoming communications between cell groups. Significant interactions are then identified after performing 100 random permutations of group labels of cells and recalculating the probability of interactions. A weighted directed graph is then constructed from the significant connections between cell groups.
Spatial Transcriptomic Analysis
A 20-μm–thick section was taken on a Visium spatial gene expression slide followed by permeabilization for 12 min. cDNA libraries were synthesized using the Visium Spatial Gene Expression Reagent Kit as per the manufacturer’s recommendations (PN-1000186 and 1000190; 10x Genomics) and sequenced using the NovaSeq 6000 (Illumina). Generated reads were preprocessed using the standard Space Ranger (https://support.10xgenomics.com/spatial-gene-expression/software) pipeline to align the raw sequencing reads onto the hg38 genome and obtaine the respective h5-format and spatial image files for UW human skin. Resulting preprocessed files were loaded in Seurat (35) for further downstream data processing and analysis. Briefly, we loaded the .h5 and spatial image files using the “Load10X_Spatial” module in Seurat and created Seurat Object for data processing. Sample quality was assured by computing the mitochondrial content (MT-content <15% is retained) and filtering out the poor-quality spots using the “subset” module of Seurat. We used the “SCTransform” method (36) available in Seurat that implements the regularized negative binomial regression for normalizing the 10X spatial data. The spatial feature plot function in Seurat was used to visualize the spatial location and expression profile of candidate genes (belongs to endothelial cells) in UW human skin.
Animal Studies
Commercially available C57BL/6 mice, obtained from The Jackson Laboratory (Bar Harbor, ME), were used for the experiments. Adult (8–12-week-old male mice, ∼25 g in weight) were used for experiments. For diabetic induction, mice were injected with streptozotocin (STZ) in the intraperitoneal region (50 mg/kg, 5 days) prepared in citrate buffer (sodium citrate, 0.05 mol/L, 4.5 pH). Mice were fasted for 6 h at the time of STZ injection with free access to water. Two weeks after STZ injection, blood samples were collected by tail venopuncture of the mice and used for the estimation of blood glucose levels. Blood glucose levels were assessed using the Bayer Contour blood glucose monitoring system. Animals with fasting (6-h) blood glucose levels for two consecutive readings of >250 mg/dL (∼14 mmol) were considered diabetic. For the T2DM model, db/db (BKS.Cg-m+/+Leprdb/J) mice were purchased (stock no. 000642; The Jackson Laboratory). Adult mice (24–38-week-old db/db mice, ∼50 g in weight, 11 male/8 female) with glucose levels >350 mg/dL were used for experiments. All studies were performed in compliance with the Institutional Animal Care and Use Committee of the Indiana University. Mice were housed individually after surgery with a 12-h light/dark cycle and temperature in the institutional animal facilities and allowed access to food and water ad libitum.
Hindlimb Ischemia Surgery
Unilateral hindlimb ischemia was induced via ligation and subsequent resection of the femoral artery (37). Briefly, STZ-induced and db/db diabetic mice were anesthetized with 1–3% isoflurane and placed supine under a stereomicroscope (Zeiss OPMI) on a heated pad. Preoperative analgesia was obtained using a subcutaneous injection of buprenorphine and carprofen. A 3-cm incision was made to expose the origin of the femoral artery proximally to the bifurcation of the popliteal and saphenous artery distally. The femoral artery was then exposed and carefully separated from the femoral vein and nerve using blunt dissection. The proximal femoral artery was ligated just distal to external iliac artery, and the distal femoral artery was ligated just proximal to the popliteal and saphenous artery with 7–0 silk suture. The femoral artery was then transected just proximal to the distal ligation. The femoral artery was then carefully removed from caudal to cranial, making sure to ligate and transect any larger branches along the way. The femoral artery was then transected just distal to the proximal ligation and fully resected. Hemostasis was achieved using a combination of direct pressure and electrocautery of the smaller side branches. Finally, a single dose of bupivacaine was administered subcutaneously in the hindlimb to provide local anesthesia. Laser speckle imaging (PeriCam PSI Low Resolution; PeriMed) was conducted ∼2 h postsurgery to confirm successful blood flow occlusion and at the different time points mentioned (days 3, 7, 10, and 14 postsurgery). Limb perfusion was measured using Pimsoft software as reported (38,39).
Tissue Nanotransfection Technology 2.0
In vivo tissue nanotransfection technology 2.0 (TNT 2.0) was performed as described previously with a modification in the chip design (37,40). Briefly, the hollow microneedle array was fabricated on a double-sided polished silicon wafer using a standard semiconductor process in a cleanroom environment as described (40,41). cDNA open reading frame (ORF) clones to express gene of interest were purchased from GenScript. PLCγ2 ORFs used were 1) OMu18551D (NM_172285.1) and 2) OMu83524D (NM_172285.2) using pcDNA3.1+/C-(K)-DYK vector. VEGFA ORFs used were 1) OMu22838D (NM_001110267.1), 2) OMu22871D (NM_001110268.1), 3) OMu23283D (NM_001287058.1), and 4) OMu09529D (NM_001287056.1) using the pcDNA3.1+/C-(K)DYK vector. Targeted delivery of PLCγ2 ORF to endothelial cells was achieved through CDH5 promoter–driven expression of PLCγ2 ORF (pPromoter backbone; Applied Biological Materials Inc.) (Supplementary Fig. 8B). For in vivo knockdown experiments in the endothelial compartment, PLCγ2 shRNA vector (target sequence 5′-ACCGCAGAGCCCTTCTTATTT-3′) was purchased from VectorBuilder (identification no. VB900081-8211xwc) and subcloned to insert CDH5 promoter (NovoPro Bioscience Inc.). Scramble shRNA lentiviral control vector (identification no. VB010000-0009mxc) was purchased from VectorBuilder.
Cell Culture
Human dermal microvascular endothelial cells (HMECs) were cultured in MCDB-131 medium (#10372-019; Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS, 10 mmol/L l-glutamine (#25030-081; Thermo Fisher Scientific), and 1% antibiotic (#15240-062; Life Technologies) (38,39). Cells were seeded in 12-well plates at a density of 0.1 × 106 cells/well. When the confluency was ∼80%, transfection was achieved by human (h)PLCγ2 siRNA (#L-008339-02-0010; Dharmacon) using DharmaFECT transfection reagent (GE Dharmacon) and OptiMEM serum-free medium (Invitrogen/Thermo Fisher Scientific) (38,39). siRNA-treated and -nontreated HMECs were collected after 72 h of transfection for flow cytometric analysis for validation of the PLCγ2 inhibition.
Flow Cytometry Analyses
Cells were stained according to the manufacturer’s protocol (#424401; True-Nuclear Transcription Factor Buffer Set; BioLegend). Briefly, sham or PLCγ2 siRNA-transfected cells were incubated in 1× fixative (True-Nuclear 4X Fix Concentrate) for 30 min and pelleted. They were then permeabilized by resuspending with vigorous vortexing in 1 mL of 1× permeabilization buffer (True-Nuclear 10X Perm Buffer) and incubated at 4°C for at least 1 h. Cells were washed twice in the staining media (PBS containing 1% BSA) and then resuspended in staining media at 0.5–1 × 106 cells/100 μL. Monoclonal antibodies against PLCγ2 (#3872S, Cell Signaling Technology, Danvers, MA; with #A-11011, Goat Anti-Rabbit IgG, Alexa Fluor 568) were added and incubated for 1 h at room temperature. The cells were washed with staining media and pelleted. Finally, samples were resuspended in 100 μL staining media and analyzed by flow cytometer (BD LSR II Flow Cytometer). The fluorescence and light-scattering properties (forward scatter and side scatter) of the cells were determined. Signals from cells labeled with conjugated fluorophores were detected (42). Data analyses was done using FlowJo software version 10.7.1. Gates were set manually. Gating strategy for the fluorophores has been shown in the respective figures.
In Vitro Angiogenesis Assay
Tube formation on Matrigel was assessed as a determinant of in vitro angiogenesis (38,39). Briefly, HMECs were transfected with control or hPLCγ2 siRNA. Following 48 h of transfection, cells were treated with 100 ng of recombinant VEGF 165 protein (#293VE050; R&D Systems) for 12 h. Seeding was done at a seeding density of 1.2 × 105 cells/well in a Matrigel-coated four-well plate (CB-40234A, Corning Matrigel Membrane Matrix; Corning). In vitro angiogenesis was assessed by calculating the tube length after 4 h of seeding using fluorescence microscopy using Calcein AM staining (38,39).
Extracellular Flux and Oxygen Consumption Rate (Seahorse) Assays
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measurements were performed using a Seahorse Bioscience XF-96 instrument as described previously (39,43). In brief, 50,000 HMECs/well were seeded in 96-well Seahorse microplate plates in MCDB131 media. After attachment, cells were transfected with control or hPLCγ2 siRNA. Following 12 h of transfection, cells were treated with 100 ng of recombinant VEGF 165 protein (#293VE050; R&D Systems) for 12 h. A day before the experiment, the sensor cartridge was hydrated overnight using the calibration buffer supplied by the manufacturer (Seahorse Biosciences, Santa Clara, CA). On the day of the experiment, the cells were washed with calibration buffer twice and incubated with glucose-free Seahorse XF base medium supplemented with 25 mmol/L of glucose and 25 mmol/L glutamine for 30 min at 37°C in a CO2-free incubator. The injection ports of the sensors were filled with 20 μL of treatment or vehicle in buffer. The sensor was then placed into the XF-96 instrument and calibrated. After calibration, the calibration fluid plate was replaced with the cell plate. The measurement cycle consisted of a 2-min mix, 1-min wait, and a 2-min measurement. Four basal rate measurements were followed by sequential addition of oligomycin (8 μg/mL), carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (100 μmol/L), rotenone (100 μmol/L), and antimycin A (100 μmol/L) prepared in glucose-free Seahorse XF base medium. Each injection was followed by four measurement cycles. The consumption rates were calculated from the continuous average slope of the decreased O2 using a compartmentalization model (43). For any one treatment, the rates from five to six wells were used.
Histology, Immunohistochemistry, and Immunocytochemistry
Histology of skin was performed from 10-μm–thick paraffin sections or tissue embedded in OCT. Paraffin tissue sections were deparaffinized using xylene and rehydrated using decreasing gradient of ethanol. Antigen retrieval was done using citrate buffer, blocked with 10% normal goat serum, and incubated with primary antibodies against PLCγ2 (1:100 dilution; no. 3872S; Cell Signaling Technology or 1:100 dilution; no. ab109267; Abcam) and anti-VWF antibody (1:100 dilution; no. ab11713; Abcam) followed by appropriate fluorescence-conjugated secondary antibodies (Alexa Fluor 568–tagged anti-sheep, 1:200 dilution; and Alexa Fluor 488–tagged anti-rabbit, 1:200 dilution). Images were collected and analyzed using a Zeiss Axio Scan Z1 and Zeiss LSM 880 microscope guided by Zen blue imaging software (38,39). Immunohistochemistry images were analyzed using the ImageJ software (National Institutes of Health).
Western Blot
Cells or homogenized tissues were harvested using cell lysis buffer (#9803; Cell Signaling Technology), sonicated on ice (three pulses for 3 s), and centrifuged (14,000g for 20 min at 4°C). The protein estimation in the clear lysates was done using bicinchoninic acid assay. Proteins (25–30 μg) were separated on 4–12% Bis-Tris Gel/MOPS running buffer (cat. no. NP0321BOX and NP0001; NuPAGE) (45 min at 200 V) using the NuPAGE electrophoresis system (Invitrogen). Proteins were transferred to polyvinylidene difluoride membranes (2.15 h at 30 V). Membranes were blocked with 5% skim milk in Tris-buffered saline with 0.1% Tween 20 for 1 h at room temperature and incubated with the respective primary antibodies overnight at 4°C. The following day, the membranes were washed and incubated with the corresponding horseradish peroxidase–conjugated secondary antibody for 1 h at room temperature. The membranes were washed, and the images were acquired using HyGLO chemiluminescent detection reagent (cat. no. E-2500; Thomas Scientific, Swedesboro, NJ) and an Azure c600 Gel Imaging System by Azure Biosystems (39). β-Actin was used as loading control. ImageJ software was used for quantification of bands by densitometry analysis.
Statistical Analysis
Prism v8.0 (GraphPad Software) was used for statistical analyses. Data were assumed to be normally distributed for all analyses conducted. Data for independent experiments are presented as means ± SEM unless otherwise stated. The Student t test (two-tailed) or ANOVA was used to test the significant differences among groups. In case of ANOVA, the Tukey honestly significant difference (HSD) post hoc test was applied. A P value of <0.05 was considered significant.
Data and Resource Availability
The scRNA-seq and Visium spatial transcriptomics raw data are deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus under accession numbers GSE176417 and GSE182208. All other data generated or analyzed during this study are included in this published article (and its Supplementary Material).
Results
scRNA-seq Characterization of Cell Clusters in Human Diabetic Skin
The transcriptional profile of 36,487 single cells (11,734 diabetic and 24,753 nondiabetic) derived from three diabetic and five nondiabetic human skin samples was analyzed (Fig. 1A and Supplementary Fig. 1A–C). Unsupervised clustering identified 12 cellular clusters with distinct expression profiles (Fig. 1B–D). Using the DEG signatures, putative identities were attributed to each cluster (Fig. 1B and D). Figure 1C and D show selected signature genes in the form of feature plots (Fig. 1C) and heat map (Fig. 1D, left). Cell and gene counts for each cell type are shown in Fig. 1D (right). This included endothelial cells, T cells, fibroblasts, SMCs, NK cells, keratinocytes, myeloid cells, LECs, and melanocytes. Fibroblasts (clusters 2 and 3) and keratinocytes (clusters 6 and 10) were identified as two distinct clusters representing different states for each. Additionally, the data quality from normal and diabetic skin cells was comparable with similar numbers of detected genes and total number of detected reads (Fig. 1D, right, and Supplementary Fig. 1D). Cluster 4 (SMCs) included established characteristics of pericytes, as evident by the high expression of published markers (44,45), such as SMA (ACTA2), DES, NG2, and RGS5 (Supplementary Fig. 1E). Hence, by investigating the abundance of each cell type, cellular composition was consistent across diabetic and nondiabetic skin (Supplementary Table 1).
Cluster-Specific Differential Gene Expression in Human Diabetic Skin
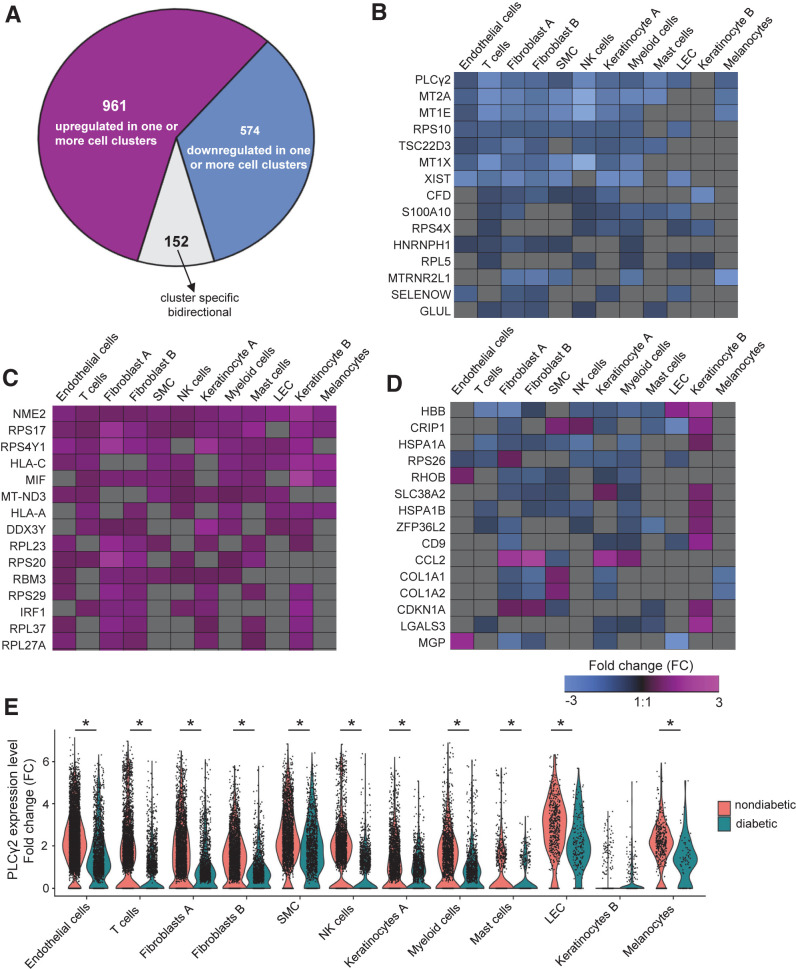
Differential gene expression analysis was performed between different cell types obtained from diabetic and nondiabetic human skin. Of the 27,393 detected genes, 6298 genes were significantly affected by diabetes in at least one cell type (false discovery rate [FDR] <0.05). A total of 1,687 genes passed the log fold-change cutoff ±0.25. Of those 1,687 genes, 1,535 exhibited the same directionality of either upregulation or downregulation in diabetic skin regardless of the cell-type identity (961 upregulated and 574 downregulated) (Fig. 2A–C, Supplementary Figs. 2 and 3, and Supplementary Tables 2 and 3). The directionality of differential expression of 152 genes was different across cell populations (Fig. 2A and D, Supplementary Figs. 2 and 3, and Supplementary Table 4). Of interest was the expression profile of the PLCγ2 gene, which was downregulated in 11 out of 12 cellular clusters identified in diabetic human skin (Fig. 2B and E). The other family of genes that was downregulated in multiple cell types in diabetic skin was the metallothionein (MT) family (MT2A, MT1E, and MT1x) (Fig. 2B). Both PLCγ2 and MTs are known downstream targets of vascular growth factors and contribute to angiogenesis (46,47). Among the upregulated genes among multiple cell types in diabetic skin, the NME/NM23 (also known as nucleoside diphosphate kinase 2 [NME2]) gene was the top candidate (Fig. 2C). NME2 was upregulated in all 12 cellular clusters in diabetic skin. The other significant gene which was upregulated in 9 out of 12 clusters was the cytokine macrophage migration inhibitory factor, which is known to contribute to metabolic inflammation in diabetes (48) (Fig. 2C). Specific sets of genes were dichotomously expressed in different cellular clusters of diabetic human skin (Fig. 2D). For example, the chemokine C-C motif chemokine ligand 2 (CCL2) was upregulated in fibroblasts, keratinocytes, and myeloid cells while at the same time downregulated in SMCs isolated from diabetic skin (Fig. 2D).
Figure 2.
Diabetes-related DEGs in distinct cell types of skin. A: Pie chart showing the number of the upregulated, downregulated, or bidirectional genes across all identified clusters. Heat map showing a subset of genes that has shared downregulation (B) or shared upregulation (C) (FDR-adjusted P < 0.05 and logFC ±0.25) in diabetic skin. D: Heat map of logFC showing a subset of diabetic-related genes (FDR-adjusted P < 0.05 and logFC ±0.25) that has bidirectional genes (down- or upregulated) in different clusters of the skin. Heat maps were created using Genesis software. E: Violin plot representing expression level of PLCγ2 in each cellular cluster in diabetic and nondiabetic skin. Each cell is represented as a dot. PLCγ2 was downregulated in 11 cellular clusters in diabetic skin (not significantly different in only keratinocyte B cluster) (*FDR-adjusted P < 0.05).
Differential Gene Expression in the Endothelial Cluster of Human Diabetic Skin
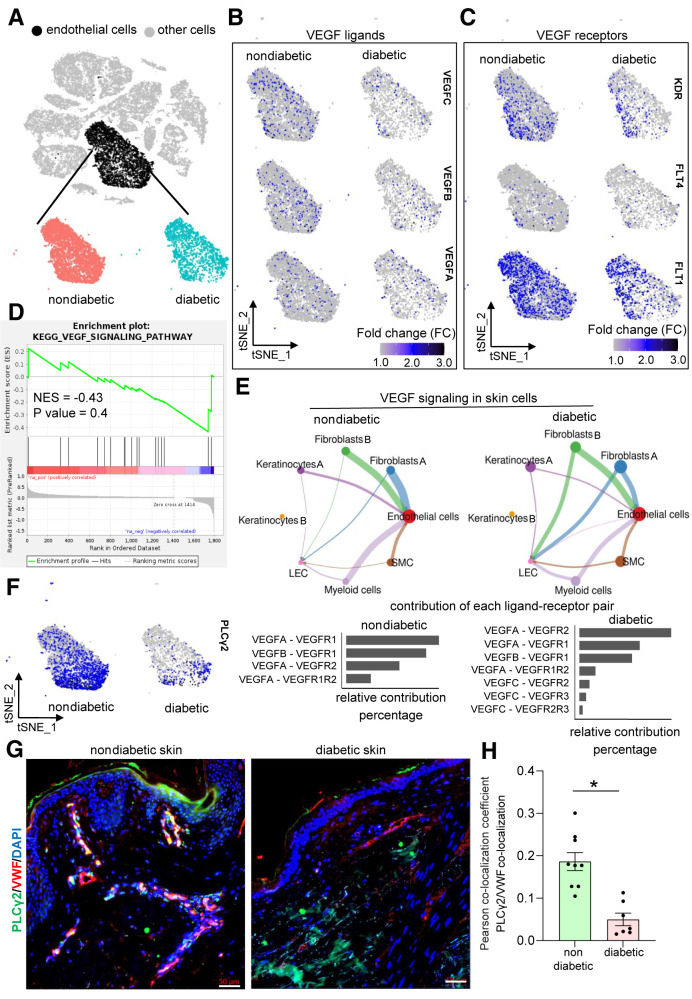
We further analyzed the gene expression changes in diabetic versus nondiabetic skin by subsetting only the endothelial cluster marked by enrichment of CHD5highVWFhighPECAM1high cells (Fig. 3A and Supplementary Fig. 4A–C). GSEA for DEGs among endothelial cells using the KEGG database resulted in identification of five upregulated and one downregulated pathway in diabetic skin endothelial cells (P < 0.05) (Supplementary Fig. 4D and E and Supplementary Table 5). The upregulated pathways included ribosome, focal adhesion, TP53, regulation of actin cytoskeleton, and chemokine signaling pathways (Supplementary Fig. 4E and F). In contrast, the cell adhesion molecule pathway was identified to be the only downregulated pathway in diabetic endothelial cells. This included classical vascular genes like PECAM1 and CD34 (Supplementary Fig. 4E and G). In addition, the genes known to augment the blood vessel functionality, like integrin β1 (ITGB1) (49) and integrin α6 (ITGA6) (50), were also downregulated in diabetic endothelial cells. Interestingly, the feature plots of VEGF ligands and receptors (Fig. 3B and C), gene expression levels (Supplementary Figs. 5 and 6), and the GSEA analysis of DEGs (normalized enrichment score −0.43; P = 0.4) (Fig. 3D) of the endothelial subcluster indicated that the VEGF signaling pathway was comparable between diabetic and nondiabetic skin.
Figure 3.
Identification and extraction of endothelial cell clusters within the skin samples of individuals with diabetes and without diabetes. A: Identification and extraction of the endothelial cell cluster in human skin by visualizing the expression level of CDH5, VWF, and PECAM1 gene expression enrichment (Supplementary Fig. 4A). B: tSNE plots showing nonsignificant difference of VEGF ligands between diabetic and nondiabetic skin endothelial cells (VEGFC, FDR-adjusted P value: 1; VEGFB, adjusted P value: 1; VEGFA, not expressed in >20% of the cells in either group). C: tSNE plots showing nonsignificant difference of VEGF receptors between diabetic and nondiabetic skin endothelial cells (KDR, FDR-adjusted P value: 1; FLT1, FDR-adjusted P value: 0.04, logFC < ±0.25; FLT4, not expressed in >20% of the cells in either group). D: GSEA showing that the VEGF signaling was not significantly altered between diabetic and nondiabetic skin endothelial cells (normalized enrichment score [NES] −0.43; P = 0.4). E: Signaling interaction analysis of VEGF pathway across different cell types in diabetic and nondiabetic skin. E, top: VEGF signaling network in nondiabetic (left) and diabetic (right) skin. E, bottom: Ligands and receptors contributed in VEGF signaling in nondiabetic (left) and diabetic (right) skin cells. F: tSNE plots showing PLCγ2 expression level between nondiabetic (left) and diabetic (right) skin endothelial cells (logFC: −0.52; FDR-adjusted P value: 5.98E-100). Immunohistochemical analysis of VWF+/PLCγ2+ colocalization in human nondiabetic and diabetic skin (G) and its colocalization coefficient (H). Scale bars, 50 µm. n = 9 and 7. *P < 0.05 (Student t test). Data are represented as the mean ± SEM.
Whether the cell–cell signaling of the VEGF pathway was affected in diabetic endothelial cells was tested. The differential CCIs between the VEGF ligands and receptors were calculated among eight different cellular clusters (e.g., endothelial cells, fibroblasts [both clusters], myeloid cells, LECs, keratinocytes [both clusters], and SMCs) in diabetic versus nondiabetic human skin using Cellchat (34). Endothelial cells were the major receiver of the VEGF signaling, seconded by LECs in both diabetic and nondiabetic skin (Fig. 3E and Supplementary Table 6). Both diabetic and nondiabetic cells relied on similar communication patterns for VEGF expression and signaling regarding originating and receiving cells (Fig. 3E). These findings led to the investigation of PLCγ2, downstream of VEGF signaling, which was downregulated in diabetic skin endothelial cells (Fig. 3F). PLCγ2 protein level was low in human diabetic endothelial cells (Fig. 3G and H).
Endothelial Cell PLCγ2 Expression Augmented Rescue of Diabetic Ischemic Limb in Response to VEGF Therapy
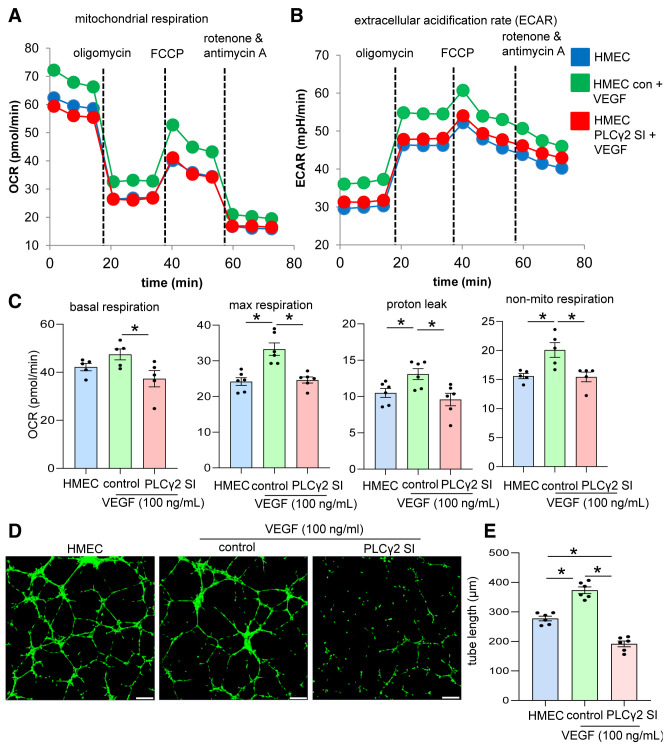
VEGF-dependent priming of endothelial cell tube formation is known to be driven through the collective action of downstream signaling molecules (51). Deregulation of any of these molecules fails to stimulate VEGF-induced endothelial cell function (51,52). Downregulation of PLCγ2 expression in diabetic endothelial cells necessitated the investigation of the significance of this gene during VEGF-induced endothelial cell functionality. The significance of PLCγ2 in VEGF-mediated increase in endothelial cell metabolism and function was assessed in cultured HMECs. First, HMECs supplemented with exogenous VEGF with or without PLCγ2 inhibition were subjected to mitochondrial function analyses. VEGF supplementation to cultured endothelial cells increases the mitochondrial function, as indicated by elevation in OCR, ECAR, and associated mitochondrial functions (Fig. 4A–C). Such VEGF-induced increase in endothelial cell metabolism was blunted in response to PLCγ2 inhibition (Fig. 4A–C and Supplementary Fig. 7). Consistently, inhibition of PLCγ2 blunted the VEGF-induced increase in Matrigel tube formation, a functional outcome (Fig. 4D and E).
Figure 4.
VEGF-mediated increase in endothelial cell functionality is PLCγ2 dependent. OCR (A) and ECAR (B) calculation of HMECs in normal conditions and after VEGF exposure (100 ng/mL) and cotreatment with either control (con) siRNA or PLCγ2 siRNA (SI) using the Seahorse XF 96-well plate reader. OCR was determined during sequential treatments with oligomycin (ATP synthase inhibitor), carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP), a protonophore that lowers the mitochondrial membrane potential to create conditions for maximal oxidative respiration, and antimycin A plus rotenone to inhibit the electron transport chain. C: Basal respiration, maximal respiration, proton leak, and nonmitochondrial (non-mito) respiration measurement in HMECs in normal conditions and supplemented with VEGF (100 ng/mL) and cotreatment with control (con) siRNA or PLCγ2 siRNA (SI). Matrigel tube formation images (D) and tube length (E) analysis in HMECs in normal conditions and after cotreatment with control (con) siRNA or PLCγ2 siRNA. Scale bars, 200 µm. n = 6. *P < 0.05 (one-way ANOVA, followed by Tukey HSD post hoc test).
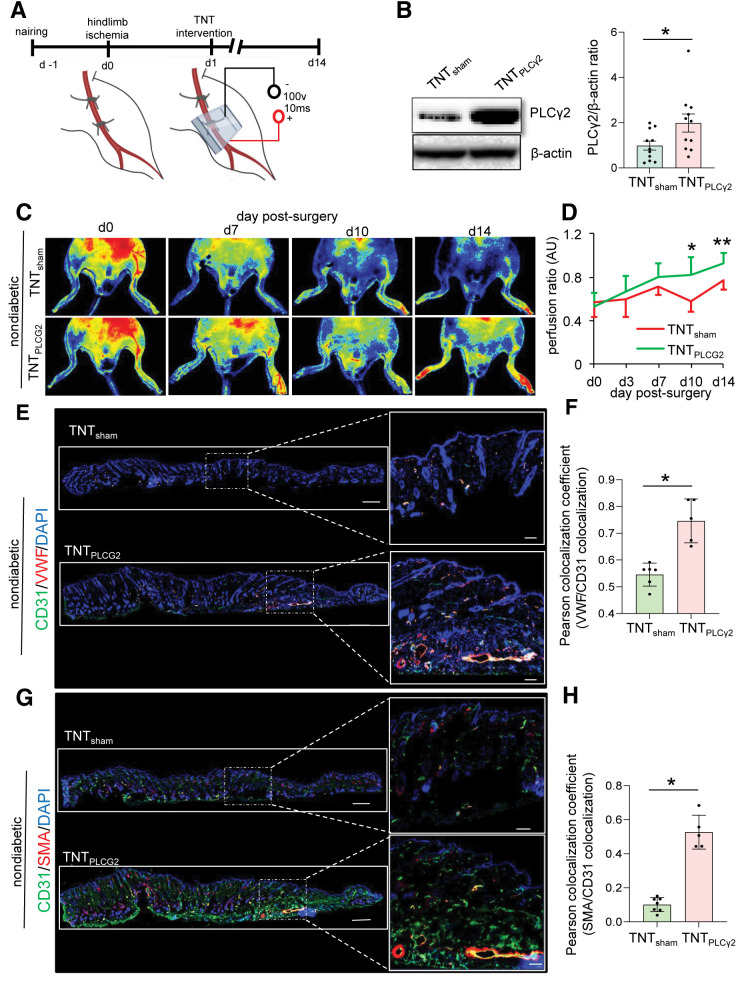
The functional significance of PLCγ2 expression in ischemic limb perfusion was tested in a series of rescue experiments, the first of which used TNT 2.0–mediated direct cytosolic delivery of PLCγ2 ORFs in normoglycemic C57BL/6 mice (Fig. 5A and B and Supplementary Fig. 8A). Compared with sham-treated ischemic limbs, PLCγ2-treated limbs showed improved perfusion as early as day 10 post-TNT (Fig. 5C and D). Marked revascularization of PLCγ2-treated ischemic tissue was observed. Such effect was manifested as higher abundance of CD31+/VWF+ and CD31+/SMA+ blood vessels compared with sham (Fig. 5E–H).
Figure 5.
TNT-mediated overexpression of PLCγ2 improves perfusion in nondiabetic ischemic limbs. A: Schematic diagram showing TNT-assisted delivery of PLCγ2 ORF in murine ischemic limbs. Schematic diagram was created with BioRender.com. B: Western blot analysis of TNT-mediated PLCγ2 overexpression in murine skin. Data represent 10 different sites from 3 mice. *P < 0.05 (Student t test). Data are represented as the mean ± SEM. PeriMed laser speckle–assisted perfusion images (C) and their analysis (D) in ischemic limbs on which TNT procedure was done with sham or PLCγ2 ORF in nondiabetic conditions. Perfusion was calculated based on the ratio of the ischemic versus normal/contralateral limb. n = 6 to 7. *P < 0.05 (Student t test); **P < 0.005. E: Immunohistochemical analysis of CD31+/VWF+. G: CD31+/SMA+ colocalization in ischemic limbs of mice treated with sham or PLCγ2 ORF at day 14 postsurgery and their colocalization coefficient (F and H) (n = 5–7). *P < 0.05, Student t test. Data are represented as the mean ± SEM. AU, arbitrary unit.
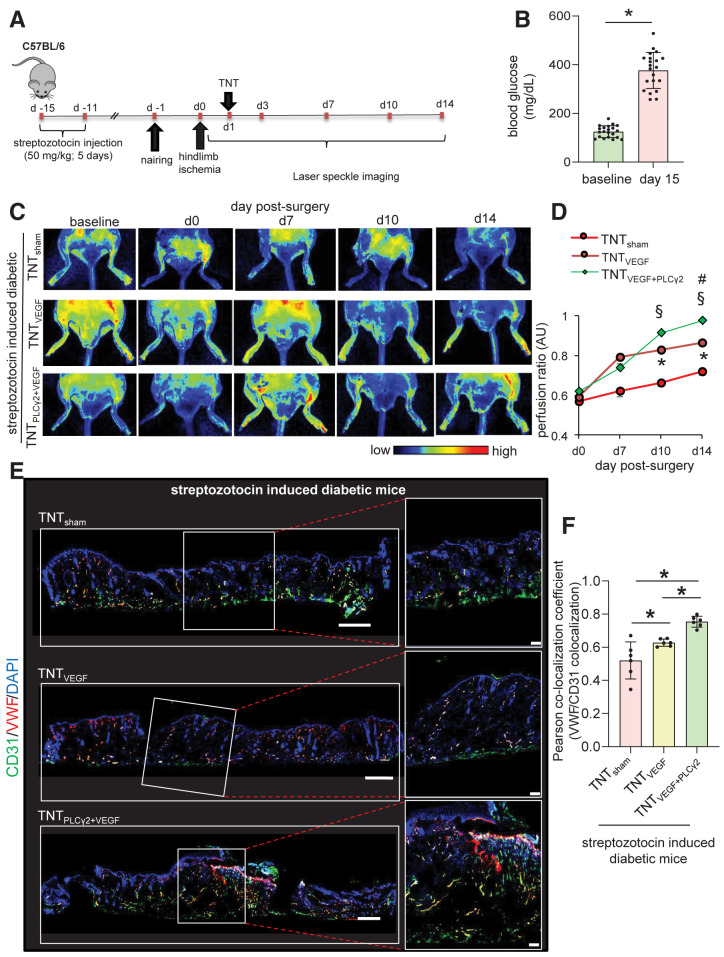
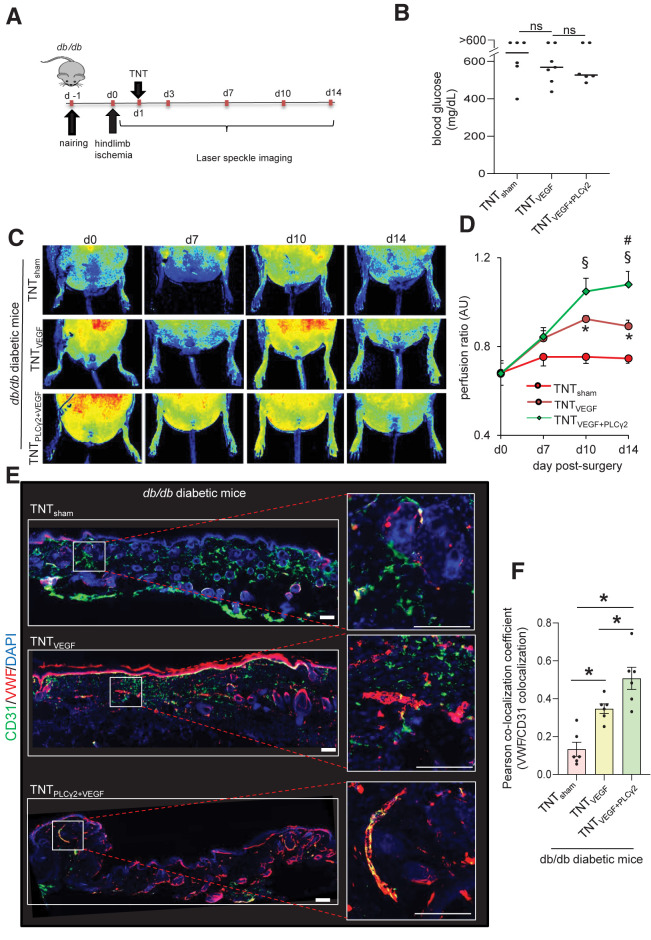
In patients with T2DM, the ability of ischemic tissue to synchronize the molecular and cellular events leading to the restoration of tissue perfusion is known to be markedly impaired (13). scRNA-seq data indicated that although VEGF signaling was not significantly altered in diabetic versus nondiabetic skin, PLCγ2 expression was blunted. Follow-up rescue studies therefore focused on understanding the significance of TNT-based VEGF therapy in the presence or absence of endothelial PLCγ2 in diabetic ischemic tissue. Induction of diabetes in C57BL6 mice was achieved via a low dose of STZ (50 mg/kg, 5 days) (Fig. 6A and B). Fifteen days post–STZ injection, unilateral hindlimb ischemia was induced via occlusion and subsequent transection of the femoral artery. Next, electrophoretic TNT was performed to deliver VEGF ORF alone or a cocktail of VEGF and CDH5 promoter–driven PLCγ2 ORF to the ischemic limb on day 1 post–hindlimb ischemia surgery (Fig. 6A). The targeted overexpression of PLCγ2 to endothelial cells was validated using flow cytometry (Supplementary Figs. 8B and 9A–D). Such successful PLCγ2 expression in endothelial cells of diabetic mice augmented VEGF therapy to increase perfusion to the diabetic ischemic limbs (Fig. 6C and D). Additionally, shRNA-mediated depletion of PLCγ2 in endothelial cells abrogated the effect of VEGF therapy to increase in perfusion of diabetic ischemic limb (Supplementary Fig. 10). Higher perfusion at the ischemic tissue site of diabetic mice subjected to VEGF plus endothelial PLCγ2 was supported by histological studies demonstrating increased abundance of CD31+/VWF+ and CD31+/SMA+ vasculature (Fig. 6E and F and Supplementary Fig. 9E and F). Similar results were obtained using a murine model of T2DM, db/db mice, in which endothelial PLCγ2 augmented VEGF therapy to increase perfusion to the ischemic limbs (Fig. 7A–E). These findings recognize PLCγ2 as a factor that can increase the efficiency of VEGF therapy in diabetic ischemic tissue.
Figure 6.
Endothelial cell–targeted delivery of PLCG2 augments VEGF therapy rescue of STZ-induced diabetic ischemic limb. A: Schematic diagram showing TNT-mediated delivery of VEGF ORFs or/and endothelial PLCγ2 ORFs in ischemic hindlimb of STZ-induced diabetic mice. B: Blood glucose level in mice used in this study before and after injection of STZ (50 mg/kg). PeriMed laser speckle–assisted perfusion images (C) and their analysis (D) in ischemic limbs on which TNT procedure was done with sham, VEGF only, or VEGF plus CDH5 promoter–driven (endothelial) PLCγ2 cocktail. Perfusion was calculated based on the ratio of the ischemic vs. normal/contralateral limb. n = 6 to 7. *P < 0.05 TNTVEGF vs. TNTsham; §P < 0.05 TNTVEGF+PLCγ2 vs. TNTsham; #P < 0.05 TNTVEGF+PLCγ2 vs. TNTVEGF (one-way ANOVA, followed by Tukey HSD post hoc test). Immunohistochemical analysis of CD31+/VWF+ colocalization in diabetic ischemic limbs of mice treated with above-mentioned cocktails at day 14 postsurgery (E) and the colocalization coefficient (F). n = 6 to 7. *P < 0.05 (one-way ANOVA, followed by Tukey HSD post hoc test). AU, arbitrary unit.
Figure 7.
Endothelial cell–targeted delivery of PLCG2 augments VEGF therapy rescue of the T2DM (db/db) diabetic ischemic limb. A: Schematic diagram showing TNT-mediated delivery of VEGF ORFs or/and endothelial PLCγ2 ORFs in the ischemic hindlimb of db/db diabetic mice. B: Blood glucose level in db/db mice used in this study. TNTsham (n = 6; 4 male and 2 female), TNTVEGF (n = 7; 4 male and 3 female), and TNTVEGF+PLCG2 (n = 6; 3 male and 3 female). PeriMed laser speckle–assisted perfusion images (C) and their analysis (D) in ischemic limbs on which TNT procedure was done with sham, VEGF only, or VEGF plus CDH5 promoter–driven (endothelial) PLCγ2 cocktail. Perfusion was calculated based on the ratio of the ischemic vs. normal/contralateral limb. n = 6 to 7. *P < 0.05 TNTVEGF vs. TNTsham; §P < 0.05 TNTVEGF+PLCγ2 vs. TNTsham; #P < 0.05 TNTVEGF+PLCγ2 vs. TNTVEGF (one-way ANOVA, followed by Tukey HSD post hoc test). Immunohistochemical analysis of CD31+/VWF+ colocalization in diabetic ischemic limbs of db/db mice treated with above-mentioned cocktails at day 14 postsurgery (E) and the colocalization coefficient (F). n = 6. *P < 0.05 (one-way ANOVA, followed by Tukey HSD post hoc test). AU, arbitrary unit.
Discussion
PLCγ2 encodes a transmembrane signaling enzyme that works downstream of VEGF signaling (53). This signaling molecule is required for the normal differentiation and function of hematopoietic cells, including B lymphocytes, NK cells, mast cells, platelets, and osteoclasts (53–56). Recently, it has been demonstrated that PLCγ2 also participates in tissue morphogenesis (57). PLCγ2-deficient mice exhibit misconnections of blood and lymphatic vessels (58). Biochemically, PLCγ2 catalyzes the hydrolysis of phosphatidylinositol 4,5-bisphosphate into diacylglycerol and inositol 1,4,5-trisphosphate as well as second messenger molecules (59,60). Direct activation of PLCγ2 by tyrosine phosphorylation controls diverse biological processes, including chemotaxis, platelet aggregation, and adaptive immunity. This work demonstrates that under conditions of diabetes, limited PLCγ2 can neutralize the angiogenic effects of VEGF. Thus, PLCγ2 emerges as an important factor that is responsible for responsiveness to VEGF therapy.
One of the major reasons for the clinical disappointment in single-gene therapy using VEGF (monotherapy) of patients with PAD is the inability to respond to changes in ligand concentration due to impaired receptor signaling (61–63). Previous studies have tried delivering two angiogenic or tissue-protective molecules together to circumvent the low-efficiency hurdle of single-gene therapy (61). Combined gene delivery of VEGF165 with angiopoietin-1 (64), fibroblast growth factor 2 (basic) (bFGF) (65), fibroblast growth factor 4 (FGF4) (66), urokinase plasminogen activator (uPA) (67), stromal cell–derived factor 1α (SDF-1α) (68), platelet-derived growth factor subunit B (PDGF-B) (69), granulocyte colony-stimulating factor (G-CSF) (70), or hepatocyte growth factor (HGF) (71) was able to augment increase of angiogenic response of VEGF therapy. This work demonstrates that combined gene therapy using PLCγ2 and VEGF is effective to enhance vasculogenic efficacy to rescue from ischemic insult. One of the important concerns of VEGF gene therapy remains its strong linkage with dose-dependent microvascular permeability and peripheral edema (19). An ∼60% of patients recruited in various clinical trials developed moderate to severe edema (19). Conversely, lower levels of VEGF result in regression of newly formed blood vessels (72). Hence, development of new delivery systems that can efficiently deliver gene products without tissue damage is warranted for clinical use (19). Administration of exogenous VEGF by a number of delivery routes (transvascular, intradermal, and subfascial) using recombinant VEGF (73) along with various viral and nonviral gene delivery methods using liposomes (74), adenoviral vectors (75), adeno-associated virus vectors (76), and sonoporation (77) has been evaluated to demonstrate the therapeutic potential of exogenous VEGF (72). Application of novel techniques like nonviral gene therapy is of high translational value (78). In vivo electroporation is based on the principle of enhanced plasma membrane permeability of the tissue upon application of short electric pulses of high voltage, resulting in enhanced nucleic acid uptake (79). The nonviral electroporation-mediated technique is safe and effective for prolonged transgene expression (72,80,81). Electrotransfer offers an alternative nonviral platform for gene delivery of plasmid VEGF to enhance ischemic tissue survival. Recent studies have demonstrated the efficacy of intradermal delivery of plasmid VEGF165 (82) by electrotransfer to enhance wound healing in rat and mice models. To that end, our recently reported nonviral TNT approach seemed capable of allowing direct cytosolic delivery of angiogenic factors by applying a highly intense and focused electric field through arrayed nanochannels (37,40). TNT benignly nanoporates the juxtaposing tissue cell membranes and electrophoretically drives gene products into the cells in a more focused and ample manner, minimizing tissue injury (6,61). Notably, TNT-assisted delivery of a vasculogenic cocktail is not associated with tissue edema that commonly develops during vascular therapies (83). Taken together, our study demonstrates that TNT-mediated delivery of endothelial PLCγ2, as part of combination gene therapy, is effective in diabetic ischemic limb rescue.
The present work addresses the significance of endothelial PLCγ2 in augmenting the effects of VEGF therapy in ischemic tissue. CCI analyses recognized endothelial cells as the primary receiver of the VEGF signaling in both diabetic as well as nondiabetic skin. In this context, it is important to appreciate the importance of the interaction between endothelial and mural (pericytes and SMCs) cells in the blood vessel wall (44,45). Any imbalance in the interaction between these two cellular components is likely to cause vascular defect (44). Our observation that PLCγ2 levels were also downregulated in nonendothelial compartments, such as pericytes and fibroblasts in diabetic skin, points toward the need to understand the dynamic mechanisms at play between these two compartments. Three members of the MT family—MT2A, MT1E, and MT1x—were downregulated in at least eight compartments of diabetic skin. Induction of MTs in wound margins promotes reepithelization and proliferation (84). In addition, dermal expression of MTs promotes keratinocyte migration by attenuating lipid peroxidation (85,86). It is evident in a diabetic hindlimb ischemia model that the endothelial overexpression of MT improves mobilization and angiogenic function of endothelial progenitor cells by enhancing VEGF signaling (87). Thus, it will be of interest to understand how PLCγ2 and MTs may interact mechanistically to improve the effects of VEGF therapy in diabetic ischemic limb rescue.
Article Information
Funding. This work was supported by National Institute of General Medical Sciences grant GM108014 and also, in part, by grant GM077185 to C.K.S. This work was also supported in part by National Institute of Diabetes and Digestive and Kidney Diseases grants DK128845 and DK125835 to C.K.S. and DK076566 to S.R. and by U.S. Department of Defense grants W81XWH-22-1-0146 to K.S. and W81XWH-21-1-0033 to C.K.S. Research programs led by C.K.S. as well as S.R. were supported by the Lilly Endowment Indiana Collaborative Initiative for Talent Enrichment (INCITE) program. The research program of C.K.S. is also supported by John Templeton Foundation grant 61742.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. C.K.S. and K.S. conceived and designed the work with important contributions from Y.R., A.S.A., and P.V., S.R., C.K.S., and K.S. designed the clinical study and supervised sample collection and clinical annotation, with important help from E.H., S.M., M.P.M., K.E.W., and G.M.G. C.K.S. and K.S. designed and supervised scRNA-seq experiments with important contributions from Y.R., A.S.A., P.V., S.L., M.K., R.S., and J.W. Sectioning and library generation for Visium spatial transcriptomics was done by Y.R., and data analysis was done by S.L., R.S., and K.S. Molecular and immunohistochemical analyses and data interpretation were done by Y.R., P.V., S.S.V., S.K.M., S.M., S.Ka., S.Kh., and K.S. Animal experiments and data analyses were done by Y.R., P.V., S.S.V., E.H., M.K., P.R.G., S.Ka., and K.S. S.R., K.S., and C.K.S. wrote the manuscript. All authors read or provided comments on the manuscript. K.S. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Y.R., A.S.A., P.V., and S.S.V. contributed equally to this work.
This article contains supplementary material online at https://doi.org/10.2337/figshare.19184360.
References
- 1. Spreen MI, Gremmels H, Teraa M, et al.; PADI and JUVENTAS Study Groups . Diabetes is associated with decreased limb survival in patients with critical limb ischemia: pooled data from two randomized controlled trials. Diabetes Care 2016;39:2058–2064 [DOI] [PubMed] [Google Scholar]
- 2. Wang SK, Murphy MP. Getting a leg up on cell therapy for critical limb ischemia. Circ Res 2017;120:1227–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lüscher TF, Creager MA, Beckman JA, Cosentino F. Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: part II. Circulation 2003;108:1655–1661 [DOI] [PubMed] [Google Scholar]
- 4. Abbott RD, Brand FN, Kannel WB. Epidemiology of some peripheral arterial findings in diabetic men and women: experiences from the Framingham Study. Am J Med 1990;88:376–381 [DOI] [PubMed] [Google Scholar]
- 5. Jude EB, Oyibo SO, Chalmers N, Boulton AJ. Peripheral arterial disease in diabetic and nondiabetic patients: a comparison of severity and outcome. Diabetes Care 2001;24:1433–1437 [DOI] [PubMed] [Google Scholar]
- 6. Avogaro A, Albiero M, Menegazzo L, de Kreutzenberg S, Fadini GP. Endothelial dysfunction in diabetes: the role of reparatory mechanisms. Diabetes Care 2011;34(Suppl. 2):S285–S290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kolluru GK, Bir SC, Kevil CG. Endothelial dysfunction and diabetes: effects on angiogenesis, vascular remodeling, and wound healing. Int J Vasc Med 2012;2012:918267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goligorsky MS. Endothelial cell dysfunction: can’t live with it, how to live without it. Am J Physiol Renal Physiol 2005;288:F871–F880 [DOI] [PubMed] [Google Scholar]
- 9. Laing T, Hanson R, Chan F, Bouchier-Hayes D. The role of endothelial dysfunction in the pathogenesis of impaired diabetic wound healing: a novel therapeutic target? Med Hypotheses 2007;69:1029–1031 [DOI] [PubMed] [Google Scholar]
- 10. Roustit M, Cracowski JL. Assessment of endothelial and neurovascular function in human skin microcirculation. Trends Pharmacol Sci 2013;34:373–384 [DOI] [PubMed] [Google Scholar]
- 11. Rajendran P, Rengarajan T, Thangavel J, et al. The vascular endothelium and human diseases. Int J Biol Sci 2013;9:1057–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hadi HA, Suwaidi JA. Endothelial dysfunction in diabetes mellitus. Vasc Health Risk Manag 2007;3:853–876 [PMC free article] [PubMed] [Google Scholar]
- 13. Howangyin KY, Silvestre JS. Diabetes mellitus and ischemic diseases: molecular mechanisms of vascular repair dysfunction. Arterioscler Thromb Vasc Biol 2014;34:1126–1135 [DOI] [PubMed] [Google Scholar]
- 14. Li Y, Hazarika S, Xie D, Pippen AM, Kontos CD, Annex BH. In mice with type 2 diabetes, a vascular endothelial growth factor (VEGF)-activating transcription factor modulates VEGF signaling and induces therapeutic angiogenesis after hindlimb ischemia. Diabetes 2007;56:656–665 [DOI] [PubMed] [Google Scholar]
- 15. Schiekofer S, Galasso G, Sato K, Kraus BJ, Walsh K. Impaired revascularization in a mouse model of type 2 diabetes is associated with dysregulation of a complex angiogenic-regulatory network. Arterioscler Thromb Vasc Biol 2005;25:1603–1609 [DOI] [PubMed] [Google Scholar]
- 16. Rivard A, Silver M, Chen D, et al. Rescue of diabetes-related impairment of angiogenesis by intramuscular gene therapy with adeno-VEGF. Am J Pathol 1999;154:355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iyer SR, Annex BH. Therapeutic angiogenesis for peripheral artery disease: lessons learned in translational science. JACC Basic Transl Sci 2017;2:503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Isner JM, Pieczek A, Schainfeld R, et al. Clinical evidence of angiogenesis after arterial gene transfer of phVEGF165 in patient with ischaemic limb. Lancet 1996;348:370–374 [DOI] [PubMed] [Google Scholar]
- 19. Sanada F, Taniyama Y, Muratsu J, et al. Gene-therapeutic strategies targeting angiogenesis in peripheral artery disease. Medicines (Basel) 2018;5:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kusumanto YH, van Weel V, Mulder NH, et al. Treatment with intramuscular vascular endothelial growth factor gene compared with placebo for patients with diabetes mellitus and critical limb ischemia: a double-blind randomized trial. Hum Gene Ther 2006;17:683–691 [DOI] [PubMed] [Google Scholar]
- 21. Rajagopalan S, Trachtenberg J, Mohler E, et al. Phase I study of direct administration of a replication deficient adenovirus vector containing the vascular endothelial growth factor cDNA (CI-1023) to patients with claudication. Am J Cardiol 2002;90:512–516 [DOI] [PubMed] [Google Scholar]
- 22. Mäkinen K, Manninen H, Hedman M, et al. Increased vascularity detected by digital subtraction angiography after VEGF gene transfer to human lower limb artery: a randomized, placebo-controlled, double-blinded phase II study. Mol Ther 2002;6:127–133 [DOI] [PubMed] [Google Scholar]
- 23. Baumgartner I, Pieczek A, Manor O, et al. Constitutive expression of phVEGF165 after intramuscular gene transfer promotes collateral vessel development in patients with critical limb ischemia. Circulation 1998;97:1114–1123 [DOI] [PubMed] [Google Scholar]
- 24. Kao KJ. Effects of leukocyte depletion and UVB irradiation on alloantigenicity of major histocompatibility complex antigens in platelet concentrates: a comparative study. Blood 1992;80:2931–2937 [PubMed] [Google Scholar]
- 25. Macosko EZ, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015;161:1202–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 2015;33:495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abouhashem AS, Singh K, Azzazy HME, Sen CK. Is low alveolar type II cell SOD3 in the lungs of elderly linked to the observed severity of COVID-19? Antioxid Redox Signal 2020;33:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aran D, Looney AP, Liu L, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019;20:163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci 2019;28:1947–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000;28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics 2002;18:207–208 [DOI] [PubMed] [Google Scholar]
- 34. Jin S, Guerrero-Juarez CF, Zhang L, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun 2021;12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–1902.e1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 2019;20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gallego-Perez D, Pal D, Ghatak S, et al. Topical tissue nano-transfection mediates non-viral stroma reprogramming and rescue. Nat Nanotechnol 2017;12:974–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Singh K, Sinha M, Pal D, et al. Cutaneous epithelial to mesenchymal transition activator ZEB1 regulates wound angiogenesis and closure in a glycemic status-dependent manner. Diabetes 2019;68:2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Singh K, Pal D, Sinha M, et al. Epigenetic modification of microRNA-200b contributes to diabetic vasculopathy. Mol Ther 2017;25:2689–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou X, Brown BA, Siegel AP, et al. Exosome-mediated crosstalk between keratinocytes and macrophages in cutaneous wound healing. ACS Nano 2020;14:12732–12748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xuan Y, Ghatak S, Clark A, et al. Fabrication and use of silicon hollow-needle arrays to achieve tissue nanotransfection in mouse tissue in vivo. Nat Protoc 2021;16:5707–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sinha M, Sen CK, Singh K, et al. Direct conversion of injury-site myeloid cells to fibroblast-like cells of granulation tissue. Nat Commun 2018;9:936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gordillo GM, Biswas A, Singh K, et al. Mitochondria as target for tumor management of hemangioendothelioma. Antioxid Redox Signal 2021;34:137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Armulik A, Abramsson A, Betsholtz C. Endothelial/pericyte interactions. Circ Res 2005;97:512–523 [DOI] [PubMed] [Google Scholar]
- 45. Gerhardt H, Betsholtz C. Endothelial-pericyte interactions in angiogenesis. Cell Tissue Res 2003;314:15–23 [DOI] [PubMed] [Google Scholar]
- 46. Hoffmann BR, Wagner JR, Prisco AR, Janiak A, Greene AS. Vascular endothelial growth factor-A signaling in bone marrow-derived endothelial progenitor cells exposed to hypoxic stress. Physiol Genomics 2013;45:1021–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miyashita H, Sato Y. Metallothionein 1 is a downstream target of vascular endothelial zinc finger 1 (VEZF1) in endothelial cells and participates in the regulation of angiogenesis. Endothelium 2005;12:163–170 [DOI] [PubMed] [Google Scholar]
- 48. Morrison MC, Kleemann R. Role of macrophage migration inhibitory factor in obesity, insulin resistance, type 2 diabetes, and associated hepatic co-morbidities: a comprehensive review of human and rodent studies. Front Immunol 2015;6:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yamamoto H, Ehling M, Kato K, et al. Integrin β1 controls VE-cadherin localization and blood vessel stability. Nat Commun 2015;6:6429. [DOI] [PubMed] [Google Scholar]
- 50. Primo L, Seano G, Roca C, et al. Increased expression of alpha6 integrin in endothelial cells unveils a proangiogenic role for basement membrane. Cancer Res 2010;70:5759–5769 [DOI] [PubMed] [Google Scholar]
- 51. Bowers SLK, Kemp SS, Aguera KN, Koller GM, Forgy JC, Davis GE. Defining an upstream VEGF (vascular endothelial growth factor) priming signature for downstream factor-induced endothelial cell-pericyte tube network coassembly. Arterioscler Thromb Vasc Biol 2020;40:2891–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee CS, Ghim J, Song P, Suh PG, Ryu SH. Loss of phospholipase D2 impairs VEGF-induced angiogenesis. BMB Rep 2016;49:191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jiang L, Xie Y, Wei L, et al. Identification of the vascular endothelial growth factor signalling pathway by quantitative proteomic analysis of rat condylar cartilage. FEBS Open Bio 2016;7:44–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mao D, Epple H, Uthgenannt B, Novack DV, Faccio R. PLCgamma2 regulates osteoclastogenesis via its interaction with ITAM proteins and GAB2. J Clin Invest 2006;116:2869–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang D, Feng J, Wen R, et al. Phospholipase Cgamma2 is essential in the functions of B cell and several Fc receptors. Immunity 2000;13:25–35 [DOI] [PubMed] [Google Scholar]
- 56. Hashimoto A, Takeda K, Inaba M, et al. Cutting edge: essential role of phospholipase C-gamma 2 in B cell development and function. J Immunol 2000;165:1738–1742 [DOI] [PubMed] [Google Scholar]
- 57. Ichise H, Ichise T, Ohtani O, Yoshida N. Phospholipase Cgamma2 is necessary for separation of blood and lymphatic vasculature in mice. Development 2009;136:191–195 [DOI] [PubMed] [Google Scholar]
- 58. Ichise H, Ichise T, Yoshida N. Phospholipase Cγ2 is required for luminal expansion of the epididymal duct during postnatal development in mice. PLoS One 2016;11:e0150521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rhee SG, Bae YS. Regulation of phosphoinositide-specific phospholipase C isozymes. J Biol Chem 1997;272:15045–15048 [DOI] [PubMed] [Google Scholar]
- 60. Li Z, Zhao R, Yang W, et al. PLCG2 as a potential indicator of tumor microenvironment remodeling in soft tissue sarcoma. Medicine (Baltimore) 2021;100:e25008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Makarevich PI, Dergilev KV, Tsokolaeva ZI, et al. Angiogenic and pleiotropic effects of VEGF165 and HGF combined gene therapy in a rat model of myocardial infarction. PLoS One 2018;13:e0197566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu FT, Stefanini MO, Mac Gabhann F, Kontos CD, Annex BH, Popel AS. VEGF and soluble VEGF receptor-1 (sFlt-1) distributions in peripheral arterial disease: an in silico model. Am J Physiol Heart Circ Physiol 2010;298:H2174–H2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Collinson DJ, Donnelly R. Therapeutic angiogenesis in peripheral arterial disease: can biotechnology produce an effective collateral circulation? Eur J Vasc Endovasc Surg 2004;28:9–23 [DOI] [PubMed] [Google Scholar]
- 64. Liu X, Chen Y, Zhang F, et al. Synergistically therapeutic effects of VEGF165 and angiopoietin-1 on ischemic rat myocardium. Scand Cardiovasc J 2007;41:95–101 [DOI] [PubMed] [Google Scholar]
- 65. Lee JS, Kim JM, Kim KL, et al. Combined administration of naked DNA vectors encoding VEGF and bFGF enhances tissue perfusion and arteriogenesis in ischemic hindlimb. Biochem Biophys Res Commun 2007;360:752–758 [DOI] [PubMed] [Google Scholar]
- 66. Jazwa A, Tomczyk M, Taha HM, et al. Arteriogenic therapy based on simultaneous delivery of VEGF-A and FGF4 genes improves the recovery from acute limb ischemia. Vasc Cell 2013;5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Traktuev DO, Tsokolaeva ZI, Shevelev AA, et al. Urokinase gene transfer augments angiogenesis in ischemic skeletal and myocardial muscle. Mol Ther 2007;15:1939–1946 [DOI] [PubMed] [Google Scholar]
- 68. Yu JX, Huang XF, Lv WM, et al. Combination of stromal-derived factor-1alpha and vascular endothelial growth factor gene-modified endothelial progenitor cells is more effective for ischemic neovascularization. J Vasc Surg 2009;50:608–616 [DOI] [PubMed] [Google Scholar]
- 69. Shi R, Lian W, Han S, et al. Nanosphere-mediated co-delivery of VEGF-A and PDGF-B genes for accelerating diabetic foot ulcers healing in rats. Gene Ther 2018;25:425–438 [DOI] [PubMed] [Google Scholar]
- 70. Sacramento CB, da Silva FH, Nardi NB, et al. Synergistic effect of vascular endothelial growth factor and granulocyte colony-stimulating factor double gene therapy in mouse limb ischemia. J Gene Med 2010;12:310–319 [DOI] [PubMed] [Google Scholar]
- 71. Makarevich P, Tsokolaeva Z, Shevelev A, et al. Combined transfer of human VEGF165 and HGF genes renders potent angiogenic effect in ischemic skeletal muscle. PLoS One 2012;7:e38776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Basu G, Downey H, Guo S, et al. Prevention of distal flap necrosis in a rat random skin flap model by gene electro transfer delivering VEGF(165) plasmid. J Gene Med 2014;16:55–65 [DOI] [PubMed] [Google Scholar]
- 73. Kryger Z, Zhang F, Dogan T, Cheng C, Lineaweaver WC, Buncke HJ. The effects of VEGF on survival of a random flap in the rat: examination of various routes of administration. Br J Plast Surg 2000;53:234–239 [DOI] [PubMed] [Google Scholar]
- 74. Liu PY, Tong W, Liu K, et al. Liposome-mediated transfer of vascular endothelial growth factor cDNA augments survival of random-pattern skin flaps in the rat. Wound Repair Regen 2004;12:80–85 [DOI] [PubMed] [Google Scholar]
- 75. Giunta RE, Holzbach T, Taskov C, et al. AdVEGF165 gene transfer increases survival in overdimensioned skin flaps. J Gene Med 2005;7:297–306 [DOI] [PubMed] [Google Scholar]
- 76. Deodato B, Arsic N, Zentilin L, et al. Recombinant AAV vector encoding human VEGF165 enhances wound healing. Gene Ther 2002;9:777–785 [DOI] [PubMed] [Google Scholar]
- 77. Yoon CS, Jung HS, Kwon MJ, et al. Sonoporation of the minicircle-VEGF(165) for wound healing of diabetic mice. Pharm Res 2009;26:794–801 [DOI] [PubMed] [Google Scholar]
- 78. Seyed Jafari SM, Blank F, Ramser HE, et al. Efficacy of combined in-vivo electroporation-mediated gene transfer of VEGF, HGF, and IL-10 on skin flap survival, monitored by label-free optical imaging: a feasibility study. Front Surg 2021;8:639661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sersa G, Stabuc B, Cemazar M, Miklavcic D, Rudolf Z. Electrochemotherapy with cisplatin: clinical experience in malignant melanoma patients. Clin Cancer Res 2000;6:863–867 [PubMed] [Google Scholar]
- 80. Seyed Jafari SM, Shafighi M, Beltraminelli H, Geiser T, Hunger RE, Gazdhar A. Improvement of flap necrosis in a rat random skin flap model by in vivo electroporation-mediated HGF gene transfer. Plast Reconstr Surg 2017;139:1116e–1127e [DOI] [PubMed] [Google Scholar]
- 81. Gehl J. Electroporation: theory and methods, perspectives for drug delivery, gene therapy and research. Acta Physiol Scand 2003;177:437–447 [DOI] [PubMed] [Google Scholar]
- 82. Ferraro B, Cruz YL, Coppola D, Heller R. Intradermal delivery of plasmid VEGF(165) by electroporation promotes wound healing. Mol Ther 2009;17:651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lemmerman LR, Balch MHH, Moore JT, et al. Nanotransfection-based vasculogenic cell reprogramming drives functional recovery in a mouse model of ischemic stroke. Sci Adv 2021;7:eabd4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lansdown AB. Metallothioneins: potential therapeutic aids for wound healing in the skin. Wound Repair Regen 2002;10:130–132 [DOI] [PubMed] [Google Scholar]
- 85. Ågren MS, Chafranska L, Eriksen JO, et al. Spatial expression of metallothionein, matrix metalloproteinase-1 and Ki-67 in human epidermal wounds treated with zinc and determined by quantitative immunohistochemistry: a randomised double-blind trial. Eur J Cell Biol 2021;100:151147. [DOI] [PubMed] [Google Scholar]
- 86. Yu D, Qin F, Sun Y. [Effects of metallothionein (MT) on burned skin of rats]. Zhonghua Zheng Xing Shao Shang Wai Ke Za Zhi 1999;15:92–94 [PubMed] [Google Scholar]
- 87. Wang K, Dai X, He J, et al. Endothelial overexpression of metallothionein prevents diabetes-induced impairment in ischemia angiogenesis through preservation of HIF-1α/SDF-1/VEGF signaling in endothelial progenitor cells. Diabetes 2020;69:1779–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]